Abstract
AmpG is a transmembrane protein with permease activity that transports meuropeptide from the periplasm to the cytoplasm, which is essential for the induction of the ampC encoding β-lactamase. To obtain new insights into the relationship between AmpG structure and function, comparative genomics analysis, secondary and tertiary structure modeling, site-directed mutational analyses and genetic complementation experiments were performed in this study. AmpGs from different genera of bacteria (Escherichia coli, Vibrio cholerae and Acinetobacter baumannii) could complement AmpG function in Pseudomonas aeruginosa. The minimal inhibitory concentration (MIC) to ampicillin is 512 μg/ml for wild type strain PAO1, while it is 32 μg/ml for an ampG deletion mutant strain (PAO1ΔampG) with a corresponding decrease in the activity of the ampC-encoded β-lactamase. Site-directed mutagenesis of conserved AmpG residues (G29, A129, Q131 and A197) resulted in a loss of function, resulting in a loss of resistance to ampicillin in PAO1ΔampG. The G29A, G29V, A129T, A129V, A129D, A197S and A197D mutants had lower resistance to ampicillin and significantly decreased activity of the AmpC β-lactamase. The G29A, G29V, A129V, A197S and A197D mutants had decreased ampG mRNA transcript levels. The A129T and A129D mutants had normal ampG mRNA transcript levels, but the function of the protein was drastically reduced. Our experimental results demonstrate that the conserved amino acids played essential roles in maintaining the function of AmpG. Combined with the AmpG structural information, these critical amino acids can be targeted for the development of new anti-bacterial agents.
Introduction
With the widespread use of antibiotics, bacterial resistance has become a major problem for global public health. Resistance to anti-bacterial agents is progressing at a rapid rate, while the speed of new anti-bacterial agent development is relatively slow, resulting in fewer and fewer alternative anti-microbial agents for use in clinical settings [1]. β-lactam anti-bacterial agents, including penicillin and cephalosporins, are the most widely used antibiotics in clinic, but with the rapid emergence of resistance, treatment failure and recurrent infections have become a serious threat to human health [2]. It has become imperative to develop more effective countermeasures to control antibiotic resistance in clinical settings.
Bacterial resistance to β-lactam anti-bacterial agents is mainly due to the production of β-lactamases by bacteria. The AmpC type β-lactamase belongs to a class of serine cephalosporinases produced by certain gram-negative bacteria and cannot be inhibited by clavulanic acid [3]. This type of β-lactamase has a broader substrate spectrum than extended spectrum β-lactamases (ESBLs), which can be extended to all types of β-lactam anti-bacterial agents except carbapenems [1]. AmpC type β-lactamases, but not ESBLs can hydrolyze cefoxitin. There are several genes involved in the regulation of ampC expression, including ampD, ampR, ampG and ampE [4, 5]. The gene ampG encodes an inner membrane permease for the transportation of muropeptides through the cell membrane. The muropeptides are peptidoglycan catabolites that, upon entry into the cytoplasm, bind to AmpR to activate its transcriptional activator function and induce the production of AmpC type β-lactamase [6–8]. Kong KF et al. reported that P. aeruginosa appeared to have two ampG paralogs, ampG (PA4393) and ampP (PA4218) [9]. In a later study by Zhang et al. [10], PA4393 was demonstrated to have AmpG permease function, while PA4218 encoded a protein that does not have permease activity. In P. aeruginosa strains PAO1 and PAK, inactivation of the ampG genes drastically repressed the intrinsic β-lactam resistance, while ampGh1 deletion had no effect on the resistance. Inactivation of nagZ or ampG fully restored the susceptibility and basal ampC expression in ampD or dacB mutants, but only ampG inactivation fully blocked ampC induction, resulting in reduction in the MIC of the potent AmpC inducer imipenem from 2 to 0.38 μg/ml [11]. Overall, ampG acts as a “gatekeeper” and plays an important role in the expression of the AmpC type β-lactamase.
The use of AmpG inhibitors together with β-lactam anti-bacterial agents would undoubtedly restore bacterial susceptibility and extend the usefulness of common β-lactams in clinic. Greater knowledge on the structure of the target protein will likely lead to useful information for development of an effective inhibitor. In this work, we investigated several conserved residues based on the predicted structure of the AmpG protein to locate potentially functional domains. This work can aid the design and synthesis of chemicals that can be utilized as inhibitors targeting the transport function of the AmpG protein. This approach may open new avenues for the development of antibacterial agents to treat infectious diseases.
Materials and Methods
Bacterial strains and plasmids
The strains and plasmids used or constructed in this work are listed in Table 1. Pseudomonas aeruginosa PAO1 (P. aeruginosa PAO1) and plasmid pUCP24 [10] were obtained from the Laboratory of Microbial Genetics, University of Florida, Gainesville, USA. Escherichia coli 7 (E. coli 7), Vibrio cholerae 03 (V. cholerae 03) and Acinetobacter baumannii 2089 (A. baumannii 2089) are wild strains isolated from the First Affiliated Hospital of Wenzhou Medial University, China.
Table 1. Bacterial strains and plasmids used in this work.
| Strain or plasmid | Relevant characteristic (s) | Reference/source |
|---|---|---|
| Strains | ||
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 (argF-lacZYA)U169 80dlacZ | [31] |
| PAO1 | reference strain; genome completely sequenced | [32] |
| PAO1ΔampG | PAO1 ampG deletion (PA4393) | [10] |
| E. coli 7 | this work | |
| A. baumannii 2089 | this work | |
| V. cholerae 03 | this work | |
| Plasmids | ||
| pMD18-ampGmut | pMD18 vector carrying mutated ampG from PAO1 | this work |
| pUCP24 | pUC18-derived broad-host-range vector; Gmr | [33] |
| pUCP24-ampGEC | ampG gene from E. coli 7 cloned into pUCP24; Gmr | this work |
| pUCP24-ampGAB | ampG gene from A. baumannii 2089 cloned into pUCP24; Gmr | this work |
| pUCP24-ampGVC | ampG gene from V. cholerae 03 cloned into pUCP24; Gmr | this work |
| pUCP24-ampGPA | ampG gene from PAO1 cloned into pUCP24; Gmr | this work |
| pUCP24-ampGmut | pUCP24 vector carrying ampG from PAO1 with the indicated point mutations | |
| pUCP24-ampGPA-A129T | this work | |
| pUCP24-ampGPA-A129S | this work | |
| pUCP24-ampGPA-A129P | this work | |
| pUCP24-ampGPA-A129G | this work | |
| pUCP24-ampGPA-A129V | this work | |
| pUCP24-ampGPA-A129D | this work | |
| pUCP24-ampGPA-Q131E | this work | |
| pUCP24-ampGPA-Q131P | this work | |
| pUCP24-ampGPA-Q131R | this work | |
| pUCP24-ampGPA-Q131H | this work | |
| pUCP24-ampGPA-G29R | this work | |
| pUCP24-ampGPA-G29C | this work | |
| pUCP24-ampGPA-G29D | this work | |
| pUCP24-ampGPA-G29A | this work | |
| pUCP24-ampGPA-G29V | this work | |
| pUCP24-ampGPA-A197T | this work | |
| pUCP24-ampGPA-A197S | this work | |
| pUCP24-ampGPA-A197G | this work | |
| pUCP24-ampGPA-A197D | this work |
Genetic complementation assays
The ampG genes of P. aeruginosa PAO1, E. coli 7, V. cholerae 03 and A. baumannii 2089 were amplified from corresponding genomic DNA templates. A BamHI restriction site was added at the 5’ end of the sense primers (PAampG-F, ECampG-F, ABampG-F and VCampG-F) and a HindIII (or SalI) restriction site was added at the 5’ end of the anti-sense primer (PAampG-R, ECampG-R, ABampG-R and VCampG-R) (Table 2). The ampG polymerase chain reaction (PCR) products were first cloned into the pMD18-T vector (TaKaRa, Dalian, China). The recombinant plasmid (pMD18-ampG) was identified initially by PCR and was then verified by sequencing. The verified pMD18-ampG plasmid was digested with the restriction enzymes BamHI and HindIII (or SalI). The ampG fragment was recovered and then ligated into the pUCP24 vector digested with the same restriction enzymes (BamHI and HindIII/SalI). The recombinant plasmid pUCP24-ampG was transformed into E. coli JM109, and the recombinant was further identified and verified by PCR. The plasmid pUCP24-ampG was extracted and introduced into PAO1ΔampG as described previously [10]. PAO1ΔampG carrying vector pUCP24 was used as a negative control.
Table 2. Primers used in this study.
| Primer | Sequencea, b | Purpose |
|---|---|---|
| PAampG-F | 5’GGGATCCCAACGCGCACGCTTGCGCGAGGA 3’(BamHI) | Cloning of ampG of PAO1 |
| PAampG-R | 5’GAAGCTTTCAGTGCTGCTCGGCGTTCTGGT3’(HindIII) | |
| ECampG-F | 5’GGGATCCATGTCCAGTCAATATTTACG 3’(BamHI) | Cloning of ampG of E.coli 7 |
| ECampG-R | 5’GAAGCTTTTACGTCAGATGCGTTTTTCG 3’(HindIII) | |
| ABampG-F | 5’GGGATCCATGTTCCGACAGCAAAATCTTTA 3’(BamHI) | Cloning of ampG of A. baumannii 2089 |
| ABampG-R | 5’GGTCGACTTATACAGTTTTAGCATCTTTCCA 3(SalI) | |
| VCampG-F | 5’GGGATCCTAGGTACAAGTAGTTGGGGCCAGG 3’(BamHI) | Cloning of ampG of V. cholerae 03 |
| VCampG-R | 5’GGTCGACCAGTCCTATTTTAGTAGGTCATTTT 3’(HindIII) | |
| PAampG-G29R | 5’AACACCAGCATGGTCGGCAGgcgGGCGGCGAAGCCGAGCA 3’ | G29R |
| PAampG-G29C | 5’AACACCAGCATGGTCGGCAGgcaGGCGGCGAAGCCGAGCA3’ | G29C |
| PAampG-G29D | 5’AACACCAGCATGGTCGGCAGgtcGGCGGCGAAGCCGAGCA3’ | G29N |
| PAampG-G29A | 5’AACACCAGCATGGTCGGCAGggcGGCGGCGAAGCCGAGCA3’ | G29A |
| PAampG-G29V | 5’AACACCAGCATGGTCGGCAGgacGGCGGCGAAGCCGAGCA3’ | G29V |
| PAampG-29-R | 5’CTGCCGACCATGCTGGTGTTCAACACCCTGTCGGTGTGGC3’ | |
| PAampG-A129T | 5’TCGATCGCGATGTCCTGGGTggtGGAGGCGAACGCCACCA3’ | A129T |
| PAampG-A129S | 5’TCGATCGCGATGTCCTGGGTggaGGAGGCGAACGCCACCA3’ | A129S |
| PAampG-A129P | 5’TCGATCGCGATGTCCTGGGTgggGGAGGCGAACGCCACCA3’ | A129P |
| PAampG-A129G | 5’TCGATCGCGATGTCCTGGGTgccGGAGGCGAACGCCACCA3’ | A129G |
| PAampG-A129V | 5’TCGATCGCGATGTCCTGGGTgatGGAGGCGAACGCCACCA3’ | A129G |
| PAampG-A129D | 5’TCGATCGCGATGTCCTGGGTgtcGGAGGCGAACGCCACCA3’ | A129N |
| PAampG-129-R | 5’ACCCAGGACATCGCGATCGACGCCTACCGCCTGGAAATCG3’ | |
| PAampG-Q131E | 5’TAGGCGTCGATCGCGATGTCctcGGTGGCGGAGGCGAACG3’ | Q131E |
| PAampG-Q131P | 5’TAGGCGTCGATCGCGATGTCcggGGTGGCGGAGGCGAACG3’ | Q131P |
| PAampG-Q131R | 5’TAGGCGTCGATCGCGATGTCccgGGTGGCGGAGGCGAACG3’ | Q131R |
| PAampG-Q131H | 5’TAGGCGTCGATCGCGATGTCatgGGTGGCGGAGGCGAACG3’ | Q131H |
| PAampG-131-R | 5’GACATCGCGATCGACGCCTACCGCCTGGAAATCGCCGAGG3’ | |
| PAampG-A197T | 5’AGCCCAGGCAGGATCAGCAGggtGAACAACGCATAGGTCA3’ | A197T |
| PAampG-A197S | 5’AGCCCAGGCAGGATCAGCAGggaGAACAACGCATAGGTCA3’ | A197S |
| PAampG-A197G | 5’AGCCCAGGCAGGATCAGCAGgccGAACAACGCATAGGTCA3’ | A197G |
| PAampG-A197D | 5’AGCCCAGGCAGGATCAGCAGgtcGAACAACGCATAGGTCA3’ | A197N |
| PAampG-197-R | 5’CTGCTGATCCTGCCTGGGCTGGTCACCAGCCTGCTGATCC3’ | |
| PAampG-FqPCR | 5’GCGGTCTCGGTGATGGTGCT 3’ | qPCR |
| PAampG-RqPCR | 5’CGCTGGACGAACTCGGTGAT 3’ | |
| EC-FqPCR | 5’TGATGGACCGCTACACGC 3’ | qPCR |
| EC-RqPCR | 5’ACCCAGAACGCTGATTGC 3’ | |
| AB-FqPCR | 5’ACAGGCGCAACTCAAGAT 3’ | qPCR |
| AB-RqPCR | 5’CCCAATAAAGCAGCAACA 3’ | |
| VC-FqPCR | 5’TGACTGGCTGGCTGAAAG 3’ | qPCR |
| VC-RqPCR | 5’CGATGGATTGGCAGAAAA 3’ | |
| RpoD-FqPCR | 5’CCGTTGCTGAATATCCGGAA 3’ | qPCR |
| RpoD-RqPCR | 5’CAAATTTTTCGCGAGCCAGT 3’ |
a: The underlined sequences are restriction endonuclease sites.
b: The nucleotide sequence corresponding to the mutated amino acids are in lowercase.
Site-directed mutagenesis of the conserved amino acids in ampG gene
The mutant nucleotide was introduced into the 3’ end primer of the 5’ end fragment as well as the 5’ end primer of the 3’ end fragment where a 10 bp overlap was located. In total, 19 pairs of primers were designed using the online software Premier 5.0 for the four conserved amino acids from the ampG gene for site-directed mutagenesis (Table 2). The common sense primer (PAampG-F) was flanked by the BamHI restriction site, and the common anti-sense primer (PAampG-R) was flanked by the HindIII restriction site (Table 2). For the recombinant PCR of each mutated site, a pair of the common primers was used with the corresponding 5’ and 3’ end mutated fragments as the templates. The PCRs were performed under the following conditions: an initial 5 min denaturation at 95°C, followed by 35 amplification cycles consisting of a 45 sec denaturation step at 95°C, a 55 sec annealing step at 55°C and a 45 sec extension step at 72°C, followed by a 10 min final extension at 72°C (ExTaq, TaKaRa, Dalian, China). The PCR products (ampGmut) were purified and inserted into pMD18-T vectors. The recombinant pMD18-ampGmut plasmids were initially identified by PCR and were then verified by sequencing. The verified pMD18-ampGmut recombinant plasmids were digested with BamHI and HindIII. The ampGmut fragments were recovered and ligated into pUCP24 digested with the same restriction enzymes (BamHI and HindIII). The recombinant pUCP24-ampGmut plasmids were then transformed into PAO1ΔampG for the complementation studies.
Antibiotic susceptibility testing
The minimal inhibitory concentration (MIC) of the antimicrobial agents was determined by the agar dilution method for the control and recombinant strains in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The antimicrobial agents were obtained from the National Institute for the Control of Pharmaceutical and Biological Products (NICPBP) and pharmaceutical companies in China. E. coli ATCC 25922 was used as a quality control for the MIC tests.
Detection of β-lactamase activity
The β-lactamase detection procedures were performed as described by Zhang Y et al. [10]. P. aeruginosa cells were induced for 1 h with 4 μg/ml cefoxitin (Calbiochem, San Diego, USA) and for 2 h with 50 μg/ml cefoxitin. Crude cell extracts were prepared by sonication, and the protein content of the crude extracts was determined by using the BCA Protein Assay Reagent (Pierce, USA) and bovine serum albumin as the standard [12]. The β-lactamase activity was quantified with an UV spectrophotometer using 100 μM of nitrocefin as the substrate [10]. The activity of the β-lactamase was defined as the nanomoles of nitrocefin hydrolyzed at 30°C per min by one milligram of the protein. All the induction experiments were performed in triplicate and the results represent an average of three experiments.
qRT-PCR analysis of ampG mRNA concentration in ampG mutants
To validate the transcription level of the mutant ampGs in vivo, qRT-PCR was performed using the StepOne™ RT-PCR System (Applied Biosystems, USA) [13]. Total RNAs were extracted from the control and recombinants cultured with ampicillin (100 μg/ml) using the Rneasy Plant Mini Kit (Qiagen, Germany) and were then treated with Dnase I using the Rnase-Free Dnase Set (Qiagen, Germany). cDNA was obtained by reverse transcription using the PrimeScript RT-PCR Kit (TaKaRa, Dalian). After qPCR analysis, relative quantification of the targets in each sample were calculated using rpoD as the internal control [14].
Sequence comparison and phylogenetic analysis of ampG
The ampG sequences from different bacterial species were compared by BlastX from NCBI, and the open reading frames of the ampG genes were identified by MEGA5.05 to analyze the similarities of the ampG nucleotide sequences and the AmpG amino acid sequences. Phylogenetic trees were constructed using the maximum likelihood method, and the resulting trees were tested with bootstrap values of 100 replicates using PhyML3.0 [15].
Prediction of the AmpG secondary structure
The AmpG secondary structure was predicted using a program at http://www.predictprotein.org/, and a Kyte-Doolittle algorithm from Lasergene 7 (DNASTAR, Madison, WI) [9] was used to predict AmpG transmembrane (TM) helices. The parameters of TOPPRED-transmembrane topology prediction were as follows: Full window size: 21, Core window size: 11, Wedge window size: 5, Using hydrophobicity file: GES-scale, Cutoff for certain transmembrane segments: 1.00, Cutoff for putative transmembrane segments: 0.60, Critical distance between 2 transmembrane segments: 2, Critical loop length: 60.
AmpG tertiary structure modeling
The folding templates were generated using BLAST Search (DS Server) from Discovery Studio [16] and FFAS03 (http://ffas.burnham.org/ffas-cgi/cgi/document.pl) [17]. The structure model of AmpG was further established using I-TASSER according to homology modeling (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) [18]. Five model structures, including 1b3u (PROTEIN PHOSPHATASE PP2A), 3o7q (L-fucose-proton symporter), 1pw4 (Glycerol-3-phosphate transporter), 2bku and 4aps (GTP-BINDING NUCLEAR PROTEIN RAN) were obtained. DS Ramachandran plot and PDBsum (www.ebi.ac.uk/thornton-srv/databases/pdbsum.Generate.html) were applied for final model evaluation [19].
Results
Diversity of the ampG gene
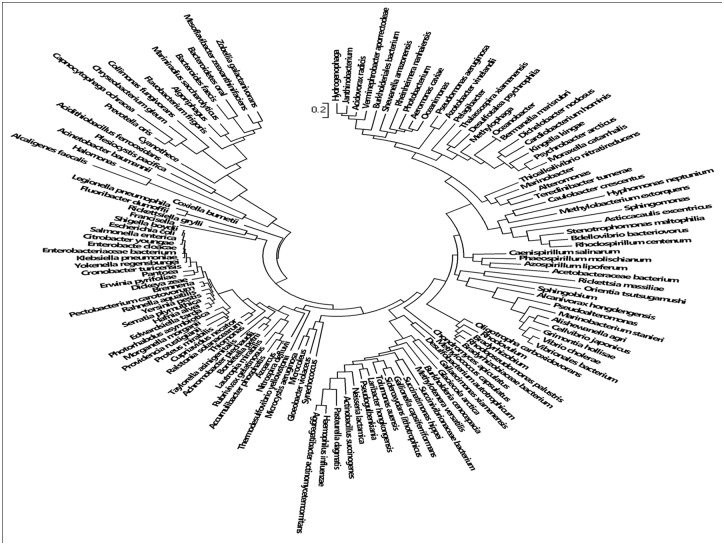
A total of 2245 AmpG protein related sequences were collected by searching the NCBI Protein database using AmpG as the key word. The AmpG proteins were widely distributed over different bacterial species, including 134 known genera (Fig 1, S1 Table). Using the P. aeruginosa PAO1 AmpG as a reference, the amino acid sequence similarity analysis showed that AmpG of Azotobacter vinelandii DJ had the highest identity of 61.9% while that of Capnocytophaga showed the lowest similarity of 22.7% (S1 Table). The similarities of the three AmpGs used for the genetic complementation analyses to P. aeruginosa were 53.3% (E. coli 7), 36.0% (A. baumannii 2089) and 33.0% (V. cholerae 03).
Fig 1. Phylogenetic tree of the ampG gene.
The AmpG sequences from 134 species were used to generate the tree.
The TM topology of AmpG
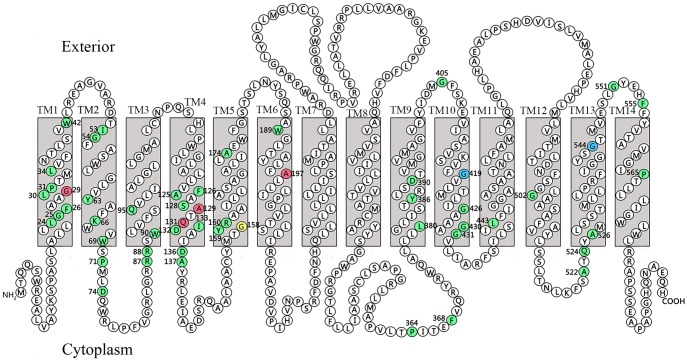
The length and number of TM helices in the various AmpG proteins differ significantly. The longest AmpG is from P. aeruginosa (CDR92618) [9], which consists of 598 amino acids (AA) and 14 predicted TM helices (Fig 2). The shortest AmpG is from Microcoleus, which only has 401 AA and 11 TM helices (S1 Table). Most AmpG proteins contain 12 or 14 TM helices. The four AmpGs used for genetic complementary analysis have 14 (PAO1, E. coli 7), 11 (A. baumannii 2089) and 10 TM helices (V. cholerae 03) (S1 Table).
Fig 2. Secondary structure of AmpG from PAO1.
The conserved amino acids are shown in green. Amino acids 29G, 129A, 131Q and 197A that are shown in red are conserved amino acids that were subjected to site-directed mutagenesis. Amino acids 419G (in blue), 544G (in blue) and 158G (in yellow) correspond to C151, G268 and G373 in AmpG of E. coli SN0301, respectively.
Conservation of AmpG
A total of 134 AmpGs from 134 genera were chosen as the representatives for conservation analysis. Using AmpG from PAO1 as a reference, the amino acids that appeared in more than 80% of the 134 representative AmpGs were considered as conserved [9]. Excluding TM 7 and 8 in P. aeruginosa (TM 7 and 8 do not exist in many AmpGs), 10 or more TM helices were identified in nearly half (66/134) AmpG proteins (S2 Table). The results of the conserved amino acid analysis showed that there were 51 conserved amino acids all clustered in the 12 TM helices. In P. aeruginosa PAO1, these 12 conserved amino acid clusters (Table 3) were distributed over 12 of the 14 TM helices with TM 7 and 8 as the exceptions. Seven of the 12 clusters locate to TMs 1, 2, 3, 4, 9, 10 and 13. These conserved amino acid residues could be critical for maintaining the function of AmpG.
Table 3. The 12 conserved amino acid clusters.
| Sequence | TM | |
|---|---|---|
| Class 1 | L24G25F26AAG29L30P31TML34VFNTLSVW42 | Segment no.1 |
| Class 2 | I53G54FASWLGLVY63AFK66WVW69SP71MLD74 | Segment no.2 |
| Class 3 | R87R88SW90LVFSQ95 | Segment no.3 |
| Class 4 | A125F126AS128A129TQ131D132I133AID136A137 | Segment no.4 |
| Class 5 | Y159R160AAILLASAGALILA174 | Segment no.5 |
| Class 6 | W189GLTYALFA197 | Segment no.6 |
| Class 7 | P364ITEF368VQRYRWQALLLL380GLISTY386RLSD390 | Segment no.9 |
| Class 8 | G405FSKEVIASVSKVFG419VLMTLIG426AAAG430G431 | Segment no.10 |
| Class 9 | SIL443FIGGAAS | Segment no.11 |
| Class 10 | DNFSGG502LAASAFVA | Segment no.12 |
| Class 11 | A522TQ524YA526MLSSTMLLLPRFIGGYSG544TMVESLG551 | Segment no.13 |
| Class 12 | F555FYVTAVMGIP565 | Segment no.14 |
Note: The residues with the superscripted numbers are the conserved amino acids.
Functional analysis of AmpG proteins cloned from different genera of bacteria
In our previous work, we used homologous recombination technology to obtain a ampG knockout in P. aeruginosa strain PAO1 (PAO1ΔampG) [10]. The MIC for ampicillin decreased from 512 μg/ml (PAO1) to 32 μg/ml (PAO1ΔampG) [10]. This result indicated that the ampG gene is essential for the induction of the AmpC type β-lactamase. Although, the ampG genes in different bacteria differ from each other in length and number of transmembrane regions, they share many conserved domains in the transmembrane regions. To detect the activity of various AmpGs from different origins, we performed genetic complementation analysis of ampGs cloned from E. coli 7 (491 AA, 14 TM helices), V. cholerae 03 (462 AA, 10 TM helices), A. baumannii 2089 (415 AA, 11 TM helices) as well as ampG of P. aeruginosa PAO1 (594 AA, 14 TM helices). The plasmid pUCP24 was used as the ampG carrier vector, and PAO1ΔampG was the host strain. The results showed that all of the cloned ampG genes complemented the function of the deleted ampG gene, and the MIC for ampicillin was almost fully restored to the level of the P. aeruginosa PAO1 parental strain. The β-lactamase activity of the genetically complemented recombinants also showed similar results (Table 4). Overall, these results show that three genes from different bacterial species have similar functions and can compensate the function of the deleted ampG gene in PAO1ΔampG, regardless of their structural differences. However, the AmpGs from E. coli and A. baumannii only showed approximately one third AmpC induction compared to P. aeruginosa or V. cholerae AmpG. The reason for this difference is unknown.
Table 4. Summary of ampicillin MIC, AmpC β-lactamase activity and qRT-PCR of ampG RNA results.
| Strain | MIC(μg/ml) | β-Lactamase activitya | qPCR(2-ΔΔCt) | |
|---|---|---|---|---|
| Non-induced | Induced | |||
| ATCC 25922 | 4 | 0 | 0 | - |
| PAO1 | 512 | 112.40±7.2 | 1981.64±56 | 1.00±0.0 |
| PAO1ΔampG | 32 | 35.49±2.5 | 44.82±3.3 | 0.08±0.0 |
| PAO1ΔampG-ampGPA | 1024 | 37.87±1.8 | 5019.31±98.2 | 8835.75±45.4 |
| PAO1ΔampG-ampGEC | 512 | 118.50±7.0 | 1622.63±34 | - |
| PAO1ΔampG-ampGAB | 1024 | 49.48±4.2 | 1680.35±28 | - |
| PAO1ΔampG-ampGVC | 512 | 24.40±1.5 | 5063.64±67.3 | - |
| PAO1ΔampG-UCP24-ampGPA-A129T | 64 | 309.64±10.6 | 139.37±6.6 | 10363.27±128.3 |
| PAO1ΔampG-pUCP24-ampGPA-A129S | 1024 | 205.27±7.8 | 9556.45±132 | 6758.90±47.2 |
| PAO1ΔampG-UCP24-ampGPA-A129P | 1024 | 895.45±66.8 | 12450.43±524 | 8803.84±56.9 |
| PAO1ΔampG-pUCP24-ampGPA-A129G | 512 | 262.83±8.9 | 5342.68±88.7 | 9738.85±123.3 |
| PAO1ΔampG-pUCP24-ampGPA-A129V | 64 | 200.74±14 | 232.28±26 | 0.52±0.0 |
| PAO1ΔampG-pUCP24-ampGPA-A129D | 64 | 233.23±12.5 | 576.69±35 | 6218.63±45.6 |
| PAO1ΔampG-pUCP24-ampGPA-Q131E | 1024 | 282.83±10 | 12559.25±213 | 6097.70±78.5 |
| PAO1ΔampG-pUCP24-ampGPA-Q131P | 1024 | 101.28±5.8 | 12518.16±432.8 | 6433.10±104.6 |
| PAO1ΔampG-pUCP24-ampGPA-Q131R | 1024 | 186.91±3 | 12393.96±541 | 9838.86±218.9 |
| PAO1ΔampG-pUCP24-ampGPA-Q131H | 1024 | 187.31±2.1 | 11109.25±426 | 13156.64±321.2 |
| PAO1ΔampG- pUCP24-ampGPA-G29R | 512 | 179.91±3.8 | 5142.63±132 | 4140.62±34.0 |
| PAO1ΔampG- pUCP24-ampGPA-G29C | 512 | 117.52±4.6 | 4928.87±265.4 | 4054.96±62.6 |
| PAO1ΔampG- pUCP24-ampGPA-G29D | 1024 | 223.10±1.6 | 4750.43±192 | 9054.04±102.7 |
| PAO1ΔampG- pUCP24-ampGPA-G29A | 64 | 40.15±4 | 236.33±12.3 | 0.74±0.0 |
| PAO1ΔampG- pUCP24-ampGPA-G29V | 64 | 17.30±1 | 209.16±36 | 0.32±0.0 |
| PAO1ΔampG-pUCP24-ampGPA-A197T | 1024 | 163.03±21 | 3453.21±43.2 | 5123.37±36.9 |
| PAO1ΔampG-pUCP24-ampGPA-A197S | 64 | 32.21±2.2 | 145.28±5.7 | 0.63±0.0 |
| PAO1ΔampG-pUCP24-ampGPA-A197G | 1024 | 170.69±6 | 5023.37±246 | 7865.3±44.8 |
| PAO1ΔampG-pUCP24-ampGPA-A197D | 128 | 46.28±7.8 | 406.78±11.2 | 0.12±0.0 |
a: Nanomoles of nitrocefin hydrolyzed per minute per milligram of protein.
_, not tested.
Tertiary structure prediction of AmpG of P. aeruginosa
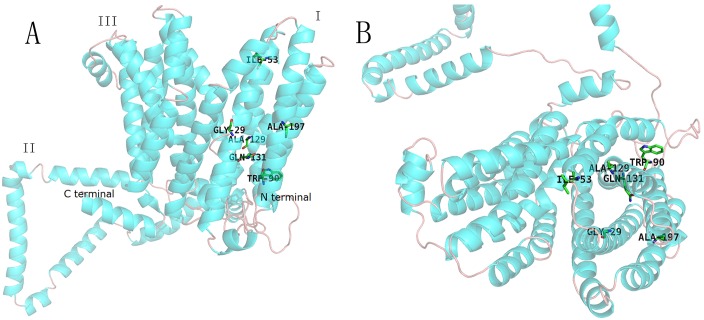
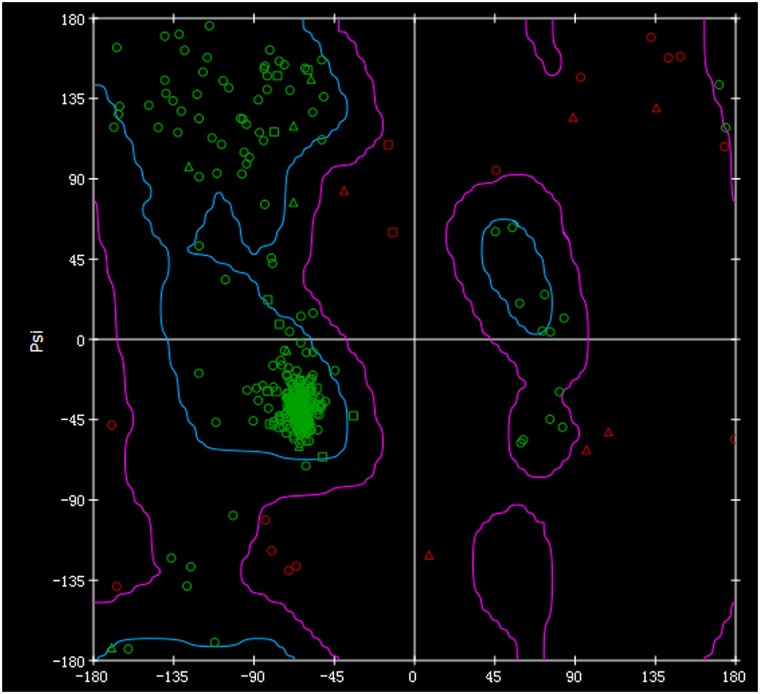
As the crystal structure of AmpG has not yet been resolved, we used DS Blast, FFAS03 and I-TASSER to predict the structure of AmpG from P. aeruginosa PAO1. The folding templates 1b3u, 3o7q, 1pw4, 2bku and 4aps were derived from human PROTEIN PHOSPHATASE PP2A, L-fucose-proton symporter of E. coli K-12, Glycerol-3-phosphate transporter of E. coli and GTP-BINDING NUCLEAR PROTEIN RAN of Saccharomyces cerevisiae, respectively. Using combined I-TASSER with two functional modules of Align Sequence to Templates and DS's Build Homology Models, 3D models of AmpG protein were created. According to the matching degree of secondary structure, the optimal structure was then chosen. The predicted 3D structure of P. aeruginosa PAO1 AmpG mainly consists of three parts (Fig 3). The first and third parts each consist of 6 transmembrane α helices that form an opening inside of the cell. The second part located between these two parts consist of 167 AA forming α helices and loop structures outside of the membrane. Both the N-terminus and C-terminus are located close to each other in the cytoplasm. Geometric evaluations of the modeled 3D structures of P. aeruginosa AmpG were performed using Discovery Studio by calculating the Ramachandran plot. The plot showed that 90.2% of residues were found in the favored regions, 8.8% were in the allowed regions and 1.0% were in the outlier region (Fig 4).
Fig 3. The modeled 3D structures of AmpG from PAO1.
Twelve TM helices and one internal membrane region are illustrated. The first and third parts consist of six transmembrane α helices. The region between the first and third parts (named the second part) is a sequence of 167 AA in length. The mutated amino acids at positions 29, 53, 90, 129, 131 and 197 are illustrated in the third part (A). The picture on the right illustrates the protein as viewed from outside of the membrane (B).
Fig 4. Ramachandran plots for the modeled 3D structures of AmpG from PAO1.
The Ramachandran plot indicates low energy conformations for φ (phi) and ψ (psi). The conventional terms are used to represent the torsion angles on either side of the alpha carbons in the peptides. A triangle indicates glycine, a square indicates proline and all other types of amino acids are indicated as circles. The regions circled with gray lines represent the most favorable combinations of phi-psi values.
Effect of conserved amino acid mutations on the function of AmpG
Based on the results shown above, 51 amino acids distributed in 12 clusters are conserved. To analyze the correlation of these conserved amino acids with the function of AmpG, 4 conserved amino acids (G29, A129, Q131 and A197) from the transmembrane regions were chosen for mutational analysis. These four amino acid residues are located in TM 1 (G29), 4 (A129, Q131) and 6 (A197) assembly of the first part of the protein (Fig 2). According to the chemical properties of the amino acids, 4 to 6 different amino acid substitutions were designed for each of the conserved amino acids. A total of 19 mutated ampG genes were generated (Table 1), and these gene clones were transformed into recipient PAO1ΔampG cells. The MICs for ampicillin and the β-lactamase activities were detected (Table 4). The transcription levels of the ampG mutants were examined by qRT-PCR. As the results show, most mutants conferred strong resistance to ampicillin, similar to the original P. aeruginosa PAO1 strain. The G29A, G29V, A129T, A129V, A129D, A197S, and A197D mutants conferred significantly lower resistance to ampicillin and had significantly lower β-lactamase activities (Table 4).
Similar to PAO1ΔampG, the expression level of ampG containing the G29A, G29V, A129V, A197S and A197D mutations decreased significantly (Table 4), suggesting that amino acids at position 29, 129 and 197 have strong effects on the expression of ampG. Conversely, the expression levels of the A129T and A129D mutants were close to PAO1ΔampG-ampGPAO1 and the rest of the mutants. However, the MICs for ampicillin and the β-lactamase activities of the A129T and A129D mutants were similar to PAO1ΔampG, suggesting that the mutants with A129T and A129D did not affect ampG expression but rather AmpG function was drastically decreased.
Discussion
The gene ampG is encoded on the chromosome and is widely distributed in many genera of bacteria. AmpG is a transmembrane protein that acts as a permease and transports muropeptide from the periplasm into the cytoplasm. The muropeptide derivatives are essential for the induction of AmpC type β-lactamase expression [20]. It was previously reported that the main material transported by AmpG is GlcNAc-anhMurNAc, and muropeptides that lack GlcNAc or anhMurNAc are not transported [21]. Dietz D et al. indicated that AmpG is responsible for the transportation of N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid- tetrapeptide aldehyde from the periplasm into the cytoplasm, which is generated from the degradation of murein by hydrolases [22]. The degraded products of murein are transferred to AmpD by AmpE and are then hydrolyzed by AmpD or are first degraded by β-N-acetyl-glucosaminidase to N-acetyl-muramyl tripeptide in the cytoplasm and then hydrolyzed by AmpD. When N-acetyl-muramyl tripeptide binds with AmpR, it may activate AmpR and induce transcription of ampC [23]. Deletion or mutation of ampG may cause the cell membrane to lack AmpG completely or may result in a structural abnormality in AmpG. Lack of or presence of a mutant form of AmpG may result in a defect in transport of N-acetyl-muramyl tripeptide into the cytoplasm, thereby reducing AmpC type β-lactamase induction [24–26].
Although AmpG was found to be widely distributed throughout gram-negative bacteria, it does vary in size and in the number of TM helices. However, secondary structure prediction showed that a large part of the TM helices are conserved in most bacteria. The AmpG protein in one type of E. coli contained 14 TM helices, and there were four hydrophobic segments within the cytoplasm. In this work, AmpG proteins from different genera of the bacteria (P. aeruginosa PAO1, 14, V. cholerae 03, 10, A. baumannii 2089, 11 and E. coli 7, 14) were used to perform genetic complementation experiments. The AmpG proteins in these bacteria have different numbers of TM helices. The results presented here showed that all of the AmpG proteins could function properly in the recipient PAO1ΔampG. Similar to the wild type, the ampG complemented PAO1ΔampG strains regained high level ampicillin MICs and expressed AmpC type β-lactamase with normal activity. Similar experiments have been performed by Huang et al., who showed that the AmpG of E. coli can replace a mutated ampG or ampN in Stenotrophomonas maltophilia and restore β-lactamase production [27]. In this work, although the donor AmpG proteins had different numbers of TM helices, and similarity between the TM helices of four AmpGs were only 33–53%, they all contain 10 (V. cholerae 03), 11 (baumannii 2089) or 14 (E. coli 7 and P. aeruginosa PAO1) clusters of conserved amino acid residues. Schmidt et al. [24] used nitrosoguanidine (NTG) to induce mutations in E. coli SN0301 (carrying ampC and ampR of E. cloacae) and obtained three non-functional ampG gene mutants, including ampG1 (G151-D151), ampG3 (G268-D268) and ampG5 (G373-D373). The three mutated amino acids correspond to positions 158 (G), 419 (G) and 544 (G) in AmpG of P. aeruginosa PAO1, and each amino acid position is in the conserved amino acid regions located in TM 5, 10 and 13 in AmpG of P. aeruginosa PAO1, respectively. Mutation of ampG can lead to the functional loss of the AmpG protein, resulting in a failure to induce β-lactamase or a significant reduction in AmpC type of β-lactamase induction. It is suggested that the conserved amino acids play an important role in maintaining normal AmpG function.
A single amino acid might be enough to determine the substrate specificity and transportation activity of AmpG. The expression levels of ampG mRNA in the G29A, G29V, A129V, A197S and A197D mutants were significantly decreased in PAO1ΔampG compared to the wild type ampG. Conversely, when A129 was mutated to D or T, the transcriptional levels of ampG was unchanged, but the ampicillin MIC levels and AmpC type β-lactamase activities were drastically reduced. These results indicate that although the mutations did not affect the transcription of ampG, the AmpG structure might have changed and resulted in a loss of function. Structural and biochemical tests of the protein LacY showed that co-transport of protons involve a complex network of salt bridges and hydrogen bonds (H-bond) [28]. Some amino acid residues are essential for the protein functions of binding, transferring and releasing of protons. The function of glutamate at position 269 is to bind with a proton and the lactose substrate and subsequently induces proton transfer to the glutamic acid at position 325. This process drives a conformational change of the protein into an inward opening. The substrate is released into the cell, then the proton is released from the glutamic acid at position 325, the protein then changes conformation to the outward opening position, and another cycle begins [28, 29]. In this study, it was predicted that the N- and C-terminus of P. aeruginosa PAO1 AmpG are located in the cytoplasm and that they are spatially close to each other. The binding site for the substrate is in the center of the cavity located in the outer end of the transmembrane channel formed by the transmembrane regions. When the substrate binds to the cavity, it will lead to a conformational change of the protein so that the inner end of the channel opens and drives the entrance of the substrate into the cytoplasm [30]. The amino acids utilized in this work (G29, A129, A197) were all conserved and were located in the first part of the transmembrane regions. Mutation of these residues might cause a steric effect and/or electrostatic force that prevents the transmembrane protein from binding to the substrate or hinders the conformational change of AmpG when it binds to the substrate. Thus, the normal function of AmpG is diminished. When the muropeptides (such as N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid-tetrapeptide aldehyde), which are the initial substrate of the inducer for the AmpC type β-lactamase, cannot be transported by AmpG to cytoplasm, the AmpC type β-lactamase level in the cell would be diminished as a consequence.
With the rapid emergence of antibiotic resistance, curing infectious diseases is a serious challenge in the clinical setting. It is critically important to study the regulatory mechanism of the expression of AmpC type β-lactamase at a cellular level. Agents that interfere with the function of AmpG may hinder the transportation of the muropeptide and decrease the level of AmpC type β-lactamase in the cell, which in turn increases the sensitivity of the cell to commonly used anti-bacterial agents. This study suggests a new research avenue: utilize AmpG as a drug target and conduct rational design and inhibitor screens with the goal of overcoming bacterial resistance to β-lactam antibiotics for clinical therapy of infectious diseases.
Supporting Information
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was funded by grants from the Natural Science Foundation of Zhejiang Province (LY14C060005), the Science and Technology Foundation of National Health and Family Planning Commission of China (WKJ2012-2-032), the National Natural Science Foundation of China (81401702, 81501808, 81501780), the Science and Technology Foundation of Wenling City (W201195, W201075, 2016C31BE0029), and the NIH (R21-AI119043). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Komolafe OO. Antibiotic resistance in bacteia—an emerging public health problem. 2003; 15(2): 63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RR, Hota B, Ahmad I, Scott RD 2nd, Foster SD, Abbasi F, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009; 49(8): 1175–84. 10.1086/605630 [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactmases and its correlation with molecular ateucture. Antimicrob Agents chemother. 1995; 39 (6): 1211–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidtke AJ, Hanson ND. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008; 52(11): 3922–7. 10.1128/AAC.00341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langaee T, Gagnon L, Huletsky A. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC beta-lactamase expression. Antimicrob Agents Chemother. 2000; 44(3): 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang TC, Chen TF, Tsai JJP, Hu RM . AmpG is required for BlaXc beta-lactamase expression in Xanthomonas campestris Pv. campestris str. 17. FEMS Microbiology Letters. 2013; 340(2): 101–8. 10.1111/1574-6968.12071 [DOI] [PubMed] [Google Scholar]
- 7.Hanson ND, Moland T, Hossain A, Neville SA, Gosbell IB, Thomson KS. Unusual Salmonella enterica serotype Typhimurium isolate producing CMY-7, SHV-9 and OXA-30 beta-lactamases. Journal of Antimicrobial Chemotherapy. 2002; 49(6): 1011–14. [DOI] [PubMed] [Google Scholar]
- 8.Korfmann G S C. AmpG is essential for high level expression of AmpC beta-lactam ase in Enterobacter cloacae. A ntim icrob A gents Chem other. 1989; 33(11): 1946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong KF, Aguila A, Schneper L, Mathee K. Pseudomonas aeruginosa β-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol. 2010; 10(10): 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Bao QY, Gagnon LA, Huletsky A, Oliver A, Jin SG, Langaee T. ampG gene of Pseudomonas aeruginosa and its role in beta-lactamase expression. Antimicrob Agents Chemother. 2010; 54(11): 4772–9. 10.1128/AAC.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamorano L, Reeve TM, Juan C, Moyá B, Cabot G, Vocadlo DJ, et al. AmpG inactivation restores susceptibility of pan-beta-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother. 2011; 55(5): 1990–1996. 10.1128/AAC.01688-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trepanier S, Prince A, Huletsky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997; 41(11): 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Yang Y, Chen W, Ding L, Li P, Zhao X, et al. Identification of differentially expressed proteins of Arthrospira (Spirulina) plantensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci. 2013; 11(1): 6 10.1186/1477-5956-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25(4): 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 15.Ying JC, Wang HF, Bao BK, Zhang Y, Zhang JF, Zhang C, et al. Molecular Variation and Horizontal Gene Transfer of the Homocysteine Methyltransferase Gene mmuM and its Distribution in Clinical Pathogens. International Journal of Biological Sciences. 2015; 11(1): 11–21. 10.7150/ijbs.10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klepeis JL, Floudas CA. Analysis and prediction of loop segments in protein structures. Computers and Chemical Engineering. 2005; 29(3): 423–36. [Google Scholar]
- 17.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server forprofile–profile sequence alignments. Nucleic Acids Res. 2005; 33 (Web Server issue): W284–8. 10.1093/nar/gki418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambrish R, Alper K, Yang Z. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols (Print). 2010; 5(4): 725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski RA. “PDBsum: summaries and analyses of PDB structures,” Nucleic Acids Research. 2001; 29(1): 221–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simner PJ, Zhanel GG, Pitout J, Tailor F, McCracken M, Mulvey MR, et al. Prevalence and characterization of extended-spectrum β-lactamase- and AmpC β-lactamase-producing Escherichia coli: results of the CANWARD 2007–2009 study. Diagn Microbiol Infect Dis. 2011; 69(3): 326–34. 10.1016/j.diagmicrobio.2010.10.029 [DOI] [PubMed] [Google Scholar]
- 21.Cheng Q, Park JT. Substrate Specificity of the AmpG Permease Required for Recycling of Cell Wall Anhydro-Muropeptides. Journal of Bacteriology. 2002; 184(23): 6434–36. 10.1128/JB.184.23.6434-6436.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietz H, Pfeifle D, Wiedemann B. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob Agents Chemother. 1997; 41(10): 2113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev. 2008; 72(2): 211–27. 10.1128/MMBR.00027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt H, Korfmann G, Barth H, Martin HH..The signal transducer encoded by ampG is essential for induction of chromosomal AmpC beta-lactamase in Escherichia coli by beta-lactam antibiotics ‘and 'unspecific' inducers. Microbiology. 1995; 141 (Pt 5): 1085–92. [DOI] [PubMed] [Google Scholar]
- 25.Garcia DL, Dillard JP. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J Bacteriol. 2008; 190(11): 3799–807. 10.1128/JB.01194-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV..Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2009; 191(7): 2012–22. 10.1128/JB.01547-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YW, Cheng WL, Hu RM, Lin YT, Chung TC, Yang TC. AmpN-AmpG operon is essential for expression of L1 and L2 beta-lactamases in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2010; 54(6): 2583–9. 10.1128/AAC.01283-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaback HR. Structure and mechanism of the lactose permease. C R Biol. 2005; 328(6): 557–67. 10.1016/j.crvi.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 29.Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001; 2(8): 610–20. 10.1038/35085077 [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957; 180(4577): 134–6. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook JE. Molecular cloning: a laboratory manual, p.1.21–1.101. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 1989. [Google Scholar]
- 32.Stover CK.Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature.2000; 406:959–64. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- 33.West SE. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene.1994; 148:81–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.