Abstract
In contrast to the highly conserved mitogenomic structure and organisation in most animals (including rotifers), the two previously sequenced monogonont rotifer mitogenomes were fragmented into two chromosomes similar in size, each of which possessed one major non-coding region (mNCR) of about 4–5 Kbp. To further explore this phenomenon, we have sequenced and analysed the mitogenome of one of the most studied monogonont rotifers, Brachionus calyciflorus. It is also composed of two circular chromosomes, but the chromosome-I is extremely large (27 535 bp; 3 mNCRs), whereas the chromosome-II is relatively small (9 833 bp; 1 mNCR). With the total size of 37 368 bp, it is one of the largest metazoan mitogenomes ever reported. In comparison to other monogononts, gene distribution between the two chromosomes and gene order are different and the number of mNCRs is doubled. Atp8 was not found (common in rotifers), and Cytb was present in two copies (the first report in rotifers). A high number (99) of SNPs indicates fast evolution of the Cytb-1 copy. The four mNCRs (5.3–5.5 Kb) were relatively similar. Publication of this sequence shall contribute to the understanding of the evolutionary history of the unique mitogenomic organisation in this group of rotifers.
Introduction
Metazoan mitogenomes are one of the most conservative genomic regions in terms of gene content and organization: commonly a single, closed, circular molecule ranging from about 15 to 20 kb, containing 37 genes (13 protein-coding, 2 rRNAs, and 22 tRNAs). Metazoan mtDNA is usually very compact: there are generally no introns, little or no intergenic DNA, and most genes show clear signatures of selection for small size [1,2]. Nevertheless, fragmentation of the “standard” single-circle mtDNA into multichromosomal genomes, which is common in higher plants [3,4], has occurred independently in some metazoans as well: nematodes [5], mesozoans [6], cnidaria [7], insects [8], and some monogonont rotifers [9,10].
Rotifers are a model system for studies of the evolution of sex [11], as well as aquatic food webs, predator-prey dynamics, population dynamics, speciation, ecotoxicology and evolutionary adaptation [12,13]. The taxonomy of Syndermata (or Rotifera sensu lato) is still unresolved, but the clade is usually divided into Seisonidea, Acanthocephala, Bdelloidea and Monogononta classes [14–16]. Monogononta are cosmopolitan, predominantly freshwater, rotifers that normally reproduce by cyclical parthenogenesis (an alternation between ameiotic parthenogenesis and sporadic sexual episodes), although some populations lose the ability to reproduce sexually [17]. In 2008, the sequencing of the first complete mitogenome of a monogonont rotifer, Brachionus plicatilis, has revealed a unique structure: two circular chromosomes (about 11 and 12.5 Kbp), both possessing a long (mostly) non-coding region of about 4.9 Kbp [9]. Only one monogonont mitogenome, B. koreanus, has been published subsequently, and it was revealed to be structurally very similar to B. plicatilis [10]. B. rubens mitogenome is also available from the GenBank (KJ489417 and KJ489418), but currently (Nov, 2016) remains unpublished. Mitogenome sequence of one of the most abundant and most studied monogonont rotifers, Brachionus calyciflorus, is currently not available. It is a cryptic species complex, occurring in freshwater ecosystems, that represents an excellent system for inferring the evolutionary processes of phylogeographic structure due to its patchy nature of habitats, lack of hybridization and cyclical parthenogenesis [18,19]. In order to further explore the phenomenon of fragmented mitogenomes in monogonont rotifers, we have sequenced and analysed the mitogenome of Brachionus calyciflorus.
Materials and Methods
Samples and DNA extraction
Plankton sample was collected using plankton nets (64 microns) from an outdoor pond (N 31°25′46.45”, E 120°16’54.38”) at the Freshwater Fisheries Research Center of the Chinese Fisheries Science Research Institute South. As this is an experimental pond belonging to our institute, and the field studies did not involve endangered or protected species, no permissions were required for sampling. Samples were collected in a 1 L beaker, filtered using a 165 micron mesh to remove large cladocerans, copepods and organic waste, and washed 2–3 times. A single rotifer was chosen under a microscope using a pipette, morphologically identified as Brachionus calyciflorus [20,21] and cultured at 26°C in an incubator with artificial illumination. Culture medium (100 mL) contained the water from the pond (55 micron mesh-filtered and autoclaved), 0.1 mL/L of a custom-made composite bacterial broth (probiotics, Bacillus spp. and Lactobacillus spp., and sterilised commercial fermented organic fertiliser) and 0.06 mL/L fresh Chlorella spp. (cultivated in a standard BG-11 medium to OD680≈0.8). In approximately 7 to 10 days, when the number of individual rotifers in the population reached about 100 (1 rotifer/mL, log phase), rotifers were mesh-filtered again (55 microns), checked for the presence of other animal species under a microscope, and moved into a fresh medium (200 mL). This batch (No. 2) was cultured until the concentration reached about 4 rotifers/mL (log phase), rotifers again collected as described, placed in a fresh 500 mL medium (batch No. 3), and further cultured until the concentration reached about 10 rotifers/mL (log phase). At the end of this step, rotifers were harvested using plankton net (55 micron mesh). To minimize the amount of Chlorella in the sample, we have rinsed the collected rotifers with distilled water several times and then placed them in a fresh distilled water medium for 24h to allow them to digest and excrete any remaining algae. Eventually, rotifers were harvested again, placed in 100% ethanol and preserved at -20°C. About 150 mg (wet weight) of rotifers was obtained for the DNA extraction.
PCR amplification, sequencing and annotation
DNA was extracted using Aidlab DNA extraction kit (Aidlab Biotechnologies, Beijing), and 18S and Cox1 fragments amplified (Table 1) and sequenced to corroborate the accuracy of the morphological identification [16,18,22]. Degenerate primers were designed to match the generally conserved regions of mtDNA genes and used to amplify and sequence short fragments of 12s, Cox1, Cox3, Cytb, Nad1 and Nad4-5 genes. Specific primers were then designed based on these sequences and used to amplify long-chain products in several PCR reactions. These primers were designed to amplify products with overlaps of about 100 bp. Reaction volume of 50 μL contained 1.0 μL of LongAmp® Taq polymerase (New England BioLabs, Inc.), 10 μL of 5x LongAmp® Taq Reaction Buffer, 1.0 μL dNTP mix (10 mM), 5.0 μL DNA template, 1.5 μL each primer (10x), and 30μL PCR-grade H2O. PCR conditions were optimized for each reaction, with the annealing temperature adjusted to suite the specific primer pair, extension time set to 1 min per Kb of expected product size, and cycles (35 on average) adjusted depending upon the amplification efficiency of the primers. PCR products were sequenced directly by the dideoxynucleotide procedure, using an ABI 3730 automatic sequencer. When that was not possible, the products were cloned into a pMD18-T vector (TaKaRa) and then sequenced. Sequences were assembled in a stepwise manner, ensuring that the overlaps are identical, hence confirming that no numts or fragments of Chlorella DNA were mistakenly incorporated during the mitogenome sequence assembly. All obtained fragments were BLASTed [23] to confirm that the amplicon is the actual target sequence. DNAstar v7.1 (Dnastar Inc., USA) was used to assemble the sequences and locate the putative ORFs for protein-coding genes (PCGs). Then we used BLAST and Blastx to compare the putative ORFs with published nucleotide and amino acid sequences of related species, and manually determine the actual initiation codon and termination codon positions. Annotation of tRNAs was performed using tRNAscan [24] and ARWEN [25] and the results checked manually. MEGA7 [26] was used for similarity analyses and to conduct sequence alignments and OGDRAW to visualize the genome [27].
Table 1. Primers used for PCR amplification of the Brachionus calyciflorus mitochondrial genome.
| Primer | Sequence (5’-3’) | Gene/size |
|---|---|---|
| chromosome-I | ||
| LCCYTBF | GTACTYCCTWGAGRTCAADTGTC | Cytb-1 |
| LCCYTBR | CCTARTTYATTAGRAATAGAKCG | 473 |
| 1LCF1 | TGCAGAGAATTTTATTCCTGC | Cytb-12S |
| 1LCR1 | TGTATGATGGTGGATCATCC | 6478 |
| LC12SF | AAGAMAAGGWTTAGATAYCC | 12S |
| LC12SR | CADAGGTGWCGGGYGATTTGT | 367 |
| 1LCF2 | GCTTAATTCGGATTTGAAAG | 12S-Nad1 |
| 1LCR2 | ATGTGTCTCTGCTAAAGTAC | 1860 |
| LCND1F | TAGYGCAGWCTATTTDTTACGA | Nad1 |
| LCND1R | AATCYTGACWCTAGTYCAGACTC | 211 |
| 1LCF3 | GATCTTTTAGGATGTCTTCAC | Nad1-16S |
| 1LCR4 | GAGCTAGTTAGAATTTTCAC | 1943 |
| LC16SF | GTACYCTGADTGTGCTRAGGYAGC | 16S |
| LC16SR | CGGTTTRAADTCAGAYCATRTA | 411 |
| 1LCF5 | ATTACTACCTCGATGTTGG | 16S-Cox3 |
| 2LCR4 | GAAAAGTGGTGGTTATACAG | 7693 |
| LCCOX3F | ACARGATTCCWTGGTAYACAKGT | Cox3 |
| LCCOX3R | CAGAYAACAWCTACGARGTRTCA | 146 |
| 2LCF5 | GTAGGTACTATTTTTTTATTCTATG | Cox3- Cox1 |
| 2LCR7 | TCTCTTTGTTGTACGAGAAC | 7611 |
| LCCOX1F | ACCRGTTGAAYTGTGTWTCCTCC | Cox1 |
| LCCOX1R | GCTDCAAYATAAHAAGYATKATG | 761 |
| 2LCF1 | CTTCAGTACCTTTACTTTGAG | Cox1- Cytb2 |
| 1LCR5 | CGAGTTAAAGTAGGATTACC | 1217 |
| chromosome-II | ||
| LCND4F | CCTHTAGYTCATGWGGRGGMTCC | Nad4-Nad5 |
| LCND5R | GGAYTTGDATAACRCATVAGT | 1676 |
| 2LCF3.1 | CTTCAACTCTAGTTACAGCAGGC | Nad5- Nad3 |
| LCDoR | CATATATTGGTATAGGGGTTG | 1859 |
| 2LCF3.4 | CTTTAGAGTGACTATCTTGG | Nad3- Nad6 |
| 2LCR1.6 | GCAGATTGCACAAGTATAACCA | 5873 |
| 2LCF1-7 | ATGACAATCTTATTAAGCTT | Nad6-Nad4 |
| 2LCR1.1 | CAAACAACAGTAACATAAACTAAG | 1496 |
| 18S | ||
| LC18SF | ACCTGGTTGATCCTGCCAG | 18S |
| LC18SR | GATCCTTCTGCAGGTTCAC | 1800 |
Results and Discussion
Genome size and organisation
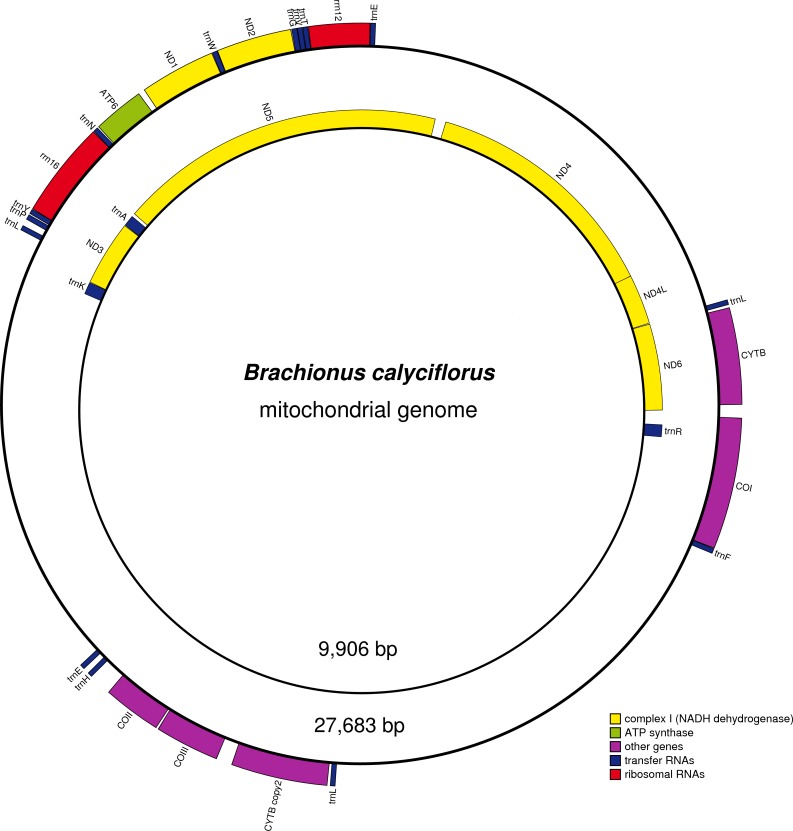
The mitogenome of B. calyciflorus is, just as the other three available monogonont mitogenomes, also composed of two circular chromosomes. However, chromosome-I (mtDNA-I, GenBank accession number KX822781) is extremely large—27 535 bp, whereas chromosome-II (mtDNA-II, KX822782) is smaller than in other monogononts—9 833 bp (Fig 1). The corresponding sizes in other monogononts are: 10 421 / 11 923 bp in B. koreanus [10], 11 153 / 12 672 bp in B. plicatilis [9], and 11 398 / 12 820 in B. rubens. With the total size of 37 368 bp, B. calyciflorus mitogenome is among the largest ever reported in metazoans [28]; the only metazoan mitogenomes comparable in size were found in some molluscs: 30 to 40 Kb in Placopecten magellanicus [29] and 47 to 50 Kb in Scapharca broughtonii [30]. Variation in the length of non-coding regions (NCRs) generally accounts for much of the size variability in mitogenomes, mostly due to segmental duplications [28]. For example, there appears to be no functional constraint on the size of the intergenic regions in the angiosperm mitogenomes, which are thus the largest and least gene-dense eukaryotic mitochondrial genomes [4]. This is also the case in B. calyciflorus mitogenome, where the exceptional size is a result of the existence of four major NCRs (mNCRs): one in the chromosome-II (5441 bp), and three in the chromosome-I (5386, 5391 and 5369 bp). We have not studied whether the size of the chromosomes varies among individual specimens, but the chromosome size and gene content among B. plicatilis individuals were relatively constant [9].
Fig 1. Maps of the two B. calyciflorus mitochondrial chromosomes.
Whereas the gene order in vertebrates is almost invariant, in invertebrates it is relatively variable. However, this is mostly due to the variation in number and location of tRNAs, whereas the order of PCGs and rRNAs is very stable [29]. The gene order in B. koreanus and B. plicatilis was almost identical, save for one rearrangement between tRNAArg and tRNAIle [10]. However, B. calyciflorus mitogenome is an outlier among the monogonont mitogenomes in terms of gene order as well. Furthermore, even the gene content on the two chromosomes was notably different from the remaining three available monogonont mitogenomes. Whereas Cytb, 12s, Nad2, Nad1, Atp6 and 16s genes are present (in that order) on the first chromosome of all four monogonont genomes, B. calyciflorus chromosome-I also contains Cox2, Cox3, an extra copy of Cytb and Cox1 (Table 2, Fig 1). Gene content (tRNAs notwithstanding) and order on chromosome-II are also almost identical in the remaining three mitogenomes: Cox1, Nad6, Nad4l, Nad5, Cox2, Cox3 and Nad3. In B. calyciflorus, however, it contains only Nad6, Nad4l, Nad4, Nad5 and Nad3 (in that order). Gene order (including the PCGs) is also very different from the one previously reported in an Acanthocephalan, Seison sp. [15].
Table 2. Structural features and organisation of the Brachionus calyciflorus mitochondrial genome.
| No. | Gene | Position | Size | IGN | Codon | Anti- | ||
|---|---|---|---|---|---|---|---|---|
| Start | End | bp | bp | Start | Stop | Codon | ||
| chromosome-I | ||||||||
| 1 | Cytb-1 | 1 | 1140 | 1140 | ATG | TAA | ||
| 2 | tRNALeu-1 | 1169 | 1232 | 64 | 28 | TAG | ||
| 3 | mNCR-1 | 1233 | 6618 | 5386 | ||||
| 4 | tRNAGlu-1 | 6739 | 6803 | 65 | 120 | TTC | ||
| 5 | 12S rRNA | 6804 | 7525 | 722 | ||||
| 6 | tRNAThr | 7526 | 7589 | 64 | TGT | |||
| 7 | tRNAVal | 7590 | 7653 | 64 | TAC | |||
| 8 | tRNAGly | 7652 | 7719 | 68 | -2 | TCC | ||
| 9 | Nad2 | 7723 | 8613 | 891 | 3 | ATG | TAA | |
| 10 | tRNATrp | 8612 | 8675 | 64 | -2 | TCA | ||
| 11 | Nad1 | 8676 | 9573 | 898 | ATG | T— | ||
| 12 | tRNAGln | 9575 | 9639 | 65 | 1 | TTG | ||
| 13 | Atp6 | 9640 | 10251 | 612 | ATG | TAA | ||
| 14 | tRNAAsp | 10262 | 10325 | 64 | 10 | GTC | ||
| 15 | 16S rRNA | 10326 | 11469 | 1144 | ||||
| 16 | tRNATyr | 11470 | 11532 | 63 | GTA | |||
| 17 | tRNAPro | 11542 | 11608 | 67 | 9 | TGG | ||
| 18 | tRNAMet | 11609 | 11674 | 66 | CAT | |||
| 19 | tRNALeu-2 | 11684 | 11747 | 64 | 9 | TAA | ||
| 20 | mNCR-2 | 11748 | 17138 | 5391 | ||||
| 21 | tRNAGlu-2 | 17139 | 17203 | 65 | TTC | |||
| 22 | tRNAHis | 17266 | 17328 | 63 | 62 | GTG | ||
| 23 | Cox2 | 17587 | 18282 | 696 | 258 | GTG | TAA | |
| 24 | Cox3 | 18302 | 19066 | 765 | 19 | ATA | TAA | |
| 25 | tRNACys | 19097 | 19157 | 61 | 30 | GCA | ||
| 26 | Cytb-2 | 19245 | 20384 | 1140 | 87 | ATG | TAA | |
| 27 | tRNALeu-3 | 20413 | 20476 | 64 | 28 | TAG | ||
| 28 | mNCR-3 | 20477 | 25845 | 5369 | ||||
| 29 | tRNAPhe | 25846 | 25909 | 64 | 75 | GAA | ||
| 30 | tRNASer | 25910 | 25974 | 65 | TGA | |||
| 31 | Cox1 | 25985 | 27535 | 1551 | 10 | ATG | TAA | |
| chromosome-II | ||||||||
| 1 | Nad6 | 1 | 462 | 462 | ATG | TAG | ||
| 2 | Nad4L | 464 | 733 | 270 | 1 | ATG | TAA | |
| 3 | Nad4 | 730 | 2031 | 1302 | -4 | ATA | TAA | |
| 4 | Nad5 | 2085 | 3777 | 1693 | 53 | ATG | TAA | |
| 5 | tRNAAsn | 3778 | 3841 | 64 | GTT | |||
| 6 | tRNAAla | 3845 | 3906 | 62 | 3 | TGC | ||
| 7 | Nad3 | 3907 | 4258 | 352 | ATG | T— | ||
| 8 | tRNALys | 4259 | 4325 | 67 | TTT | |||
| 9 | mNCR-1 | 4326 | 9766 | 5441 | ||||
| 10 | tRNAArg | 9767 | 9833 | 67 | ACG | |||
| 11 | tRNAIle | 9842 | 9906 | 65 | 8 | GAT | ||
IGN is the number of inter-genetic nucleotides, where a positive value indicates a non-coding region, whereas a negative value indicates a gene overlap.
Genome characteristics
Despite the unusually large size and unusual organisation, the mitogenome contained most of the standard 37 mtDNA genes. Only Atp8 was not found among the standard 13 mitochondrial protein-coding genes, but Cytb was present in two copies. The absence of Atp8 is common in rotifers [15,28,31]. Duplicated protein coding genes are common in mitogenomes of some metazoan groups [32], but with the exception of mitogenomes of some birds, characterized by a tandemly duplicated region encompassing a part of the Cytb, three tRNAs, Nad6 and the control region [33], the duplication of Cytb appears to be a relatively rare event in metazoans. Even though the two Cytb copies were very similar (about 99.1%), a high number (99) of SNPs indicates a relatively fast evolution of one of the copies, probably facilitated by the functional redundancy. Similarity and phylogeny analyses including available orthologs (not shown) indicate that Cytb-1 is the faster-evolving copy.
Apart from the already discussed genes, chromosome-I and II also contained 18 and five tRNAs, respectively. Generally, tRNAs are the gene category with the highest ‘dispensability’ in the mitochondrial genome [28], but all of the main 20 tRNAs were found in the B. calyciflorus mitogenome. However, only 18 were found using the available annotation programs: 15 on the chromosome-I and three on the chromosome-II. Only 15 of these were unique: tRNAGlu was found in two and tRNALeu in three copies. Five more tRNAs were found after the detailed manual search of non-coding regions: tRNAMet, tRNACys and tRNASer on chromosome-I, and tRNAAsn and tRNAIle on chromosome-II. Whereas many metazoan genomes have clusters of five to seven tRNA genes, in monogonont rotifers, including B. calyciflorus, tRNA genes appear to be dispersed, with no clusters larger than three [9,10].
Four major NCRs notwithstanding, non-coding regions ranged from 1 to 258 bp in length in chromosome-I (13 ncrs) and from 1 to 53 bp in chromosome-II (3 ncrs). Two putative gene overlaps were found in chromosome-I (both 2 bp): tRNAVal—tRNAGly and Nad2—tRNATrp; and one in chromosome-II (4 bp): Nad4l and Nad4. However, it is likely that Nad2 used the unfinished T—stop codon instead of the complete TAA, in which case there would be no overlap with tRNATrp. Regarding the Nad4l and Nad4 overlap, if we consider the possibility of the unfinished T—stop codon in this sequence…ATT(ATAA)CT…, overlap could be only two bases…ATT(AT)AACT…, or there could even be no overlap at all, but in that case Nad4l would have to be two bases shorter:…ATT)(ATAACT… Comparable to B. plicatilis, all genes on both chromosomes would be transcribed from the same DNA strand, and all four mNCRs are in the same orientation relative to the direction of transcription. Overall, three different start codons were found: ATG (10 genes), ATA (2) and GTG (1); and two stop codons: TAA (12) and TAG (1) (Table 2). All these codons are typical for the invertebrate, and rotiferan, mitochondrial DNA [15,34]. The unfinished T—codon was not counted separately, as we presumed that it would be completed (TAA) by the posttranscriptional polyadenylation [35,36]. The orthodox initiation codon ATG was also used in 8 / 9 of the twelve protein-coding genes in B. plicatilis / B. koreanus mitogenomes, respectively [9,10]. These two species used incomplete stop codons in Nad1, Nad3 and Atp6 genes. In B. calyciflorus, it was only Nad1 and Nad3 (Atp6 used TAA). ATA was the start codon for Nad4 and Cox3 in all three species. Hence, there appears to be a relatively high level of conservation in the start/stop codon usage among the three monogonont species.
Base composition was very similar between the two chromosomes, both of which exhibited a strong A+T bias: 68.7% in chromosome-I and 69.6% in chromosome-II. To test whether this is a consequence of the large proportion of non-coding regions, we have checked whether the base composition in mNCRs is different from the rest of the genome. A+T content was only somewhat lower in the mNCRs (66%) in comparison to the complete genome (69%), mainly due to the decrease in the amount of T: 37.5% in the genome vs. 33.0% in the mNCRs. In comparison, the base composition of the mNCRs in B. plicatilis was significantly different from the rest of the mitogenome, also due to a decrease in the amount of T in favour of other bases [9]. However, the overall A+T bias appears to be typical not only for monogonont mitogenomes (B. plicatilis = 60.7–71.7% and B. koreanus = 68.81%) [9,10], but also for syndermatan mitogenomes [15,16].
Major non-coding regions
There were three major non-coding regions (mNCRs) in the first chromosome: mNCR-I-1 (5386 bp), mNCR-I-2 (5391 bp) and mNCR-I-3 (5369 bp); and only one in the second chromosome: mNCR-II (5441 bp). Sequences were very similar: the overall average genetic distance was only 0.02. The smallest pairwise genetic distance was found between NCR-I-2 and NCR-II (0.002), and the highest between NCR-I-2 and NCR-I-3 (0.032). All four sequences were almost identical at the 5’-end, whereas the largest differences were observed at the 3’-end; notably, a deletion of about 20–25 bases at the end of NCR-I-3 and a T-rich extension of about 46 bases at the end of NCR-II. Whereas NCRs of angiosperm mitochondria contain (usually) non-functional polypeptides that are translated from open reading frames created as by-products of genome alteration [4], we did not find putative ORFs in either of the B. calyciflorus mNCRs.
Generally, there is no correlation between the size of mitogenome and gene content, as the differences in size of mitochondrial genomes are mostly caused by marked variations in the length and organisation of intergenic regions. In B. calyciflorus, non-coding regions comprised more than half (59.75%) of the total genome: 61.09% of the chromosome-I and 56.0% of the chromosome-II. This is comparable to other extremely large metazoan mitogenomes; specifically, some molluscan mitogenomes also show transpositions of certain regions that can more than double the genome size [29,30]. The role of these regions is still debated, some are thought to be mobile and ‘selfish’, whereas others might play a role in recombination or serve as replication and transcription regulatory signals [28,37].
Conclusions
The organisation of all three previously sequenced monogonont mitogenomes was almost identical: they had two circular chromosomes of a relatively similar size, with almost identical gene content and order (among the three species, not between the two chromosomes), and one major non-coding region on each chromosome. B. calyciflorus mitogenome is also organised into two circular chromosomes, but different in other aspects: the first chromosome possesses three mNCRs and it is almost three times larger than the second chromosome. Gene content (and order) of the two chromosomes is also quite different from other monogononts.
Even though monogonont rotifers are a model system in several research fields, very few mitogenomes have been sequenced so far. This is highly likely to be a consequence of a high plasticity of mitogenomic structure and organisation, as well as the existence of a varying number of very long, almost identical non-coding regions, which can make the PCR amplification and mitogenome sequence assembly a very difficult and time-consuming process. Publication of this sequence shall help streamline the future attempts to sequence monogonont mitogenomes, and ultimately contribute to the understanding of the evolutionary history of the unique mitogenomic organisation in this group of rotifers.
Data Availability
GenBank accession numbers of the two sequences are KX822781 and KX822782.
Funding Statement
This study was supported by the Jiangsu Agriculture Science and Technology Innovation Fund [JASTIF; grant no. CX(13)2039] and Three New Projects of Agricultural Aquaculture Program of Jiangsu Province (grant no. D2015-14). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rand DM. “Why genomes in pieces?” revisited: Sucking lice do their own thing in mtDNA circle game. Genome Res. Cold Spring Harbor Laboratory Press; 2009;19: 700–702. 10.1101/gr.091132.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Z, Cuthbert JM, Taylor DR, Sloan DB. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci U S A. 2015;112: 10185–91. 10.1073/pnas.1421397112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitazaki K, Kubo T, Kitazaki K, Kubo T. Cost of Having the Largest Mitochondrial Genome: Evolutionary Mechanism of Plant Mitochondrial Genome. J Bot. 2010;2010: 1–12. [Google Scholar]
- 5.Armstrong MR, Blok VC, Phillips MS. A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics. 2000;154: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe KI, Bessho Y, Kawasaki M, Hori H. Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J Mol Biol. 1999;286: 645–50. 10.1006/jmbi.1998.2523 [DOI] [PubMed] [Google Scholar]
- 7.Pont-Kingdon G, Vassort CG, Warrior R, Okimoto R, Beagley CT, Wolstenholme DR. Mitochondrial DNA of Hydra attenuata (Cnidaria): a sequence that includes an end of one linear molecule and the genes for l-rRNA, tRNA(f-Met), tRNA(Trp), COII, and ATPase8. J Mol Evol. 2000;51: 404–415. [DOI] [PubMed] [Google Scholar]
- 8.Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009;19: 904–912. 10.1101/gr.083188.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 2008;25: 1129–1137. 10.1093/molbev/msn058 [DOI] [PubMed] [Google Scholar]
- 10.Hwang D-S, Suga K, Sakakura Y, Park HG, Hagiwara A, Rhee J-S, et al. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondrial DNA. 2014;25: 29–30. 10.3109/19401736.2013.775274 [DOI] [PubMed] [Google Scholar]
- 11.Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11: 41–46. [DOI] [PubMed] [Google Scholar]
- 12.Serra M, Galiana A, Gomez A. Speciation in monogonont rotifers. Hydrobiologia. 1997;358: 63–70. [Google Scholar]
- 13.Declerck SAJ, Papakostas S. Monogonont rotifers as model systems for the study of micro-evolutionary adaptation and its eco-evolutionary implications. Hydrobiologia. 2016; [Google Scholar]
- 14.Sørensen M V., Giribet G. A modern approach to rotiferan phylogeny: Combining morphological and molecular data. Mol Phylogenet Evol. 2006;40: 585–608. 10.1016/j.ympev.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 15.Sielaff M, Schmidt H, Struck TH, Rosenkranz D, Mark Welch DB, Hankeln T, et al. Phylogeny of Syndermata (syn. Rotifera): Mitochondrial gene order verifies epizoic Seisonidea as sister to endoparasitic Acanthocephala within monophyletic Hemirotifera. Mol Phylogenet Evol. 2016;96: 79–92. 10.1016/j.ympev.2015.11.017 [DOI] [PubMed] [Google Scholar]
- 16.Lasek-Nesselquist E. A mitogenomic re-evaluation of the Bdelloid phylogeny and relationships among the Syndermata. PLoS One. 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer CP, Schmidt J, Wiedlroither A, Riss S. Loss of sexual reproduction and dwarfing in a small metazoan. PLoS One. 2010;5: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert JJ, Walsh EJ. Brachionus calyciflorus is a species complex: Mating behavior and genetic differentiation among four geographically isolated strains. Hydrobiologia. 2005;546: 257–265. [Google Scholar]
- 19.Xiang X ling, Xi Y long, Wen X li, Zhang G, Wang J xia, Hu K. Patterns and processes in the genetic differentiation of the Brachionus calyciflorus complex, a passively dispersing freshwater zooplankton. Mol Phylogenet Evol. 2011;59: 386–398. 10.1016/j.ympev.2011.02.011 [DOI] [PubMed] [Google Scholar]
- 20.Segers H. Rotifera: Monogononta. Freshwater Invertebrates of the Malaysian Region. 2002. pp. 106–120.
- 21.Wallace RL, Snell TW. Rotifera. Ecology and classification of North American freshwater invertebrates. 2010. pp. 173–235.
- 22.Vasileiadou K, Papakostas S, Triantafyllidis A, Kappas I, Abatzopoulos TJ. A multiplex PCR method for rapid identification of Brachionus rotifers. Mar Biotechnol (NY). 2009;11: 53–61. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997. pp. 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33: W686–9. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laslett D, Canbäck B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24: 172–175. 10.1093/bioinformatics/btm573 [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb). 2008;101: 301–32062. [DOI] [PubMed] [Google Scholar]
- 29.Smith DR, Snyder M. Complete mitochondrial DNA sequence of the scallop Placopecten magellanicus: Evidence of transposition leading to an uncharacteristically large mitochondrial genome. J Mol Evol. 2007;65: 380–391. 10.1007/s00239-007-9016-x [DOI] [PubMed] [Google Scholar]
- 30.Liu YG, Kurokawa T, Sekino M, Tanabe T, Watanabe K. Complete mitochondrial DNA sequence of the ark shell Scapharca broughtonii: An ultra-large metazoan mitochondrial genome. Comp Biochem Physiol—Part D Genomics Proteomics. 2013;8: 72–81. 10.1016/j.cbd.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 31.Steinauer ML, Nickol BB, Broughton R, Ortí G. First sequenced mitochondrial genome from the phylum acanthocephala (Leptorhynchoides thecatus) and its phylogenetic position within metazoa. J Mol Evol. 2005;60: 706–715. 10.1007/s00239-004-0159-8 [DOI] [PubMed] [Google Scholar]
- 32.Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M. A resourceful genome: Updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014;30: 555–564. 10.1016/j.tig.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 33.Sammler S, Bleidorn C, Tiedemann R. Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination. BMC Genomics. 2011;12: 35 10.1186/1471-2164-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolstenholme DR. Animal Mitochondrial DNA: Structure and Evolution. International Review of Cytology. 1992. pp. 173–216. [DOI] [PubMed] [Google Scholar]
- 35.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290: 470–474. [DOI] [PubMed] [Google Scholar]
- 36.Schuster G, Stern D. RNA Polyadenylation and Decay in Mitochondria and Chloroplasts. Progress in Molecular Biology and Translational Science. 2009. pp. 393–422. 10.1016/S0079-6603(08)00810-6 [DOI] [PubMed] [Google Scholar]
- 37.Burger G, Gray MW, Lang BF. Mitochondrial genomes: Anything goes. Trends in Genetics. 2003. pp. 709–716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GenBank accession numbers of the two sequences are KX822781 and KX822782.