Abstract
Canonical signal transduction via heterotrimeric G proteins is spatiotemporally restricted, i.e. triggered exclusively at the plasma membrane, only by agonist activation of G protein-coupled receptors via a finite process that is terminated within a few hundred milliseconds. Recently, a rapidly emerging paradigm has revealed a non-canonical pathway for activation of heterotrimeric G proteins via the non-receptor guanidine-nucleotide exchange factor, GIV/Girdin. Biochemical, biophysical and functional studies evaluating this pathway have unraveled its unique properties and distinctive spatiotemporal features. As in the case of any new pathway/paradigm, these studies first required an in-depth optimization of tools/techniques and protocols, governed by rationale and fundamentals unique to the pathway, and more specifically to the large multimodular GIV protein. Here we provide the most up-to-date overview of protocols that have generated most of what we know today about non-canonical G protein activation by GIV and its relevance in health and disease.
Keywords: GIV/Girdin, Trimeric G proteins, Immunoblotting, Immunoprecipitation, In cellulo GST-pull down, Immunofluorescence, FRET
Introduction
A vast majority of environmental cues are transmitted to the interior of a eukaryotic cell via a complex network of two major signaling hubs: 1) Receptor tyrosine kinases (RTKs) and 2) heterotrimeric G proteins (henceforth referred to as trimeric G proteins). Canonical signal transduction via trimeric G proteins has been studied extensively and is known to be spatially and temporally restricted i.e. triggered exclusively at the plasma membrane (PM) by agonist activation of G-protein-coupled receptors (GPCRs) via a process that completes within a few hundred milliseconds. Recently, a rapidly emerging body of work has revealed an alternative (non-canonical) pathway for activation of trimeric G proteins by the Gα-protein Interacting Vesicle associated protein (GIV, a.k.a. Girdin; gene name CCDC88A) that has distinctive temporal and spatial features. GIV is the preeminent member of a growing family of proteins, collectively known as guanine nucleotide exchange modulators [GEMs (Ghosh, 2015c)], that binds Gα-subunits and accelerates nucleotide exchange on, i.e., activate Gαi subunits via an evolutionarily conserved short (~ 30 aa) motif. But unlike any other modulator of G proteins, GIV has a unique modular makeup (see Fig 1, top) that allows it to couple to diverse classes of ligand-activated receptors, such as growth factor receptor tyrosine kinases (RTKs), G protein coupled receptors (GPCRs), integrins, toll-like receptors, and the family of TGF-β receptors, many of which are believed to relay signals exclusively via tyrosine-based signals. Published work has revealed that such coupling allows diverse types of stimuli to activate Gαi proteins via GIV, and has established a non-canonical mechanism for convergent and coordinated G protein signaling [reviewed in (Ghosh, 2015a; Ghosh, 2015c; Ghosh, 2016)]. The unique combination of modules and motifs in GIV (see Fig 1, top) which enables it to receive tyrosine-based signal inputs and relay them via G protein intermediates, positioning GIV at the intersection of two of the largest signaling hubs in eukaryotes. Consistent with its ability to integrate signals downstream of multiple receptors, both at the PM and on other subcellular organelles, GIV modulates diverse cellular processes including cell migration, survival, autophagy, secretion, cell polarity, endocytosis, exocytosis and cell adhesion [reviewed in (Aznar et al, 2016b)]. Because it straddles the two signaling hubs that are most frequently targeted for their therapeutic significance, the role of tyrosine-based G-protein signals triggered by GIV has been studied and confirmed in diverse cell systems and disease models, e.g. cancer progression, organ fibrosis, insulin resistance/type II diabetes, vascular injury etc. [reviewed in (Aznar et al, 2016b; Ghosh, 2015c; Ghosh, 2016)], thus making it an interesting target for fundamental as well as translational studies. These studies have not only revealed GIV's pathophysiologic importance (Ghosh, 2016), but also its diagnostic and therapeutic potential in a variety of disease states [reviewed in (Aznar et al, 2016b; Ghosh, 2015a; Ghosh, 2015c)].
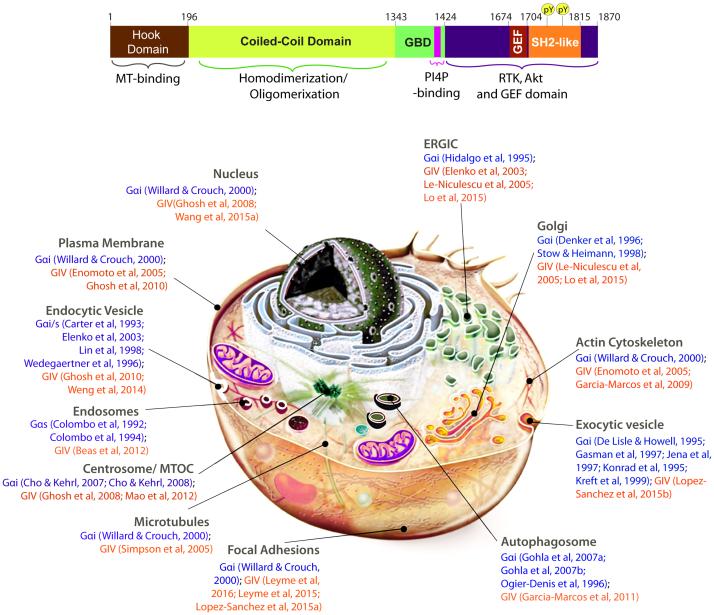
Figure 1. GIV, a multimodular cytosolic protein and GEF for Gαi that localizes to and functions at various subcellular locations.
Top: Bar diagram showing the various known functional modules of full-length GIV. Residue numbers marking the boundaries of each domain are shown. The C-terminal domain features multiple short patches of functional motifs and modules that enable GIV to bind other proteins and transduce signals. Among them, a C-terminally located GEF motif (Garcia-Marcos et al, 2010; Garcia-Marcos et al, 2009) binds and activates Gαi proteins by triggering nucleotide exchange. Just downstream of the GEF motif is a SH2-like domain (Lin et al, 2014) which recognizes and folds upon binding phosphotyrosine ligands on the cytoplasmic tails of multiple RTKs. GIV is also a substrate of multiple TKs (Lin et al, 2011). Phosphorylation of GIV at two tyrosines (pY1764 and pY1798) by multiple receptor and non-receptor protein tyrosine kinases generates two docking sites for SH2-adaptor containing proteins. Bottom: Schematic of a cell with various subcellular organelles and locations where both G proteins and GIV are known to locate. Numbers denote citations that provide evidence for such localizations. Published work demonstrates that GIV and its GEF function is functional at most, if not all of these locations.
However, due to its large size (1870 AA; Mr ≈ 220 kDa), susceptibility to proteolytic degradation, existence of multiple isoforms and localization to multiple subcellular locations, GIV can be technically challenging to work with. This article contains several detailed and optimized protocols to investigate GIV and its cellular functions using various biochemical and biophysical techniques. We provide a comprehensive collection of protocols ranging from an inexpensive transfection method to Forster Resonance Energy Transfer (FRET) assays along with troubleshooting guidelines. We also summarize various siRNA/shRNA sequences used (by us and other groups) to deplete GIV, list antibodies for use in immunoblotting, immunoprecipitation and immunocytochemical studies to observe the subcellular compartmentalization of GIV, provide the sequence of primers and probes for the measurement of GIV mRNA expression, and provide detailed rationale for the design of fusion-tagged constructs of GIV. The comprehensive sets of protocols, information on available reagents and tools, and troubleshooting tips provided here will serve as a one-stop source of valuable information for new researchers from diverse fields interested in studying GIV and its role in cellular processes of their interest.
Strategic Planning: General guidelines for studying GIV’s functions in cells Choice of cell lines for studying GIV's functions in cells
GIV is a ubiquitously expressed protein; however, not every tissue or cell type expresses it at equivalent levels or as a full-length transcript (Le-Niculescu et al, 2005). Among all tissues tested, full-length GIV is found to be predominantly expressed in the brain, testes and ovary and virtually undetectable in liver (Enomoto et al, 2005). Among cell lines, COS-7 express GIV at highest levels (Fig 2A), whereas MCF7 cells have very low levels of GIV (Lopez-Sanchez et al, 2014). Thus, it is important to first assess if the cell model to be studied indeed expresses full-length GIV. Methods for detection of full-length GIV at the mRNA and protein level are discussed in Basic Protocol 2.
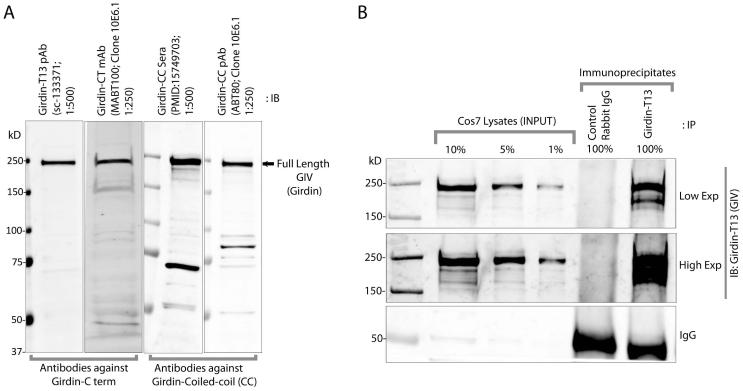
Figure 2. Detection of GIV/Girdin by immunoblotting and immunoprecipitation.
A: Equal aliquots of lysates (~ 60 μg) of COS-7 cells were analyzed for GIV expression by immunoblotting (IB) with a variety of commercially available monoclonal and polyclonal antibodies raised against either the coiled-coil (N-terminal) or C-terminal (CT) domains of GIV at the indicated dilutions. Full-length GIV is detected at ~250 kD by all antibodies. In addition, some coiled-coil antibodies detect a few shorter products in a variety of cell lines, some of which are isoforms of GIV. B. Equal aliquots of lysates (~ 2.0 mg) of COS-7 were incubated with ~1.5 μg control or anti-Girdin T13 (SCBT) antibody and protein A beads. Immune complexes were eluted and analyzed for GIV by IB.
Besides the level of GIV expression, it is also important to take into consideration the levels of accessory signaling components that are essential to assess the full impact of GIV on signal transduction. Because GIV straddles tyrosine-based (growth factor-initiated) and G protein-based signaling mechanisms, it is important to consider whether the cell lines chosen to study GIV's functions are appropriate and acceptable for studying both signaling mechanisms. Such careful consideration will ensure that key readouts which reflect the rich cross-talk between the two mechanisms enabled by GIV are accounted for and analyzed. For example, canonical G protein signaling has been extensively studied in HEK 293 cells; however, studying GIV's roles in these cells risks neglecting the rich crosstalk between RTKs and G protein pathways because they express low endogenous levels of multiple RTKs, including, EGFR and the related ErbB RTKs (Stern et al, 2007) and PDGFR (Freedman et al, 2002). Similarly, COS-7 and MCF-7 cell lines also have low levels of endogenous EGFR (Hatton et al, 2015) and should be avoided when the role of GIV as a signal transducer within the EGFR pathway is being studied. Therefore, while the HEK 293 and COS-7 cell lines are appropriate model systems for overexpressing tagged-proteins for use in in vitro protein-protein interaction assays, they are incomplete systems (when used as is) for evaluating the effect of GIV on signal transduction or cell behavior. We recommend using human cervical cancer cells (HeLa cells) as a model system to study the role of GIV in linking the tyrosine-based signaling pathways (and the prototype RTK, EGFR) and the G protein pathways for the following reasons: (i) they express intermediate levels of GIV and G proteins (i.e., neither too high, nor too low) and rely on this pathway for growth factor-induced migration and proliferation (Garcia-Marcos et al, 2009; Ghosh et al, 2010); (ii) they express physiologic levels of EGFRs (~50,000 EGFRs/cell) (Ley & Ellem, 1992); (iii) they have a complete set of adaptors/intermediates at near physiologic levels to effectively relay signals downstream of the RTK (Capuani et al, 2015); (iv) EGFR trafficking in HeLa cells has been characterized in detail (Ceresa & Bahr, 2006; Dinneen & Ceresa, 2004); (v) they are amenable to genetic manipulation for the generation of stable cell lines, either depleted of endogenous GIV or G proteins, or expressing various GIV/G protein mutants. All of these characteristics make HeLa cells an excellent model for GIV-related studies.
Genetic manipulation of cells
As in the case of most proteins, gene depletion has been used by us and others as the first step in evaluating GIV's role in any given process. Various methods used for GIV depletion are summarized in Table 1. However, because GIV has multiple modules, and serves key functions in virtually every subcellular cellular compartment (Fig 1, bottom), depletion of GIV alone may be associated with defects that are too broad to allow meaningful interpretation of findings. For example, because GIV is essential for vesicular transport from the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) to the Golgi (Lo et al, 2015) as well as exocytosis (Lopez-Sanchez et al, 2015b), failure of protein secretion could reflect a defect in both steps of the process. Expression of domain-deleted mutants is also a risky approach that is likely to provide dominant negative phenotypes that can be too broad to yield meaningful conclusions. For example, deletion of the N-terminal Hook-like domain is expected to not only abolish GIV's ability to bind microtubules (Simpson et al, 2005), but also abolish GIV's ability to interact with membrane-associated active [GTP-bound] Arf1 GTPases (Lo et al, 2015). Similarly, deletion of the C-terminal ~200 aa is likely to abolish a whole host of interactions, including G proteins (Garcia-Marcos et al, 2009), EGFR and other RTKs (Lin et al, 2014), PI3K (Lin et al, 2011), Akt (Enomoto et al, 2005), IRS1 (Ma et al, 2015d), Exo-70 (Lopez-Sanchez et al, 2015b) and actin (Enomoto et al, 2005). Thus, whenever possible, all attempts should be made to identify point mutants that cannot interact with the binding partner of interest, and validated extensively to confirm their specificity.
Table 1.
Methods to deplete GIV
| Name | Sequence | Species | Method of Introduction |
Vector | Citations | Target |
|---|---|---|---|---|---|---|
|
GIV
shRNA 3' UTR |
5'- CCGGGCTTTCATTA CC AGCTCTGAACTCGA GTTCAGAGCTGGTA ATGAAAGCTTTTTTG -3' |
Monkey (COS- 7), Human (HeLa, MDA- MB-231, Hs578T), |
Lentivirus | pLKO.1 | (Lopez-Sanchez et al, 2015b; Ma et al, 2015d) |
Human |
|
GIV
shRNA1 |
5'- GAAGGAGAGGCAAC TGGAT-3' |
Human (MDA- MB-231, MCF-7, HUVEC, BTSC, MCF10A,LN229, SH-SY5Y) Monkey(COS-7, Vero) Rat (PC- 12) |
Lentivirus | pLKO.1-puro, pSIREN-RetroQ, pcPURU6b |
(Bhandari et al, 2015; Enomoto et al, 2009; Gu et al, 2014; Jiang et al, 2008; Kitamura et al, 2008; Leyme et al, 2016; Leyme et al, 2015; Mao et al, 2012; Midde et al, 2015; Muramatsu et al, 2015; Natsume et al, 2012; Ohara et al, 2012) |
Rat, Human, Mouse |
|
GIV
shRNA2 |
5'- AAGAAGGCTTAGGC AGGAATT-3' |
Human (MDA- MB-231, MCF-7, VSMC) |
Lentivirus | pLKO.1-puro | (Leyme et al, 2016; Leyme et al, 2015; Miyake et al, 2011) |
Rat, Human, Mouse |
|
GIV
shRNA3 |
5'- GGAACAAACAAGAT TAGAA-3' |
Monkey (COS-7, Vero), Rat(PC- 12), Human(BTSC, MCF10A, SH- SY5Y) |
retroviral | pcPURU6b, pSIREN-RetroQ |
(Enomoto et al, 2009; Muramatsu et al, 2015; Natsume et al, 2012; Ohara et al, 2012) |
Rat, Human, Mouse |
|
GIV
shRNA4 |
5'- AGCTGGAACTTCTT CATGA-3' |
N/A | Retrovirus | pcPURU6b | (Enomoto et al, 2009) | |
|
GIV
shRNA5 |
5'- CCGGAGGCAAGAGT TGAGGAATTAACTC GAGTTAATTCCTCAA CTCTTGCCTTTTTTG -3' |
Human (DLD1) | Lentivirus | LvUCTP | (Zhang et al, 2014) | Human |
|
GIV
shRNA6 |
5'- GATCCCCGTCAATA ATGATGCCTCACTTC AAGAGAGTGAGGCA TCATTATTGACTTTT T-3' |
Human (U251) | Lipofectamin e |
pGCsi- H1/Neo/GFP |
(Ni et al, 2015) | Human |
|
GIV/GIR
DIN siRNA1 |
5'- AACCAGGTCATGCT CCAAATT-3' |
Monkey (Vero, COS-7), Human (THP-1, DLD1, HeLa, LX- 2, HUVEC, MDA-MB- 231,HREC, KYSE, MG-63), Rabbit (SMC), Canine (MDCK) |
Oligofectami ne |
N/A | (Beas et al, 2012; Dunkel et al, 2012; Enomoto et al, 2005; Garcia-Marcos et al, 2009; Garcia-Marcos et al, 2012; Ghosh et al, 2008; Hu et al, 2015; Ichimiya et al, 2015; Ito et al, 2013; Jiang et al, 2008; Kitamura et al, 2008; Lin et al, 2014; Lin et al, 2011; Lopez-Sanchez et al, 2014; Lopez-Sanchez et al, 2013; Ma et al, 2015a; Mao et al, 2012; Midde et al, 2015; Sasaki et al, 2015; Shibata et al, 2013) |
Human |
|
GIV/GIR
DIN siRNA2 |
5' GAGGCAGACAGUGU CAUUATT-3' |
Human (A549, H520,) |
Oligofectami ne |
N/A | (Pan et al, 2015) | Human |
|
GIV/GIR
DIN siRNA3 |
5'- GCAACAAGCUUACC UCAAUTT-3 |
Mouse (podocytes) |
PTD-DRBD | N/A | (Wang et al, 2015) | Human |
|
GIV/GIR
DIN siRNA4 |
5'- AAGAAGGCTTAGGC AGGAATT-3' |
Human (KYSE, PC-3, HeLa, MG-63), Canine (MDCK) |
Oligofectami ne |
N/A | (Hu et al, 2015; Shibata et al, 2013; Tomiyama et al, 2015) |
Human, Dog |
|
GIV/GIR
DIN siRNA5 |
5'- GGACCAACCUGGAU GAAUATT-3' |
Rabbit (Smooth Muscle Cell) |
Lipofectamin e RNAMAX |
N/A | (Miyachi et al, 2014; Miyachi et al, 2015) |
Rabbit |
|
GIV/GIR
DIN siRNA6 |
5'- GGCAGAACAUCCAC UAGCATT-3' |
Rabbit (Smooth Muscle Cell) |
Lipofectamin e RNAMAX |
N/A | (Miyachi et al, 2014; Miyachi et al, 2015) |
Rabbit |
|
GIV/GIR
DIN siRNA7 |
5'- CCAGAAUGUACCGA GAUGAUU-3' |
Human (DLD1) | Lipofectamin e |
N/A | (Zhang et al, 2014) | Human |
|
GIV/GIR
DIN siRNA8 |
5'- CUUCAUUAGUUCUG CGGGAUU-3' |
Human (DLD1) | Lipofectamin e |
N/A | (Zhang et al, 2014) | Human |
|
GIV/GIR
DIN siRNA9 |
5'- CAAGAGUUGAGGAA UUAAAUU-3' |
Human (DLD1) | Lipofectamin e |
N/A | (Zhang et al, 2014) | Human |
|
GIV/GIR
DIN siRNA1 0 |
5'- GGACCAACCUUGAU GAAUAUU-3' |
Human (DLD1) | Lipofectamin e |
N/A | (Zhang et al, 2014) | Human |
|
GIV/GIR
DIN siRNA1 1 |
5'- GAAGGAGAGGCAAC TGGAT-3' |
Human (U-87) | GenePorter | N/A | (Gu et al, 2014) | Human |
Basic Protocol 1: Detection of full-length GIV by Immunoblotting
Perhaps the first major challenge in studying GIV is being able to detect the full-length protein consistently and reliably in samples. Although its calculated molecular weight is ~220 kDa, full-length GIV typically runs at ~250 kDa (Fig 2A), and is often accompanied by more than just one band close to that size (appearing sometimes like a ladder). Because it is a large protein with multiple possible isoforms and is known to undergo numerous post-translational modifications (see www.phosphosite.org), it is believed that most of those bands are differentially modified forms of GIV. Gene depletion studies have indeed confirmed such to be the case in some commonly studied cell lines, e.g., HeLa and COS-7. Listed below are several key steps during immunoblotting that significantly improve the detection of full-length GIV--
Use the standard wet transfer protocol (Ni et al, 2016) adhering to the following conditions/buffers optimized specifically for the detection of full-length GIV protein.
Materials
Running and Transfer Buffers (see buffer recipes in Reagents and Solutions).
Bovine serum albumin (BSA; Sigma) or dry milk powder (Store bought brand)
PVDF membrane; 0.45 μm pore size (Millipore)
Blotting paper, extra thick (Sigma).
Primary antibodies (Table 2)
IRDye® Infrared dye conjugated secondary antibodies (Li-COR)
Phosphate buffered saline with 0.05% Tween 20 (PBS/Tween)
Minigel system (BioRad)
2000 volt power supply (BioRad).
Orbital shaker (Multiple suppliers).
LiCOR Odyssey® CLx Imaging system (Li-COR Odyssey®)
Table 2.
Antibodies tested to successfully immunoprecipitate and/or immunoblot endogenous or exogenously expressed GIV
| Antibody/Cat# | Source | Type | Application |
|---|---|---|---|
|
GIV-CT (Girdin T-13) /sc-
133371 |
Santa Cruz Biotechnology (SCBT) |
Rabbit Polyclonal | IB, IP, IF, PLA |
| GIV CC/ ABT80, ABT168 | Millipore | Rabbit Polyclonal | IB, IP, IF, PLA |
| GIV CC / MABT100 | Millipore | Mouse Monoclonal | IB |
| GIV pY1765 / SP158 | Spring Bioscience | Rabbit Monoclonal | IB, IP, IF, PLA |
CT = Carboxyl terminus; CC = coiled-coil; IB = immunoblotting; IF immunofluorescence; IP = immunoprecipitation; PLA = proximity ligation assay
Procedure
- Whenever possible, resolve samples on an 8% SDS-PAGE gel. Use Tris/Glycine/SDS based running buffer.
- ○ In case of protein-protein interaction assays, where the binding partner of interest is smaller and the samples need to be resolved on a higher percent gel, use a 4-20% gradient gel.
- ○ Using fresh lysates; freeze-thaw cycles greatly impair the detection of GIV, likely due to activation of proteases.
For transfer, use 25 mM Tris/190 mM glycine buffer with 20% methanol (10% or lower significantly reduces the efficacy of transfer). Add 0.01-0.04% SDS to the transfer buffer to increase efficiency of transferring GIV to the membrane.
- Use PVDF membranes. This is preferred over Nitrocellulose to improve the detection of full-length GIV.
- ○ This is because PVDF has a protein binding capacity of 170 to 200 μg/cm2 whereas nitrocellulose has a protein binding capacity of 80 to 100 μg/cm2. Because PVDF has a higher protein binding capacity, it also offers higher sensitivity, in that it allows detection of low levels of proteins. Consequently, the use of PVDF membrane is expected to produce higher background noise, warranting diligent washing after incubation with primary and secondary antibodies.
- Transfer overnight (35 V for 16 h at RT, and then @ 120 V for 15 min before removing the membrane) with precautions to avoid overheating of the transfer buffer.
- ○ If the PVDF membrane looks dry after the transfer, make sure to dip it in methanol to re-prime the membrane before proceeding to immunoblotting.
- Although there are several anti-GIV antibodies available commercially (summarized in Table 2), we recommend the following for immunoblotting --
- Girdin-T13 (Catalog # sc-133371; Santa Cruz Biotechnology; 1:500 in 5% milk / PBS-T). This rabbit polyclonal antibody detects the C-terminal 18 aa of full-length GIV, and by far, gives the cleanest signal (highly specific; Fig 2A). However, due to the small epitope against which it was raised, it is less sensitive for detecting smaller amounts of GIV (or GIV molecules that lose their C-termini due to proteolysis during lysis). Alternatives include a mouse monoclonal antibody generated by Millipore (MABT100; 1:250 in 5% milk/PBS-T) (Fig 2A).
- Girdin coiled-coil antibodies (Catalog# ABT80 and ABT168; Millipore;1:500 in 5% milk/PBS-T) which were raised against a ~400 aa stretch within the coiled-coil domain of rat Girdin (Fig 2A). This antibody is highly sensitive, in that it is able to detect lower levels of expression, but may show cross-reacting bands in some cell lines. This antibody is preferred for the detection of small amounts of GIV in co-immunoprecipitates.
- For all phospho-specific antibodies, including anti-pY1764 GIV (Spring Bio), we recommend using 5% BSA/PBS-T (instead of milk) for blocking and for preparing the dilution of primary and secondary antibodies.
- Incubate blots with primary antibodies overnight at 4°C.
- ○ When testing expression in new cell line or tissue, analyze the entire length of the membrane (and not just the region around the expected molecular weight). This is essential to look for signals that may indicate proteolytic breakdown and/or spliced isoforms and assess for other cross-reacting proteins.
- Wash membrane using excess PBST × 4 times (15 min × 1, and 5 min × 3).
- ○ Ensure that wash trays are clean.
- ○ Ensure that at no point in time forceps or tweezers, or edges of the trays, or bare skin touches the membrane directly, especially in the region of the blot where proteins have been transferred. Because of the highly sensitive nature of the infrared detection imaging dyes (see below), every imperfection, big or small will be detected.
- Incubate membrane in secondary antibodies. Use IRDye conjugated secondary antibodies at 1:10,000 dilution, at room temperature for 45 - 60 min.
- ○ Infrared fluorescent dyes and near-infrared (NIR) fluorescence imaging deliver similar sensitivity as chemiluminiscence. It is also much more sensitive than visible fluorescence due to low background autofluorescence in the near-infrared region and, therefore, higher signal-to-noise ratios. Furthermore, infrared detection is static, which allows a wider linear detection range than chemiluminescence without a loss of signal. Another advantage of infrared imaging is the ability to simultaneously detect two proteins on the same blot at the same molecular weight; this minimizes the need for stripping and re-probing leading to an increase in detection efficiency.
- Use Li-COR Odyssey for consistent and reproducible digital images, and for dual color imaging without the hassles and unpredictability of film (Mathews et al, 2009).
- ○ Li-COR protein detection system enables determination of band intensity in immunoblots over a greater dynamic range than that of conventional chemiluminescence. Li-COR also allows visualization of all the data at the time of the first scan, without sacrificing sensitivity or detection of fainter higher molecular weight GIV bands.
- ○ For the detection of GIV and other proteins by Li-COR Odyssey, Immobilon FL (Immobilon-FL PVDF, 0.45 μm; Millipore) membrane outperforms the other membranes tested.
Basic Protocol 2: Detection of full length GIV mRNA by quantitative real time polymerase chain reaction (qRT-PCR)
To analyze GIV transcripts, we recommend that primers are designed to specifically amplify GIV's C-terminus. This strategy allows detection of GIV isoforms that contain the most critical C-terminal region which appears to be necessary and sufficient to couple G proteins to RTKs and trigger G protein signaling (Ma et al, 2015a). Either Taqman assay detection system or SYBR Green detection system can be used. There are two qRT-PCR methods that can be used to detect GIV transcripts: 1.Relative Standard Curve method; 2. Comparative Ct method.
Materials
RNeasy kit (QIAGEN)
SuperScript® II Reverse Transcriptase
RNase H
MicroAmp® Optical 96-Well Reaction Plate (Applied Biosystems™)
MicroAmp® Optical 96-Well Optical Adhesive Film (Applied Biosystems™)
TaqMan® Fast Universal PCR Master Mix (2X) (Applied Biosystems™) or Fast SYBR® Green Master Mix (2X)
CCDC88A (GIV) Assay Mix : Hs01554973_m1(see Table4) for Taqman assay detection or GIV primers designed by Primer Express® Software v3.0.1 (Applied Biosystems™) (see Table 3) for SYBR green detection.
GAPDH Assay mix: Hs99999905_m1(see Table 4) for Taqman assay detection or GADPH primers designed by Primer Express® Software v3.0.1 (Applied Biosystems™) (see Table 3) for SYBR green detection
Forward and reverse primers diluted to working concentration (10μM working stocks are sufficient for most assays)
PCR microtubes with attached caps size 0.2 mL
Microcentrifuge
DNA/cDNA template— cDNA reaction diluted 1:10 to detect a medium to highly expressed targets or 1:2 to 1:5 for rare transcripts or 10 ng to 100 ng gDNA
Sterile filter pipette tips
PCR grade water
Applied Biosystems StepOnePlus™
Table 4.
Primers for detection of GIV mRNA
| Gene Name | Primer Sequence | NCBI Ref Seq(s) | Amplicon length |
PCR efficiency |
|---|---|---|---|---|
| CCDC88A (Mouse) | Fwd: 5’-GTGATCTCTACTGCTGAAGG-3’ Rev: 5’-TGTTGCTCCCTAGACCTGCT-3’ |
NC_000077.6 | 185 bp | 96.6% |
| CCDC88A (Human) | Fwd: 5’- ATCTCAACTGCCGAAGGAACT-3’ Rev: 5’-TGTTGCTCCCTAGACCTGCT-3’ |
NG_031944.1 | 182 bp | 97.7% |
| GAPDH (Human) | Fwd: 5’-TCAGTTGTAGGCAAGCTGCGACGT-3’ Rev: 5’-AAGCCAGAGGCTGGTACCTAGAAC-3’ |
NG_007073.2 | 185 bp | 98.2% |
| GAPDH (Mouse) | Fwd: 5’- CTGCAGCCTCGTCCCGTAGAC A - 3’ Rev: 5’-TGCCGTGAGTGGAGTCATACTGGA- 3’’ |
NC_000072.6 | 181 bp | 97.1% |
Table 3.
Sequence of probes for Taqman gene expression assay for detection of GIV
| Gene Name | Assay number | NCBI Ref Seq(s) | Amplicon length |
|---|---|---|---|
| GAPDH | Hs99999905_m1 | NM_002046.3 | 122 |
| CCDC88A | Hs01554973_m1 |
NM_001135597.1 NM_001254943.1 NM_018084.4 |
130 bp |
Procedure
Isolate mRNA using commercially available kits as per the manufacturer's protocol [e.g., RNeasy kit (QIAGEN)].
Synthesize first-strand cDNA using Superscript II reverse transcriptase (Invitrogen), followed by ribonuclease H treatment (Invitrogen) prior to performing quantitative real-time PCR (Kusser et al, 2006).
Use 500ng of mRNA for reverse transcription.
Include a no-template control which omits any DNA or RNA template from a reaction and a no-reverse transcriptase control in each experiment as negative controls.
Run reactions on a real-time PCR system, e.g., ABI StepOnePlus; (Applied Biosystems).
- Detect gene expression with SYBR green (Invitrogen).
- ○ For detection of GIV mRNA with high degrees of accuracy, reproducibility and sensitivity (up to 1-10 copies), we recommend using the Taqman®-detection method. This method uses fluorogenic probes specific to target gene to detect target as it accumulates during PCR. Table 3 summarizes the Taqman probe that has been successfully validated for use on patient-derived samples (Barbazan et al, 2016).
Primers for detection of GIV by SYBR green are listed in Table 4. These primers were designed using Primer Express® Software v3.0.1 (Applied Biosystems™).
- Determine relative quantitation of gene expression by normalizing to GAPDH using relative standard curve method or Comparative CT Method (ΔΔCt) [see Guide to Quantitative PCR by Applied Biosystems™).
- ○ For relative standard curve method, use larger PCR products (~500 bp) that include GIV or GADPH sequence used in qPCR and its series dilution (1:10) as standards (see Table 5).
- ○ When using SYBR green detection for Comparative CT Method (ΔΔCt), we recommends that you determine the PCR efficiencies of GIV and GAPDH are approximately equal and above 95% (see Table 4) and use melt curve to validate the primers.
Table 5.
GIV and GAPDH standards serial dilutions
| GIV/GADPH Standard | Volume | Standard Concentration (pg/ul) |
|---|---|---|
| Std. 1 | 50ul | 100 |
| Std. 2 | 50ul | 10 |
| Std. 3 | 50ul | 1 |
| Std. 4 | 50ul | 0.1 |
| Std. 5 | 50ul | 0.01 |
Basic protocol 3: Immunoprecipitation of endogenous or exogenously expressed GIV
This protocol describes the method for immunoprecipitation of full-length GIV from cell or tissue lysates (Fig 2B). This approach is routinely used to identify GIV-interacting proteins or to detect post-translational modifications on GIV. The antibodies listed in Table 2 have been tested to successfully immunoprecipitate endogenous and overexpressed GIV. If the goal is to detect an interaction between GIV and a partner, we recommend performing the immunoprecipitation of the partner and monitoring co-immunoprecipitated GIV by immunoblotting.
Materials
Cell culture set up, media and buffers
Cell scraper
Microcentrifuge tubes
30-G needle
Antibodies (Table 2)
Protein A/G Sepharose beads (GE Healthcare)
1X Phosphate Buffered Saline (PBS) (see Reagents and Solutions)
IP Lysis Buffer (see Reagents and Solutions)
1X PBS-T Wash Buffer (see Reagents and Solutions)
5X Laemmli Sample buffer (see Reagents and Solutions)
Procedure
A. Cell Lysis
- Prepare the IP Lysis buffer (see recipe provided in Reagents and Solutions) and keep it on ice.
- ○ Add DTT, sodium orthovanadate, protease and phosphatase inhibitors fresh.
- ○ The use of relatively high concentration of protease inhibitors is essential to enhance the detection of full-length GIV in samples.
- ○ If looking for pSer modifications, add 5-10 mM NaF, 5-10 mM sodium pyrophosphate as well.
Remove the cells from the incubator and place them on ice.
Wash with cold 1X PBS, aspirate off, scrape cells very gently in 1 ml of 1X PBS.
Collect cells into a pre-chilled microcentrifuge eppendorf tube and briefly spin them down at low speed (<1500 × g for 10-15 secs) in a cold microcentrifuge.
Aspirate PBS supernatant ensuring not to suction cells.
- Add 500 μl of lysis buffer and quickly vortex tubes to resuspend pellets in the lysis buffer.
- ○ It is crucial to keep everything on ice as much as possible. GIV has a fragile C-terminus and gets proteolyzed easily at higher temperatures.
Pass the lysates through a 30G needle ~10 times at 4°C, and clarify lysates by centrifuging at 12,500Xg for 10 min at 4°C.
Take the clarified supernatant out in a fresh eppendorf tube and estimate protein concentration by a standard protein quantification assay (Bradford, 1976).
B. Immunoprecipitation
- 9. Incubate cell lysates (~1-2 mg of total protein) with 2 μg of antibody (see Table 2 for GIV antibodies) for at least 4 h at 4°C rocking on a rotator.
- ○ Include a negative control – cell lysate incubated with control IgG.
- 10. After incubation with antibody, add 25 μl of packed, equilibrated Protein A /G Sepharose beads (GE Healthcare) and incubate for 60 min at 4°C. For rabbit primaries use Protein A, and for mouse primaries use Protein G Sepharose beads.
- ○ Equilibrate Protein A/G sepharose beads by washing in cold lysis buffer three times.
Wash beads 4 times, each time with 1 ml PBS-T buffer. Centrifugation at 4°C, and no more than ~ 1500 × g to avoid crushing the beads, which may result in undesirably high non-specific background.
- 12. Elute the bound proteins in 40 μl of Laemmli's sample buffer by heating to 95°C for 5 min.
- ○ Do not boil if you are looking for interaction of GIV with transmembrane receptors, e.g., G protein-coupled receptors (GPCRs), glucose transporters (GLUT), and integrins. In those cases, elute with Laemmli's buffer at room temperature or at 37°C for 10-15 min. Boiling in these specific cases causes irreversible damage to the proteins and colloidal aggregate formation that cannot be resolved by SDS PAGE.
- ○ It is recommended that the beads be eluted twice (20 ul each time and pool the eluate) to maximally recover the bead-bound immune complexes.
13. Analyze samples by immunoblotting. Use the standard wet transfer protocol adhering to the conditions/buffers optimized for GIV (see Basic Protocol 2).
Note: See COMMENTARY section for special considerations when co-immunoprecipitation of protein complexes are desired or when targeted complexes are bound to the actin cytoskeleton.
Basic Protocol 4: Transfection of GIV constructs into mammalian cells
Owing to the large size of its cDNA (5610 bp), the efficiency of exogenous expression of full-length GIV tends to be sub-optimal (only ~2-3 folds over endogenous) and poses challenges in some experiments such as generation of stable cell lines and for studying protein-protein interactions. The method detailed here offers an efficient and inexpensive way for expression of GIV using polyethylenimine (PEI). We tested and compared efficiency of homemade PEI solution with that of commercially available transfection reagents (Genejuice, Mirus, Lipofectamine) and found PEI to be the most efficient (a comparison with GeneJuice® transfection reagent (EMD Millipore) is shown in Fig 3A). Here, we describe a detailed method for transfection of COS-7 cells with GIV-FLAG construct using PEI. The same protocol can be applied to and optimized for several different cell lines and constructs of GIV. Because the expression levels achieved with PER is often well above physiologic, transient transfection with PEI is not recommended for any functional assays or for studying cell phenotypes because it is likely to be associated with overexpression-related artifacts. PEI is also not suitable for transient transfections during FRET analyses (see Basic Protocol 7).
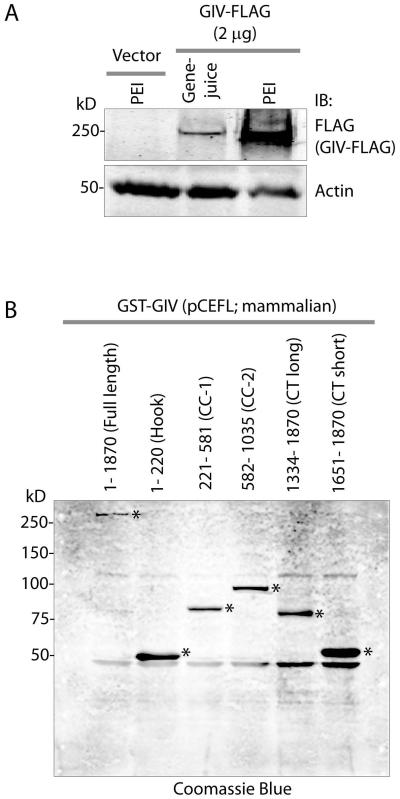
Figure 3. Exogenous expression of GIV/Girdin in mammalian cells.
A: COS-7 cells were transiently transfected with control vector or GIV-FLAG constructs using two indicated transfection reagents and lysed at 48 h after transfection. Equal aliquots of those lysates (~ 60 μg) were analyzed for GIV expression by IB with a commercial anti-FLAG antibody (Sigma; 1:500 dilution). Exogenous expression of full-length GIV is detected at ~250 kD exclusively in lanes transfected with GIV-FLAG. The level of expression is significantly higher (~ 7.5 fold by band densitometry) when transfection was carried out with PEI (middle lane) compared to Genejuice (right lane). B. COS-7 cells transfected with various pCEFL GST-GIV constructs were lysed after 48 h. Equal aliquots of lysates (~ 30 μg) were separated by SDS PAGE and analyzed for GST-GIV by Coomassie blue.
Materials
Polyethylenimine (PEI) (Polysciences Inc.; Catalog # 23966)
Syringe and 0.22 μm filter
Cell culture set up, media and buffers
1X PBS (See Reagents and Solutions)
Microcentrifuge tubes, 1.5 ml.
Procedure
A. Preparation of PEI stock solution
- Dissolve PEI in deionized H2O to a final concentration of 1 mg/ml. Adjust pH to 7.0. It could take a few hours to completely dissolve PEI.
- ○ Heating the solution to 60°C with gentle stirring on a magnetic stirrer helps expedite the process.
- Filter through a sterile 0.22 μm syringe filter.
- ○ This step is very important not only for sterility but also because the presence of un-dissolved PEI particles may precipitate DNA leading to poor efficiency of transfection.
Make aliquots and store at −80°C. Once frozen, the aliquots at −80°C are stable for 2 years. Once thawed, the tube can be kept at 4°C for a maximum of 1 month.
B. Transfection of cells with PEI
4. Maintain cells in complete medium (e.g., in the case of HeLa or COS-7 cells, maintain in DMEM supplemented with 10% FBS and 1X penicillin-streptomycin-glutamine).
- 5. Plate cells for transfection so that the cells are ~90% confluent the next day.
- ○ Although 90% confluency is desirable in the case of COS-7, optimization may be necessary in the case of other cell lines.
6. Next day, prepare a mix of DNA:PEI (ratio 1 μg : 1.5 μl) in 100μl of serum free media. Vortex and incubate the master mix at room temperature for 20 min. Although each construct should be tested individually to determine the optimal amount of DNA for transfection, ~ 1 μg DNA is a good starting point for a single well of a 6-well plate; ~ 4-5 μg DNA is optimal for a 10 cm dish.
7. Gently add the mix to the cells maintained in complete medium drop-wise and incubate for 5 h. Before adding the transfection mix, aspirate off the medium and add fresh complete medium to the cells.
8. Once incubation is complete, aspirate off the medium, wash cells with 1X PBS three times and incubate cells with fresh complete media for 48 h before harvesting.
Note: The detailed guidelines provided above apply to HeLa and COS-7 cell lines, and may need optimization when used for other cell lines. A troubleshooting guide is provided in COMMENTARY section (see Table 6).
Table 6.
Troubleshooting guide for poor transfection efficiency
| Problem encountered | Suggestion |
|---|---|
| Cell incompatibility | Check if PEI has been previously tested with your cell line. If not, change cell line if possible. We have successfully transfected GIV constructs with PEI in HEK293, HeLa, COS-7, NIH3T3, DLD1 and MDCK cell lines using the Basic Protocol 4. |
| DNA:PEI Ratio | We suggest using 1-2 μl of PEI per μg of DNA transfected. May need to be optimize for your cells. |
| Poor PEI quality | PEI could be kept at 4°C for 1 month maximum. However, if a decline of transfection efficiency is observed, we recommend taking out a new aliquot from −80°C. |
| High cell death | - Prolonged incubation with PEI could be toxic for the cells. We recommend an incubation of mix DNA/PEI for a maximum of 5h. - May need to adjust DNA/PEI ratio or decrease amount of DNA transfected. |
Basic protocol 5: In cellulo GST pull down assay using GST-tagged GIV from mammalian cells
This protocol describes an in cellulo pull-down assay using GST-tagged GIV (full-length or individual domains of GIV; Fig 3B) expressed and purified from mammalian cells. The assay provides an alternative or a complimentary approach to immunoprecipitation studies (Basic protocol 3) to help identify protein-protein interactions. The GST-tagged constructs of GIV (cloned into pCEFL-vector, in which expression is driven by a EF1 promoter) express at a high level in mammalian cells and offer the advantage of a one-step GST pull down assay. The lysates of cells expressing GST-GIV fusion proteins are incubated with Glutathione Sepharose 4B beads to pull down the fusion protein together with the interacting proteins. The proteins bound on beads can then be analyzed by immunoblotting or mass spectrometry.
Note: Purification of recombinant His- and GST tagged C-terminus of GIV (AA 1660-1870; harboring the GEF motif, putative SH2-like domain and the Akt-Actin binding region) from bacteria has been described in detail previously (Garcia-Marcos et al, 2011).
Materials
Cell culture set up, media and buffers
Cell scraper
Microcentrifuge tubes
30-G needle
Glutathione Sepharose beads (GE Healthcare Life Sciences)
1X Phosphate Buffered Saline (PBS) (see Reagents and Solutions)
GST pull down Lysis Buffer (see Reagents and Solutions)
1X PBS-T Wash Buffer (see Reagents and Solutions)
5X Laemmli Sample buffer (see Reagents and Solutions)
Procedure
Lyse cells transfected with the desired construct in 500 μl of cold GST pulldown Lysis buffer using the lysis protocol described in Basic Protocol 3.
- Incubate lysates (~1-2 mg protein) with 25 μl of packed, equilibrated Glutathione-Sepharose beads for 4 h at 4°C rocking on a rotator.
- ○ Equilibrate Glutathione-Sepharose beads by washing in cold GST pulldown lysis buffer three times.
After incubation is over, wash beads 3 times with 1 ml of 1XPBS-T wash buffer. Centrifugation (5000xg) should be done at 4°C.
- Elute the bound proteins in 40 μl of Laemmli sample buffer by heating to 95°C for 5 min.
- ○ Elution should be repeated twice (20 ul each time) and pooled to maximally recover the bead-bound protein complexes.
Analyze by immunoblotting (see Basic protocol 1).
Basic Protocol 6: Whole-cell Immunofluorescence
GIV is a cytosolic protein, and has been found at various locations within cells (summarized in Fig 4). This protocol describes the method to follow sub-cellular localization of GIV – under basal and/or receptor-stimulated conditions via whole-cell immunofluorescence (IF) microscopy. The various commercially available antibodies tested for IF and detection pattern for GIV are summarized (Fig 4).
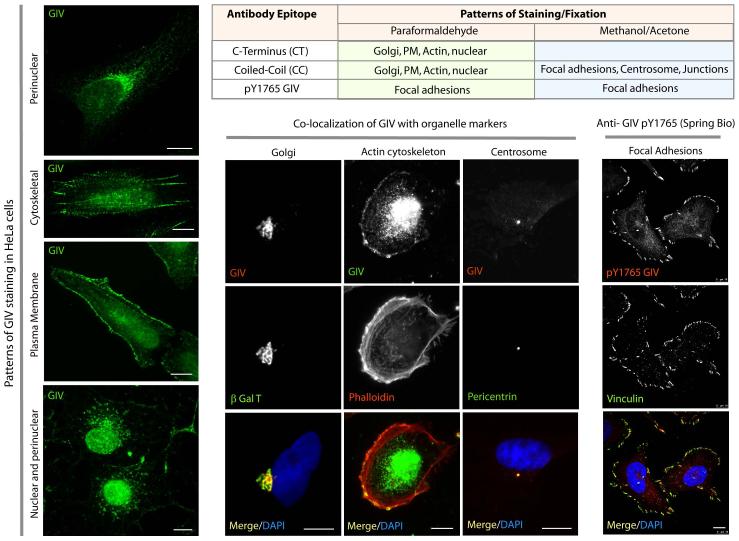
Figure 4. Subcellular distribution of GIV, as determined by whole cell immunofluorescence.
HeLa cells grown on cover slips were fixed and co-stained for GIV with a variety of other markers (as indicated) and analyzed by confocal immunofluorescence. Table 2 lists the commercially available anti-GIV antibodies, and the patterns of staining seen using each antibody.
Materials
■ 10X PBS
■ 1X PBS
■ 3% Paraformaldehyde (PFA) in 1X PBS
■ 0.1 M Glycine in 1X PBS (Quenching solution)
■ 0.2% Triton X-100 (TX-100) in 1X PBS (Permeabilizing solution)
■ 1% BSA and 0.1% TX-100 in 1X PBS (Blocking Buffer)
■ Multi-well plates, Coverslips, Glass slides
■ Nail polish (sealant)
■ Primary and secondary antibodies
■ DAPI (4 ',6-diamino-2-fenilindol) (Life Technologies)
■ ProLong® (Life Technologies)
■ Confocal or fluorescence microscope.
Procedure
A. Sample Preparation
Plate cells in multi-well plates containing sterile coverslips so that they will reach approximately 80% confluence by the time immunofluorescence is going to be carried out (e.g., 7 × 104 cells/well for a 12-well plate).
Incubate cells for 16 hours in the CO2 incubator to allow cells to attach to the coverslip.
B. Fixation
- 3. Incubate cells with 3% PFA for 25 minutes at room temperature.
- ○ Fixation of cultured cells is generally achieved by replacing the culture medium with the fixative solution. However, the sudden change in surface tension following removal of the culture medium can damage some cell types. Thus, we recommend that the fixative be first directly added to the culture medium. For example, when 3% PFA is the desired fixative, adding the same volume of 6% PFA as the volume of culture medium will finally result in a 3% PFA solution, which is strong enough to pre-fix the cells. After 2 minutes, the pre-fixation culture medium should be replaced with a fresh volume of 3% PFA. The pre-fixation step makes the cells more rigid so they can withstand any potential deleterious effects created by changes in surface tension.
- ○ Fixation can result in hydrophobic cross-linking of tissue proteins. The time, temperature, pH, and fixative used will determine the degree of cross-linking. Once the fixation protocol has been optimized, the same procedure should be used consistently.
- ○ Fixation is best (fastest) when carried out at room temperature. However, in experiments that require stimulation with ligands (e.g., EGF, insulin, etc.) and analysis of specific time points after such activation, it is recommended that cells are incubated with 3% PFA for 10 minutes on ice, followed by 15-20 minutes at room temperature.
4. Aspirate fixation solution and add 0.1 M glycine in 1X PBS for 10 minutes at room temperature to quench excess aldehyde groups.
5. Aspirate quenching solution and incubate cells with 0.2% TX-100 in 1X PBS for 20 minutes to permeabilize cells.
C. Immunofluorescence Preparation
6. Incubate cells with 1X PBS containing 1% BSA and 0.1% TX-100 (blocking buffer) for 30 minutes at room temperature.
7. Aspirate off blocking solution and incubate cells in primary antibody in blocking solution for 60 minutes at room temperature.
8. Wash monolayer 3 times for 5 minutes with 1X PBS.
- 9. Incubate cells in secondary antibody in blocking solution for 60 minutes at room temperature.
- ○ Nuclei can be detected by incubating cells in the dark with 0.1 - 1 μg/ml DAPI in PBS for 10-30 min.
10. Wash monolayer 3 times for 5 minutes with 1X PBS.
11. Aspirate washing solution and mount the samples with ProLong® Gold and analyze using a confocal microscope.
Note: See COMMENTARY section for troubleshooting tips, and for different modes of fixation recommended for the visualization of different subcellular pools of GIV.
Basic protocol 7: Förster Resonance Energy Transfer (FRET) studies to assess the spatiotemporal dynamics of GIV-associated protein complexes and GIV-dependent G protein signaling
This protocol describes the fundamentals of designing and carrying out FRET-based studies for assessing the spatiotemporal dynamics of GIV-dependent signaling in live cells. These assays have the potential to provide spatial information (like immunofluorescence studies) and temporal information (like protein-protein interaction assays carried out after ligand stimulation) in real time and are able to capture single-cell dynamics of signal transduction like no other assay. Despite these advantages, major disadvantages include the need to overexpress protein probes above physiologic levels and the toxicity that comes with expressing fluorescent proteins. In addition, a lot of consideration is warranted in designing constructs that are appropriate for use in these assays. Below we discuss some of these issues that are to be specifically considered when using GIV or G protein probes in FRET-based assays.
Materials
■ Mammalian cells (e.g. HeLa, Cos7)
■ Plasmid(s) of interest for transient expression
■ 35 mm FluoroDish (World Precision Instruments Inc.; Catalog # FD35-100)
■ DMEM (Corning; Catalog # 10-013-CV) supplemented with 10% FBS (HyClone; Catalog # SH30071.03) and 1X penicillin-streptomycin-glutamine (Gibco; Catalog #10378-016)
■ Imaging media: DMEM without Phenol Red (Corning; Catalot #17-205-CV)
■ Trans-IT®-LT1 tansfection reagent (Mirus Bio LLC)
- ■ Microscope: Inverted confocal laser scanning microscope (Olympus FV1000/3000 or equivalent) equipped with:
- ○ Oil immersed objective (60x, 1.49 N.A);
- ○ Excitation lasers for donor (CFP) in the range of 405 to 440 nm (HeNe) and for direct excitation of acceptor (YFP) a 514-515 nm laser (Argon-ion);
- ○ Detection via gated spectral detection or suitable bandpass filter
- ■ Software for imaging and analysis purposes:
- ○ In-built software (Olympus Fluoview or equivalent)
- ○ ImageJ software with RiFRET plugin installed (free download platform: https://imagej.nih.gov/ij/ (Roszik et al, 2009; Schneider et al, 2012)).
Procedure
A. Choice and design of FRET probes
- When choosing FRET probes or designing new probes, consider the following the general principles of fluorescent tag proteins:
- ○ Fusion of fluorescent proteins to target protein can perturb the native function of the target protein. Extreme caution must be taken while introducing the bulky fluorescent protein (for example, the 28 kDa GFP has 238 amino acid residues) at a location that does not sterically interfere with these binding interactions. As a rule, multiple tag positions should be validated for known localizations, functions and interactions of any given protein. In the case of GIV, no fluorescent-tagged full-length construct (either N- or C-terminally tagged, with or without linker domains) expressed well or localized appropriately in cells. When a fluorescent protein tagged GIV-CT fragment (last ~210 aa) was developed, N-terminal, but not C-terminal tag position maintained GIV's ability to bind and activate Gαi protein (see Table 7).
- Independently validate each FRET probe.
- ○ The most robust method to test this involves replacing the gene that encodes the wild-type protein with the gene that encodes the fusion protein, and then evaluating the mutant cells for any phenotypic changes compared to wild-type cells. In the case of CFP-GIV-CT probes, extensive biochemical and biophysical validation included the confirmation that the C-terminal fragment of GIV fulfilled several of the known functions of full-length GIV, in that it could bind ligand-activated EGFR, bind Gαi in cells, undergo tyrosine phosphoregulation, activate downstream pathways (Akt phosphorylation) and trigger cell invasion in matrigel (Midde et al, 2015).
- Whenever possible, pair a newly designed probe with a previously validated probe with appropriate positive (where FRET is expected) and negative (FRET is not expected) controls so that only one probe is tested at any given time. For example, in studies evaluating newly generated fluorescent CFP-GIV-CT probes, all other constructs used as FRET probes were previously validated:
- ○ C-terminally tagged EGFR-YFP plasmid DNA construct, which was used (Midde et al, 2015) to study when and where GIV binds ligand activated EGFR and to investigate the temporospatial dynamics of the assembly of EGFR-GIV-Gαi ternary complexes in cells (Midde et al, 2015) was a generous gift from Zhixiang Wang, University of Alberta (Pennock & Wang, 2008). A GEF-deficient GIV mutant was used as negative control.
- ○ EGFR-CFP and Grb2-YFP, which were used as positive controls in the above assays (Midde et al, 2015) were generous gifts from Larry Samelson, National Institute of Health (NIH) (Yamazaki et al, 2002).
- ○ CFP-tagged Insulin receptor β, which was used to interrogate the obligatory role of GIV in coupling Gαi to ligand-activated Insulin receptors (Midde et al, 2015) was a generous gift from Ingo Leibiger (Karolinska Institute) (Uhles et al, 2003).
- ○ Internally tagged Gαi1-YFP, Gαi3-YFP, Gαi3-CFP, NT tagged CFP-Gβ1, and Gγ2 constructs, which have been extensively used by us to study the dynamics of non-canonical G protein activation by GIV-family of GEFs (Aznar et al, 2015; Lo et al, 2015; Ma et al, 2015a; Midde et al, 2015), were originally developed and validated by the groups of Bunemann and Gilman for studies on canonical G protein signaling that is activated by GPCRs (Bunemann et al, 2003; Gibson & Gilman, 2006).
- ○ The 3rd generation cAMP FRET biosensor, mTurq-EPAC-Venus (tEPACvv) was a generous gift from Kees Jalink (Klarenbeek et al, 2011). tEPACvv encompasses mTurquoise as donor and mVenus-mVenus as dual acceptor which has superior quantum yield and longer life-time, making it the most advanced probe to monitor sub-micromolar changes in cellular cAMP concentration.
Table 7.
Various epitope tagged functional constructs of GIV
| Construct | Expression vector (mammalian/bacterial) |
Tag/position | Important observations |
Cited References |
|---|---|---|---|---|
| Full-length GIV |
p3XFLAG-CMV-14 and pcDNA3.1 expression vectors for transient or stable intracellular C-terminal 3XFLAG or Myc tag, respectively. |
C-terminal FLAG and Myc tags |
N-terminal tags mislocalize protein in cells, alter protein functions. Bulky C-terminal tags (GFP etc.), even with the placement of flexible linkers, express poorly. |
GIV-FLAG in p3XFLAG-CMV-14 (ADDGENE ID# 65947) (Beas et al, 2012; Bhandari et al, 2015; Denker et al, 1996; Garcia-Marcos et al, 2011; Garcia-Marcos et al, 2010; Garcia-Marcos et al, 2009; Ghosh et al, 2008; Lin et al, 2011; Lo et al, 2015; Lopez-Sanchez et al, 2013; Lopez-Sanchez et al, 2015a; Ma et al, 2015d) GIV-myc in pcDNA 3.1 (Bhandari et al, 2015) |
| GIV fragments | In the following vector backbones: pET28b (His-tagged, bacterial), pGEX-4T (GST-tagged, bacterial), pCEFL-GST (GST-tagged, mammalian), or pcDNA3.1-CFP (CFP-tagged, mammalian) |
N-terminally placed His6-, GST- or CFP- tag. |
Placement of each tag in the C-terminus significantly reduces binding of GIV-CT to G protein (reason unclear). |
pER28b (ADDGENE ID# 69864) (Bhandari et al, 2015; Garcia-Marcos et al, 2011; Garcia-Marcos et al, 2010; Garcia-Marcos et al, 2009) pGEX-4T (ADDGENE ID# 69863) (Garcia-Marcos et al, 2011; Garcia-Marcos et al, 2010; Ogier-Denis et al, 1996) pCEFL (Bhandari et al, 2015; Lo et al, 2015; Midde et al, 2015) pcDNA3.1 CFP (ADDGENE ID# 69532) (Midde et , 2015) |
B. Transfection of FRET probes
4. Grow desired cells in sterile 35 mm FluoroDish (or MatTek glass bottom dishes); maintain in 10% FBS and antibiotics.
5. Transfect cells at 60-70% confluency.
6. Use Trans-IT®-LT1 tansfection reagent (Mirus Bio LLC) for these transfections using manufacturer’s protocol. Maintain cells in complete media (10% FBS and antibiotics) after transfection. TransIT-LT1 is a low toxicity, serum-compatible transfection reagent that eliminates the need for any culture medium change.
- 7. Transfect the optimal amount of various donor and acceptor plasmid constructs (0.1 - 1 μg, depending on the construct). To minimize complexities arising from molecular crowding, FRET probes should be overexpressed to levels ~1.5 - 2 fold higher than the endogenous proteins, as determined by immunoblotting.
- ○ Expression of each plasmid should be checked individually to ensure that the exogenously expressed fluorescent protein localizes as expected for the endogenous protein.
- 8. Choose cells carefully for analysis; include only those that express equimolar amounts of donor and acceptor probes.
- ○ Because stoichiometry of FRET probes has a significant impact on FRET efficiency, only cells that expressed equimolar amounts of donor and acceptor probes (as determined by computing the fluorescence signal/intensity by a photon counting histogram; PCH) should be included in FRET analyses. The PCH analysis constitutes a tested tool for extracting quantities from fluorescence fluctuation data, i.e., the measured photon counts per molecule and the average number of molecules within the observation volume (Chen et al, 2005). The ratio of transfected plasmids may need to be titrated until many cells show equimolar expression of fluorescent protein probes.
- ○ To consistently achieve equimolar stochiometry in FRET based assays, we recommend the use of cleavage site sequence plasmids that are engineered to enable regulated expression of multiple proteins in an equimolar ratio. These plasmids encode multiple proteins in tandem, each separated by high efficiency cleavage sites of a 2A peptide derived from porcine teschovirus-1 (Chng et al, 2015; Daniels et al, 2014; Kim et al, 2011). Studies using these engineered plasmids in human cell Lines, drosophila, zebrafish and mice have validated their use for the reliable co-expression of multiple constructs in diverse settings (Chng et al, 2015; Daniels et al, 2014; Goedhart et al, 2011; Kim et al, 2011; Martin et al, 2006).
9. Titrate expression levels of the FRET probes in order to express just enough for visualization and analyses, but avoid overexpression-related misfolding, mislocalization and other artifacts.
C. Preparation of cells for imaging
10. Starve cells overnight (~16-18 h) in serum-free DMEM (Gibco) [for COS-7 cells] or in 0.2% FBS [for HeLa cells] for all experiments in which acute responses to ligand stimulation is being monitored.
11. Switch the media to DMEM without phenol red (imaging media) prior to live cell imaging.
12. Allow cells to acclimatize to the imaging media for sufficient time (~2-3 h) prior to imaging.
D. Imaging of cells expressing fluorescent FRET probes
- 13. Use an inverted confocal laser scanning microscope equiped with an 60x high NA (1.49) objective for live cell FRET imaging.
- ○ The microscope should be stabilized on a vibration proof platform, caged in temperature controlled (37°C) and CO2 (5%) supplemented chamber.
- 14. Allow cells to acclimatize; avoid temperature variations during imaging.
- ○ Live-cell imaging chambers are produced in a wide variety of configurations that often utilize entirely different technologies for maintaining the specimen at a constant temperature. Whatever the method used for heating the chamber, cells should be allowed to acclimatize to these conditions prior to the start of imaging.
- ○ Fluctuations in temperature produced as a result of air conditioners and central heating units in the laboratory, intense illumination sources on the microscope, as well as unevenly heated objectives and stages are usually the primary culprits in focus drift. In addition, differential expansion and contraction rates in materials used to construct the culture chamber and/or microscope optical train can result in a change of the distance between the objective front lens and the glass-bottomed dish, leading to a loss of focus. With the highest magnification objectives, a change of just one degree Celsius is often sufficient to produce a shift of between 0.5 and 1.0 micrometers in the focal plane.
15. Use a 60x 1.49 N.A oil immersed objective to minimize chromatic aberration and enhance resolution for 405-605 nm imaging.
16. Acquire data through the method of sensitized emission using in-built softwares, such as the Olympus Fluoview.
- 17. To excite CFP, use either a 405 or 440 nm laser diode. To excite YFP use a 515 nm Argon-ion laser.
- ○ In our studies CFP was excited using a 405 nm laser diode because in a recently published work (Broussard et al, 2013) Claire Brown's group has described in great detail why 405 nm excitation is preferred for CFP, when all the controls are performed. Those guidelines should be strictly followed in designing controls in our study.
18. Minimize bleed through by adjusting the bandwidth of spectral emission through gating.
19. Exclude light that is out of the plane of focus. To accomplish this, enhanced CFP emission should be collected from 425-500 nm and YFP emission should be collected through 535-600 nm and passed through a confocal pinhole of 1 airy unit (A.U) before being sent to photomultiplier tube.
- 20. Image every field of view sequentially through CFPex/CFPem, CFPex/YFPem and YFPex/YFPem (three excitation and emission combinations) and save as donor, transfer and acceptor image files, respectively, through an inbuilt wizard.
- ○ FRET images should be obtained by pixel-by-pixel ratiometric intensity method and efficiency of transfer should be calculated by the ratio of intensity in transfer channel to the quenched (corrected) intensity in the donor channel. Cells transfected with CFP and YFP alone should be imaged under all three previously mentioned excitation and emission combinations, and those imaged should be used to correct for cross-talk. Furthermore, untransfected cells and a field of view with-out cells should be imaged to correct for background, autofluorescence and light scattering.
21. Arrange the appropriate laser output, scan speed and –average to minimize fluorescent photobleacing.
22. Create suitable detector gain and –offset to minimize signal to noise ratio’s (SNR).
23. Use a RiFRET plugin in Image J software (Roszik et al, 2009) to analyze the FRET images and calculate the efficiency of energy transfer.
24. Avoid inhomogeneities from samples by carrying out fluorescence microscopy studies on single cells in mesoscopic regime as previously rationalized (Borejdo et al, 2012; Midde et al, 2013; Midde. K, 2014).
25. Whenever possible, include specific well validated mutants of the same construct as negative controls.
Note: See COMMENTARY section for troubleshooting tips, and special considerations for FRET-based assays to measure activation of G proteins by GIV in living cells.
Concluding remarks
In this article, we have attempted to provide a comprehensive overview of multiple experimental techniques optimized to study GIV/Girdin in a cellular context. This collection of general guidelines, specific protocols, important controls, various available resources, troubleshooting tips and special considerations/advice for both technical issues and interpretation of results (wherever applicable) will serve as an invaluable resource for investigators who wish to study GIV and its role in various cellular processes and will help them optimize the systems of their choice.
REAGENTS AND SOLUTIONS
Use Milli-Q purified water in all recipes
■ 1X Phosphate-Buffered Saline (PBS): 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, 0.05% Tween 20, pH 7.4.
■ IP Lysis Buffer: 25 mM HEPES pH 7.2, 125 mM potassium acetate, 5 mM magnesium acetate, 2 mM DTT, 0.4% Triton X-100, 5X protease inhibitors (Complete; EDTA-free, Roche Diagnostics), 2x Phosphatase inhibitors (Cocktails 1 and 2; Sigma), 0.5mM Sodium orthovanadate
■ 1X PBS-T wash buffer: 1X PBS, 0.1% (v/v) Tween20, 5 mM EDTA, 10 mM MgCl2, 2mM DTT, 0.5mM Sodium orthovanadate
■ 5x Laemli sample buffer: 156 mM Tris pH 6.8, 25% β-mercapto-ethanol (BME), 5% SDS, 25% Glycerol, 0.025% Bromophenol Blue
■ RIPA buffer: 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 0.5-1.0 % sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM Na3VO4, 5X protease inhibitors (Complete EDTA-free, Roche Diagnostics), 2x Phosphatase inhibitors (Cocktails 1 and 2; Sigma)
■ Pulldown Lysis Buffer: 50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.4% [vol:vol] Nonidet P-40, 10 mM MgCl2, 5 mM EDTA, 30 μM GDP, 2 mM DTT, 5X protease inhibitor mixture (Complete EDTA-free, Roche Diagnostics), 0.5mM Sodium orthovanadate, 2X Phosphatase inhibitors (Cocktails 1 and 2; Sigma)
■ 1X Running buffer: 25 mM Tris, 192 mM glycine, 0.1% sodium dodecyl sulfate
■ 1X Transfer Buffer: 25 mM Tris, 192 mM glycine, 20% (v/v) methanol, 0.01–0.04% Sodium dodecyl sulfate.
COMMENTARY
Background Information
GIV is a large (~1870 aa) multimodular protein with many functions at various locations within cells. It is predicted to have multiple functional interactions, post-translational modifications, and spliced isoforms. Significant parts of GIV are unstructured and highly susceptible to proteolytic cleavage. As the premiere member of a novel family of non-receptor GEFs for trimeric G proteins, one that couples G proteins to diverse families of receptors (most of which are known to transduce signals via tyrosine kinases and phosphoproteins), studies on GIV have helped in the emergence of a the tyrosine-based G protein signaling paradigm. Several studies have revealed how deregulated GIV-dependent G protein signaling influences a variety of pathophysiologic processes, thereby introducing GIV to new fields of study and new researchers with diverse interests. The protocols outlined here are intended to help those researchers evaluate GIV's roles in their cell lines and model systems with ease.
Critical Parameters
Protein-Protein Interactions
When protein-protein interactions are being studied, immunoprecipitation of GIV is not the approach of choice in all cases. For example, it is a lot easier to analyze GIV-Gαi interactions by immunoprecipitating the G protein subunit. Thus, it is recommended that immunoprecipitation be tried both ways to assess which is more effective in detecting protein complexes.
If protein complexes being studied are expected to be firmly associated with the actin cytoskeleton, it is recommended that reversible cross-linkers should be used to stabilize protein complexes in situ prior to lysis using harsher buffers, such as radioimmunoprecipitation assay (RIPA) buffer (see recipe), which effectively extracts proteins and multiprotein complexes off of the cytoskeleton.
Transient expression of GIV
Due to the inefficiency and heterogeneous nature of plasmid uptake in cells (wherein few cells take up and express GIV at supraphysiologic levels, while most express none), transient transfection of GIV should be limited to in vitro biochemical assays in which lysates of transfected mammalian cells are used as source of full-length protein. Although PEI provides a way to express GIV more homogeneously and efficiently than most other reagents, it is not recommended for immunofluorescence microscopy (or for any other phenotypic assays) where expression of GIV several fold (> 5-10 fold) above physiologic levels may lead to artifacts. We recommend using cells that have been depleted of endogenous GIV (by shRNA; see Table 1) to generate cell lines stably expressing GIV-WT or other mutant constructs of GIV. Also, when designing epitope-tagged GIV constructs, position of tags should be given special consideration (see Table 7).
Detecting subcellular localization of GIV using whole-cell immnuofluorescence
GIV is localized at various subcellular locations (Fig 1, bottom; Fig 4). To confirm GIV's localization in any compartment, control and GIV-depleted cells should be stained side-by-side. Whenever possible, subcellular fractionation and biochemical validation of such association is warranted. For visualization of GIV on actin cytoskeleton, the PM, the Golgi and endosomes, PFA is the preferred fixative. For visualization of GIV at focal adhesions, centrosome, microtubules and cell-cell junctions, Methanol/Acetone are preferred fixatives.
Special considerations when analyzing G protein activation using FRET-based assays
Tyrosine-based, non-canonical G protein signaling is governed by a new set of rules that differ from GPCR-dependent canonical signaling. These rules have been extensively reviewed (Aznar et al, 2016b), and stated briefly here. First, the rules of receptor engagement are different. Unlike the canonical GPCR/G protein pathway, in which the G proteins engage exclusively with ligand-activated receptor (GPCR)-GEFs, the non-receptor GIV-GEF engages with a diverse array of receptors, including GPCRs, integrins and RTKs (Aznar et al, 2016a; Garcia-Marcos et al, 2009; Ghosh et al, 2010) and, thereby, enables transactivation of G proteins in response to a wide variety of stimuli. Second, the temporal and spatial aspects of tyrosine-based G protein signaling are distinct, and are less restricted than canonical G protein signaling. Canonical signal transduction via trimeric G proteins is spatially and temporally restricted, i.e. triggered exclusively at the plasma membrane (PM), only by agonist activation of GPCRs via a process that completes within a few hundred milliseconds. Tyrosine-based G protein signaling, on the other hand, starts later (~ 5 min and lasts hours) and can occur both at the PM and on internal membranes discontinuous with the PM. Thus, FRET imaging studies evaluating GIV-dependent G protein activation have distinct features that require special considerations. Below are a few of them--
- When the simultaneous expression of 3 constructs is desired (e.g., Gαi1-YFP, CFP-Gβ1, and Gγ2 for use in G protein activation assays), CFP-Gβ1 and Gγ2 are transfected first, followed by Gαi1-YFP (~8-12 h later).
- ○ This is because the Gαi1-YFP plasmid is highly efficient for protein expression, and when all three are co-transfected simultaneously the Gαi1-YFP plasmid suppresses the expression of the CFP-Gβ1 and Gγ2 plasmids.
As outlined in Basic Protocol 7, only those cells expressing equimolar amounts of YFP and CFP should be considered for imaging.
- A variety of ligands can be assessed for their ability to trigger G protein activation, both at the PM and on internal membranes/organelles.
- ○ Before using each ligand, the cell line should be assessed for the presence of each receptor that is being interrogated (see Strategic Planning).
When conducting confocal FRET imaging, the focal plane should be chosen carefully to sample adequate stretches of the PM as well as the internal organelle (e.g., the Golgi; see Fig 5).
Unlike canonical G protein signaling, GIV-dependent G protein signaling is delayed [i.e., occurs ~ 5 min after EGF stimulation (Midde et al, 2015)]. Therefore, the cells should be imaged at appropriate intervals to avoid photobleaching during constant imaging.
To pinpoint a role of GIV and its GEF function, GIV-depleted cells and cells expressing the GEF-deficient GIV-F1685A mutant should be assessed side by side with appropriate controls.
- For reproducible quantitative long-term imaging of Gi activation by FRET, we recommend the use of newer generation of biosensors with high dynamic range built using fluorescent proteins with enhanced photophysical properties and minimal photobleaching, while allowing for the expression of each FRET probe at a set molar ratio.
- ○ Recently such probes were engineered by the groups of Geodhart and Gadella (van Unen et al, 2016) using the brightest and most photostable CFP variant, mTurquoise2, as donor fused to Gαi subunit, and cp173Venus fused to the Gγ2 subunit as acceptor. These new biosensors allow for the expression of the Gαi FRET biosensors together with Gβ1 and Gγ2 from a single plasmid, providing an easy way to ensure balanced expression levels of all three subunits with reduced variation in mammalian cells. This new generation of FRET biosensors for Gαi1, Gαi2 and Gαi3 activation were validated for use in live-cell assays for Gαi activation within the canonical GPCR/G protein pathway.
Figure 5. Measurement of GIV-dependent Gi activation by FRET imaging.
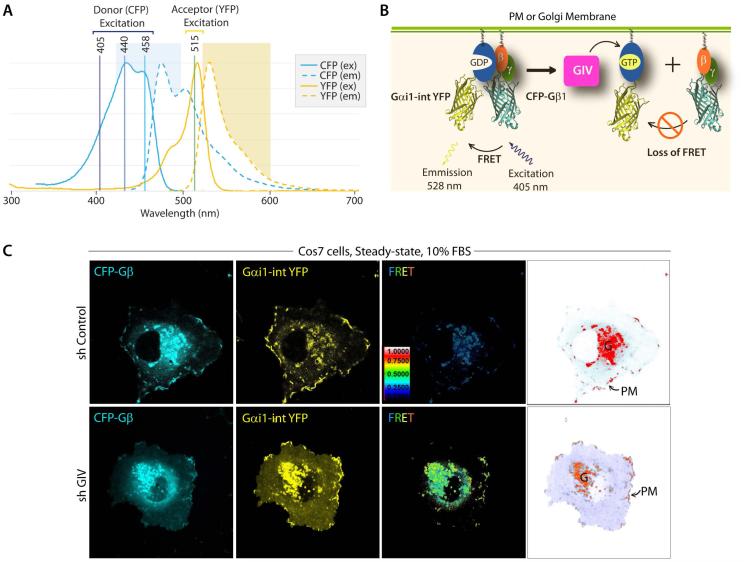
A. Illustration of spectral overlap integral between the fluorescence spectrum of cyan-fluorescent protein (CFP), and the absorption spectrum of yellow-fluorescent protein (YFP). CFP and YFP are mutants of GFP. The 3 lasers that are commonly used for excitation of donor (CFP) and direct activation of acceptor (YFP) are indicated. The 405 nm laser selectively excites CFP with little or no excitation of YFP. B. Schematic for the internally tagged Gαi1-(int)YFP, CFP-Gβ1 and untagged Gγ2 constructs used as paired FRET probes. When associated as a Gαi1-βγ heterotrimer, FRET can be observed. Activation of Gi by GIV-GEF (magenta box) triggers the dissociation of Gαi1-(int)YFP from CFP-Gβ1γ2 heterodimers, resulting in loss of FRET. Thus, high FRET indicates that a majority of Gi1 is inactive, whereas a loss of FRET (ligand-dependent) or low FRET (at steady-state) indicates that a significant pool of Gi1 is active. C. Control (sh Control) and GIV-depleted (sh GIV) COS7 cells were cotransfected with Gαi1-(int)YFP, CFP-Gβ1 and Gγ2 (untagged) and live cells were analyzed by FRET imaging at steady-state, in the presence of 10% serum. From left to right, representative freeze-frame CFP, YFP and FRET images are shown. FRET image panels display intensities of acceptor emission due to efficient energy transfer in each pixel. Right most panel shows the different membranes (in red) that are imaged simultaneously to analyze activation of Gi at both sites-- 1) internal membranes in the perinuclear region, which was largely determined to be the Golgi (G), and 2) peripheral PM ruffles. In control cells, FRET is low (i.e., Gi1 is active) at both locations, whereas in GIV-depleted cells FRET is high at both locations. These observations have been described in details by Lo I et al (Lo et al, 2015).
Troubleshooting
Detection of full length GIV by Immunoblotting
Immunodetection of full length GIV may pose the greatest challenge due to proteolysis of GIV during lysis; this will give rise to bands that are shorter than expected size. This problem is frequently encountered in cells or tissues in which lysosomes are particularly abundant, i.e., phagocytic cells, secretory cells, sperms, liver cells, etc. Such proteolytic breakdown can be overcome at least in part or full by strictly adhering to all steps of lysis (e.g., use of 5X protease inhibitors in lysis buffer, optimized transfer conditions, etc.) outlined in Basic Protocols 1 and 3, minimizing the time taken to process samples, and avoiding temperature rise above 4°C.
Transfection with PEI
Transfection with PEI may require some optimization especially the PEI/DNA ratio to use. We recommend to start with a ratio 1.5 μl PEI/1 μg DNA. PEI transfection is useful for certain assays such as co-immunoprecipitation assays and generation of stable cell lines. Table 6 lists the troubleshooting guide to optimize your transfection protocol.
Whole Cell Immunofluorescence
Certain commercially available anti-GIV primary antibodies may not be compatible with PFA fixation. Also, some subcellular localizations of GIV cannot be visualized easily with PFA fixation (e.g., GIV at centrosomes, or at cell-cell junctions; see Fig 4). Therefore, alternative methods of fixation should be tried in all cases:
(i) Methanol: Add ice cold 100% methanol and place at −20°C for 10 min. Wash with PBS or PBS 1% BSA. Note: Methanol will also permeabilize, but not in all cases; some epitopes are very sensitive to methanol as it can disrupt epitope structure. If this is occurring, try acetone.
(ii) Acetone: Add ice cold 100% acetone and place at −20°C for 5-10 min. Wash with PBS or PBS 1% BSA. Note: Acetone will also permeabilize, and therefore, no further permeabilization step is required.
FRET assays
Although the FRET-based assays are technically challenging, they have the advantage of being able to provide real time spatial and temporal information in single living cells, something no other assay can do. All general considerations for FRET assays apply also to studies on GIV, and researchers who intend to use this approach are advised to refer to (Snapp & Hegde, 2006) during troubleshooting for key guidelines on some of the fundamentals of FRET imaging, e.g., how to determine which protein will be the donor and acceptor, selection of donor and acceptor fluorophores, identification of relevant positive and negative controls, identification of optimal photobleaching conditions for the acceptor, and quantification of FRET measurements. To mitigate the concerns of overexpression-related artifacts, alternatives like direct stochastic optical reconstruction microscopy (dSTORM) imaging (Midde et al, 2015) and antibody-based FRET (Snapp & Hegde, 2006) should be considered; both techniques utilize antibodies on fixed cells to analyze the proximity of endogenous proteins.
Anticipated Results
Basic protocols 1 and 2 will help researchers detect and quantify the expression of full length GIV protein and mRNA, respectively in a consistent, reliable and reproducible manner. Basic protocols 3 and 5 will help in the identification of novel interacting partners or posttranslational modifications (using specific antibodies for Western blotting or by analyzing the IP sample by mass spectrometry). Basic protocol 4 will help achieve high levels of expression of full length GIV in cells for use in biochemical studies. Basic protocol 6 will allow researchers determine sub-cellular localization of GIV as new functions of this protein continues to be unraveled at new locations within cells. Basic protocol 7 will help researchers design FRET-based experiments that will provide insights into the spatiotemporal dynamics of GIV's interactions and G protein activation within each receptor/ligand system.
Time Considerations
Once all necessary reagents are available, most of the protocols listed above can be successfully completed in 2-7 days. However, extra time should be factored in for optimization of protocols if a new cell line or signaling pathway is being investigated.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA100768, CA160911 and DK099226 (to P.G). P.G. was also supported by the Burroughs Wellcome Fund (CAMS award), the American Cancer Society (ACS-IRG 70-002) and by the UC San Diego Moores Cancer Center (P30CA23100). D.B was supported by NIH BUILD Research Stimulation Grant (UL1MD009601) and CSUPERB New Investigator grant (GF00631143). I.L-S was supported by a fellowship from the American Heart Association (AHA #14POST20050025), N.K by pre-doctoral trainings awards from the NCI and NIDDK (T32DK0070202 and T32CA067754), and L.S was supported by a pre-doctoral training award from the NIDDK (T32DK0070202).
Footnotes
Author Contributions: All authors participated in writing and editing specific protocols and figures featured in this article. D.B and P.G compiled all parts and assembled the manuscript text and figures. Shared corresponding authorship between D.B and P.G reflects their equal contribution in thorough vetting of each protocol featured in this article.
REFERENCES
- Aznar N, Kalogriopoulos N, Midde K, Lo IC, Ghosh P. Heterotrimeric G Protein Signaling via GIV/Girdin: Breaking the rules of engagement, space and time. BioEssays. 2016a doi: 10.1002/bies.201500133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar N, Kalogriopoulos N, Midde KK, Ghosh P. Heterotrimeric G protein signaling via GIV/Girdin: Breaking the rules of engagement, space, and time. Bioessays. 2016b;38:379–393. doi: 10.1002/bies.201500133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar N, Midde KK, Dunkel Y, Lopez-Sanchez I, Pavlova Y, Marivin A, Barbazan J, Murray F, Nitsche U, Janssen KP, Willert K, Goel A, Abal M, Garcia-Marcos M, Ghosh P. Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. Elife. 2015;4:e07091. doi: 10.7554/eLife.07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazan J, Dunkel Y, Li H, Nitsche U, Janssen KP, Messer K, Ghosh P. Prognostic Impact of Modulators of G proteins in Circulating Tumor Cells from Patients with Metastatic Colorectal Cancer. Sci Rep. 2016;6:22112. doi: 10.1038/srep22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beas AO, Taupin V, Teodorof C, Nguyen LT, Garcia-Marcos M, Farquhar MG. Galphas promotes EEA1 endosome maturation and shuts down proliferative signaling through interaction with GIV (Girdin) Mol Biol Cell. 2012;23:4623–4634. doi: 10.1091/mbc.E12-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Lopez-Sanchez I, To A, Lo IC, Aznar N, Leyme A, Gupta V, Niesman I, Maddox AL, Garcia-Marcos M, Farquhar MG, Ghosh P. Cyclin-dependent kinase 5 activates guanine nucleotide exchange factor GIV/Girdin to orchestrate migration-proliferation dichotomy. Proc Natl Acad Sci U S A. 2015;112:E4874–4883. doi: 10.1073/pnas.1514157112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borejdo J, Rich R, Midde K. Mesoscopic analysis of motion and conformation of cross-bridges. Biophys Rev. 2012;4:299–311. doi: 10.1007/s12551-012-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broussard JA, Rappaz B, Webb DJ, Brown CM. Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt. Nature protocols. 2013;8:265–281. doi: 10.1038/nprot.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci U S A. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuani F, Conte A, Argenzio E, Marchetti L, Priami C, Polo S, Di Fiore PP, Sigismund S, Ciliberto A. Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells. Nat Commun. 2015;6:7999. doi: 10.1038/ncomms8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceresa BP, Bahr SJ. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem. 2006;281:1099–1106. doi: 10.1074/jbc.M504175200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wei LN, Muller JD. Unraveling protein-protein interactions in living cells with fluorescence fluctuation brightness analysis. Biophys J. 2005;88:4366–4377. doi: 10.1529/biophysj.105.059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng J, Wang T, Nian R, Lau A, Hoi KM, Ho SC, Gagnon P, Bi X, Yang Y. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. MAbs. 2015;7:403–412. doi: 10.1080/19420862.2015.1008351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Rossano AJ, Macleod GT, Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PloS one. 2014;9:e100637. doi: 10.1371/journal.pone.0100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker SP, McCaffery JM, Palade GE, Insel PA, Farquhar MG. Differential distribution of alpha subunits and beta gamma subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. The Journal of cell biology. 1996;133:1027–1040. doi: 10.1083/jcb.133.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneen JL, Ceresa BP. Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res. 2004;294:509–522. doi: 10.1016/j.yexcr.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Dunkel Y, Ong A, Notani D, Mittal Y, Lam M, Mi X, Ghosh P. STAT3 protein up-regulates Galpha-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. J Biol Chem. 2012;287:41667–41683. doi: 10.1074/jbc.M112.390781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M, Tatsumi H, Taya S, Tsuboi D, Kuroda K, Kaneko N, Sawamoto K, Miyamoto R, Jijiwa M, Murakumo Y, Sokabe M, Seki T, Kaibuchi K, Takahashi M. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K, Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Developmental cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Freedman NJ, Kim LK, Murray JP, Exum ST, Brian L, Wu JH, Peppel K. Phosphorylation of the platelet-derived growth factor receptor-beta and epidermal growth factor receptor by G protein-coupled receptor kinase-2. Mechanisms for selectivity of desensitization. J Biol Chem. 2002;277:48261–48269. doi: 10.1074/jbc.M204431200. [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J Biol Chem. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc Natl Acad Sci U S A. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Kietrsunthorn PS, Pavlova Y, Adia MA, Ghosh P, Farquhar MG. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc Natl Acad Sci U S A. 2012;109:1961–1966. doi: 10.1073/pnas.1120538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. G protein coupled growth factor receptor tyrosine kinase: no longer an oxymoron. Cell Cycle. 2015a;14:2561–2565. doi: 10.1080/15384101.2015.1066538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Heterotrimeric G proteins as emerging targets for network based therapy in cancer: End of a long futile campaign striking heads of a Hydra. Aging (Albany NY) 2015c;7:469–474. doi: 10.18632/aging.100781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. The untapped potential of tyrosine-based G protein signaling. Pharmacol Res. 2016;105:99–107. doi: 10.1016/j.phrs.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Beas AO, Bornheimer SJ, Garcia-Marcos M, Forry EP, Johannson C, Ear J, Jung BH, Cabrera B, Carethers JM, Farquhar MG. A G{alpha}i-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol Biol Cell. 2010;21:2338–2354. doi: 10.1091/mbc.E10-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. The Journal of cell biology. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SK, Gilman AG. Gialpha and Gbeta subunits both define selectivity of G protein activation by alpha2-adrenergic receptors. Proc Natl Acad Sci U S A. 2006;103:212–217. doi: 10.1073/pnas.0509763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Adjobo-Hermans MJ, Elzenaar I, Hink MA, Gadella TW., Jr. Quantitative co-expression of proteins at the single cell level--application to a multimeric FRET sensor. PloS one. 2011;6:e27321. doi: 10.1371/journal.pone.0027321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Wang L, He J, Liu X, Zhang H, Li W, Fu L, Ma Y. Girdin, an actin-binding protein, is critical for migration, adhesion, and invasion of human glioblastoma cells. J Neurochem. 2014;131:457–469. doi: 10.1111/jnc.12831. [DOI] [PubMed] [Google Scholar]
- Hatton N, Lintz E, Mahankali M, Henkels KM, Gomez-Cambronero J. Phosphatidic Acid Increases Epidermal Growth Factor Receptor Expression by Stabilizing mRNA Decay and by Inhibiting Lysosomal and Proteasomal Degradation of the Internalized Receptor. Mol Cell Biol. 2015;35:3131–3144. doi: 10.1128/MCB.00286-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JT, Li Y, Yu B, Gao GJ, Zhou T, Li S. Girdin/GIV is upregulated by cyclic tension, propagates mechanical signal transduction, and is required for the cellular proliferation and migration of MG-63 cells. Biochem Biophys Res Commun. 2015;464:493–499. doi: 10.1016/j.bbrc.2015.06.165. [DOI] [PubMed] [Google Scholar]
- Ichimiya H, Maeda K, Enomoto A, Weng L, Takahashi M, Murohara T. Girdin/GIV regulates transendothelial permeability by controlling VE-cadherin trafficking through the small GTPase, R-Ras. Biochem Biophys Res Commun. 2015;461:260–267. doi: 10.1016/j.bbrc.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Ito T, Komeima K, Yasuma T, Enomoto A, Asai N, Asai M, Iwase S, Takahashi M, Terasaki H. Girdin and its phosphorylation dynamically regulate neonatal vascular development and pathological neovascularization in the retina. Am J Pathol. 2013;182:586–596. doi: 10.1016/j.ajpath.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Jiang P, Enomoto A, Jijiwa M, Kato T, Hasegawa T, Ishida M, Sato T, Asai N, Murakumo Y, Takahashi M. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer research. 2008;68:1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Asai N, Enomoto A, Maeda K, Kato T, Ishida M, Jiang P, Watanabe T, Usukura J, Kondo T, Costantini F, Murohara T, Takahashi M. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat Cell Biol. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, Jalink K. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PloS one. 2011;6:e19170. doi: 10.1371/journal.pone.0019170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusser W, Javorschi S, Gleeson MA. Real-Time RT-PCR: cDNA Synthesis. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4114. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- Ley KD, Ellem KA. UVC modulation of epidermal growth factor receptor number in HeLa S3 cells. Carcinogenesis. 1992;13:183–187. doi: 10.1093/carcin/13.2.183. [DOI] [PubMed] [Google Scholar]
- Leyme A, Marivin A, Garcia-Marcos M. GIV/Girdin (Galpha-interacting, Vesicle-associated Protein/Girdin) Creates a Positive Feedback Loop That Potentiates Outside-in Integrin Signaling in Cancer Cells. J Biol Chem. 2016;291:8269–8282. doi: 10.1074/jbc.M115.691550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyme A, Marivin A, Perez-Gutierrez L, Nguyen LT, Garcia-Marcos M. Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. The Journal of cell biology. 2015;210:1165–1184. doi: 10.1083/jcb.201506041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Ear J, Midde K, Lopez-Sanchez I, Aznar N, Garcia-Marcos M, Kufareva I, Abagyan R, Ghosh P. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol Biol Cell. 2014;25:3654–3671. doi: 10.1091/mbc.E14-05-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Ear J, Pavlova Y, Mittal Y, Kufareva I, Ghassemian M, Abagyan R, Garcia-Marcos M, Ghosh P. Tyrosine phosphorylation of the Galpha-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci Signal. 2011;4:ra64. doi: 10.1126/scisignal.2002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo IC, Gupta V, Midde KK, Taupin V, Lopez-Sanchez I, Kufareva I, Abagyan R, Randazzo PA, Farquhar MG, Ghosh P. Activation of Galphai at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling. Developmental cell. 2015;33:189–203. doi: 10.1016/j.devcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Dunkel Y, Roh YS, Mittal Y, De Minicis S, Muranyi A, Singh S, Shanmugam K, Aroonsakool N, Murray F, Ho SB, Seki E, Brenner DA, Ghosh P. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat Commun. 2014;5:4451. doi: 10.1038/ncomms5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Garcia-Marcos M, Mittal Y, Aznar N, Farquhar MG, Ghosh P. Protein kinase C-theta (PKCtheta) phosphorylates and inhibits the guanine exchange factor, GIV/Girdin. Proc Natl Acad Sci U S A. 2013;110:5510–5515. doi: 10.1073/pnas.1303392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Kalogriopoulos N, Lo IC, Kabir F, Midde KK, Wang H, Ghosh P. Focal adhesions are foci for tyrosine-based signal transduction via GIV/Girdin and G proteins. Mol Biol Cell. 2015a;26:4313–4324. doi: 10.1091/mbc.E15-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Ma GS, Pedram S, Kalogriopoulos N, Ghosh P. GIV/girdin binds exocyst subunit-Exo70 and regulates exocytosis of GLUT4 storage vesicles. Biochem Biophys Res Commun. 2015b;468:287–293. doi: 10.1016/j.bbrc.2015.10.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma GS, Aznar N, Kalogriopoulos N, Midde KK, Lopez-Sanchez I, Sato E, Dunkel Y, Gallo RL, Ghosh P. Therapeutic effects of cell-permeant peptides that activate G proteins downstream of growth factors. Proc Natl Acad Sci U S A. 2015a;112:E2602–2610. doi: 10.1073/pnas.1505543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma GS, Lopez-Sanchez I, Aznar N, Kalogriopoulos N, Pedram S, Midde K, Ciaraldi TP, Henry RR, Ghosh P. Activation of G proteins by GIV-GEF is a pivot point for insulin resistance and sensitivity. Mol Biol Cell. 2015d;26:4209–4223. doi: 10.1091/mbc.E15-08-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JZ, Jiang P, Cui SP, Ren YL, Zhao J, Yin XH, Enomoto A, Liu HJ, Hou L, Takahashi M, Zhang B. Girdin locates in centrosome and midbody and plays an important role in cell division. Cancer Sci. 2012;103:1780–1787. doi: 10.1111/j.1349-7006.2012.02378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Albagli O, Poggi MC, Boulukos KE, Pognonec P. Development of a new bicistronic retroviral vector with strong IRES activity. BMC Biotechnol. 2006;6:4. doi: 10.1186/1472-6750-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews ST, Plaisance EP, Kim T. Imaging systems for westerns: chemiluminescence vs. infrared detection. Methods Mol Biol. 2009;536:499–513. doi: 10.1007/978-1-59745-542-8_51. [DOI] [PubMed] [Google Scholar]
- Midde K, Rich R, Marandos P, Fudala R, Li A, Gryczynski I, Borejdo J. Comparison of orientation and rotational motion of skeletal muscle cross-bridges containing phosphorylated and dephosphorylated myosin regulatory light chain. J Biol Chem. 2013;288:7012–7023. doi: 10.1074/jbc.M112.434209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midde KK, Aznar N, Laederich MB, Ma GS, Kunkel MT, Newton AC, Ghosh P. Multimodular biosensors reveal a novel platform for activation of G proteins by growth factor receptors. Proc Natl Acad Sci U S A. 2015;112:E937–946. doi: 10.1073/pnas.1420140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midde KRR, Saxena A, Gryczynski I, Borejdo J, Das HK. Membrane topology of human presenilin-1 in SK-N-SH cells determined by Fluorescence Correlation Spectroscopy and Fluorescent Energy Transfer. Cell Biochemistry and Biophysics. 2014 doi: 10.1007/s12013-014-9999-z. in press. [DOI] [PubMed] [Google Scholar]
- Miyachi H, Mii S, Enomoto A, Murakumo Y, Kato T, Asai N, Komori K, Takahashi M. Role of Girdin in intimal hyperplasia in vein grafts and efficacy of atelocollagen-mediated application of small interfering RNA for vein graft failure. J Vasc Surg. 2014;60:479–489. doi: 10.1016/j.jvs.2013.06.080. e475. [DOI] [PubMed] [Google Scholar]
- Miyachi H, Takahashi M, Komori K. A Novel Approach against Vascular Intimal Hyperplasia Through the Suppression of Girdin. Ann Vasc Dis. 2015;8:69–73. doi: 10.3400/avd.ra.14-00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake H, Maeda K, Asai N, Shibata R, Ichimiya H, Isotani-Sakakibara M, Yamamura Y, Kato K, Enomoto A, Takahashi M, Murohara T. The actin-binding protein Girdin and its Akt-mediated phosphorylation regulate neointima formation after vascular injury. Circ Res. 2011;108:1170–1179. doi: 10.1161/CIRCRESAHA.110.236174. [DOI] [PubMed] [Google Scholar]
- Muramatsu A, Enomoto A, Kato T, Weng L, Kuroda K, Asai N, Asai M, Mii S, Takahashi M. Potential involvement of kinesin-1 in the regulation of subcellular localization of Girdin. Biochem Biophys Res Commun. 2015;463:999–1005. doi: 10.1016/j.bbrc.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Natsume A, Kato T, Kinjo S, Enomoto A, Toda H, Shimato S, Ohka F, Motomura K, Kondo Y, Miyata T, Takahashi M, Wakabayashi T. Girdin maintains the stemness of glioblastoma stem cells. Oncogene. 2012;31:2715–2724. doi: 10.1038/onc.2011.466. [DOI] [PubMed] [Google Scholar]
- Ni D, Xu P, Gallagher S. Immunoblotting and Immunodetection. Curr Protoc Mol Biol. 2016;114:10 18 11–10 18 37. doi: 10.1002/0471142727.mb1008s114. [DOI] [PubMed] [Google Scholar]
- Ni W, Fang Y, Tong L, Tong Z, Yi F, Qiu J, Wang R, Tong X. Girdin regulates the migration and invasion of glioma cells via the PI3K-Akt signaling pathway. Mol Med Rep. 2015;12:5086–5092. doi: 10.3892/mmr.2015.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- Ohara K, Enomoto A, Kato T, Hashimoto T, Isotani-Sakakibara M, Asai N, Ishida-Takagishi M, Weng L, Nakayama M, Watanabe T, Kato K, Kaibuchi K, Murakumo Y, Hirooka Y, Goto H, Takahashi M. Involvement of Girdin in the determination of cell polarity during cell migration. PloS one. 2012;7:e36681. doi: 10.1371/journal.pone.0036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Shen J, Cao J, Zhou Y, Shang L, Jin S, Cao S, Che D, Liu F, Yu Y. Interleukin-17 promotes angiogenesis by stimulating VEGF production of cancer cells via the STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci Rep. 2015;5:16053. doi: 10.1038/srep16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28:3020–3037. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszik J, Lisboa D, Szollosi J, Vereb G. Evaluation of intensity-based ratiometric FRET in image cytometry--approaches and a software solution. Cytometry A. 2009;75:761–767. doi: 10.1002/cyto.a.20747. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kakuwa T, Akimoto K, Koga H, Ohno S. Regulation of epithelial cell polarity by PAR-3 depends on Girdin transcription and Girdin-Galphai3 signaling. Journal of cell science. 2015;128:2244–2258. doi: 10.1242/jcs.160879. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Matsuo Y, Shamoto T, Hirokawa T, Tsuboi K, Takahashi H, Ishiguro H, Kimura M, Takeyama H, Inagaki H. Girdin, a regulator of cell motility, is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol Rep. 2013;29:2127–2132. doi: 10.3892/or.2013.2406. [DOI] [PubMed] [Google Scholar]
- Simpson F, Martin S, Evans TM, Kerr M, James DE, Parton RG, Teasdale RD, Wicking C. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 2005;6:442–458. doi: 10.1111/j.1600-0854.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS. Rational design and evaluation of FRET experiments to measure protein proximities in cells. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb1709s32. Chapter 17: Unit 17 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern KA, Visser Smit GD, Place TL, Winistorfer S, Piper RC, Lill NL. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol Cell Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama L, Sezaki T, Matsuo M, Ueda K, Kioka N. Loss of Dlg5 expression promotes the migration and invasion of prostate cancer cells via Girdin phosphorylation. Oncogene. 2015;34:1141–1149. doi: 10.1038/onc.2014.31. [DOI] [PubMed] [Google Scholar]
- Uhles S, Moede T, Leibiger B, Berggren PO, Leibiger IB. Isoform-specific insulin receptor signaling involves different plasma membrane domains. The Journal of cell biology. 2003;163:1327–1337. doi: 10.1083/jcb.200306093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Unen J, Stumpf AD, Schmid B, Reinhard NR, Hordijk PL, Hoffmann C, Gadella TW, Jr., Goedhart J. A New Generation of FRET Sensors for Robust Measurement of Galphai1, Galphai2 and Galphai3 Activation Kinetics in Single Cells. PloS one. 2016;11:e0146789. doi: 10.1371/journal.pone.0146789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Misaki T, Taupin V, Eguchi A, Ghosh P, Farquhar MG. GIV/girdin links vascular endothelial growth factor signaling to Akt survival signaling in podocytes independent of nephrin. J Am Soc Nephrol. 2015;26:314–327. doi: 10.1681/ASN.2013090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. Journal of cell science. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Li AJ, Han Y, Yin L, Lin MB. Inhibition of Girdin enhances chemosensitivity of colorectal cancer cells to oxaliplatin. World journal of gastroenterology : WJG. 2014;20:8229–8236. doi: 10.3748/wjg.v20.i25.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]