Abstract
Klotho functions as an aging suppressor, which, in mice, extends lifespan when overexpressed and accelerates development of aging-like phenotypes when disrupted. Klotho is mainly expressed in brain and kidney and is secreted into the serum and CSF. We have previously shown that Klotho is reduced in brains of old monkeys, rats, and mice. We further reported the ability of Klotho to enhance oligodendrocyte differentiation and myelination. Here, we examined the signaling pathways induced by Klotho in MO3.13, a human oligodendrocytic hybrid cell line. We show that exogenous Klotho affects the ERK and Akt signaling pathways, decreases the proliferative abilities and enhances differentiation of MO3.13 cells. Furthermore, microarray analysis of Klotho-treated MO3.13 cells reveals a massive change in gene expression with 80 % of the differentially expressed genes being downregulated. Using gene set enrichment analysis, we predicted potential transcription factors involved in regulating Klotho-treated MO3.13 cells and found that these cells are highly enriched in the gene sets, that are similarly observed in cancer, cardiovascular disease, stress, aging, and hormone-related chemical and genetic perturbations. Since Klotho is downregulated in all brain tumors tested to date, enhancing Klotho has therapeutic potential for treating brain and other malignancies.
Keywords: Aging, Cancer, Myelin, Signaling pathways, ERK, Apoptosis, Glia
Introduction
The anti-aging protein Klotho was named after the goddess who spins the thread of life (Kuro-o et al. 1997). The absence of Klotho shortens lifespan in both mice and Caenorhabditis elegans (Chateau et al. 2010; Kuro-o et al. 1997). In mice, knockout of Klotho induces a dramatic phenotype recapitulating many of the disorders commonly associated with human aging (Kuro-o et al. 1997), including vascular calcification, infertility, emphysema, osteoporosis, skin atrophy and hair loss, thymic involution, osteopenia, motor neuron degeneration, and cognitive impairment (Kuro-o 2009). In contrast, overexpression of Klotho in mice extends lifespan ~20–30 % and suppresses insulin signaling (Kurosu et al. 2005). We found that Klotho is significantly downregulated in the aged brain (Duce et al. 2008) likely due to the methylation of its promoter (King et al. 2012b). Interestingly, the Klotho promoter has also been found to be methylated in tumors (Gan et al. 2011; Lee et al. 2010; Pan et al. 2011; Rubinek et al. 2012; Wang et al. 2011), which would explain Klotho downregulation in many cancers. Further, restoration of Klotho results in inhibition of cancer cells growth in many cancers (Xie et al. 2013; Shu et al. 2013; Chen et al. 2012; Chang et al. 2012).
Klotho is a type I transmembrane protein which is mainly expressed in brain, kidney, and reproductive organs (Masuda et al. 2005) and exists also in a shed form that is detectable in serum and CSF (Chen et al. 2007; Matsumura et al. 1998). Klotho has pleiotropic functions which include regulation of FGF23 signaling, suppression of the insulin/insulin-like growth factor 1 (IGF-1) signaling, regulation of calcium homeostasis and nitric oxide production, suppression of Wnt signaling and oxidative stress, and inhibition of cancer development. For extensive review on Klotho, see German et al. (2012).
Klotho is a potential tumor suppressor and acts as an inhibitor of the IGF-1 pathway and activator of the FGF pathway in human breast cancer (Wolf et al. 2008). Klotho expression is downregulated in pancreatic adenocarcinoma. Overexpression of Klotho, or treatment with soluble Klotho, reduces growth of pancreatic cancer cells in vitro and in vivo (Abramovitz et al. 2011). Klotho expression also is associated with epithelial ovarian cancer progression, and it may serve as an independent marker for cancer prognosis (Lu et al. 2008). In addition, Klotho inhibits transforming growth factor-β1 (TGF-β1) signaling and suppresses renal fibrosis and cancer metastasis in mice (Doi et al. 2011). Other than TGF-β1 signaling, secreted Klotho has been shown to inhibit Wnt and IGF-1 signaling, which are pathways that are known to promote endothelial to mesenchymal transition (EMT). Thus, secreted Klotho may function as an endogenous anti-EMT factor by inhibiting multiple growth factor signaling pathways such as TGF-β1, Wnt, and IGF-1 (Doi et al. 2011).
We have previously demonstrated a novel function of Klotho in oligodendrocyte maturation and developmental myelination of the CNS (Chen et al. 2013). Klotho may function as a humoral factor secreted by neurons or choroid plexus to promote myelination in neurodevelopment, and have a regulatory role in maintaining or supporting oligodendrocyte and oligodendrocyte precursor cell (OPC) function in the adult CNS (Chen et al. 2013). To further understand the molecular mechanisms driving Klotho’s effects as a differentiation factor and a tumor suppressor in a CNS cell type, we performed a cDNA microarray study on cultured human oligodendrocytic hybrid MO3.13 cells (McLaurin et al. 1995) with or without the addition of the shed form of Klotho. We found that Klotho acts as a suppressor for numerous pathways in these cells. The gene signature profiles of Klotho-treated MO3.13 cells were similar to those induced by chemical and genetic perturbations seen in cancer, cardiovascular disease, stress, aging, and hormone-related treatments, suggesting that Klotho may be an inhibitory part of a common pathway responsible for the development of age-related disorders in humans.
Materials and Methods
Materials
The recombinant mouse Klotho protein containing the extracellular domain of mouse Klotho (Ala 35- Lys 982) was from R&D Systems (Minneapolis, MN, USA). All other chemicals were from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise specified.
Cell Culture and Protein Sample Collection
MO3.13 cells (McLaurin et al. 1995) were maintained in Dulbecco’s modified Eagle medium supplemented with 10 % fetal bovine serum and antibiotics at 37 °C and 5 % CO2 on 6-well plates (BD Falcon, San Jose, CA, USA). When cells reached 70–80 % confluence, they were washed twice with HBSS (Invitrogen, Carlsbad, CA, USA) and incubated with serum-free Dulbecco’s modified Eagle medium (DMEM) with or without recombinant mouse Klotho at a concentration of 0.4 μg/mL at 37 °C for the time indicated in the experiments. The medium was collected and centrifuged at 16,000×g for 1 min to remove detached cells. The cells on the plate were washed twice with ice-cold phosphate buffered saline (PBS) and lysed with RIPA buffer (150 mM NaCl, 1 % Triton X-100, 0.5 % deoxycholate, 0.1 % sodium dodecyl sulfate (SDS), 50 mM Tris, pH 7.5) containing protease and phosphatase inhibitor cocktails (Roche). The cell lysate was centrifuged at 16,000×g for 15 min, and the supernatant was collected for SDS-polyacrylamide gel electrophoresis (PAGE). For MO3.13 cells differentiation studies, cultures were maintained in high-glucose DMEM OPC culture medium (4 mML-glutamine, 1 mM sodium pyruvate, 0.1 % BSA, 50 μg/mL insulin, 30 nM sodium selenite, 10 nM D-biotin, and 10 nM hydrocortisone) with or without recombinant Klotho at 0.4 μg/mL for 7 days. The cell lysates were collected as described above for SDS-PAGE.
Western Blotting
Protein concentrations were measured using the Micro BCA Protein Assay Reagent Kit (Pierce, Rockford, IL, USA) according to the manufacturer’s protocol. For SDS-PAGE, cell lysates containing the same amount of total protein were boiled for 5 min and loaded on 4–20 % pre-cast Tris-HCl gels (Bio-Rad, Hercules, CA, USA). Proteins were transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). All antibodies were diluted in TBST (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1 % (v/v) Tween 20) containing 5 % (w/v) nonfat dry milk (Carnation, Glendale, CA, USA). Secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse, anti-rat, or anti-rabbit (1:5,000; Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA). Enhanced chemiluminescence (ECL) was detected using Immobilon Western Chemiluminescent Substrate (Millipore). Autoradiography was done using Kodak Scientific Imaging Film X-OMAT™ AR (Eastman Kodak, Rochester, NY, USA).
The antibodies for STAT3, p-STAT3, and the Akt and ERK pathways were from the phospho Akt and ERK pathway kit (Cell Signaling, Danvers, MA, USA) and were used according to the manufacturer’s protocol. For protein phosphorylation, we used the antibodies against phosphorylated tyrosine (p-Tyr) monoclonal antibody (1:1,000; Cell Signaling), serine and threonine (p-Ser/Thr) monoclonal antibody (1:1,000; BD Transduction Laboratories, San Jose, CA, USA), serine (p-Ser) monoclonal antibody (clone 4A3, and 1:1,000, Biomol, Hamburg, Germany), and p-Ser polyclonal antibody (#61-8100, 1:1,000; Zymed–Life Technologies, Grand Island, NY, USA). For the proliferation assay, we used the anti-PCNA clone PC10 (1:1,000; Upstate, Temecula, CA, USA) and anti-GAPDH monoclonal antibody (1:2,000; Santa Cruz Biotechnology, Dallas, TX, USA) as loading control. For MO3.13 cell differentiation studies, the primary antibodies used were anti-MAG mouse monoclonal antibody (clone B11F7, 1:1,000), anti-CNP mouse monoclonal antibody (1:1,000, Sigma-Aldrich), anti-OSP rabbit polyclonal antibody (1:1,000, Abcam), and anti-PLP mouse monoclonal antibody (1:1000, Millipore).
MTT Cell Proliferation Assay
MO3.13 cells were seeded into 96-well plates in 100 μL DMEM with 10 % fetal bovine serum (FBS) at 4,000 cells per well. After 24 h, the medium was replaced with fresh DMEM with 10 % FBS or serum-free DMEM and cells were treated with recombinant Klotho at 0.4 μg/mL for 48 h. Following treatment, 15 μL of 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye (Promega) were added to cells, incubated for 3 h at 37 °C, then quenched with 100 μL of solubilization solution from the Promega kit for 30 min at room temperature. Absorbance was read at 560 nm.
Crystal Violet Staining
The staining was performed as described previously (Chen et al. 2013). Cells were seeded onto 12-well plates in DMEM with 10 % FBS at 15,000 cells per well. After 24 h, cells were treated with recombinant Klotho at 0.4 μg/mL for 24, and 48 h. Medium was removed from cells, cells were washed with PBS and fixed with 70 % ethanol. Cells were then incubated with 500 μL of 1 % Crystal Violet solution (Sigma-Aldrich, C-3886) at room temperature for 30 min, and then washed thoroughly with water and air dried. The dye was then solubilized with 70 % ethanol, plates were shaken for 30 min at room temperature, and the absorbance read at 560 nm; reference was at 750 nm.
Apoptosis Assay
The MO3.13 cells were seeded into 96-well plates in 100 μL DMEM with 10 % FBS to reach 70–80 % confluence on day of treatment. After 24 h, the medium was replaced with fresh DMEM with 10 % FBS or serum-free DMEM and cells were treated with recombinant Klotho at 0.4 μg/mL for 48 h. Following treatment, 50 μL of caspase-glo 3/7 reagent (Promega) was added to cells, which were then incubated for 30 min and luminescence was read. Each condition was used in four repeats.
Microarray Study
All microarray experiments were performed at Boston University Microarray Resource Facility exactly as described in GeneChip® Whole Transcript (WT) Sense Target Labeling Assay Manual (Affymetrix, Santa Clara, CA, USA). MO3.13 cells were cultured in six-well plates to 70–80 % confluency. The cells were treated with or without recombinant mouse Klotho for 2.5 h, and the total RNA (1 μg) was isolated using QIAGEN’s RNeasy kit (Qiagen, Valencia, CA, USA) and sample integrity was verified using RNA 6000 Nano Assay RNA chips run in Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The labeled fragmented cDNA (~5 μg) was hybridized to the Exon Arrays for 16 h in GeneChip Hybridization oven 640 at 45 °C with rotation (60 rpm). The hybridized samples were washed and stained using Affymetrix fluidics station 450. The first stain with streptavidin-R-phycoerythrin was followed by signal amplification using a biotinylated goat antistreptavidin antibody followed by another Streptavidin Phycoerythrin staining (Hybridization, Washing and Staining Kit, Affymetrix, Santa Clara, CA, USA). Microarrays were immediately scanned using Affymetrix GeneArray Scanner 3000 7G Plus (Affymetrix). The resulting CEL files were summarized using Affymetrix Expression Console (version 1.1). Expression level estimates for each probe set were derived using IterPLIER (Affymetrix). Affymetric Human Exon 1.0 ST Array mRNA data from each individual sample have been deposited with the NCBI Gene Expression Omnibus (http://wwww/ncbi.nlm.nih.gov/geo/ under accession number GSE51536).
Differential gene expression was detected using CyberT (Baldi and Long 2001). Genes that vary significantly between the two experimental conditions were identified using a t test. A false-discovery rate (FDR) for each of the resulting p values was calculated to account for multiple hypotheses testing and a critical FDR value of 5 % was used to construct the list of genes that are differentially expressed between the two conditions. The 100 probe sets with p values <0.05 were selected for further analysis. Ingenuity Pathway Analysis (Ingenuity Systems) was used to explore biological characteristics that were enriched among these genes as well as previously reported interactions between them.
qRT-PCR
Total RNA was isolated as described above. A reverse transcription was performed with 2 μg of total RNA from each sample. Primers for selected genes were designed by SABiosciences (customized quantitative reverse transcriptase PCR (qRT-PCR) array with five controls as follows: actin, beta (ACTB), beta-2-microglobulin (B2M), ribosomal protein L13a (RPL13A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and genomic DNA contamination control). The qRT-PCR experiments were performed with Brilliant SYBR green (Stratagene) detection on a Bio-rad C1000 Thermal Cycler. The whole cDNA from 2 μg of total RNA was used for one 96 plate. Triplicates of 25 μL reactions containing primer and cDNA template were used for quantitation. A PCR reaction was carried out as follows: 1 cycle of 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min. This was followed by a dissociation curve beginning at 55 °C and increasing by 0.2 °C every 3 s, with SYBR green fluorescence measured at every interval. Differential gene expression was detected using CyberT (Baldi and Long 2001) which is a Bayesian approach to the traditional Student’s t test. Because of the small sample size, we were unable to identify significantly differentially expressed genes after correcting for the false discovery rate, so we selected the 100 probe sets with smallest CyberT p values (which ranged from 7.87E-06 to 0.018) for further analysis. Ingenuity Pathway Analysis (Ingenuity Systems) was used to explore biological characteristics that were enriched among these genes as well as previously reported interactions between them.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states. In the current study, GSEA was performed according to Subramanian et al. (2005). The Molecular Signatures Database (MSigDB) is a collection of annotated gene sets for use with GSEA software. In total, 2392 curated gene sets were explored from the MSigDB (version 3.1), which include 639 canonical pathways (CP) gene sets, 1,186 chemical and genetic perturbation (CGP) gene sets, and 500 transcription factors targets (TFT) sets. We downloaded the GSEA software and the MSigDB gene sets from the GSEA website (http://www.broadinstitute.org/gsea/msigdb/index.jsp). Klotho-treated MO3.13 cells versus untreated control MO3.13 cells gene expression profiles were ranked based on p value. The ranked gene list was utilized as input for the CP gene sets, CGP gene sets, and TFT gene sets, respectively. The primary result of the gene set enrichment analysis is the enrichment score (ES), which reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. GSEA calculates the ES by walking down the ranked list of genes, increasing a running sum statistic when a gene is in the gene set, and decreasing it when it is not. The magnitude of the increment depends on the correlation of the gene with the phenotype. The ES is the maximum deviation from zero encountered in walking the list. A positive ES indicates gene set enrichment at the top of the ranked list; a negative ES indicates gene set enrichment at the bottom of the ranked list. While, the normalized enrichment score (NES) is the primary statistic for examining gene set enrichment results. By normalizing the enrichment score, GSEA accounts for differences in gene set size and in correlations between gene sets and the expression dataset; therefore, the NES can be used to compare analysis results across gene sets. The FDR is the estimated probability that a gene set with a given NES represents a false-positive finding. For example, an FDR of 25 % indicates that the result is likely to be valid three out of four times. The GSEA analysis report highlights enrichment gene sets with an FDR of less than 25 % as those most likely to generate interesting hypotheses and drive further research, but provides analysis results for all analyzed gene sets. In general, given the lack of coherence in most expression datasets and the relatively small number of gene sets being analyzed, an FDR cutoff of 25 % is appropriate. In the current study, the significant gene sets were selected based on the NES and FDR, accordingly.
Luciferase Assay
The YY1 luciferase reporter plasmid (OC-luc) was kindly provided by Dr. Janet Stein (University of Massachusetts Medical School, Worcester, MA, USA), and Pax3 reporter plasmid (pluc-TKCD-19) was kindly provided by Dr. Frank Rauscher III (Wistar Institute, Philadelphia, PA, USA). The NFAT/AP-1 3x luc plasmid was from Addgene (Cambridge, MA, USA). All other reporter plasmids were from SABiosciences (Qiagen). MO3.13 cells were plated into 96-well plates at a density of 0.65×106 cells/plate for transfection on the following day. Two hundred nanograms of luciferase reporter DNA (with 5 ng Renilla luciferase) were transfected into cells using Lipofectamine 2000 (Invitrogen), and cells were treated with or without recombinant mouse Klotho in medium indicated in the experiments. Twenty hours post-transfection, cells were assayed for luciferase activity using the Dual-Luciferase system (Promega) as described (Oh et al. 2010).
Statistical Analysis
For microarray study, genes that vary significantly between the two experimental conditions were identified using the traditional Student’s t test. An FDR for each of the resulting p values was calculated to account for multiple hypotheses testing and a critical FDR value of 5 % was used to construct the list of genes that are differentially expressed between the two conditions. For qRT-PCR data analysis, differential gene expression was detected using CyberT which is a Bayesian approach to the traditional Student’s t test. For GSEA analysis, Klotho-treated versus untreated control gene expression profiles were ranked based on p value. We then use the ES, the NES, and FDR to analyze the data as described above in the GSEA section. For western blotting, MTT cell proliferation assay, Crystal Violet staining, apoptosis assay, and luciferase assay, the significance was calculated using the traditional Student’s t test.
Results
Phosphorylation Analysis of Klotho-Treated MO3.13 Cells
To investigate whether cultured MO3.13 cells respond to Klotho, we treated the cells with Klotho and protein phosphorylation was assayed by western blot using antibodies to p-Tyr, p-Ser/Thr, and p-Ser. Treatment of MO3.13 oligodendroglioma cells with exogenous Klotho results in changes in protein phosphorylation with some proteins having a reduced phosphorylation, while others an increased phosphorylation (Fig. 1a) suggesting that the cells contain receptor(s) responsive to Klotho, and that signal is transduced into the cells. Endogenously expressed Klotho protein was not detectable in MO3.13 cells by western blot analysis similarly to other cancer cells (Wolf et al. 2008).
Fig. 1.
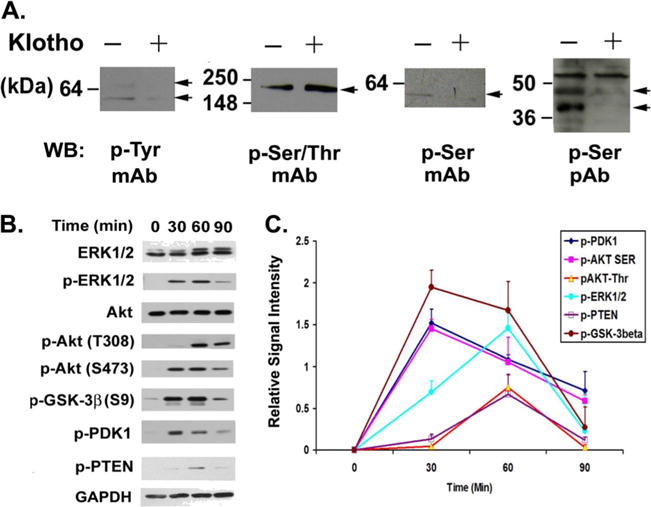
Phosphorylation kinetic analysis of Klotho-treated MO3.13 cells shows involvement of ERK and AKT pathways. A Protein phosphorylation analysis of Klotho-treated MO3.13 cells. MO3.13 cells were treated with Klotho for 2.5 h and the cell lysates were analyzed by SDS-PAGE and western blotting for protein phosphorylation using the indicated monoclonal or polyclonal antibodies against phosphorylated tyrosine (p-Tyr), serine/threonine (p-Ser/Thr), or serine (p-Ser) with (+) or without (−) Klotho treatment. B MO3.13 cells were treated with Klotho for the durations indicated and the cell lysates were analyzed by SDS-PAGE and western blotting for protein phosphorylation of the indicated proteins. C Densitometry of the kinetics of the signal intensity of the bands in A. Results are the average of three to five independent experiments. Error bars indicate standard deviation
Klotho has been reported to affect intracellular signaling through Akt and ERK1/2 pathways in HEK293 and breast cancer cells (Kurosu et al. 2006; Wolf et al. 2008). To examine the potential signaling pathways induced in Klotho-treated MO3.13 cells, we analyzed the kinetics of protein phosphorylation from 0 to 90 min in the ERK1/2 and Akt pathways. Western blot results indicate that proteins in the Akt pathway including PTEN (Ser380), PDK1 (Ser241), Akt, and GSK-3β (Ser9) are phosphorylated upon Klotho treatment (Fig. 1b). Note that the phosphorylation of Akt at S473 occurs at the 30-min timepoint before the phosphorylation of Akt T308 (Fig. 1b, c), consistent with the report that S473 phosphorylation precedes and promotes T308 phosphorylation (Scheid et al. 2002). ERK1/2 proteins are also phosphorylated in response to Klotho treatment. The peak phosphorylation of Akt and PDK1 is at 30 min, while the maximal phosphorylation of ERK1/2, PTEN, and GSK-3β is at 60 min (Fig. 1c). These results suggest that the Akt and ERK/12 pathways are both involved in Klotho-induced signaling in MO3.13 cells.
Microarray Analysis of Klotho-Treated Mo3.13 Cells
To decipher the downstream signaling pathways induced by Klotho on MO3.13 cells, we performed a microarray analysis using RNA from cultured MO3.13 cells treated with or without Klotho for 2.5 h. We pursued MO3.13 rather than primary oligodendrocytes for the microarray study because the diversity of maturational stages of primary cells would likely increase the variability of the microarray results. Also, contamination with other glia would complicate the analysis compared to MO3.13 cells, which are a cell line. We also wanted to study the anticancer activity of Klotho and chose the MO3.13 cells for this purpose since it is a oligodendrocytic hybrid cell line. We used the Affymetrix human Exon 1.0 ST arrays, which provide the most comprehensive coverage of the genome. Of the top 100 differentially expressed genes, 80 % were downregulated by Klotho treatment (Fig. 2a). Ingenuity Pathway Analysis of the top 100 genes showed the networking of Akt, ERK, MAP kinases, and NFκB pathways (Fig. 2b). Two additional networks are shown in Supplemental material Fig. S1a and S1b.
Fig. 2.
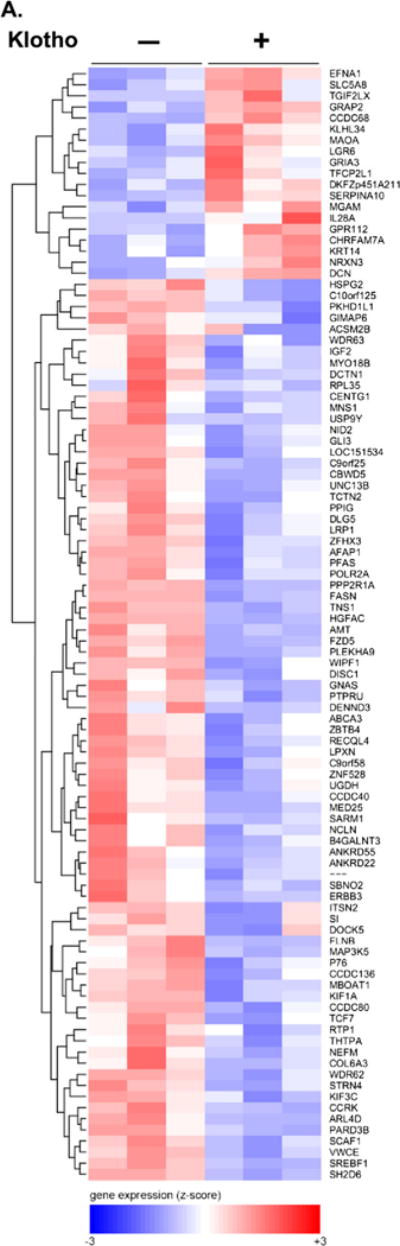
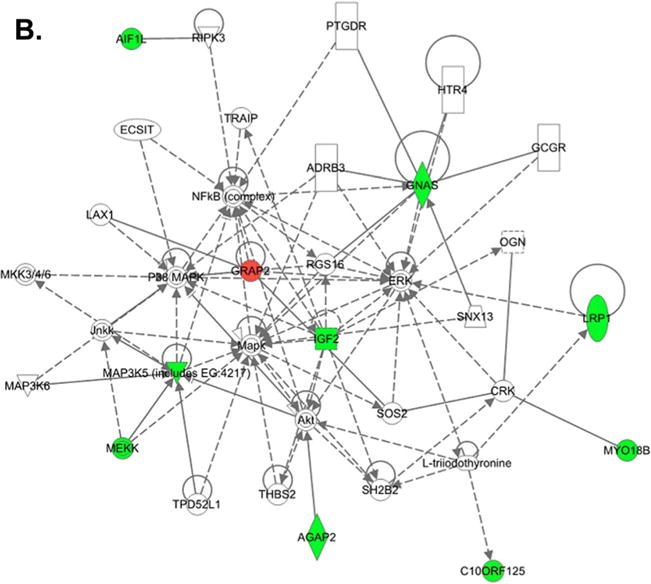
Microarray analysis of Klotho-treated MO3.13 cells. a Cluster heat map analysis of the top 100 regulated genes of Klotho treated (+) and untreated (−) MO3.13 cells. Note that 80 % of the top genes were downregulated by Klotho treatment. b Network analysis of the genes regulated by Klotho-treated MO3.13 cells shows the networking of Akt, ERK, MAP kinases, and NFκB pathways. Green downregulated genes, red upregulated genes
qRT-PCR Confirmation of Microarray Data
To confirm the microarray results, we performed qRT-PCR analysis of 91 genes (see Supplemental material Table 1S) from the top 100 regulated genes using RNA from cultured MO3.13 cells treated with Klotho for 1, 2.5, and 3.5 h. The real-time qRT-PCR confirmation of the 91 regulated gene sets revealed that 79 % of the upregulated genes, and 90 % of the downregulated genes were consistent using both methods (Supplemental material Table 1S, at 2.5 h timepoint). Figure 3 shows the selected qRT-PCR results of three independent comparisons of RNA from Klotho-treated and untreated MO3.13 cells. Complete qPCR results at three different timepoints are in Supplemental materials Table 1S. These results demonstrate that the qRT-PCR analysis of the chosen regulated genes is consistent with and confirms the results obtained from the microarray experiment.
Fig. 3.
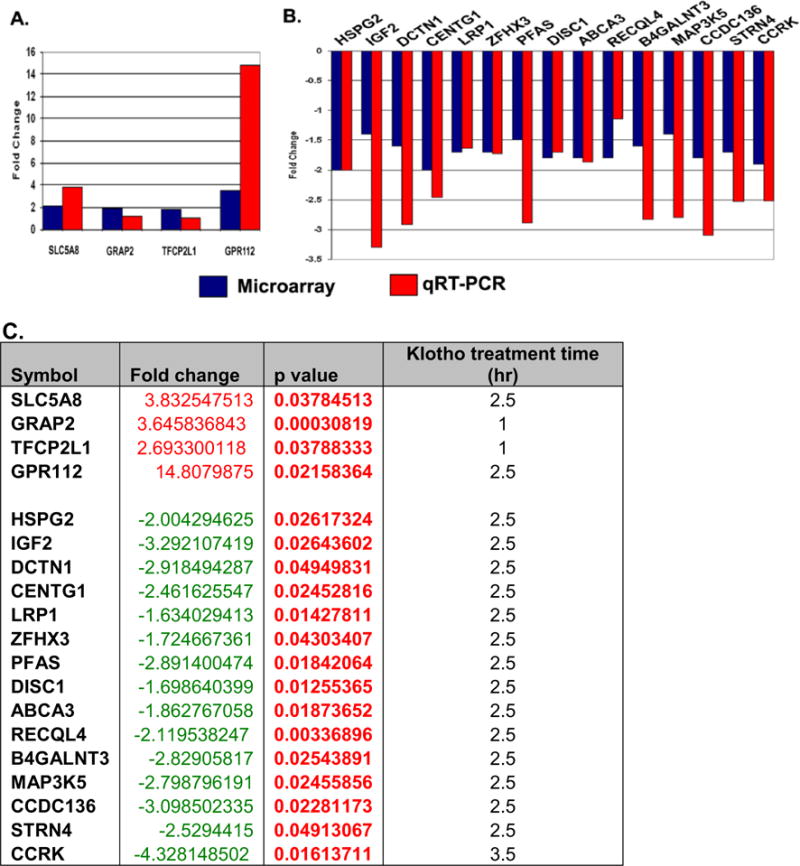
Real-time RT-PCR confirmation of microarray data from Klotho-treated MO3.13 cells. qRT-PCR confirmation of the regulated gene sets from microarray data of Klotho-treated MO3 cells. Ninety-one regulated genes from the microarray study were tested for validation by qRT-PCR. Seventy-nine percent of the upregulated (a) genes and 90 % of the downregulated (b) genes were consistent using both methods. All results are statistically significant at p<0.05. The fold change, p value, and Klotho treatment time for the regulated genes are listed in c
Cell Proliferation, Apoptosis, and Differentiation Analyses of Klotho-Treated MO3.13 Cells
To determine whether Klotho has antiproliferative effects on MO3.13 cells, we used two independent methods: an MTT cell proliferation assay and Crystal Violet staining. Using the MTT assay, cells seeded in 10 % FBS DMEM and treated with Klotho are about 15 % less confluent compared to untreated cells in the same medium condition (Fig. 4a). To enhance this effect by minimizing the growth factors normally found in FBS, cells were treated with Klotho in serum free DMEM as well. These cells are only 52 % as confluent compared to the untreated cells. Crystal Violet staining is another method that reflects the number of viable cells. The inhibitory effect of Klotho on MO3.13 cells after 48 h in serum-free DMEM was demonstrated by Crystal Violet staining (Fig. 4b) and supports the results obtained by the MTT assay also measured after 48 h. Similar results were obtained by western blot analysis using the cell proliferation marker PCNA (Fig. 4c, d). Klotho’s inhibition of proliferation was more pronounced in the serum-free conditions. Using the caspase-glo luminescence assay to measure apoptosis in Klotho-treated cells, Klotho significantly increased the amount of apoptosis by 6 % in cells seeded in 10 % FBS and by 26.5 % in cells growing in serum-free medium (p<0.05) (Fig. 4e).
Fig. 4.
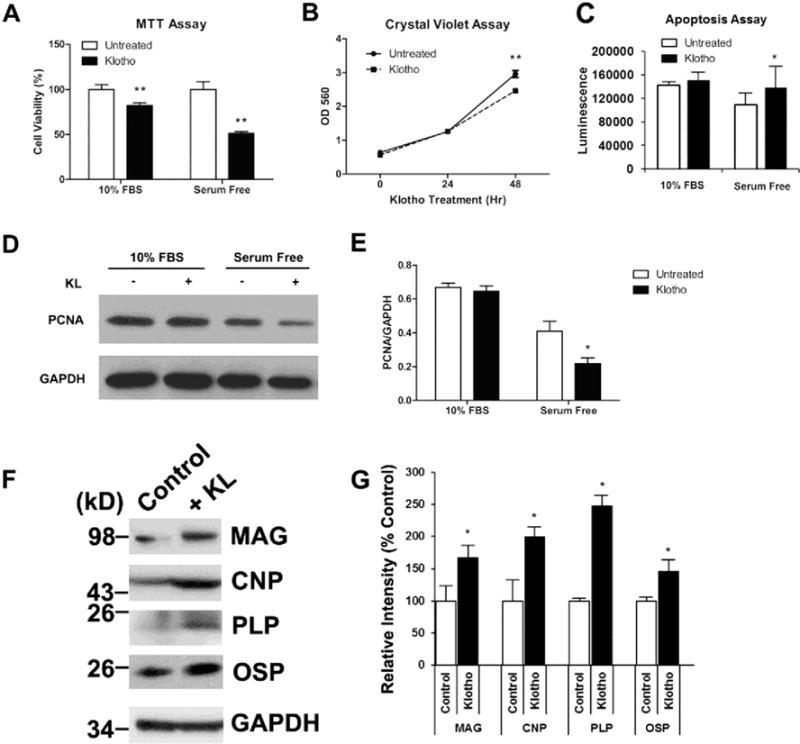
Klotho inhibits MO3.13 cell proliferation, increases apoptosis, and enhances differentiation. a MO3.13 cells were treated with Klotho for 48 h in either DMEM containing 10 % FBS or serum-free DMEM and assayed for cell proliferation using the MTT assay. Cells treated with Klotho are 15 and 48 % less confluent when grown in DMEM containing 10 % FBS or serum-free DMEM, respectively. b Cells treated with Klotho for 48 h in DMEM containing 10 % FBS show a significant decrease in cell number as measured by the Crystal Violet assay. *p< 0.001 indicate statistical significance by t test. Error bars indicate standard deviation. Results are from three to five independent experiments. c MO3.13 cells were treated with the same conditions as in a for 48 h and assayed using the cell apoptosis assay. Cells treated with Klotho have a significant increase in apoptosis when grown in serum-free DMEM. d Western blot analysis of the same experiments in a using an antibody against the cell proliferation marker PCNA with GAPDH as loading control. e Statistical analysis of densitometric results from d. *p<0.05 indicate statistical significance by t test. Results from a–d represent the average of three to five independent experiments. f MO3.13 cells were treated with Klotho for 7 days in DMEM OPC culture medium as described in “Materials and Methods” section. The cell lysates were collected for SDS-PAGE using antibodies against mature oligodendrocyte markers indicated, with GAPDH as control. g Statistical analysis of densitometric results from f. *p<0.05 indicate statistical significance by t test. Results are from three independent experiments
To determine whether Klotho has an effect on MO3.13 cells differentiation, as we have observed in OPCs (Chen et al. 2013), we treated MO3.13 with or without Klotho for 1 week in medium that can promote rat primary oligodendrocyte differentiation according to Buntinx et al. (2003) and Boscia et al. (2012) and our previous protocol (Chen et al. 2013). The results showed that Klotho enhances in MO3.13 cells the expression of maturation markers by two- to threefold after 7 days but not after 3 days of treatment. The oligodendrocyte maturation markers tested included MAG, CNP, PLP, and OSP (Fig. 4f, g). MBP was not detectable by WB in MO3.13 cells at up to 7 days in culture. Taken together, these results suggest that Klotho inhibits the proliferation, increases apoptosis, and enhances differentiation of MO3.13 cells.
Systems Biological Approach to Identify Potential Transcriptional Factors Involved in Klotho Regulation of MO3.13 Cells
We found that Akt and ERK1/2 pathways are involved in Klotho signaling of MO3.13 cells and primary rat oligodendrocytes (Chen et al. 2013). We then applied the knowledge-based approach—GSEA (Subramanian et al. 2005)—to identify potential transcription factors involved in Klotho regulation of MO3.13 cells. The GSEA method was used to search 500 transcription factor gene sets using microarray profile data of the significant differential signature genes from Klotho-treated MO3.13 cells. The result from the GSEA analysis predicted the following transcription factors as potential transcriptional regulators of the Klotho-treated MO3.13 cells: AP-1, AR, CEBPB, E2F, E2F1, EVI1, GATA, GATA1, GR, LXR, MYB, NFAT, NFκB, Oct-1, Pax3, PR, SRY, STAT3, STAT5A, STAT6, and YY1. Among the transcription factors predicted to be involved in oligodendrocyte biology by GSEA analysis, 12 reporters (AP-1, CEBPB, E2F, GATA, GR, LXR, NFAT, NFκB, Pax3, PR, STAT3, and YY1) were available and were used for validation using either Western blotting or the luciferase reporter assay. The results showed that 9 out of 12 transcription factors were regulated by Klotho except GR, PR, and NFAT, which were either unaffected or had no detectable reporter activities in MO3.13 cells. Western blotting confirmed that STAT3 phosphorylation was increased upon Klotho treatment of MO3.13 cells (Fig. 5a). In addition, we examined the transcription factors previously reported to be regulated by Klotho including PPAR, PI3K MAPK/FOXO, SRE MAPK/ERK, p53, Pax6, VDR, Wnt, and SP1 by luciferase reporter system. Altogether, we found that 10 reporter activities were inhibited by Klotho (SP1, NFκB, Wnt, VDR, Pax6, Pax3, E2F, p53, AP1 MAPK/JNK, and C/EBP), and 6 reporter activities were enhanced by Klotho (SRE MAPK/ERK, YY1, PI3K/AKT FOXO, LXR, GATA, and PPAR) (Fig. 5b). The results were consistent in MO3.13 cells and in OPCs. In both cell types, Klotho inhibits C/EBP, AP1 MAPK/JNK, NF-κB, and Pax3, and promotes LXR, STAT3, and SRE MAPK/ERK (Fig. 5b and Chen et al. (2013). Here, we confirm previously described Klotho regulatory pathways and provide new pathways for future investigation into Klotho functions. See Table 2S in Supplemental materials, which lists the transcription factor pathway reporters used in this study. These results demonstrate that GSEA is a useful method in predicting transcription factors involvement using microarray data comparisons and point to additional pathways Klotho may regulate.
Fig. 5.
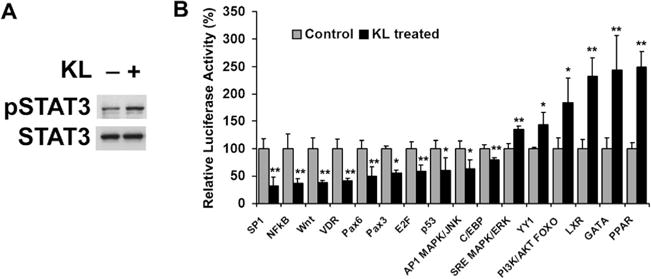
Transcription factors involved in Klotho regulation of MO3.13 cells. a Western blot analysis of STAT3 phosphorylation upon Klotho treatment in MO3.13 cells. b Luciferase assay of MO3.13 cells transfected with Renilla luciferase and luciferase reporters as indicated. Cells were treated with (+black) or without (−gray) Klotho for 24 h and tested using the luciferase assay. The luminescent signals were normalized to Renilla luciferase. Relative luciferase activity was calculated relative to control (no Klotho treatment), which was given the value of 100 %. *p<0.05 and **p<0.01 indicate statistical significance by t test. Error bars indicate standard deviation
Comparison of Gene Signatures from Klotho-Treated MO3.13 Cells with Chemical and Genetic Perturbation Database
CGP gene set enrichment analysis was applied on the significant differential signature genes from the Klotho-treated MO3.13 cells through comparison to the Molecular Signatures database from The Broad Institute (Cambridge, MA, USA). We searched 1,186 chemical and genetic perturbation gene sets, and found that signature genes from Klotho-treated MO3.13 cells were highly enriched in the signature gene sets which were also observed in cancer, cardiovascular disease, stress, aging, and hormone-related chemical and genetic perturbations (Fig. 6a, b), strongly suggesting that Klotho is involved in some common pathways, which also have functional regulations on aging, stress, and cancer.
Fig. 6.
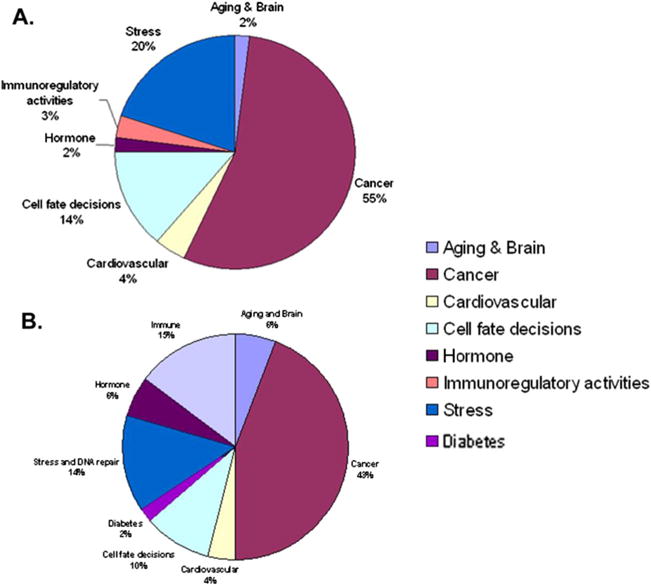
Comparison of microarray data from Klotho-treated MO3.13 cells with chemical and genetic perturbation database (CGP). CGP gene sets enrichment analysis was applied on the significant differential signature genes from the Klotho-treated MO3.13 cells compared to the molecular signature database from the Broad Institute. The top 100 “hits” from the comparison of the upregulated genes (a) and downregulated genes (b) were categorized, and the frequency of each category was plotted. The results of the comparison of the upregulated and downregulated genes with CGP database are shown in Tables 2 and 3 in the Supplemental material
Expression of Klotho in Cancer
To examine whether Klotho expression is reduced in all cancers, we searched the Oncomine database (http://www.oncomine.org). We found that Klotho gene expression is significantly downregulated in almost all cancer types including brain malignancies (Fig. 7), bladder carcinoma, lung, breast, prostate, skin, male germ cell, salivary gland, pancreatic, T-cell, head and neck, and ovarian and bone marrow tumors. The only two exceptions are thyroid and B-cell cancers where Klotho levels were not observed to change.
Fig. 7.
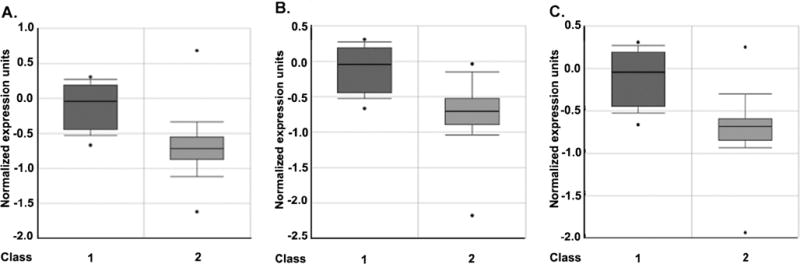
Oncomine database analysis of association of Klotho gene expression with brain cancers. a Klotho is downregulated in glioblastoma. Class 1, normal (n=23); class 2, tumor (n=77); p=2.1×10−9. b Klotho is downregulated in oligodendroglioma. Class 1, normal (n=23); class 2, oligodendroglioma (n=50); p=2.8×10−9. c Klotho is downregulated in astrocytoma. Class 1, normal (n=23); class 2, astrocytoma (n=26); p=2×10−7
Discussion
In this study, we examined the effects of Klotho on MO3.13 cells, a human oligodendrocytic hybrid cell line, and aimed to decipher the mechanisms of Klotho action. Microarray analysis of Klotho-treated MO3.13 cells compared to untreated cells reveals a massive change in gene expression with 80 % of the differentially expressed genes being downregulated. We provide evidence that Klotho affects the ERK and Akt signaling pathways and decreases the proliferative abilities of these cells. Furthermore, using systems biological approach, we predicted potential transcription factors involved in Klotho regulation of MO3.13 cells, and confirmed their involvement using the Luciferase reporter assay.
Using microarray analysis, we determined that the effects of Klotho on MO3.13 cells were mostly inhibitory. This observation is consistent with the known functions of Klotho including inhibition of IGF-1/insulin and Wnt signaling, negative regulation of vitamin D synthesis, and tumor suppression. One of the genes downregulated by Klotho is IGF2 as confirmed by both microarray and qRT-PCR methods in this study. We have previously shown that insulin can stimulate the cleavage and release of the extracellular domain of Klotho (Chen et al. 2007), and IGF2 can also increase Klotho shedding (Tucker, Chen and Abraham, unpublished data). Klotho has been reported to block insulin and IGF-1 receptor phosphorylation of the insulin receptor substrate (Torres et al. 2007; Unger 2006). Our finding suggests that Klotho provides feedback through an unknown process to turn off insulin signaling as well as inhibit IGF2 gene expression. Interestingly, IGF2 has been shown to be involved in the proliferation of medulloblastoma cells, the most frequent childhood brain tumor (Hartmann et al. 2005). Thus, upregulation of Klotho would be expected to have tumor suppressing effects. Klotho also downregulated DISC1, a susceptibility gene for major mental illness. DISC1 plays a role in brain development via modulation of Akt signaling pathway (Kim et al. 2009). The DISC1 gene appeared in the center of a pathway network analysis together with SP1 and TNF (Supplemental material Fig. S1a). Suppression of DISC1 expression reduces neural progenitor proliferation via regulation of GSK-3β signaling, leading to cell cycle exit and differentiation (Mao et al. 2009). Determining the mechanism by which Klotho modulates DISC1 expression may contribute to our understanding of the etiology of psychiatric disorders. Finally, Klotho also downregulated MAP3K5, a biomarker for highly malignant prostate cancer (Pressinotti et al. 2009).
We performed microarray and protein phosphorylation studies in order to decipher the initial effects of Klotho on MO3.13 cells. In both assays, the effects were examined within 3 h. of exogenous addition of Klotho. However, Klotho effects on MO3.13 cells proliferation and apoptosis are only significant after 48 h (Fig. 4a–e). For OPC cell differentiation, at least 3 days were required in order to detect major myelin-related genes (e.g., PLP and MBP) (Chen et al. 2013). However, in both OPCs and MO3.13 cells, we can detect oligodendrocyte maturation markers after 7 days in culture (Chen et al. (2013) and Fig. 4f, g). Klotho inhibits IGF-1 and insulin signaling in cancer cells in culture (Wolf et al. 2008) and in vivo in Klotho overexpressing mice (Kurosu et al. 2005), however, we observed an increase rather than a decrease in both ERK and PI3K pathways. We speculate that initially Klotho activates ERK and Akt pathways but later on other pathways are induced to inhibit proliferation and promote differentiation. We do detect a significant increase in apoptosis after 48 h; however, the majority of cells survived, differentiated and expressed mature oligodendrocyte markers after 1 week in culture. Klotho effects are much greater under serum-free conditions (Fig. 4), when cell proliferation is already inhibited due to the lack of growth factors. Thus, it appears that Klotho’s effects on MO3.13 cells are mainly to promote differentiation. The summary of Klotho’s effects on MO3.13 cells is listed in Table 1. Klotho activates Akt and ERK pathways within 1 h, likely regulates the phosphorylation of other proteins within 2.5 h and downregulates many genes and pathways within 3.5 h. At 24 h, Klotho does not yet affect cell proliferation; however, several transcription factor pathways are up- and downregulated. At 48 h, we observed decreased proliferation and increased apoptosis. At 1 week of treatment, Klotho enhances differentiation. Thus, we propose that the initial effect of Klotho on MO3.13 cells is to activate phosphorylation of Akt via an, as of yet, unidentified receptor, followed by activation of ERK. The downstream effects are the inhibition of numerous signaling pathways, which in turn regulate in the nucleus transcription factors such as LXR, PPAR, FOXO, YY1, and GATA to inhibit Wnt, NFκB, p53, and C/EBP pathways. The results are increased apoptosis, inhibition of proliferation, and enhanced differentiation in the MO3.13 cells. However, in other cells such as MCF7 breast cancer cells (Wolf et al. 2008), apoptosis is the dominant effect. The mechanism by which Klotho can lead to such different effects in different cell types could be due to various cell-specific Klotho receptors that remain to be identified.
Table 1.
Summary of Klotho effects on MO3.13 cells
| Time | Assay/analysis | Effects | Figure or table |
|---|---|---|---|
| 1 h | Western blot, qPCR | Enhancement of Akt and ERK signaling | Fig. 1, Table S1 |
| 2.5 h | Microarray, qPCR, Ingenuity pathway analysis, GSEA | 80 % of the differentially expressed genes downregulated. High enrichment in the gene set that is also observed in cancer, cardiovascular disease, stress, aging and hormone-related chemical and genetic perturbations | Figs. 2, 3, and 6; S1, Table S1, S3, and S4 |
| 3.5 h | qPCR | Similar to 2.5 h | Fig. 3c and Table S1 |
| 24 h | Crystal Violet, Luciferase | Cell proliferation not affected. Inhibited: SP1, NFκB, Wnt, VDR, Pax6, Pax3, E2F, p53, AP1 MAPK/JNK, and C/EBP signaling pathways Activated: SRE MAPK/ERK, YY1, PI3K/AKT FOXO, LXR, GATA, and PPAR signaling pathways | Figs. 4 and 5 |
| 48 h | Western blot, Crystal Violet, MTT, apoptosis | Decrease in proliferative abilities and increased apoptosis | Fig. 4 |
| 1 Week | Western blot | Enhanced differentiation | Fig. 4 |
GSEA gene set enrichment analysis, crystal Violet staining assay for cell proliferation/cell number, Luciferase luciferase transcription factor pathway reporter assay, MTT cell proliferation/cell viability assay, apoptosis Caspase-glo 3/7 assay
Using the knowledge-based GSEA approach, we identified a list of transcription factors potentially involved in Klotho regulation of MO3.13 cells. Among those transcription factors, a paired homeodomain protein (Pax3) is important for development of myelinating glia and the myelination process (Wegner 2000). An inverse correlation was observed between expression of Pax3 and MBP, suggesting that it represses MBP transcription (Kioussi et al. 1995). We found that Klotho inhibited the Pax3 reporter in MO3.13 cells (Fig. 5b) and in rat primary OPC cells and increased MBP expression in OPC (Chen et al. 2013). Our data suggest that Klotho may affect oligodendrocyte differentiation through the Pax3 transcription factor pathway.
The transcription factor STAT3 has been previously reported to be involved in oligodendrocyte differentiation and survival (Dell’Albani et al. 1998; Massa et al. 2000). Platelet-derived growth factor and ciliary neurotrophic factor enhance oligodendrocyte progenitor cells survival through JAK/STAT signaling pathways (Dell’Albani et al. 1998) while the protein tyrosine phosphatase SHP-1 regulates oligodendrocyte differentiation through STAT3 in response to IL-6 family of cytokines (Massa et al. 2000). STAT3 is also important in IL-6 cytokine-mediated brain inflammation (Rummel et al. 2004). In addition, STAT3 has been found to play dual tumor suppressive and oncogenic roles in the pathogenesis of glioblastomas depending on the genetic background of the tumor. The same group also identified a PTEN-regulated STAT3 brain tumor suppressor pathway (de la Iglesia et al. 2008). In the present study, we found that Klotho increased STAT3 and PTEN phosphorylation in MO3.13 cells (Figs. 3 and 5a), as well as induced the differentiation markers MAG, CNP, PLP, and OSP, suggesting a role of Klotho in oligodendrocyte differentiation, protection against inflammation, and brain tumor suppression.
The transcription factor YY1 is essential for oligodendrocyte progenitor differentiation (He et al. 2007). We found that Klotho slightly increased YY1 activity in MO3.13 cells under after 3 h incubation with Klotho (Fig. 5b). Interestingly, it has been shown that the multifunctional regulator YY1 represses 1,25-dihydroxyvitamin D3 (vitamin D)-induced transactivation of the bone tissue-specific osteocalcin gene (Guo et al. 1997). One of the important functions of Klotho is to repress vitamin D biosynthesis. However, whether Klotho regulates vitamin D metabolism through YY1, by sole inhibition of its synthetic enzyme 1,10 alpha hydroxylase, or by a combination of both pathways requires further investigation.
Among the CGP gene sets, we found that the signature genes upregulated by Klotho-treated MO3.13 cells were similar to those signature genes upregulated by the anticancer drug tamoxifen treatment (Table 3 in Supplemental material). Furthermore, a recent observation indicates that Klotho represses p53 (de Oliveira 2006). We found that many of the Klotho upregulated genes are also present in the p53 downregulated gene sets (Table 3 in Supplemental material). Similarly, many cancer-related CGP gene sets are analogous to downregulated genes of Klotho-treated MO3.13 cells (Table 4 in Supplemental material). Klotho has additionally been reported to suppress TNF-α-induced cell adhesion molecules and attenuate NF-κB activation in endothelial cells (Maekawa et al. 2009). We also found NF-κB in the list of transcription factors regulating signature genes by Klotho through GSEA analysis, and Klotho inhibited NF-κB reporter activities in OPCs (Chen et al. 2013). In addition, we found NF-κB, p53, and C/EBP in the pathway network analysis (Fig. 1b and Supplemental material S1b), and all three transcription factors were regulated by Klotho as examined in the Luciferase reporter assay (Fig. 5b). NF-κB is a crucial pronflammatory transcription factor that regulates the expression of over 500 genes involved in cellular transformation, survival, proliferation, invasion, angiogenesis, metastasis, and inflammation (Min et al. 2008). Inhibition of NF-κB activation represents a potential therapeutic strategy for treatment of many diseases (Karin et al. 2004). Thus, our findings are consistent with the role of Klotho as a tumor suppressor. Multiple pathways critical in the development of cancer are also found to be repressed by Klotho in MO3.13 cells, including IGF-1, Wnt, and p53. Dysregulation of Klotho expression would thus be anticipated to create an environment where malignant transformation can occur more easily by weakening normal cellular processes that protect against cancer.
Here, we present evidence that the Akt and ERK1/2 pathways are involved in Klotho signaling in MO3.13 cells. This finding is consistent with our previous findings of Klotho effects in primary OPC cells (Chen et al. 2013). These results suggest that MO3.13 cells contain receptor(s) to Klotho and that receptor tyrosine kinases are likely candidates. At this point, we do not know which receptor(s) is responsible for Klotho’s effects on MO3.13 cells. Whether the effects reported here are through the Wnt signaling receptor or the insulin/IGF and FGF pathways are currently under investigation.
In this study, we found that Klotho may play a dual role in human oligodendroglioma MO3.13 cells—to promote differentiation and to inhibit cancer development. The MO3.13 cell line is an immortal human-human hybrid cell line that expresses phenotypic characteristics of primary oligodendrocytes, and was created by fusing a cancer of skeletal muscle with adult human oligodendrocytes (McLaurin et al. 1995). Thus, it is not surprising that MO3.13 cells respond to Klotho treatment as both oligodendrocyte and cancer cells. The gene profiles of Klotho-treated MO3.13 cells were similar to those induced by chemical and genetic perturbations seen in cancer, cardiovascular disease, stress, aging, and hormone-related treatments, suggesting that Klotho may be involved in a common pathway responsible for the development of age-related disorders in humans. Understanding the functions of Klotho in MO3.13 cells may provide a common strategy in protecting the brain against age-dependent changes such as tumor development, maintenance of myelin integrity (Chen et al. 2013), and neurodegeneration. Furthermore, in a high throughput screen of 150,000 small molecules that would enhance Klotho expression, we have identified a number of promising compounds that elevate Klotho expression at the RNA, protein, and functional levels (Abraham et al. 2012; King et al. 2012a). When optimized, such compounds could be tested in animal models of various cancers including the currently untreatable brain malignancies, multiple sclerosis, and neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank Drs. Yuriy Alekseyev and Marc Lenburg at Boston University Microarray Core Facility for help with the microarray analysis. We thank Chun-Tsin Hsu for assistance with the qRT-PCR experiment and Dr. Christina Khodr for helpful discussions. This work was supported by NIH-NIA grant AG-00001 to CRA, and an Ellison Foundation Award to CC.
Abbreviations
- PCR
Polymerase chain reaction
- DMEM
Dulbecco’s modified Eagle’s Medium
- PBS
Phosphate buffered saline
- FBS
Fetal bovine serum
- BSA
Bovine serum albumin
- SDS-
Sodium dodecyl sulfate polyacrylamide gel
- PAGE
electrophoresis
- MAG
Myelin-associated glycoprotein
- MBP
Myelin basic protein
- CNP
2′,3′cyclic nucleotide-3′-phosphodiesterase
- OPC
Oligodendrocytic precursor cells
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12031-014-0336-1) contains supplementary material, which is available to authorized users.
Contributor Information
Ci-Di Chen, Department of Biochemistry, Boston University School of Medicine, 72 E. Concord Street, K304, Boston, MA 02118, USA.
Hu Li, Email: cabraham@bu.edu, Department Biomedical Engineering, Boston University School of Medicine, Boston, MA, USA; Center for Individualized Medicine, Department of Molecular, Pharmacology and Experimental Therapeutics, Mayo Clinic, Rochester, MN 55905, USA.
Jennifer Liang, Department of Biology, Boston University School of Medicine, Boston, MA, USA.
Kathryn Hixson, Department of Biochemistry, Boston University School of Medicine, 72 E. Concord Street, K304, Boston, MA 02118, USA.
Ella Zeldich, Department of Biochemistry, Boston University School of Medicine, 72 E. Concord Street, K304, Boston, MA 02118, USA.
Carmela R. Abraham, Department of Biochemistry, Boston University School of Medicine, 72 E. Concord Street, K304, Boston, MA 02118, USA Department of Pharmacology and Experimental Therapeutics, Boston University School of Medicine, Boston, MA, USA.
References
- Abraham CR, Chen C, Cuny GD, Glicksman MA, Zeldich E. Small-molecule Klotho enhancers as novel treatment of neurodegeneration. Future Med Chem. 2012;4:1671–1679. doi: 10.4155/fmc.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitz L, Rubinek T, Ligumsky H, Bose S, Barshack I, Avivi C, Kaufman B, Wolf I. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin Cancer Res. 2011;17:4254–4266. doi: 10.1158/1078-0432.CCR-10-2749. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long A. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Boscia F, D’Avanzo C, Pannaccione A, et al. Silencing or knocking out the Na(+)/Ca(2+) exchanger-3 (NCX3) impairs oligodendrocyte differentiation. Cell Death Differ. 2012;19:562–572. doi: 10.1038/cdd.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntinx M, Vanderlocht J, Hellings N, Vandenabeele F, Lambrichts I, Raus J, Ameloot M, Stinissen P, Steels P. Characterization of three human oligodendroglial cell lines as a model to study oligodendrocyte injury: morphology and oligodendrocyte-specific gene expression. J Neurocytol. 2003;32:25–38. doi: 10.1023/a:1027324230923. [DOI] [PubMed] [Google Scholar]
- Chang B, Kim J, Jeong D, et al. Klotho inhibits the capacity of cell migration and invasion in cervical cancer. Oncol Rep. 2012;28:1022–1028. doi: 10.3892/or.2012.1865. [DOI] [PubMed] [Google Scholar]
- Chateau MT, Araiz C, Descamps S, Galas S. Klotho interferes with a novel FGF-signalling pathway and insulin/Igf-like signalling to improve longevity and stress resistance in Caenorhabditis elegans. Aging (Albany NY) 2010;2:567–581. doi: 10.18632/aging.100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci U S A. 2007;104:19796–19801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Ma X, Liu S, Zhao W, Wu J. Inhibition of lung cancer cells growth, motility and induction of apoptosis by Klotho, a novel secreted Wnt antagonist, in a dose-dependent manner. Cancer Biol Ther. 2012;13:1221–1228. doi: 10.4161/cbt.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Sloane JA, Li H, et al. The antiaging protein Klotho enhances oligodendrocyte maturation and myelination of the CNS. J Neurosci: Off J Soc Neurosci. 2013;33:1927–1939. doi: 10.1523/JNEUROSCI.2080-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira RM. Klotho RNAi induces premature senescence of human cells via a p53/p21 dependent pathway. FEBS Lett. 2006;580:5753–5758. doi: 10.1016/j.febslet.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Dell’Albani P, Kahn MA, Cole R, Condorelli DF, Giuffrida-Stella AM, de Vellis J. Oligodendroglial survival factors, PDGF-AA and CNTF, activate similar JAK/STAT signaling pathways. J Neurosci Res. 1998;54:191–205. doi: 10.1002/(SICI)1097-4547(19981015)54:2<191::AID-JNR7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Doi S, Zou Y, Togao O, et al. Klotho inhibits transforming growth factor-beta1 (TGF-beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem. 2011;286:8655–8665. doi: 10.1074/jbc.M110.174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56:106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- Gan LH, Pan J, Chen SJ, Zhong J, Wang LJ. DNA methylation of ZIC1 and KLOTHO gene promoters in colorectal carcinomas and its clinicopathological significance. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2011;40:309–314. doi: 10.3785/j.issn.1008-9292.2011.03.014. [DOI] [PubMed] [Google Scholar]
- German DC, Khobahy I, Pastor J, Kuro OM, Liu X. Nuclear localization of Klotho in brain: an anti-aging protein. Neurobiol Aging. 2012;33(1483):e1425–e1430. doi: 10.1016/j.neurobiolaging.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Aslam F, van Wijnen AJ, et al. YY1 regulates vitamin D receptor/retinoid X receptor mediated transactivation of the vitamin D responsive osteocalcin gene. Proc Natl Acad Sci U S A. 1997;94:121–126. doi: 10.1073/pnas.94.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann W, Koch A, Brune H, et al. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave K, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GD, Chen C, Huang MM, et al. Identification of novel small molecules that elevate Klotho expression. Biochem J. 2012a;441:453–461. doi: 10.1042/BJ20101909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GD, Rosene DL, Abraham CR. Promoter methylation and age-related downregulation of Klotho in rhesus monkey. Age (Dordr) 2012b;34:1405–1419. doi: 10.1007/s11357-011-9315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Gross MK, Gruss P. Pax3: a paired domain gene as a regulator in PNS myelination. Neuron. 1995;15:553–562. doi: 10.1016/0896-6273(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Klotho and aging. Biochim Biophys Acta. 2009;1790:1049–1058. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, et al. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jeong DJ, Kim J, et al. The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Mol Cancer. 2010;9:109. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H. Klotho expression in epithelial ovarian cancer and its association with insulin-like growth factors and disease progression. Cancer Invest. 2008;26:185–192. doi: 10.1080/07357900701638343. [DOI] [PubMed] [Google Scholar]
- Maekawa Y, Ishikawa K, Yasuda O, et al. Klotho suppresses TNF-alpha-induced expression of adhesion molecules in the endothelium and attenuates NF-kappaB activation. Endocrine. 2009;35:341–346. doi: 10.1007/s12020-009-9181-3. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa PT, Saha S, Wu C, Jarosinski KW. Expression and function of the protein tyrosine phosphatase SHP-1 in oligodendrocytes. Glia. 2000;29:376–385. [PubMed] [Google Scholar]
- Masuda H, Chikuda H, Suga T, Kawaguchi H, Kuro-o M. Regulation of multiple ageing-like phenotypes by inducible klotho gene expression in klotho mutant mice. Mech Ageing Dev. 2005;126:1274–1283. doi: 10.1016/j.mad.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun. 1998;242:626–630. doi: 10.1006/bbrc.1997.8019. [DOI] [PubMed] [Google Scholar]
- McLaurin J, Trudel GC, Shaw IT, Antel JP, Cashman NR. A human glial hybrid cell line differentially expressing genes subserving oligodendrocyte and astrocyte phenotype. J Neurobiol. 1995;26:283–293. doi: 10.1002/neu.480260212. [DOI] [PubMed] [Google Scholar]
- Min C, Eddy SF, Sherr DH, Sonenshein GE. NF-kappaB and epithelial to mesenchymal transition of cancer. J Cell Biochem. 2008;104:733–744. doi: 10.1002/jcb.21695. [DOI] [PubMed] [Google Scholar]
- Oh SY, Chen CD, Abraham CR. Cell-type dependent modulation of Notch signaling by the amyloid precursor protein. J Neurochem. 2010;113:262–274. doi: 10.1111/j.1471-4159.2010.06603.x. [DOI] [PubMed] [Google Scholar]
- Pan J, Zhong J, Gan LH, Chen SJ, Jin HC, Wang X, Wang LJ. Klotho, an anti-senescence related gene, is frequently inactivated through promoter hypermethylation in colorectal cancer. Tumour Biol. 2011;32:729–735. doi: 10.1007/s13277-011-0174-5. [DOI] [PubMed] [Google Scholar]
- Pressinotti NC, Klocker H, Schafer G, et al. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low-and high-risk prostate cancer. Mol Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinek T, Shulman M, Israeli S, et al. Epigenetic silencing of the tumor suppressor klotho in human breast cancer. Breast Cancer Res Treat. 2012;133:649–657. doi: 10.1007/s10549-011-1824-4. [DOI] [PubMed] [Google Scholar]
- Rummel C, Hubschle T, Gerstberger R, Roth J. Nuclear translocation of the transcription factor STAT3 in the guinea pig brain during systemic or localized inflammation. J Physiol. 2004;557:671–687. doi: 10.1113/jphysiol.2003.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid MP, Marignani PA, Woodgett JR. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol. 2002;22:6247–6260. doi: 10.1128/MCB.22.17.6247-6260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu G, Xie B, Ren F, Liu DC, Zhou J, Li Q, Chen J, Yuan L. Restoration of klotho expression induces apoptosis and autophagy in hepatocellular carcinoma cells. Cell Oncol (Dordr) 2013;36:121–129. doi: 10.1007/s13402-012-0118-0. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha V, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres PU, Prie D, Molina-Bletry V, Beck L, Silve C, Friedlander G. Klotho: an antiaging protein involved in mineral and vitamin D metabolism. Kidney Int. 2007;71:730–737. doi: 10.1038/sj.ki.5002163. [DOI] [PubMed] [Google Scholar]
- Unger RH. Klotho-induced insulin resistance: a blessing in disguise? Nat Med. 2006;12:56–57. doi: 10.1038/nm0106-56. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang X, Jie P, et al. Klotho is silenced through promoter hypermethylation in gastric cancer. Am J Cancer Res. 2011;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- Wegner M. Transcriptional control in myelinating glia: the basic recipe. Glia. 2000;29:118–123. [PubMed] [Google Scholar]
- Wolf I, Levanon-Cohen S, Bose S, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008 doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- Xie B, Zhou J, Shu G, Liu DC, Chen J, Yuan L. Restoration of klotho gene expression induces apoptosis and autophagy in gastric cancer cells: tumor suppressive role of klotho in gastric cancer. Cancer Cell Int. 2013;13:18. doi: 10.1186/1475-2867-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


