Abstract
Although significant progress has been made in developing therapeutics against Zaire ebolavirus, these therapies do not protect against other Ebola species such as Sudan ebolavirus (SUDV). Here, we describe an RNA interference therapeutic comprising siRNA targeting the SUDV VP35 gene encapsulated in lipid nanoparticle (LNP) technology with increased potency beyond formulations used in TKM-Ebola clinical trials. Twenty-five rhesus monkeys were challenged with a lethal dose of SUDV. Twenty animals received siRNA-LNP beginning at 1, 2, 3, 4 or 5 days post-challenge. VP35-targeting siRNA-LNP treatment resulted in up to 100% survival, even when initiated when fever, viraemia and disease signs were evident. Treatment effectively controlled viral replication, mediating up to 4 log10 reductions after dosing. Mirroring clinical findings, a correlation between high viral loads and fatal outcome was observed, emphasizing the importance of stratifying efficacy according to viral load. In summary, strong survival benefit and rapid control of SUDV replication by VP35-targeting LNP confirm its therapeutic potential in combatting this lethal disease.
Severe and sporadic outbreaks of ebolaviruses occur throughout the African continent. The first epidemic in history of Zaire ebolavirus (EBOV) occurred from December 2013 to January 2016, affecting 28,638 people, with 11,316 fatalities1. The novel location of this latest outbreak in West Africa and the dissemination of infected individuals worldwide have raised concerns for future outbreaks, as there are currently no approved treatments for this deadly disease. Since 2010, Sudan ebolavirus (SUDV) has been responsible for three outbreaks and, until 2014, it had caused the largest outbreak of Ebola haemorrhagic fever on record, with 425 confirmed cases in Uganda in 2000 (ref. 2). Research into vaccines3–6 and therapeutics against SUDV in non-human primate (NHP) models has lagged behind those for EBOV. It is unclear whether any of the therapies currently in development for EBOV infection, such as monoclonal antibodies and nucleos(t)ide analogues7–11, possess broad-spectrum activity against SUDV in the rigorous NHP models of SUDV infection. Recent reports have shown cross-protective efficacy of monoclonal antibodies in guinea pig and mouse models12–16, with no data in the NHP model to date.
RNA interference (RNAi)-based therapeutics offer significant advantages for broad-spectrum-activity drug development through the incorporation of multiple siRNAs targeting clinically relevant species of Ebolavirus into a cocktail. A significant barrier to the in vivo application of RNAi has been the lack of availability of efficient delivery vehicles that promote cellular uptake17; however, use of a lipid nanoparticle (LNP) platform not only protects siRNAs from nuclease degradation in the bloodstream, but also mediates effective delivery to hepatocytes, a major replication cell for SUDV. This technology has been used successfully to protect NHPs against EBOV and Marburg virus infection18–20. Although anti-EBOV siRNA-LNP was recently tested in a very small Phase 2 clinical trial, and appeared safe but did not show clear benefit, the extremely high viral loads (>9 log10 copies ml−1 for all 12 cases) and existing organ injury in the enrolled end-stage patients probably obscured this trial’s ability to detect statistically significant treatment efficacy21. In addition, LNP technology continues to advance beyond the formulations used in the preceding TKM-Ebola clinical trials. Using a new LNP formulation with increased potency compared with that used in the recent clinical trial described in ref. 21, we show that siRNA-LNP treatment rescues NHPs from lethal infection with SUDV when treatment is initiated at an advanced stage of disease. Treatment at a late stage of disease was also able to prevent infection of immune privileged sites, which is considered a protective benefit because EBOV survivors with viral dissemination to such sites develop serious clinical sequelae17–20. Together, these results represent a substantial step forward towards the development of a broad-spectrum siRNA-LNP therapeutic against clinically relevant filoviruses.
Results
siRNAs targeting the viral polymerase, nucleoprotein and viral proteins 35 and 24 are effective against SUDV in vitro
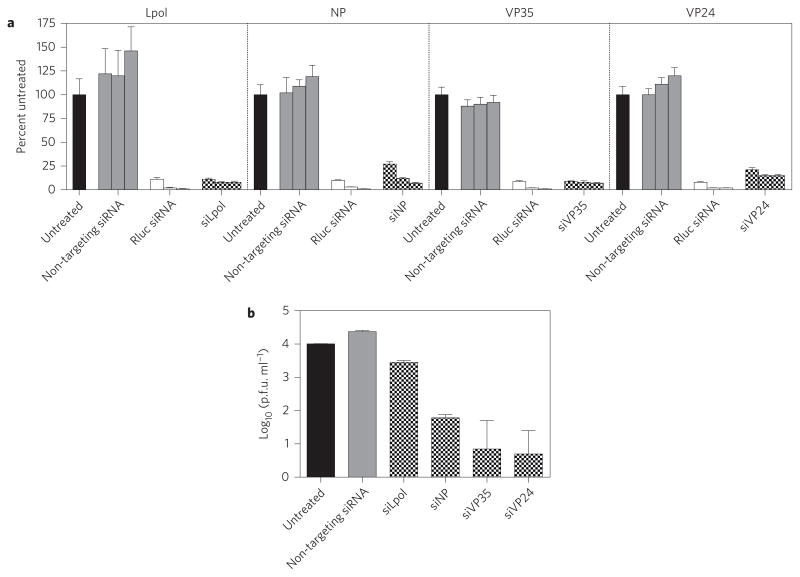
A panel of 48 siRNAs were designed against target sequences in the coding regions of the viral L polymerase (Lpol), viral proteins 24 and 35 (VP24 and VP35) and nucleoprotein (NP), which together are essential for viral replication and assembly26. VP35 is also a known innate immune antagonist preventing RIG-I/MDA5 activation27–29. Target sequences were completely conserved in the 14 genomic sequences of SUDV clinical isolates present in publically available databases (for accession numbers see Methods). siRNAs were screened for mRNA cleavage activity using a dual luciferase reporter assay against the relevant viral mRNA target in HepG2 cells in a similar manner as reported previously30 (Supplementary Fig. 1a–d). The most active siRNAs against each gene target were then chemically modified to abrogate immune stimulatory potential28,31,32. Chemically modified siRNAs possessed cleavage activity against their respective viral mRNA targets in vitro (Fig. 1a) and inhibited viral replication in SUDV-infected HepG2 cells (Fig. 1b and Supplementary Fig. 1e), with viral suppression effects ranging from 0.5 to 3 log10 units versus negative controls. siRNAs demonstrating the greatest level of antiviral activity in vitro against each gene target were selected for further evaluation in an NHP model of SUDV infection. Because the inhibition conferred by the Lpol-targeting siRNA was relatively modest, subsequent antiviral efficacy studies in infected NHPs were conducted only with siRNAs against NP, VP35 and VP24.
Figure 1. siRNAs targeting SUDV Lpol, NP, VP35 and VP24 are active against their viral mRNA targets and display potent antiviral activity in infected cells.
a, Treatment of HepG2 cells transfected with a luciferase reporter plasmid encoding Lpol, NP, VP35 or VP24 viral mRNA sequences with siRNA-LNP results in 85–93% knockdown of gene expression. Three doses of each siRNA at 0.8, 4.0 and 20 nM, respectively, were assessed. Cell lysates were quantitated for Renilla luciferase (Rluc; fused to SUDV target transgene expression) and Firefly luciferase signals. The Renilla luciferase signal was normalized to the Firefly luciferase signal and expressed as percent gene expression relative to a plasmid-only control (pDNA) assigned a value of 100%. An siRNA against Renilla luciferase and a non-virally targeting siRNA against ApoB were used as positive and negative controls, respectively. Data represent the means of technical triplicates ± s.d. of one experiment. b, siRNA-LNP treatment effectively controls viral replication in HepG2 cells infected with SUDV. Reductions ranging from 0.5 to 3 log10 units were observed compared to untreated cells or cells treated with a non-targeting luciferase siRNA negative control. Data shown are means of technical duplicates ± s.e.m. of one experiment.
Silencing of VP35 confers the most survival benefit in the lethal NHP SUDV model
Five studies, totalling 25 animals, were conducted in rhesus macaques infected with a SUDV variant isolated from the Gulu outbreak in Uganda in 2000 (refs 2,33). The initial two studies compared the efficacies of lead siRNA candidates targeting VP35, NP and VP24. The most efficacious siRNA was progressed into subsequent studies with increasing delay in treatment initiation to more stringently evaluate the ability of the siRNA to protect against lethal infection. VP35-targeting siRNA was found to be the most efficacious agent, and a third study evaluated the degree of protection conferred by VP35-targeting siRNA alone or in a cocktail with siRNA against NP. As siRNAs targeting NP have been found to confer protection in Marburg virus-infected NHP (ref. 19), it was of interest to evaluate whether a cocktail of VP35- and NP-targeting siRNAs would result in additional protection. Animals were infected with a lethal dose of SUDV by intramuscular infection (target of 1,000 p.f.u.; for back-titred calculated inoculum doses see Methods) and treated with VP35-, VP24-, NP- siRNAs or siVP35 plus siNP cocktail encapsulated in LNP, with treatment initiated at either 24, 48 or 72 h after infection. LNP potency can be enhanced by improving the individual lipid components or the composition of the lipid mixture. Using principles similar to those of Semple et al.34, the LNP formulation used in these studies was designed to be more potent than formulations used in TKM-Ebola clinical trials (Supplementary Fig. 2) and protects NHPs against EBOV Makona lethal infection20. All siRNA-LNP were administered at 0.5 mg kg−1 for a total of seven daily doses. Three untreated animals were included as controls. The results demonstrated that whereas control animals succumbed to SUDV infection on days 10, 8 and 9 post-infection in each study, respectively, 100% of the animals treated with siVP35-LNP at either 24, 48 or 72 h post-infection (six of six animals in total) survived lethal SUDV challenge. In contrast, 50% survival was observed in animals treated with siRNAs targeting VP24 or NP (one of two treated animals for each group, Fig. 2a). All animals treated with the VP35- and NP-siRNA cocktail survived challenge (two of two), but one animal showed severe signs of disease, whereas all animals treated with VP35-targeting siRNA alone had only mild clinical signs (Supplementary Table 1). This suggested that the degree of protection conferred by the siRNA cocktail was lower than treatment with VP35-siRNA alone. Overall, these results indicate that VP35 knockdown may be of more therapeutic benefit against SUDV infection in vivo than strategies silencing VP24 or NP.
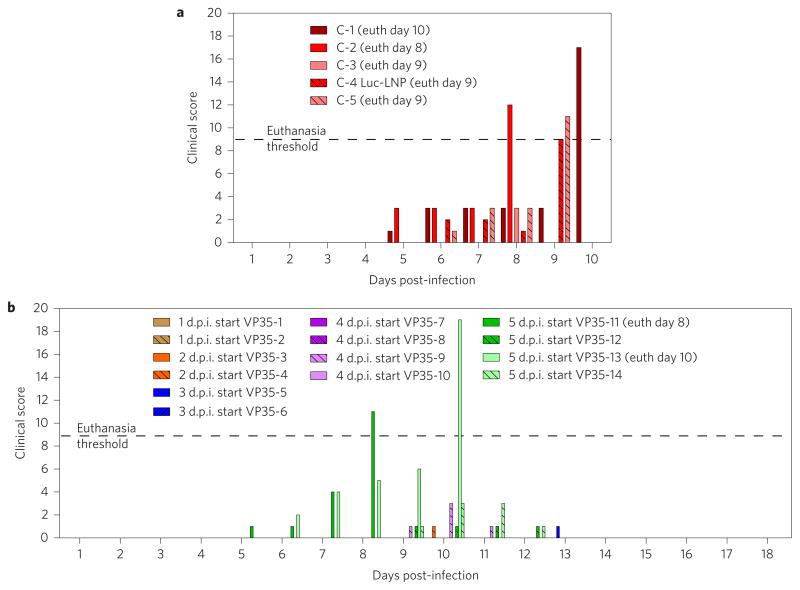
Figure 2. siRNA-LNP treatment in NHPs lethally infected with SUDV results in increased survival and effective viral control.
a, Animals were infected with a target 1,000 p.f.u. of SUDV, then treated with siRNAs targeting VP35 (n = 14), VP24 (n = 2), NP (n = 2) or a cocktail of VP35 and NP siRNAs (n = 2), beginning at 1, 2, 3, 4 or 5 days post-infection (d.p.i.). Untreated (n = 4) or Luc-LNP treated (n = 1) animals were used as negative controls. Treatment with VP35-targeting siRNA in LNP results in significant survival benefit when treatment is initiated up to 5 days post-infection. *P < 0.05, Fisher’s exact test. Treatment with siRNAs targeting VP24 or NP provides partial survival benefit (50%). Animals treated with an siRNA cocktail of siVP35 and siNP have 100% survival when treatment is initiated 3 days post-challenge; however, these animals displayed pronounced clinical signs, suggesting incomplete protection. b,c, Treatment with siVP35-LNP results in effective control of viraemia (b) and viral RNA load in the blood (c). siVP35-LNP treatment initiated at either 1, 2 or 3 days post-infection resulted in undetectable viraemia or viral RNA in four out of six animals. d,e, Viral load reductions are observed after a single dose of siVP35-LNP (d), in contrast to untreated or non-targeting siRNA-LNP treated animals (e). f, Viral RNA in tissues. When treatment was initiated at 4 and 5 days post-challenge, reductions of viral RNA in tissues ranging from 3 to 11 log10 units were observed in surviving animals, while non-survivors had similar viral loads as control animals. Data for brain, spinal cord, pancreas, urinary bladder, gonad, uterus/prostate and eye are shown for siVP35-LNP treated survivor animals only. *P < 0.05, paired t-test (one-sided) of group means for each tissue type, followed by Holm–Sidak correction for multiple comparisons. Data shown represent individual animals.
In a laboratory accident setting the timing of infection is known, and therapeutic intervention can be well informed and swift. In outbreak situations, it is generally unknown when patients become infected, and incubation periods before disease symptom onset are highly variable. In either situation, it is important to understand the efficacy of candidate therapeutic drugs when treatment is initiated after a significant delay post-infection. To this end, the efficacy of siVP35-LNP treatment was assessed in three further studies to determine the degree of protection conferred when treatment initiation was delayed up to five days post-infection. Consistent with data from untreated control animals in previous studies (Supplementary Table 1, C-1 and C-2) and with historical data, in this NHP model of SUDV infection, untreated control animals succumbed to infection on day 9 post-infection (Supplementary Table 1, C-3 and C-5). A control animal administered a non-targeting siRNA directed against a non-endogenous luciferase target encapsulated in LNP also succumbed to infection on day 9, verifying that efficacy requires the specific activity of virus-targeting siRNA. In contrast to the non-targeting siRNA control LNP group, 100% of the animals treated with siVP35-LNP survived when treatment was initiated up to 4 days post-infection (six of six total animals) and 50% survival (two of four animals) was observed when treatment initiation occurred at 5 days post-infection. These results confirm that siVP35-LNP treatment is able to confer survival benefit, even at a time when animals were febrile (seven of ten febrile in the day 3–5 groups) and displayed signs of disease (ten of ten in the day 3–5 groups) before treatment was initiated (Supplementary Table 1).
siVP35-LNP treatment effectively controls viraemia and viral RNA load
SUDV infection is an acute disease, with death in the NHP model occurring 7 to 10 days post-infection. As a direct antiviral agent, siRNA-LNP has utility in controlling viral loads and providing time for adaptive immune responses to develop that ultimately clear the infection. When siVP35-LNP treatment was initiated at either 1, 2 or 3 days post-infection, only two of six animals showed detectable viraemia by plaque assay or had detectable viral RNA quantitated by quantitative real-time PCR (qRT-PCR) throughout the entire study duration (animal VP35-3, 1.40 log10 p.f.u. ml−1 on day 10; animal VP35-4, 5.9 log10 GEq ml−1 on day 6), and these were 5 log10 units and 6.3 log10 units lower than viraemia and viral RNA loads observed in the untreated control animal from the same study (Fig. 2b,c). When siVP35-LNP treatment was initiated at 4 or 5 days post-infection, 3 to 11 log10 unit reductions in viraemia and blood viral RNA loads were observed in animals that survived, whereas viral loads in the blood and tissues of non-survivors in the 5 day treatment delay group were similar to levels attained by untreated control animals (Fig. 2f). Interestingly, after initiation of siVP35-LNP dosing, rapid viral load reductions of up to 4 log10 units were observed by the next sampling time point in all treated animals, including treated animals that eventually succumbed to infection (Fig. 2d,e). As these reductions were observed to occur earlier than the detection of a serological immune response in all treated animals (Supplementary Fig. 3), this suggests that the early viral control was directly attributable to siRNA-LNP treatment. Furthermore, treatment did not interfere with the eventual development of the adaptive immune responses that lead to infection resolution. Thus, siRNA-LNP treatment, with its rapid control of viral replication, extends the temporal window of opportunity for infected animals to develop an effective immune response to overcome lethal disease.
Treatment ameliorates disease pathology and clinical signs
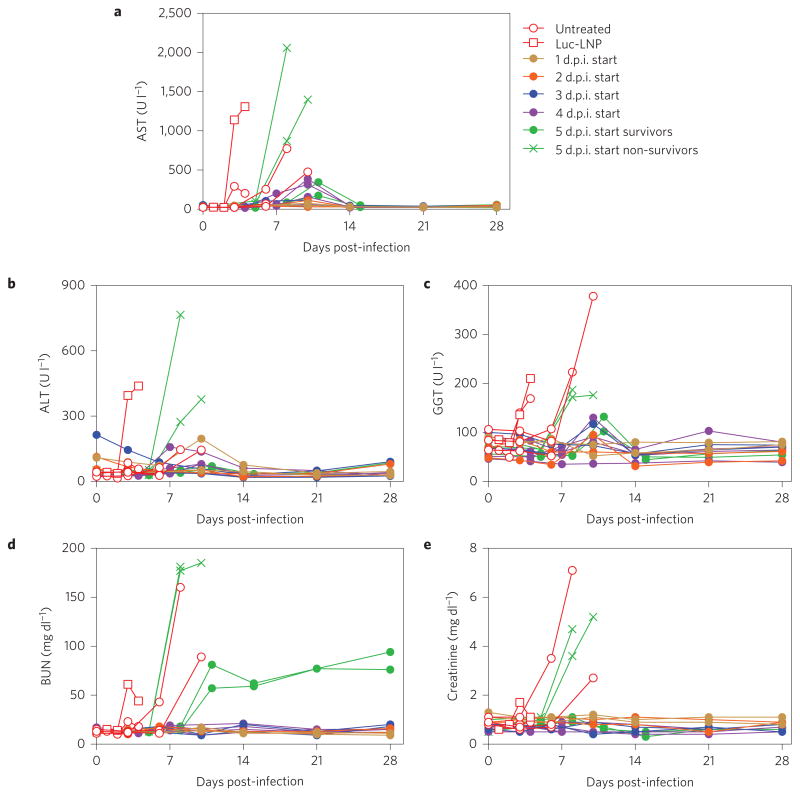
Animals that were untreated or were administered negative control siRNA-LNP exhibited disease signs such as depression, anorexia, petechial rash and haemorrhage, but animals treated with siVP35-LNP displayed only mild clinical signs when treatment was initiated up to 4 days post-infection (Fig. 3a,b). When treatment was initiated at 5 days post-infection, non-survivor animals displayed clinical signs of similar severity and time of onset as the untreated control animal, while treated animals that survived showed mild disease symptoms. These signs had resolved by the pre-determined study endpoint at day 28. In surviving animals, siVP35-LNP treatment was also able to protect against the liver and renal dysfunction typically observed with SUDV infection (Fig. 4), whereas non-survivor animals showed temporal increases in liver enzymes comparable to untreated or non-targeting siRNA-LNP control animals. By the study endpoint, serum chemistry markers had returned to baseline, with the exception of blood urea nitrogen (BUN) levels, which remained at elevated levels in survivor animals in the 5 day treatment delay group. As is typical of SUDV disease, thrombocytopenia and/or lymphopenia was observed at the time of peak viraemia in all infected animals, independent of treatment (Supplementary Fig. 4).
Figure 3. siVP35-LNP treatment ameliorates disease symptoms.
a, Untreated or the non-targeting siRNA control animals exceed euthanasia criteria during days 8–10. b, Surviving siVP35-LNP treated animals show milder clinical signs during the course of infection. Scoring changes measured from baseline included posture/activity level, attitude/behaviour, food and water intake, weight, respiration and disease manifestations such as visible rash, haemorrhage, ecchymosis or flushed skin. A score of ≥9 indicated that an animal met criteria for euthanasia.
Figure 4. siVP35-LNP treatment protects against liver and renal dysfunction induced by SUDV infection.
a–c, Surviving treated animals showed lower levels of the liver enzymes aspartate aminotransferase (AST) (a), alanine aminotransferase (ALT) (b) and gamma-glutamyl transferase (GGT) (c). d,e, Levels of blood urea nitrogen (d, BUN) and creatinine were also reduced in surviving treated animals (e).
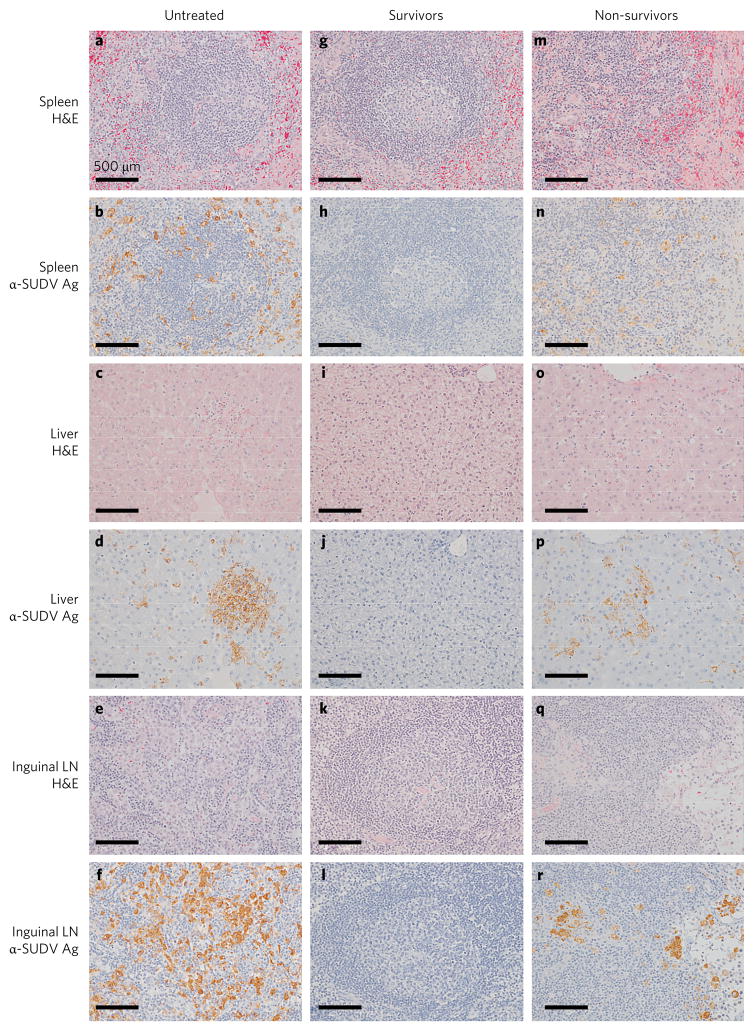
Control and treated animals that succumbed to SUDV infection displayed lesions (observed by haematoxylin and eosin (H&E) staining) in tissues consistent with SUDV infection. Significant lesions included splenic lymphoid depletion with tingible body macrophages and fragmented nuclear debris and expansion of splenic red pulp with fibrin (Fig. 5a,m), multifocal necrotizing hepatitis with sinusoidal leukocytosis (Fig. 5c,o) and mild interstitial pneumonia. No lesions (Fig. 5g,i,k) or immunoreactivity for SUDV antigen (Fig. 5h,j,l) was detected in tissue sections at the day 28 study endpoint in any siVP35-LNP-treated animal that survived challenge. Further investigation of survivor tissues demonstrated no SUDV antigen in the eyes, reproductive tissues or neural tissues (Supplementary Fig. 5). Collectively, these data suggest that treatment with siVP35-LNP confers effective and robust therapeutic benefit towards the post-exposure treatment of SUDV infection.
Figure 5. H&E lesions and immunohistochemistry using anti-SUDV antibody.
Representative control rhesus macaque tissues from untreated animal C-5, 5 day treatment delay siVP35-LNP treated survivor VP35-12 and 5 day treatment delay siVP35-LNP treated non-survivor VP35-11. a,m, Splenic lymphoid depletion. b,n, Spleen with diffuse cytoplasmic immunolabelling of scattered dendritiform mononuclear cells. c,o, Multifocal necrotizing hepatitis with sinusoidal leukocytosis. d,p, Liver with immunolabelled hepatocytes having diffuse cytoplasmic labelling. e,q, Inguinal lymph node lymphoid depletion. f,r, Immunolabelling of mononuclear cells within the subcapsular and medullary sinuses of the inguinal lymph node. g–l, Tissues lacking lesions by H&E (g,i,k) and tissues devoid of immunolabelling (h,j,l) from 5 day treatment delay post-exposure siVP35-LNP treated survivor animal VP35-12. All images were acquired at ×20 magnification. Scale bars, 500 μm. Representative images were taken from control animal C-5 (a–f), treated survivor animal VP35-12 (g–l) and treated non-survivor animal VP35-11 (m–r).
Discussion
Although much attention has focused on the recent EBOV epidemic in West Africa, outbreaks of EBOV and other Ebolavirus species will continue to occur sporadically throughout the African continent with high mortality and morbidity, and risks of global dissemination will remain high. Despite the recent accelerated development of therapeutic and vaccine candidates against EBOV triggered in part by the West African outbreak crisis, a large gap in understanding exists as to whether any of these candidates possess effective activity against other Ebolavirus species such as SUDV. There is thus an acute need to continue the development of strategies with proven activity against other Ebolavirus species, particularly of modalities that have the capacity for broad-spectrum targeting across the different species.
RNAi triggers are valued for their ability to target mRNA transcripts in a highly specific manner to mediate gene silencing. Here, the design of triggers against conserved target sites and the use of a cocktail of triggers against multiple viruses facilitate broad-spectrum targeting. Unlike small-molecule or monoclonal-antibody approaches where broad-spectrum activity requires empirical confirmation, the mechanism of action of an RNAi trigger is well-defined, and activity can be predicted by genomic sequence data that can be made available in an outbreak situation. The modular siRNA-LNP platform technology also permits the adaptation of RNAi triggers to suit, without compromising pharmacokinetic or pharmacodynamics properties (determined by the LNP component) or requiring the development of new manufacturing processes. In addition, advancements in LNP technology that increase potency and/or therapeutic indices can be applied to pre-existing RNAi triggers, generating a superior product. Thus, siRNA-LNP offers a streamlined developmental mechanism capable of product evolution.
Towards this goal of developing a broad-spectrum siRNA-LNP therapeutic, we describe here the identification of an siRNA that, when administered in vivo using LNP delivery technology with increased potency, is able to rescue NHPs lethally challenged with SUDV. VP35-targeting siRNA demonstrated a greater ability to protect against SUDV infection than siRNAs targeting Lpol, VP24 or NP (Figs 1a and 2a). VP35 is a multifunctional protein, with essential roles in viral replication (as a cofactor for Lpol activity) and nucleocapsid formation (with NP and VP24)26. VP35 and VP24 additionally inhibit innate immunity through antagonism of Type I IFN pathways27–29,35–38. However, recent transcriptional signature analysis of infections with EBOV VP24 and VP35 strains with disabled innate antagonizing domains suggests that these functions do not overlap and that VP35, in particular, plays a strong role in repressing dendritic cell maturation39,40. In addition, VP35 also inhibits mammalian RNA interference by binding to Dicer40,41. Taken together, this collective body of data suggests that siRNA knockdown of VP35 may yield greater benefit due to effects on multiple critical nodes in infection, while in parallel enhancing the RNA silencing mechanism. While siRNAs targeting VP35 and Lpol comprise the current TKM-Ebola cocktail and this cocktail shows protective benefit in EBOV-infected NHPs (ref. 20), it is unclear what the contribution of each individual siRNA is towards its efficacy. Furthermore, given the sequence diversity between SUDV and EBOV (SUDV Lpol is estimated to be ~75% similar to EBOV33), it is unclear whether there are differences in these distinct viral species regarding the contribution of Lpol to infection. It is worth noting that the most potent SUDV Lpol-targeting siRNA had strong mRNA cleavage ability but nonetheless mediated only a modest inhibition of viral replication in an infection model (Supplementary Fig. 1).
As monotherapy, the use of a single siRNA theoretically lends itself to a higher risk for the development of viral escape mutants (as opposed to a cocktail approach), although the severe and rapid disease course of infection may factor into a lower probability of escape mutant development. Sequential sequencing data to track viral mutation development from the same individual over the course of infection is sparse, but a recent report from a patient suffering relapse describes at most two non-coding changes in the genome, nine months after initial discharge42, suggesting that viral mutation rates within an individual are low. Given an intended eventual incorporation into a broad-spectrum siRNA cocktail-LNP therapeutic, activity confirmation of this individual SUDV component is a necessary step, as captured in this proof-of-concept work. In addition, siRNA-LNP may form one part of potential combination treatment strategies, and using multiple drug modalities would mitigate the risk of viral escape.
Substantial survival benefit was observed with siVP35-LNP treatment, resulting in 100% survival when treatment was initiated up to 4 days post-infection and 50% survival when treatment began at 5 days post-infection, at a point when animals were exhibiting high viral loads, fever, petechial rash and anorexia. At the time of treatment initiation, a few days before the anticipated mean time to death, blood viral RNA loads ranged from 6.3 to 10.2 log10 GEq ml−1 (Fig. 2c). Animals with viral RNA loads less than 7.0 log10 GEq ml−1 at the time of treatment start survived challenge, whereas those animals with loads greater than 8.9 log10 GEq ml−1 succumbed. Interestingly, these results mirror a threshold previously reported from analysis of survivors and fatal cases in the 2000–2001 SUDV Gulu outbreak, where it was determined that viral loads of >8 log10 copies ml−1 were ≥90% predictive of a fatal outcome2. Taken together, these findings indicate that the NHP challenge model closely approximates human SUDV infection and, importantly, suggest that for animals treated with siVP35-LNP that did not survive infection, treatment came too late: the high viral loads (and pathology) present in these animals had already determined a fatal outcome. These observations also reflect accumulating data from the recent EBOV epidemic in West Africa, where it is becoming clear that once viral loads exceed certain thresholds in patients and pathologies progress, there is very little chance for successful therapeutic intervention43,45. Consistent with this, results from the TKM-Ebola-Guinea Phase 2 clinical trial suggest that high viral loads (>9 log10 copies ml−1) and existing organ injury in subjects probably obscured the ability to detect treatment benefit21. In the 14 patients who received TKM-Ebola-Guinea, the viral geometric mean before treatment was 9.35 log10 copies ml−1 (ref. 21), a viral load level that has been associated with a fatal outcome in >90% of cases43–45. In addition to high viral loads, seven patients before treatment initiation presented with additional signs that have also been associated with a fatal outcome, such as advanced age (two patients were over 60 years old), haemorrhagic signs, hiccough and tachypnoea21. The RAPIDE-TKM trial determined efficacy by comparing the survival of treated patients to a predetermined survival probability threshold at 14 days after admission21. This predetermined threshold was based on a historical data set of patient survival when provided standard of care. However, given that the authors observe that the patients administered TKM-Ebola-Guinea probably presented with higher baseline viral loads and a greater degree of illness than what had been observed in their historical data set21, this would have made any comparisons to historical data (and determination of efficacy) difficult. Efficacy determination was also made uncertain by the grouping together of data from all treated patients, regardless of baseline viral load and clinical pathology characteristics. Thus, data from currently ongoing and any future clinical trials against Ebolaviruses should be stratified according to viral load and clinical pathology to better detect indications of efficacy.
Despite the presence of high viral loads, all animals treated with siVP35-LNP, regardless of survival, showed reductions of up to 4 log10 units in both infectious viral particle and viral RNA loads within 3 days after administration of a single dose, suggesting rapid control of viral replication. The rapid control of SUDV replication mediated by siVP35-LNP treatment confers an important advantage against this extremely acute filovirus infection, where viral titres in patients can climb quickly over the course of a few days. Although data are lacking for SUDV, EBOV infection can disseminate to secondary organs such as the eyes, heart and kidneys. Recent reports of clinical sequelae in survivors from filovirus outbreaks suggest that viral dissemination to immune-privileged organs such as the eyes and brain may result in life-long pathologies in these individuals, despite their having survived the acute disease22,23,25,46. In addition, past and current reports of filoviruses in the semen of survivors24,47–49 is concerning. Thus, effective control of viral replication in patients during the acute phase of infection by siRNA-LNP treatment may also yield long-term benefits by preventing viral spread to these organs, as reported here.
Together, the results presented here strongly support the continued development of siRNA-LNP as one weapon in the arsenal against haemorrhagic fever viruses and inclusion of SUDV VP35-targeting siRNA in a broad-spectrum siRNA cocktail for pan-Ebola antiviral activity. Demonstrated protection in this uniformly lethal NHP model represents a high bar for assessing efficacy, as animals are infected with a high viral dose that mimics the worst-case scenario of a needle-stick injury with concentrated viral material. The survival benefit and rapid control of viral replication by siVP35 treatment, in an LNP formulation with increased potency, illustrate the strong potential of this evolving technology platform in combatting highly lethal viral infections.
Methods
Dual luciferase reporter assays
A total of 48 siRNA candidates were designed to target conserved regions of SUDV Lpol, NP, VP35 and VP24 mRNA transcripts across SUDV genomic sequences present in Genbank (accession nos. AY729654, KC242783, NC_006432, KU182912, KT750754, KR063670, KC545392, KC545391, KC545390, KC545389, KC589025, JN638998, FJ968794 and EU338380). Sequences with homology to human mRNAs of 16 or more contiguous bases using BLAST (v.2.2.13) were excluded. siRNAs were evaluated in dual luciferase reporter assays for mRNA cleavage ability. The psiCHECK2 (Promega) vector was used to construct the SUDV reporter plasmids used in this study (Life Technologies and Genscript). Briefly, to construct the SUDV reporter plasmids, the mRNA sequences of SUDV VP24, VP35, VP40, NP and Lpol (Genbank accession no. AY729654) were cloned into the 3′ UTR of the Rluc gene between the SgfI and NotI restriction sites to allow for the detection of siRNA activity as represented by decreased Rluc activity. siRNAs were synthesized at Integrated DNA Technologies. Individual duplexes were encapsulated in LNP by the process of spontaneous vesicle formation as previously reported50. The resulting LNPs were dialysed against PBS and sterilized through a 0.2 μm filter before use. siRNAs targeting Renilla luciferase and ApoB8 (synthesized by Integrated DNA Technologies) were also encapsulated in LNP and were included as positive and negative controls, respectively.
HepG2 cells were obtained from the American Type Culture Collection (catalogue no. HB-8065), and no further authentication was carried out. HepG2 cells were transfected with the SUDV psiCHECK2 plasmid constructs using Lipofectamine 2000 (Life Technologies) and treated with siRNA-LNP at 0.8, 4.0 and 20 nM. Cells tested negative for mycoplasma contamination. Transfected cells were incubated for 48 h, followed by measurement of Renilla and firefly luciferase activities using a luminometer. Results were expressed as a percentage of the Renilla: firefly luciferase activity in cells transfected with the reporter plasmid only (no siRNA treatment).
In vitro infections
HepG2 cells were seeded at 6 × 104 cells per well in 24-well culture plates and incubated at 37 °C/5% CO2 overnight before treatment with 10 nM siRNA-LNP. Cells were obtained from the American Tupe Culture Collection (catalogue no. HB-8065) and no further authentication was carried out. Cells tested negative for mycoplasma contamination. Cells were incubated for 24 h before infection with 0.5 multiplicity of infection of SUDV Gulu (Genbank accession no. KU182912). Cells were incubated with virus for 1 h, then washed four times with PBS. Culture medium was replaced and cells were incubated for 48 h post-infection before collection of cell supernatants for RNA extraction by Trizol and qRT-PCR assessment.
LNP encapsulation of siRNA used in NHP studies
siRNAs were formulated into LNP by a process of stepwise ethanol dilution and spontaneous particle formation. Briefly, an ethanolic solution of lipids such as 3-N-(-methoxy poly(ethylene glycol) 2000)carbamoyl-1,2-dimyristyloxy-propylamine (PEG-C-DMA), 3-(dilinoleylmethoxy)-N,N-dimethylpropan-1-amine (DLin-MP-DMA), cholesterol and dipalmitoylphosphatidylcholine (DPPC) was mixed with a PBS solution containing nucleic acid. The resulting LNPs were subsequently concentrated and diafiltered against 20 wash volumes of PBS (pH 7.4) using a cross-flow ultrafiltration cartridge (GE Healthcare) and finally sterile-filtered through Acrodisc 0.8/0.2 mm Supor filters (Pall Corp). The degree of encapsulation (92–98%) was determined using Ribo-Green (Invitrogen) and a Varian Cary Eclipse Fluorimeter. Particle sizes (85–90 nm) and polydispersity values (<0.1) were determined using a Malvern Nano Series Zetasizer.
The viral genome sequences targeted by the lead siRNAs evaluated in in vitro and NHP studies were as follows: siNP-1 (5′-TCCTCAAACTCAAACTAAT-3′); siNP-2 (5′-CCTCAAACTCAAACTAATA-3′); siNP-3 (5′-GCTCCTCCTACAA TTCTAA-3′); siNP-4 (5′-GAGCCTACAACATGGATAA-3′); siNP-5 (5′-AGCCTA CAACATGGATAAA-3′); siNP-6 (5′-AGGGTATTTGTCAACATAT-3′); siNP-7 (5′-TGCGAATGACACAGTAATA-3′); siNP-8 (5′-GGTCTTCTGATTGTAAAGA-3′); siNP-9 (5′-GCCAAGCATGGAGAATATG-3′); siNP-10 (5′-GAATGAGATCAGCTTCCAG-3′); siNP-11 (5′-GAGGCAATCAATTATTATC-3′) siNP-12 (5′-GA GCTGCTTGTGTCAATTT-3′); siNP-13 (5′-AGCTGCTTGTGTCAATTTA-3′); siNP-14 (5′-GAGCTTAGGAGGATAATAT-3′); siVP35-1 (5′-GATGAAGA TTAAAACCTTC-3′); siVP35-2 (5′-ATGAAGATTAAAACCTTCA-3′); siVP35-3 (5′-TGAAGATTAAAACCTTCAT-3′); siVP35-4 (5′-GAAGATTAAAACCTTCATC-3′); siVP35-5 (5′-AAGATTAAAACCTTCATCA-3′); siVP35-6 (5′-ACCGCT AACAGAGGTGTTT-3′); siVP35-7 (5′-CGCTAACAGAGGTGTTTGT-3′); siVP35-8 (5′-GCCATAAATTCGGTGATAT-3′); siVP35-9 (5′-TGCAGCAACA GAAGCATAT-3′); siVP35-10 (5′-GCAGCAACAGAAGCATATT-3′); siVP35-11 (5′-TTCGGGCGACCTTACATTT-3′); siVP35-12 (5′-CGAGGAGATATACCCAAAG-3′); siVP24-1 (5′-GGCTAGGGTTTATAGTTAA-3′); siVP24-2 (5′-TCTGCAAGTAATTGTTTAG-3′); siVP24-3 (5′-CCGGTACAACTTGGTAACA-3′); siVP24-4 (5′-GGCTGGACTTGAGTTTGAT-3′); siVP24-5 (5′-CCTGTTAAATCGTCTTAAA-3′); siVP24-6 (5′-TTGGGCCTTGAGGGTAATT-3′); siVP24-7 (5′-GGGATTCTTGACCAATTAA-3′); siVP24-8 (5′-GCTGA TTGGTTACTAACAA-3′); siVP24-9 (5′-GAGCATGAGGATGTTATCT-3′); siVP24-10 (5′-GAGGATGTTATCTCTTATA-3′); siLpol-1 (5′-GAGGAAGATTAA GAAAAAG-3′); siLpol-2 (5′-AGACCAATGTGACCTAGTG-3′); siLpol-3 (5′-GA TGCATCTTGCAGTTATT-3′); siLpol-4 (5′-CGGGATTGAATTCCTACAT-3′); siLpol-5 (5′-GCTCCCTTCATCAAATATT-3′); siLpol-6 (5′-CGGCAAGGAACAA GTTAAT-3′); siLpol-7 (5′-GAGGGAACCAGAACTTTAT-3′); siLpol-8 (5′-GCA CGTGATAGCAACATTA-3′); siLpol-9 (5′-GCCCAATACTTAACATACA-3′); siLpol-10 5′-GGGCCCACGTTTAAATATT-3′); siLpol-11 (5′-GCTGGATTAACAA TTATCT-3′) and siLpol-12 (5′-GGAGTTGGTAACTGATTAT-3′).
Animal challenge
All animal studies were conducted under a protocol approved by the University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee (IACUC). IACUC approval of animal studies performed at UTMB is in compliance with the Animal Welfare Act, PHS Policy and other Federal statutes and regulations relating to animals and experiments involving animals. The facilities where this research was conducted are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adhere to principles stated in the eighth edition of the Guide for the Care and Use of Laboratory Animals, National Research Council.
Twenty-five healthy adult rhesus macaques (Macaca mulatta) of Chinese origin (4–8 kg, 13 males and 12 females, 4–7 years old) were inoculated intramuscularly (i.m.) with target 1,000 p.f.u. of SUDV Gulu strain (back-titred calculated inoculum doses of 813, 1,213, 1,225, 826 and 863 p.f.u. for each of the five studies). In total, five studies were conducted, with five animals per study. Sample sizes were based on the availability of rhesus macaques. Animals were randomized with Microsoft Excel into treatment or control groups. The most efficacious siRNA from each study was selected for further assessment in subsequent studies with increasing time delay after viral infection. The first study compared the efficacies of siRNAs targeting VP35 and VP24. siVP35-LNP and siVP24-LNP (0.5 mg kg−1) were administered to two SUDV Gulu-infected macaques, each by bolus i.v. infusion 1 day after SUDV challenge. The control animal was not treated. The second study compared the efficacy of siVP35 with an siRNA targeting NP, when treatment was initiated at 2 days post-challenge. The third, fourth and fifth studies assessed the efficacy of siVP35-LNP when treatment was initiated at 3, 4 and 5 days post-infection. The number of animals per treatment group ranged from two to four animals, and a total of seven daily doses of 0.5 mg kg−1 (siRNA dose) were administered per treatment course. All animals were given physical examinations, and blood was collected at the time of challenge and on days 3, 6, 10, 14, 21 and 28 (for 1, 2 and 3 day treatment delay studies) or on days 4, 7, 10, 14, 21 and 28 (for 4 day treatment delay study) or on days 5, 8, 10, 15, 21 and 28 (for 5 day treatment delay study) after SUDV challenge or at the time of euthanasia. In addition, all animals were monitored daily and scored for disease progression with an internal filovirus scoring protocol approved by the UTMB IACUC. The scoring changes measured from baseline included posture/activity level, attitude/behaviour, food and water intake, weight, respiration and disease manifestations such as visible rash, haemorrhage, ecchymosis or flushed skin. A score of ≥9 indicated that an animal met the criteria for euthanasia. All studies were not blinded, and all animals were included in analyses.
Detection of viraemia and viral RNA
RNA was isolated from whole blood or tissues using the Viral RNA Mini Kit or RNeasy Kit (Qiagen) using 100 μl of blood into 600 μl of buffer AVL, or 100 mg of tissue according to the manufacturer’s instructions, respectively. Primers or probe targeting the L gene of SUDV were used for qRT-PCR with the probe used here being 6-carboxyfluorescein (6FAM)-5′-CAT CCA ATC AAA GAC ATT GCG A3′-6 carboxytetramethylrhodamine (TAMRA; Life Technologies). SUDV RNA was detected using the CFX96 detection system (BioRad Laboratories) in One-step probe qRT-PCR kits (Qiagen) with the following cycle conditions: 50 °C for 10 min, 95 °C for 10 s and 40 cycles of 95 °C for 10 s and 59 °C for 30 s. Threshold cycle (CT) values representing SUDV genomes were analysed with CFX Manager Software, and data are shown as means ± s.d. of technical replicates. To create the GEq standard, RNA from SUDV stocks was extracted and the number of SUDV genomes was calculated using Avogadro’s number and the molecular weight of the SUDV genome.
Virus titration was performed by plaque assay with Vero E6 cells from all serum samples, as previously described5. Briefly, increasing tenfold dilutions of the samples were adsorbed to Vero E6 monolayers in duplicate wells (200 μl); the limit of detection was 25 p.f.u. ml−1.
Haematology, serum biochemistry and blood coagulation
Total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, haematocrit values, total haemoglobin concentrations, mean cell volumes, mean corpuscular volumes and mean corpuscular haemoglobin concentrations were analysed from blood collected in tubes containing EDTA using a laser-based haematologic analyser (Beckman Coulter). Serum samples were tested for concentrations of albumin, amylase, alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), gamma-glutamyltransferase (GGT), glucose, cholesterol, total protein, total bilirubin (T’BIL), BUN, creatinine (CRE) and C-reactive protein (CRP) using a Piccolo point-of-care analyser and Biochemistry Panel Plus analyser discs (Abaxis).
Histopathology and immunohistochemistry
Necropsy was performed on all subjects. Tissue samples of all major organs were collected for histopathological and immunohistochemical examination, immersion-fixed in 10% neutral buffered formalin and processed for histopathology as previously described5. For immunohistochemistry, specific anti-SUDV immunoreactivity was detected using an anti-SUDV VP40 protein rabbit primary antibody (Integrated BioTherapeutics) at a 1:4,000 dilution. In brief, tissue sections were processed for immunohistochemistry using the Dako Autostainer (Dako). The secondary antibody used was biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 followed by Dako LSAB2 streptavidin-HRP (Dako). Slides were developed with Dako DAB chromagen (Dako) and counterstained with haematoxylin. Non-immune rabbit IgG was used as a negative control. In Fig. 5, spleen, liver and inguinal lymph node representative images were taken at ×40 and for spleen were taken at ×20 from control animal C-5 (Fig. 5a–f), treated survivor animal VP35-12 (Fig. 5g–l) and treated non-survivor animal VP35-11 (Fig. 5m–r).
Statistical analyses
Analysis was conducted with Graphpad Prism software (version 6.04). A Fisher’s exact test was used to compare survival between untreated and treated animals. A paired t-test (one-sided) was used to compare untreated and treated group means for viral RNA levels in the various tissues assessed at necropsy.
Supplementary Material
Acknowledgments
The authors thank V. Borisevich for assistance with clinical pathology assays performed in the GNL BSL-4 laboratory, J. Heyes and K. Lam for data comparing endogenous gene silencing potency across LNP formulations, and S. Klassen for his assistance with siRNA-LNP preparation. This study was supported by the Department of Health and Human Services, National Institutes of Health grant no. U19AI109711 to T.W.G. and E.P.T., and UC7AI094660 for BSL-4 operations support of the Galveston National Laboratory.
Footnotes
Accession codes. SUDV genomic sequences that served as references for siRNA design are located in the GenBank database under accession nos. AY729654, KC242783, NC_006432, KU182912, KT750754, KR063670, KC545392, KC545391, KC545390, KC545389, KC589025, JN638998, FJ968794 and EU338380.
Author contributions
R.U.-B. and M.R. designed the siRNA and R.U.-B. conducted the dual luciferase reporter studies. R.U.-B., C.E.M., M.R., I.M. and T.W.G. designed the in vitro infection study. K.N.A. and C.E.M. performed the in vitro infection study. E.P.T., C.E.M., A.C.H.L., I.M. and T.W.G. conceived and designed the NHP studies. C.E.M., J.B.G., D.J.D. and T.W.G. performed the NHP challenge and treatment experiments and conducted clinical observations of the animals. J.B.G., K.N.A. and D.J.D. performed the clinical pathology assays. J.B.G. performed the SUDV infectivity assays. C.E.M. and K.N.A. performed the PCR assays. E.P.T., C.E.M., J.B.G., K.N.A., D.J.D., K.A.F., A.S.K, A.C.H.L. and T.W.G. analysed the data. K.A.F. performed histological and immunohistochemical analysis of the data. E.P.T., C.E.M., A.C.H.L. and T.W.G. wrote the paper. All authors had access to all of the data and approved the final version of the manuscript.
Additional information
Supplementary information is available online
Competing interests
A.L., I.M. and T.W.G. claim intellectual property regarding RNA interference for the treatment of filovirus infections. I.M. and T.W.G. are co-inventors on US patent 7,838,658 (‘siRNA silencing of filovirus gene expression’) and A.L., I.M. and T.W.G. are co-inventors on US patent 8,716,464 (‘Compositions and methods for silencing Ebola virus gene expression’). The other authors declare no competing interests. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the University of Texas Medical Branch.
References
- 1.Ebola Situation Report. World Health Organization; 2016. http://apps.who.int/iris/bitstream/10665/204172/1/ebolasitrep_20Jan2016_eng.pdf?ua=1. [Google Scholar]
- 2.Towner JS, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol. 2004;78:4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geisbert TW, et al. Vesicular stomatitis virus-based vaccines protect nonhuman primates against aerosol challenge with Ebola and Marburg viruses. Vaccine. 2008;26:6894–6900. doi: 10.1016/j.vaccine.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert AS, et al. Venezuelan equine encephalitis virus replicon particle vaccine protects nonhuman primates from intramuscular and aerosol challenge with ebolavirus. J Virol. 2013;87:4952–4964. doi: 10.1128/JVI.03361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geisbert TW, et al. Single-injection vaccine protects nonhuman primates against infection with Marburg virus and three species of Ebola virus. J Virol. 2009;83:7296–7304. doi: 10.1128/JVI.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratt WD, et al. Protection of nonhuman primates against two species of Ebola virus infection with a single complex adenovirus vector. Clin Vaccine Immunol. 2010;17:572–581. doi: 10.1128/CVI.00467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettitt J, et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med. 2013;5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 8.Qiu X, et al. Sustained protection against Ebola virus infection following treatment of infected nonhuman primates with ZMAb. Sci Rep. 2013;3:3365. doi: 10.1038/srep03365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren TK, et al. A single phosphorodiamidate morpholino oligomer targeting VP24 protects rhesus monkeys against lethal Ebola virus infection. mBio. 2015;6:e02344–14. doi: 10.1128/mBio.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynard O, et al. Identification of a new ribonucleoside inhibitor of Ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holtzberg F, et al. Pan-ebolavirus and pan-filovirus mouse monoclonal antibodies protection against Ebola and Sudan viruses. J Virol. 2015;90:266–278. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keck Z, et al. Macaque monoclonal antibodies targeting novel conserved epitopes within filovirus glycoprotein. J Virol. 2015;90:279–291. doi: 10.1128/JVI.02172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frei J, et al. Bispecific antibody affords complete post-exposure protection of mice from both Ebola (Zaire) and Sudan viruses. Sci Rep. 2016;6:19193. doi: 10.1038/srep19193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuyama W, et al. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep. 2016;6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell K, et al. Antibody treatment of Ebola and Sudan virus infection via a uniquely exposed epitope within the glycoprotein receptor-binding site. Cell Rep. 2016;15:1514–1526. doi: 10.1016/j.celrep.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haussecker D. Current issues of RNAi therapeutics delivery and development. J Control Rel. 2014;195:49–54. doi: 10.1016/j.jconrel.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 18.Geisbert TW, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thi EP, et al. Marburg virus infection in nonhuman primates: therapeutic treatment by lipid-encapsulated siRNA. Sci Transl Med. 2014;6:250ra116. doi: 10.1126/scitranslmed.3009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thi EP, et al. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunning J, et al. Experimental treatment of Ebola virus disease with TKM-130803 A single arm phase 2 clinical trial. PLoS Med. 2016;13:e1001997. doi: 10.1371/journal.pmed.1001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattia JG, et al. Early clinical sequelae of Ebola virus disease in Sierra Leone: a cross-sectional study. Lancet Infect Dis. 2015;16:331–338. doi: 10.1016/S1473-3099(15)00489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark DV, et al. Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study. Lancet Infect Dis. 2015;15:905–912. doi: 10.1016/S1473-3099(15)70152-0. [DOI] [PubMed] [Google Scholar]
- 24.Uyeki TM, et al. Ebola virus persistence in semen of male survivors. Clin Inf Dis. 2016;62:1552–1555. doi: 10.1093/cid/ciw202. [DOI] [PubMed] [Google Scholar]
- 25.Tiffany A, et al. Ebola virus disease complications as experienced by survivors in Sierra Leone. Clin Inf Dis. 2016;62:1360–1366. doi: 10.1093/cid/ciw158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prins KC, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basler CF, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ursic-Bedoya R, et al. Protection against lethal Marburg virus infection mediated by lipid encapsulated small interfering RNA. J Infect Dis. 2014;209:562–570. doi: 10.1093/infdis/jit465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. doi: 10.1016/j.virusres.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 35.Leung DW, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol. 2010;84:1169–1175. doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid SP, et al. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfmann P, Neumann G, Kawaoka Y. The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J Infect Dis. 2011;204(Suppl 3):S953–S956. doi: 10.1093/infdis/jir325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilinykh PA, et al. Different temporal effects of Ebola virus VP35 and VP24 proteins on global gene expression in human dendritic cells. J Virol. 2015;89:7567–7583. doi: 10.1128/JVI.00924-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haasnoot J, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathogens. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs M, et al. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet. 2016;388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De La Vega MA, et al. Ebola viral load at diagnosis associates with patient outcome and outbreak evolution. J Clin Invest. 2015;125:4421–4428. doi: 10.1172/JCI83162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzpatrick G, et al. The contribution of Ebola viral load at admission and other patient characteristics to mortality in a Médecins sans Frontières Ebola case management centre, Kailahun, Sierra Leone, June–October 2014. J Infect Dis. 2015;212:1752–1758. doi: 10.1093/infdis/jiv304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schieffelin JS, et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med. 2014;371:2092–2100. doi: 10.1056/NEJMoa1411680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howlett P, et al. Ebola virus disease complicated by late-onset encephalitis and polyarthritis, Sierra Leone. Emerg Infect Dis. 2016;22:150–152. doi: 10.3201/eid2201.151212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martini GA, Schmidt HA. Spermatogenic transmission of the ‘Marburg virus’ (Causes of ‘Marburg simian disease’) Klinische Wochenschrift. 1968;46:398–400. doi: 10.1007/BF01734141. [DOI] [PubMed] [Google Scholar]
- 48.Mate SE, et al. Molecular evidence of sexual transmission of Ebola virus. N Engl J Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deen GF, et al. Ebola RNA persistence in semen of Ebola virus disease survivors—preliminary report. N Engl J Med. 2015 doi: 10.1056/NEJMoa1511410. http://dx.doi.org/10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed]
- 50.Ma H, et al. Formulated minimal-length synthetic small hairpin RNAs are potent inhibitors of hepatitis C virus in mice with humanized livers. Gastroenterology. 2014;146:63–66. doi: 10.1053/j.gastro.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.