Abstract

Long wavelength ultraviolet radiation (UVA, 320–400 nm) interacts with chromophores present in human cells to induce reactive oxygen species (ROS) that damage both DNA and proteins. ROS levels are amplified, and the damaging effects of UVA are exacerbated if the cells are irradiated in the presence of UVA photosensitizers such as 6-thioguanine (6-TG), a strong UVA chromophore that is extensively incorporated into the DNA of dividing cells, or the fluoroquinolone antibiotic ciprofloxacin. Both DNA-embedded 6-TG and ciprofloxacin combine synergistically with UVA to generate high levels of ROS. Importantly, the extensive protein damage induced by these photosensitizer+UVA combinations inhibits DNA repair. DNA is maintained in intimate contact with the proteins that effect its replication, transcription, and repair, and DNA–protein cross-links (DPCs) are a recognized reaction product of ROS. Cross-linking of DNA metabolizing proteins would compromise these processes by introducing physical blocks and by depleting active proteins. We describe a sensitive and statistically rigorous method to analyze DPCs in cultured human cells. Application of this proteomics-based analysis to cells treated with 6-TG+UVA and ciprofloxacin+UVA identified proteins involved in DNA repair, replication, and gene expression among those most vulnerable to cross-linking under oxidative conditions.
Keywords: DNA−protein cross-link, SILAC, ultraviolet A (UVA), reactive oxygen species (ROS), photosensitizer, 6-thioguanine (6-TG), fluoroquinolone, ciprofloxacin, DNA damage, DNA repair
Introduction
Human genomic DNA is maintained in intimate contact with proteins that confer the structural integrity of chromosomes. Other proteins associate intermittently with DNA to affect its repair, replication, and transcription. Although generally unreactive, proteins and DNA can become covalently associated. These reactions are favored under oxidative conditions, and the production of covalent DNA–protein cross-links (DPCs) is enhanced by exposure of cells to diverse agents including chemical oxidants, ionizing radiation (IR), ultraviolet radiation (UV), reactive aldehydes, or chemotherapeutic drugs (reviewed in ref (1)). DPCs are particularly challenging for cells. Sequestration of proteins required for DNA repair, replication, or transcription is likely to impair these important functions. In addition, DPCs are large DNA adducts that block DNA replication and physically impede DNA-related processes.2 Their formation poses a risk of substantial and permanent genetic damage.
The thiopurine 6-thioguanine (6-TG) is among the therapeutic agents that promote DPC formation.3 6-Thioguanine nucleotides, the end product of the metabolism of 6-TG and of the anticancer immunosuppressants azathioprine and 6-mercaptopurine (6-MP), are substrates for incorporation into DNA. Additionally, exposure of cultured human cells to 6-TG or 6-MP depletes their antioxidant defenses and increases steady-state levels of reactive oxygen species (ROS).4,5 Patients treated with thiopurines experience skin photosensitivity6 and have a significantly increased risk of developing skin cancer.7,8 Photosensitivity is a consequence of an accumulation of 6-TG in patients’ DNA. DNA 6-TG can act both as a Type I and Type II UVA photosensitizer (reviewed in ref (9)). In the Type I mode, extremely reactive purine radical cations or purine thiyl radicals are generated following UVA activation of DNA 6-TG. As a Type II sensitizer, DNA 6-TG interacts with UVA in the presence of molecular oxygen to generate singlet oxygen (1O2), a form of ROS that is particularly damaging to proteins. Unsurprisingly, these photosensitized reactions also cause many different kinds of DNA damage. These include oxidized forms of DNA 6-TG (guanine sulfinate (GSO2) and guanine sulfonate (GSO3)10) and guanine (8-oxo-7,8-dihydroguanine11) as well as DNA single- and double-strand breaks and DNA interstrand cross-links (ICLs).4 The combination of DNA 6-TG and UVA also induces protein damage in the form of carbonyls and oxidized thiols.12 It causes oxidation-related cross-linking between the subunits of multiprotein complexes including the PCNA,13 Ku,12 RPA,14 and MCM2–7 DNA replication/repair complexes15 and between DNA and proteins.3 Importantly, protein damage induced by 6-TG+UVA is associated with a significant attenuation of DNA repair capacity.12
Ciprofloxacin is a member of the fluoroquinolone family of antibiotics that are UVA photosensitizers. Like 6-TG, ciprofloxacin is a Type II UVA photosensitizer that generates 1O2.16 UVA irradiation of cells treated with ciprofloxacin causes damage to DNA and proteins.17 Protein damage by ciprofloxacin+UVA also includes oxidation and cross-linking between subunits of DNA replication and repair complexes14,17 and is associated with impaired DNA repair.17 To our knowledge, the possible induction of DPCs by UVA-activated ciprofloxacin has not been examined.
We previously demonstrated the formation of heat- and reducing agent-resistant DPCs between oligonucleotides containing oxidized 6-TG (GSO3) and the amino or thiol groups of oligopeptides. The same study3 also presented preliminary evidence for the formation of DPCs in vivo in cultured human cells treated with 6-TG and exposed to low doses of UVA radiation. Immunoblotting identified DNA repair proteins among the cross-linked species from cells treated with 6-TG and UVA. Specifically, the PCNA DNA repair/replication protein that is known to be susceptible to oxidation was identified along with MSH2 and XPA, essential components of the DNA mismatch repair and nucleotide excision repair pathways, respectively. The presence of these important DNA repair factors in DPCs suggested that the obligatory, albeit transient association of DNA repair proteins with DNA might make them particularly vulnerable to inactivation by DNA cross-linking. We have developed a sensitive and statistically rigorous approach to identifying cross-linked proteins. Based on stable isotope labeling with amino acids in cell culture (SILAC) and mass spectrometry (MS), the method is generally applicable to DNA damaging treatments. Here we describe the application of this proteomics-based technique to analyze in detail DPC formation by 6-TG treatment and by the UVA activation of DNA 6-TG in human cells. We also report the application of the same approach to examine DPC induction by UVA activation of ciprofloxacin, a representative of a family of non DNA-embedded UVA photosensitizers.
Materials and Methods
Chemicals
6-TG and ciprofloxacin were obtained from Sigma-Aldrich.
Cells and UV Radiation
CCRF-CEM cells were routinely grown in RPMI 1640 medium (Thermo Fisher) supplemented with 10% dialyzed fetal calf serum. For SILAC, growth medium was supplemented with a combination of either 100 mg per liter of light (14N, 12C) or heavy (15N, 13C) lysine and arginine (CK Isotopes). Following growth for 7 d in this medium, full labeling of proteins was confirmed by MS (data not shown).
Cells were incubated with 6-TG for 24 h and ciprofloxacin for 1 h prior to UVA irradiation. For hydroxyurea treatment, the drug (3 mM) was included in the medium for 6 h prior to and during growth in 6-TG. DNA was extracted, and DNA 6-TG incorporation was quantified as described previously.18
Cells were UVA irradiated in phosphate buffered saline using a UVH 253 lamp (UV Light Technology Limited) with maximum emission at 365 nm and a dose rate of 0.1 kJ m–2 s–1. Neither photosensitizer treatment nor UVA irradiation reduced cell viability, whereas photosensitizer+UVA combinations were highly lethal.17
ROS were determined by FACS using CM-H2DCFDA (Invitrogen) as previously described.19
Mass Spectrometry Sample Preparation
Following treatment, 106 isotopically labeled control/treated cells were mixed and nuclei prepared by resuspension in 200 μL of 10 mM Tris-HCl pH 7.4, 2.5 mM MgCl2, 0.5% NP40, 1 mM dithiothreitol (DTT) and were harvested by centrifugation. Chromatin was released from the nuclear pellet by resuspension in 25 mM Na phosphate, pH 7.4, 5 mM MgCl2, 500 mM NaCl, 0.5% Triton, 1 mM EDTA, 1 mM DTT, 10% glycerol plus protease inhibitors. The chromatin pellet was washed three times by resuspension in the same buffer and then sheared by sequential passage through 19G, 25G, and 27G needles (20× each). Sheared chromatin samples containing 10 μg of DNA were applied to a Hybond-N+ membrane using a slot blot apparatus (GE Healthcare). DNA was cross-linked to the membrane by UVC irradiation from a Stratalinker (Stratagene) and was then washed extensively with 8 M urea (Sigma-Aldrich) and water. The areas of membrane containing the applied samples were excised.
Membrane-bound proteins were reduced with 10 mM DTT at 50 °C for 30 min and alkylated by treatment with 55 mM iodoacetamide for 30 min at room temperature in the dark. The alkylation reaction was stopped by incubation with 10 mM DTT for 10 min at room temperature. Following three washes with 10 mM triethylammonium bicarbonate (TEAB), the proteins were digested by immersing the membrane in trypsin (12.5 ng/μL) overnight at 37 °C. DTT, iodoacetamide, and trypsin were all prepared in 10 mM TEAB.
For MS analysis of total cell lysates, whole-cell extracts were prepared with RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% Triton, and protease inhibitors). Twenty micrograms of protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with colloidal coomassie (Instant Blue, Expedeon). Gel bands were excised and trypsin digested using a PerkinElmer Janus liquid handling system.20
Tryptic peptides were analyzed by liquid chromatography-mass spectrometry (LC–MS) using an Ultimate 3000 uHPLC system connected to either a Q-Exactive or Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) and acquired in data-dependent mode. The data were searched against human Uniprot (UniProt KB2012_08 taxonomy human 9606 canonical with contaminants 20 120 921) using the Andromeda search engine and MaxQuant (version 1.3.0.5).21 For MaxQuant, a false discovery rate of 0.1% was used to generate protein identification tables. The data were uploaded into Perseus version 1.4.0.11 (MaxQuant) for statistical analyses.
Immunoblotting
Chromatin extracts (20 μg of protein) were separated on 10% polyacrylamide gels (Invitrogen) and transferred to HyBond-N+ membranes. Proteins were cross-linked to the membrane with UVC. Following washing with urea and water, membranes were probed with antibodies against MSH2, MSH6, PCNA (Santa Cruz), and RPA70 (Abcam). The complexes were visualized using ECL detection agent (GE Healthcare).
Results
DNA–Protein Cross-Link Formation by 6-TG/UVA
Growth of CCRF-CEM cells in the presence of 6-TG (0.3–0.9 μM) resulted in the thiopurine replacing around 0.05–0.6% of DNA guanine. When DNA was prepared from a standard number of 6-TG treated cells by the Wizard (Promega) extraction protocol, the amount recovered declined in a 6-TG concentration-dependent manner. Irradiation of cells containing DNA 6-TG with a modest dose of UVA (50 kJ/m2) further exacerbated DNA losses. The effects of 6-TG and UVA were synergistic, and UVA alone had no detectable effect on DNA recovery. The Wizard extraction protocol involves a protein precipitation step prior to DNA harvesting and the inclusion of a proteinase K digestion step prior to DNA precipitation restored quantitative DNA yields (Supplementary Figure S1A). The reduced DNA recovery from cells treated with 6-TG or 6-TG+UVA was dependent on the presence of 6-TG in DNA. The Wizard DNA purification protocol yielded quantitative DNA recovery from 6-TG treated GM03467 Lesch-Nyhan cells without the inclusion of the protease digestion step (data not shown). These cells do not express hypoxanthine-guanine phosphoribosyltransferase and cannot scavenge 6-TG for incorporation into DNA. In addition, quantitative DNA yields were achieved without the additional protease digestion step if 6-TG incorporation into DNA was prevented by treatment of CCRF-CEM cells with 6-TG in the presence of hydroxyurea (Supplementary Figure 1A). FACS analysis (Supplementary Figure 1B) confirmed that exclusion of 6-TG from DNA also reduced UVA-induced ROS levels.
Their dependence on protease digestion suggested that DNA yields were reduced by cross-linking of DNA to protein. This possibility was investigated further using chromatin from treated cells. To selectively enrich for proteins covalently attached to DNA, chromatin extracted from 6-TG+UVA treated CCRF-CEM cells was washed extensively with high salt (500 mM NaCl) to deplete noncovalently associated proteins. Sheared, salt-washed chromatin was applied to a HyBond-N+ membrane that was then sequentially washed with 8 M urea and water. Staining with SyproRuby and SYBR Green confirmed that extensive urea washing removed all detectable membrane-associated proteins from untreated chromatin while having no noticeable impact on the amount of bound DNA. Subsequent probing of the filter with a panel of antibodies confirmed that UVA induced a 6-TG dose-dependent increase in the amount of the XPA, PCNA, MSH2,3 and RPA70 (the 70 kDa subunit of the RPA single strand DNA binding complex) DNA repair/replication proteins associated with the washed membrane (Figure 1A,B).
Figure 1.
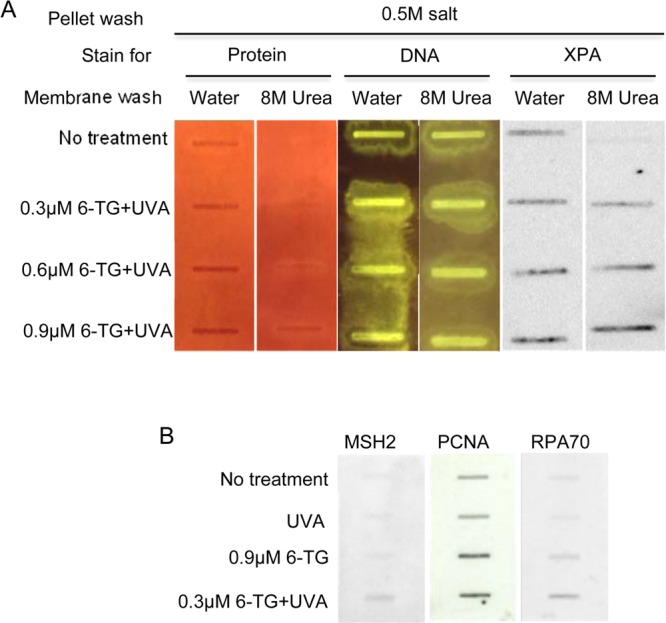
HyBond-N+ membrane binding of DNA cross-linked proteins. (A) Sheared, salt-washed chromatin from CCRF-CEM cells that were untreated or treated with 6-TG and 50 kJ/m2 UVA as indicated was applied to HyBond-N+ membranes. Membranes were washed extensively with water and 8 M urea as indicated. Membrane-associated protein and DNA was visualized by staining with Sypro Ruby and SYBR Green. XPA protein was detected by immunostaining. (B) DNA-bound protein. Sheared chromatin from untreated CCRF-CEM cells or cells treated with UVA, 6-TG or 6-TG+UVA was applied to HyBond-N+ membranes. Following extensive washing with water and 8 M urea, membranes were probed with antibodies as indicated.
MS Analysis of DNA–Protein Cross-Linking
The observation that DNA replication/repair proteins are enriched in HyBond-N+ membrane-bound chromatin prompted us to undertake a comprehensive analysis of the proteome associated with HyBond-N+-bound DNA to provide an unbiased screen for proteins involved in DPCs. The protocol is outlined in Figure 2, panel A.
Figure 2.
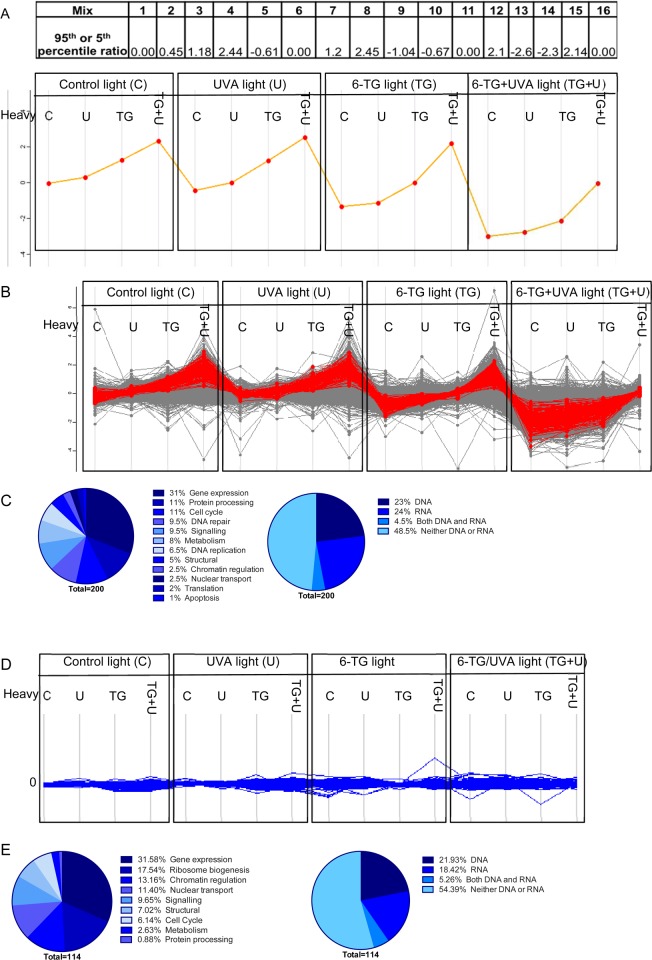
Outline of SILAC analysis. (A) CCRF-CEM cells were labeled with heavy or light arginine and lysine isotopes. Half the cells were treated with 0.9 μM 6-TG for 24 h and the other half left untreated. Half of each of these two cultures was then irradiated with 50 kJ/m2 UVA. The remaining four cultures were not irradiated. Cells were mixed in 1:1 ratios for preparation of chromatin or whole cell extracts (RIPA) as indicated. Chromatin extracts were applied to a HyBond-N+ membrane that was water and 8 M urea washed prior to in situ trypsin digestion and MS analysis. RIPA extracts were subjected to short SDS-PAGE and in-gel trypsin digestion prior to MS analysis. (B) The 16 mixes generated from different combinations of treatments.
Briefly, CCRF-CEM cells labeled with heavy or light isotopes of arginine and lysine were treated with 6-TG (0.9 μM). Half of each culture was then irradiated with UVA (50 kJ/m2). The remaining cells were mock irradiated. Chromatin was prepared from a total of 16 1:1 mixes of heavy and light isotope labeled cells that had been treated with 6-TG, UVA, 6-TG+UVA, or left untreated. The compositions of these mixes are shown in Figure 2, panel B. High salt-washed chromatin mixtures were loaded onto a HyBond-N+ membrane that was extensively washed with water and 8 M urea. Remaining membrane-associated proteins were then digested with trypsin in situ and the digests analyzed by MS.
A total of 2611 proteins were identified in two independent experiments (forward and reverse labeling analyses) of HyBond-N+ membrane bound chromatin from UVA, 6-TG, or 6-TG+UVA treated CCRF-CEM cells (Supporting Information). Because the analysis was carried out with chromatin prepared from mixtures of heavy and light labeled cells, changes in the log2 heavy/light (H:L) protein ratios reveal an enrichment of proteins associated with membrane-bound DNA. Supplementary Figure S2A presents the protein distribution for Mix 1 in which the chromatin applied to the membrane was prepared from mixtures of untreated heavy- and untreated light-labeled cells. The tight symmetrical clustering of log2 H:L ratios around the zero value in the histogram confirms the expected equal representation of heavy and light labeled proteins. Because >99.5% of log2 values for these untreated cells lie between −1 and +1, in the subsequent analysis of treated chromatin we considered values that fall outside this range (representing >2-fold enrichment) to be significant treatment-related changes that reflect DPC formation.
Comparison of mixtures of heavy 6-TG-treated/light untreated cells and heavy untreated/light 6-TG-treated cells (Mixes 3 and 9) revealed the effects of 6-TG treatment. Supplementary Figure S2B,C shows that 6-TG treatment shifted the membrane-associated protein distribution in the direction of the label in the treated cells. In the scatter plot (Figure 3A), 6-TG-induced asymmetry in log2 H:L ratios results in the majority of the data points occupying the lower right quadrant, a shift consistent with DPC formation. The effect was small, however, and only reached significance for approximately 10% (7 and 16% in two determinations) of the detected proteins, which indicated that 6-TG-induces a low level of DNA–protein cross-linking.
Figure 3.
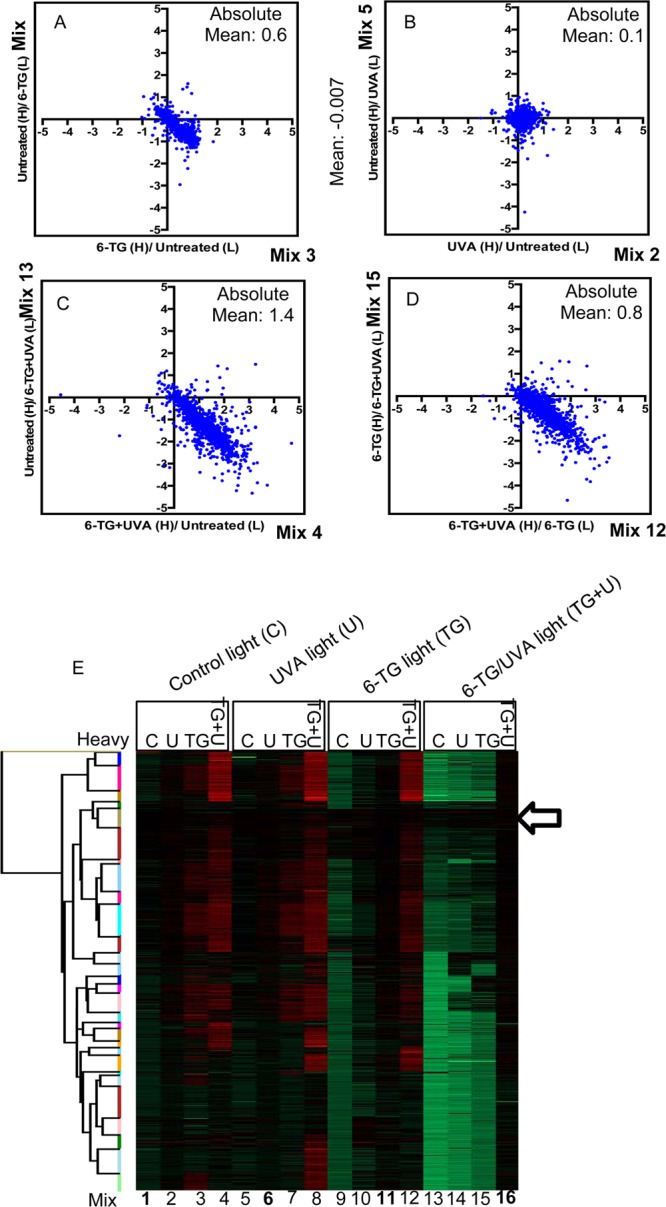
Effects of the different treatments on DPC formation. Scatterplots of SILAC log2 H:L ratios. (A) The effect of 6-TG treatment (6-TG vs untreated). Mix 3 versus Mix 9 (from Figure 2B). (B) The effect of UVA treatment (UVA vs untreated). Mix 2 versus Mix 5. (C) The effect of 6-TG+UVA treatment (6-TG+UVA vs untreated). Mix 4 versus Mix 13. (D) The additional effect of UVA on 6-TG treated cells (6-TG+UVA vs 6-TG). Mix 12 versus Mix 15. Axis values are the log2 H:L ratios for the mix indicated. The calculated absolute values for log2 H:L ratio shown confirm that 6-TG+UVA induces a significant change. They also validate synergy between 6-TG and UVA. (E) Heat map of log2 H:L ratio intensities of identified proteins. Hierarchical cluster analysis was performed in Perseus based on Euclidian distances. Arrow indicates clustered potential false positives (114 proteins). Red = increased ratio; green = decreased ratio. Mix numbers refer to those in Figure 2, panel B. Mixes of H- and L-labeled cells that received the same treatment are shown in bold.
The moderate UVA dose we used (50 kJ/m2) did not induce detectable DNA–protein cross-linking, and the log2 H:L ratios for the comparison of Mixes 2 and 5 (Figure 3B) that addresses the effect of UVA remain tightly clustered around the origin of the scatter plot.
Analysis of Mixes 4 and 13 (Figure 3C) revealed that the combination of 6-TG and UVA caused extensive DNA–protein cross-linking. By comparing Mixes 12 and 15, we specifically examined the effect of UVA on cells treated with 6-TG. In the absence of synergy between 6-TG and UVA, the log2 H:L ratios would cluster around the origin of the scatter plot as they do for samples from cells treated with UVA alone. Figure 3, panel D and Supplementary Figure S2D,E confirm that UVA induces extensive DPC formation in cells treated with 6-TG. DPC induction by 6-TG, UVA, and combined 6-TG+UVA is summarized in the heat map in Figure 3, panel E. Hierarchical cluster analysis reveals a family of proteins (arrowed) that appeared to be largely unaffected by any of the treatments and represent a set of false positives. Subsequent analysis of the cross-linking profiles of these proteins (see below) confirmed their absence of susceptibility to cross-linking by either 6-TG or 6-TG+UVA.
In summary, SILAC analysis demonstrates that 6-TG induces a low level of DPCs in CCRF-CEM cells. It also reveals that 6-TG and UVA combine synergistically to cause extensive DPC formation.
Changes in Protein Abundance Unrelated to DPC Formation
The MS analysis revealed changes in log2 H:L ratios that indicate decreases in protein yield from cells treated with 6-TG and 6-TG+UVA relative to that from untreated cells (the upper left quadrant of the scatter plots in Figure 3). While these changes are consistent with DPC formation, we considered two alternative mechanisms that might contribute to this protein underrepresentation. First, a 24-h treatment with 6-TG might significantly inhibit transcription or translation. If the inhibition is sufficiently severe, it could reduce overall cellular protein content. Second, treatment with 6-TG or 6-TG+UVA causes significant protein oxidation. An alternative (and not exclusive) possibility is that the acknowledged insolubility of oxidized proteins might contribute to diminished protein recovery. To investigate these eventualities, we compared the HyBond-N+ membrane-bound proteome with proteins in unfractionated extracts. Cultures of isotopically labeled CCRF-CEM cells that had been treated with UVA+6-TG, 6-TG alone, or UVA alone were combined with an equal number of untreated reverse labeled cells, and the mixture was divided into two equal parts. Chromatin extracted from one aliquot of cells was bound to a HyBond-N+ membrane and processed for MS as described above. The remaining cells were used to prepare a DNA-free whole cell extract using a standard (RIPA) extraction procedure. Trypsin digests of these whole cell extracts were compared with those of the corresponding HyBond-N+ proteins. Figure 4, panels A and B show the effect of 6-TG treatment (Mixes 3 and 9).
Figure 4.
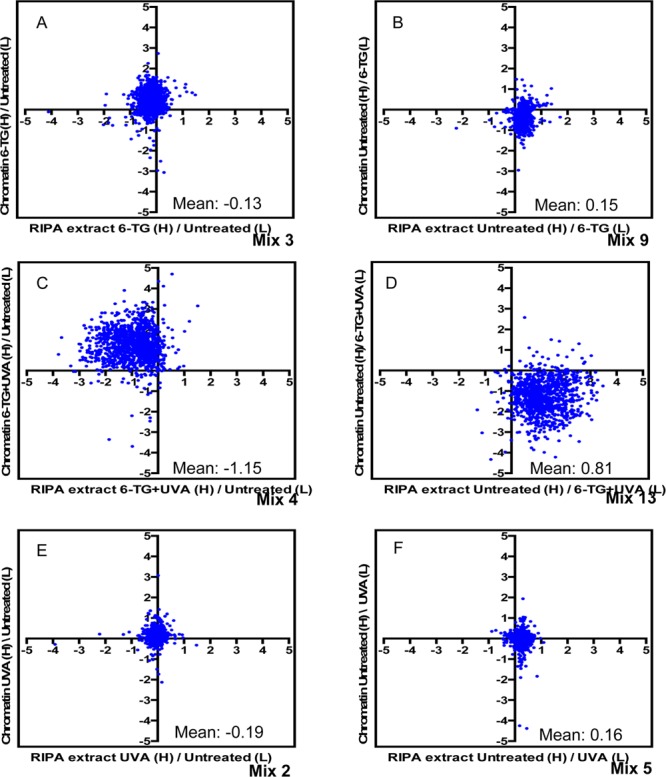
Treatment-related protein losses. Scatter plots of log2 SILAC H:L ratios comparing the effects of treatments on chromatin and RIPA (whole cell) extracts. The components of the mixes compared are shown on the axes. (A, B) The effect of 6-TG treatment. (C, D) The effect of combined 6-TG+UVA treatment. (E, F) The effect of UVA irradiation. Mean values for log2 H:L ratios RIPA extract mixes are presented on each panel.
The vertical shifts in log2 H:L ratios from the origin of the scatter plot in the direction of the label of the chromatin from 6-TG-treated cells confirm that 6-TG induces DPCs. The small changes in mean log2 H:L ratio for the RIPA extract proteins (Mix 3 = −0.31; Mix 9 = 0.15) indicate that yields are largely unaffected by 6-TG treatment. In contrast, RIPA extracts are enriched for proteins from untreated cells relative to those from cells treated with 6-TG+UVA (Figure 4C,D) yielding significant changes in mean log2 H:L values (Mix 4 = −1.15; Mix 13 = 0.81). As expected, UVA did not affect protein recovery (Mean log2 H:L ratios −0.19 and 0.16 for Mixes 2 and 5, respectively). Since neither 6-TG nor UVA alone significantly influenced protein recovery, the changes in protein abundance in extracts from cells treated with 6-TG+UVA can be ascribed to their combined effect. It follows that interference with transcription/translation during prolonged (24 h) 6-TG treatment does not have a significant impact on protein yield. We conclude that the diminished protein recovery from 6-TG+UVA treated cells most likely reflects depletion due to protein–DNA cross-linking allied to losses resulting from precipitation of proteins oxidized by 6-TG+UVA.
The observation that treatment with 6-TG+UVA causes a measurable reduction in protein recovery indicates that SILAC analysis may slightly underestimate the extent of DPC formation by this combination.
Identification of Proteins Susceptible to Cross-Linking
To identify proteins that were most susceptible to DNA cross-linking, we used R combined with ggplot222 to determine and visualize the 95th (for heavy-labeled treated cells) and fifth (for light-labeled treated cells) percentile values of log2 H:L ratio changes for each of the 16 analyses. These values are shown in Figure 5, panel A along with a plot that provides a graphic representation of the predicted maximal effect of UVA, 6-TG, or 6-TG+UVA on DNA–protein cross-linking.
Figure 5.
Cross-linking profile of CCRF-CEM chromatin proteins most vulnerable to DPC formation. (A) 95th (treated heavy-labeled) or 5th (treated light-labeled) percentile values for log2 H:L ratios are presented and plotted for each of the 16 comparisons. (B) The cross-linking profile of the 200 proteins that best fit the profile in panel A is shown superimposed on the profile for all 2611 detected proteins (in gray). (C) Protein ontology of the best fit 200 proteins in panel B, as specified by UniProt. (D) Cross-linking profiles of the 114 potential false positive proteins (Figure 3E). (E) Ontology of false positive proteins.
We used the Perseus program (maxquant) to identify 200 proteins that best fit this profile. In Figure 5, panel B, the cross-linking profile of these proteins (red) is superimposed on that of all other identified proteins (gray). Figure 5, panels A and B confirm that both 6-TG and 6-TG+UVA induce cross-linking.
Of the 200 selected proteins, 192 are either predominantly nuclear or have been detected in the nucleus (Uniprot/GeneCards). Around one-third of these proteins are involved in gene expression, and DNA repair/replication comprises the next largest category (16%, Figure 5C). DNA and RNA binding proteins are equally represented (Figure 5C) and together account for about half of the 200 selected. The significant representation of RNA binding proteins requires comment. Since 6-TG is incorporated extensively into RNA23 and UVA cross-links protein to RNA containing a photoreactive purine analog,24 some of these RNA binding proteins may be present as RNA-protein cross-links. Direct measurements (data not shown) indicated that RNA accounted for <2% of the total nucleic acid in our chromatin preparations. We therefore rule out a significant contribution from RNA-protein cross-links, and we conclude that these RNA processing proteins are most likely present in DPCs. The cross-linking profiles for the 114 proteins identified as potential false positives by cluster analysis (Figure 3E) are shown in Figure 5, panel D and their categorization in Figure 5, panel E. Cross-linking analysis confirms that none of the treatments increased their representation in DPCs. The identification of these proteins may reflect a number of nonexclusive factors. These include particularly high abundance leading to high background and lower probability of detecting treatment-related DPC formation, a high affinity for DNA, or strong direct binding to the HyBond-N+ membrane independently of DNA. Consistent with these possibilities, histones (see below) and the highly abundant intermediate filament protein vimentin (pI = 5) were among the 114 false positives.
DNA Repair and Replication Proteins
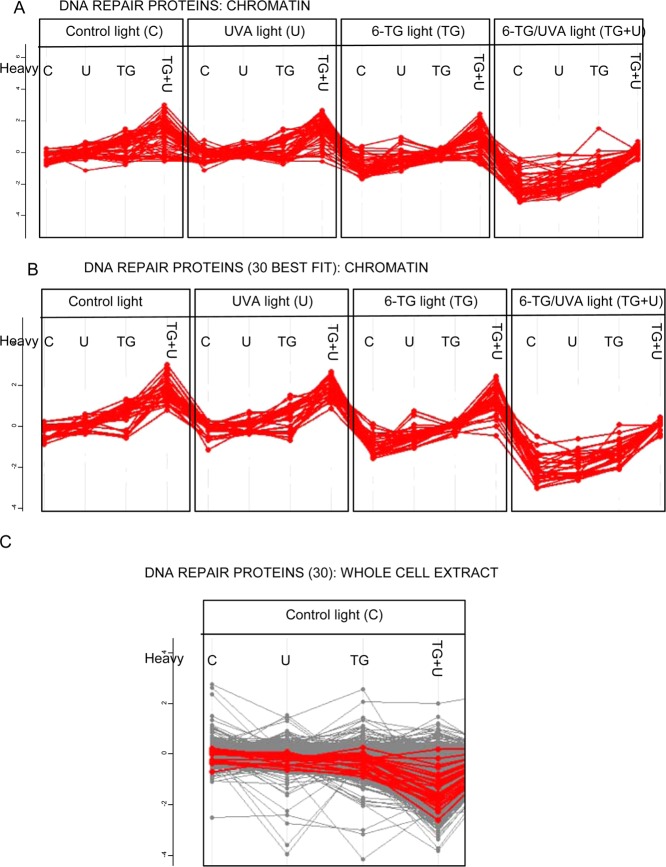
The intimate association of DNA repair and replication proteins with DNA is expected to increase their vulnerability to DNA cross-linking. Among 179 DNA repair proteins (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html), 52 were identified by our analysis. Most matched the expected profile for 6-TG and 6-TG+UVA dependent cross-linking (Figure 6A). Analysis of the 30 best fits to the generic cross-link profile (Supplementary Table S1) is shown in Figure 6, panel B. A similar analysis of whole cell (RIPA) extracts of Mixes 1–4 (Figure 2B) indicated that most of these DNA repair proteins were underrepresented to some degree in cells treated with 6-TG+UVA (Figure 6C). This observation is consistent with a significant depletion of DNA repair proteins by cross-linking to DNA.
Figure 6.
Cross-linking of DNA repair proteins. (A) Cross-linking profiles for the 52 detected designated DNA repair proteins. (B) Cross-linking profiles for the 30 most vulnerable DNA repair proteins. (C) Log2 SILAC H:L ratio plots for whole cell RIPA extracts for the 30 most vulnerable DNA repair proteins indicating that the majority are significantly depleted.
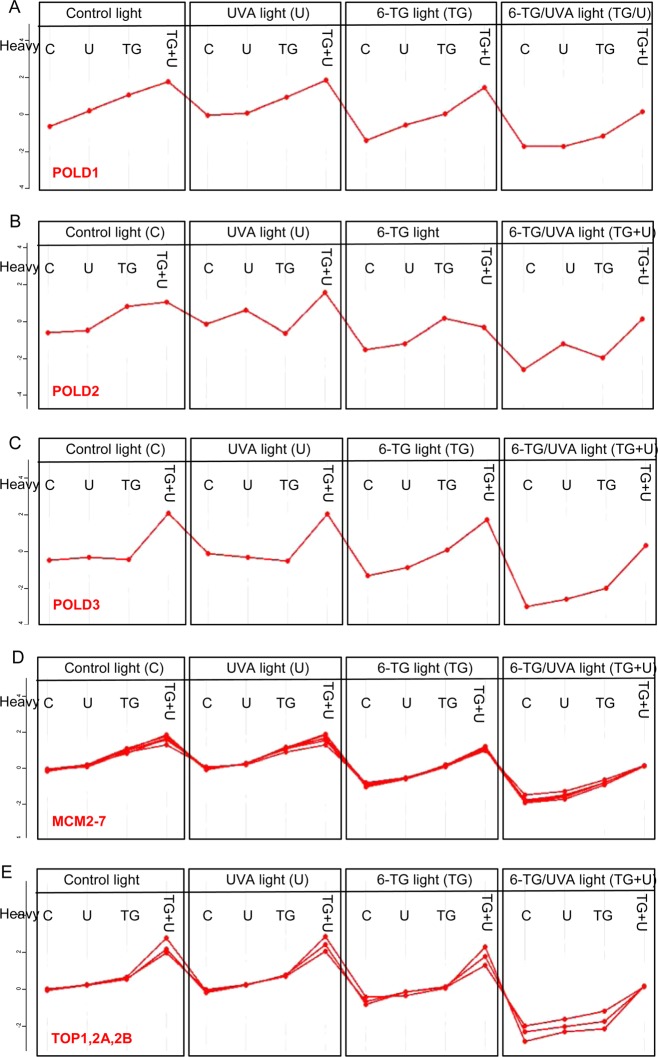
DNA polymerase δ is involved in both DNA repair and replication. It comprises three large and one small (12 kDa) subunits. Our analysis identified the three largest subunits in DPCs. We did not detect the 12 kDa subunit. The cross-linking profiles for the 125 kDa catalytic (PolD1) and the 50 kDa PolD3 auxiliary subunits (Figure 7A,C) were good fits to the generic profile of Figure 5, panel A confirming their vulnerability to DNA cross-linking. These profiles suggest that PolD1 is susceptible to cross-linking in cells treated with either 6-TG alone or 6-TG+UVA, whereas PolD3-DNA cross-linking appears to be predominantly photochemical and requires both 6-TG and UVA. Although the 66 kDa PolD2 subunit was among the chromatin proteins identified by MS analysis, its profile deviated significantly from the generic plot suggesting that it is less susceptible to cross-linking.
Figure 7.
Cross-linking of individual DNA repair and replication proteins. Cross-linking profiles for: (A) DNA polymerase delta 125 kDa, (B) 60 kDa, and (C) 55 kDa subunits; (D) MCM2–7 proteins; (E) topoisomerases 1,2A and 2B.
The MCM complex is an essential DNA replication factor. It comprises the MCM2-MCM7 proteins that assemble on DNA as a circular hexamer to initiate replication. All six MCM subunits were identified among the most susceptible proteins. Their cross-link profiles were essentially superimposable (Figure 7D) indicating a shared vulnerability to cross-linking in cells treated with 6-TG or with 6-TG+UVA.
Topoisomerases relieve supercoiling by cleaving DNA ahead of the transcription or replication apparatus. This essential function is performed by the major human topoisomerases TOP1 and TOP2A, 2B. The three topoisomerases exhibited essentially identical behavior. They were susceptible to DNA cross-linking, and like POLD3, they also appeared to be particularly susceptible to photochemical cross-linking to DNA containing 6-TG (Figure 7E).
Consistent with depletion due to DPC formation, the PolD subunits, MCM proteins, and topoisomerases were all present in significantly reduced levels in whole cell extracts following 6-TG+UVA treatment (Supplementary Figure S3).
DNA–Protein Cross-Linking by Ciprofloxacin+UVA
Ciprofloxacin is an acknowledged UVA photosensitizer. SILAC-HyBond-N+ membrane binding was used to investigate DPC formation by UVA-activated ciprofloxacin. Figure 8, panel A shows the experimental setup. Treatment with 500 μM ciprofloxacin and 50 kJ/m2 UVA induced significant DNA–protein cross-linking in CCRF-CEM cells (Figure 8B) as indicated by the clustering of most of the data for the comparison of Mixes 2 and 3 in the lower right quadrant of the scatter plot. Among the 2269 cross-linked proteins we identified, the representation of DNA and RNA binding proteins was closely similar to that generated by 6-TG+UVA (Figure 8C). Forty-one DNA repair proteins (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html) were identified in ciprofloxacin+UVA treated samples. Of these, 29 fit the profile expected for DPC induction (>2-fold change) (Supplementary Table S1, Figure 8D). Among ciprofloxacin+UVA cross-linked DNA repair proteins, there was a highly significant (p < e–10) overlap with DNA repair proteins cross-linked by 6-TG+UVA treatment (Figure 8E). Hierarchical cluster analysis (Supplementary Figure S4) confirmed ciprofloxacin+UVA induced DPC formation and again revealed a group of candidate false-positive proteins. The cross-linking profiles of the 68 candidates confirmed that they were not present in treatment-related DPCs (data not shown). Histones were highly represented in this group, and there was significant overlap with false positives identified following 6-TG+UVA treatment (Supplementary Figure S4).
Figure 8.

Cross-linking by ciprofloxacin+UVA. (A) Outline of analysis. Heavy- or light-labeled CCRF-CEM cells were treated with 500 μM ciprofloxacin (Cip) for 1 h and UVA (50 kJ/m2) as indicated. Salt-washed chromatin prepared from cells mixed in 1:1 ratios as indicated was applied to a HyBond-N+ membrane. Following washing with water and 8 M urea, membrane-associated proteins were trypsin digested in situ and analyzed by MS. The table describes the mixes that were compared. (B) Ciprofloxacin+UVA induced DPC formation. Scatter plot of Mix 2 versus Mix 3. (C) DNA and RNA binding proteins among Cirofloxacin+UVA (green) and 6-TG+UVA (blue). (D) Cross-linking profiles for 29 DNA repair proteins that best fit the most vulnerable profile. (E) Overlap between 29 DNA repair proteins cross-linked by ciprofloxacin+UVA and the 30 cross-linked by 6-TG+UVA.
Discussion
We have devised a SILAC and proteomics-based method to investigate DPC formation by photosensitizer/UVA combinations and describe its application to human cells treated under conditions that mimic and amplify the clinical effects of photosensitizing medications. DPC induction by formaldehyde has been investigated by a similar approach that employs a modified ChIP technique.25 DNA cross-linking by specific subsets of proteins has also been investigated using biotinylated double-stranded oligonucleotides,26 specific recognition sequences inserted into genomic DNA in vivo,27,28 and immunoprecipitation of cross-linked proteins or trimethylated histones.29,30 Many of these studies have employed SILAC or other labeling methods to quantify cross-linking. To our knowledge, selective enrichment of DPCs by HyBond-N+ membrane binding has not previously been combined with SILAC-LC–MS analysis. The approach identified more than 2000 cellular proteins that were cross-linked to DNA by 6-TG, by 6-TG+UVA, or by ciprofloxacin+UVA. Among the proteins most susceptible to cross-linking, most are nuclear and are involved in control of gene expression or DNA repair/replication. This distribution is consistent with incorporated DNA 6-TG or DNA-bound or -intercalated ciprofloxacin31 acting as the predominant sources of the photochemical ROS that drive DPC formation.
One important aspect of our study is that it addresses cross-linking targeted to DNA containing a reactive center (6-TG and 6-TG+UVA) as well as protein cross-linking to canonical DNA constituents (ciprofloxacin+UVA). Photoactivation of a site-specific DNA- or RNA-embedded thionucleobase, including 6-TG, has been used extensively to probe nucleic acid structure and protein–nucleic acid cross-linking.32,33 UVA-induced cross-linking of oligopeptides or purified proteins to 6-TG-containing oligonucleotides has also been used to model protein–DNA interactions in vitro,32 and we have presented preliminary evidence from 2D-DIGE and isopycnic density gradient analysis for DPC formation in cells treated with 6-TG and UVA.3 The present study describes an unbiased and quantitative investigation into DPC formation in human cells by UVA-activated photosensitizers.
A modest level of DNA substitution by 6-TG (around 0.05% of DNA guanine) in CCRF-CEM cells was sufficient to reduce DNA recovery unless the DNA was treated with a protease prior to precipitation during purification. MS analysis confirmed that 6-TG induces DPCs. Protease-reversible DNA losses during purification were exacerbated by exposure of the 6-TG-treated cells to 50 kJ/m2 UVA, a dose that had no detectable effect on DNA recovery in the absence of prior 6-TG treatment. Proteomic analysis confirmed the synergistic induction of DPCs by 6-TG and 50 kJ/m2 UVA radiation. 6-TG is an atypical photosensitizer because its incorporation into DNA introduces highly reactive DNA thiol groups through which DPCs may form preferentially. In contrast, DPCs induced by UVA-activated ciprofloxacin, which is not incorporated into DNA, must involve canonical DNA components. Our analysis revealed extensive DNA–protein cross-linking by ciprofloxacin+UVA indicating that the approach is likely to be generally applicable to DPC analysis.
Thiopurines like 6-TG perturb the cellular redox balance and increase the concentrations of intracellular ROS4,5 including superoxide anion (O2˙–),34 H2O2, and ultimately via metal-catalyzed reactions, the highly damaging hydroxyl radical (˙OH), which is a possible source of DPCs.35 We have previously shown that ICL formation in cells treated with 6-TG requires both ROS and incorporated DNA 6-TG.4 DPC induction shares these requirements, and DNA-embedded 6-TG is a key participant in DNA–protein cross-linking by 6-TG. Lesch-Nyhan cells were invulnerable to DPC induction by either 6-TG alone or 6-TG+UVA, and both treatments were without effect in CCRF-CEM cells in which replication was inhibited to prevent the accumulation of DNA 6-TG. On the basis of this requirement, we consider it unlikely that reactions between protein nucleophiles and oxidized DNA guanine36 are a significant source of DPCs mediated by 6-TG or 6-TG+UVA. We previously reported oligonucleotide–oligopeptide cross-linking involving GSO3, which is a good leaving group in nucleophilic substitution reactions, and peptide SH or NH2 groups.3 Nucleophilic attack by proteins at DNA GSO3 generated in the ROS-rich environment is a possible mechanism for 6-TG-mediated DPC formation. Cross-linking between a free protein NH2 group and a DNA 6-TG radical cation37 or a thiyl radical generated by oxidation of DNA 6-TG is a possible alternative reaction. ROS generated during 6-TG treatment cause widespread protein oxidation, and DPCs may also form via the reaction of oxidized proteins with DNA 6-TG. The more extensive photochemical cross-linking by 6-TG+UVA most likely involves Type II photosensitization. Like DNA 6-TG, ciprofloxacin is a Type II UVA photosensitizer and source of 1O2. Ciprofloxacin+UVA proved to be an effective inducer of DPCs. Since proteins are susceptible to 1O2-mediated oxidation, reactions between oxidized proteins and canonical DNA constituents are likely to be a significant factor in DPC formation by 6-TG+UVA and ciprofloxacin+UVA. Consistent with a common etiology, more than 75% of the DNA repair proteins that were identified as highly vulnerable to cross-linking by ciprofloxacin+UVA were among similar proteins identified in 6-TG+UVA DPCs.
Our analysis identified different patterns of cross-link susceptibility. Members of the group containing the large catalytic POLD1 subunit of DNA polymerase δ and the MCM proteins were efficiently cross-linked by both 6-TG and 6-TG+UVA, whereas cross-linking of proteins typified by the smaller POLD3 and the topoisomerases appeared to be largely photochemical and depended on 6-TG+UVA. These different cross-linking behaviors may reflect different cross-linking chemistries or may simply be related to the positioning of the proteins on DNA. In the case of protein multimers such as DNA pol δ□ or MCM2–7 that can also form intersubunit protein–protein cross-links, their presence in DPCs might also reflect DNA cross-linking of covalent protein–protein complexes.
Transient DPC formation is a feature of many DNA processing enzymes, and interference with the correct reversal of topoisomerase–DNA complexes is the basis of the therapeutic action of drugs such as camptothecin and etoposide. These specific enzyme-related DPCs are, however, atypical, and DPCs are likely to be structurally heterogeneous. Most studies of the induction, processing, and biological effects of DPCs have used formaldehyde, a highly reactive molecule that causes protein damage, depletes cellular reduced glutathione levels,38 and induces ICLs as well as DPCs. The possible involvement of nucleotide excision repair,39 homologous recombination,40 and the proteasome41 in DPC reversal in human cells has been suggested but is not firmly established (see ref (42) for review). Fanconi anemia cells are extremely sensitive to formaldehyde,40 and evidence from mice and from chicken cells with defective aldehyde metabolism43,44 suggests a requirement for this DNA repair pathway to repair aldehyde-induced DPCs. Whether SPRTN, the human homologue of putative DPC-specific proteases that has been identified in yeast and Xenopus laevis,45 participates in the same or in a separate repair pathway remains to be determined. Like formaldehyde, 6-TG+UVA and ciprofloxacin+UVA (and most other treatments that cause DNA–protein cross-linking) induce other DNA lesions as well as DPCs and protein damage. This pleiotropy may have hampered attempts to define DPC repair pathways in human cells. Among the putative systems for DPC repair, we identified essential NER, homologous recombinational repair, and Fanconi pathway proteins as particularly susceptible to DNA cross-linking. This observation raises the possibility that DPC repair might be compromised by depletion or oxidation of the repair proteins themselves. Because the non-DPC damage induced by UVA activated photosensitizers is unlikely to be the same as that caused by aldehydes, UVA/photosensitizer combinations may be a useful addition to studies of DPC induction and repair in human cells.
Acknowledgments
The assistance of the Clare Hall Cell Services team is gratefully acknowledged. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00717.
SI table of contents (PDF)
Protease dependence of DNA recovery and OS analysis by FACS; log2 SILAC distributions for different treatment conditions; cross-linking profiles for DNA polymerase δ, MCM proteins, and topoisomerases; heat map for ciprofloxacin+UVA cross-linked proteins and identity of false positives; DNA repair proteins most vulnerable to cross-linking by 6-TG+UVA or ciprofloxacin+UVA (PDF)
Author Present Address
# The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.
Author Contributions
‡ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Barker S.; Weinfeld M.; Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res., Rev. Mutat. Res. 2005, 589, 111–135. 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Yeo J. E.; Wickramaratne S.; Khatwani S.; Wang Y.-C.; Vervacke J.; Distefano M. D.; Tretyakova N. Y. Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS Chem. Biol. 2014, 9, 1860–1868. 10.1021/cb5001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueranger Q.; Kia A.; Frith D.; Karran P. Crosslinking of DNA repair and replication proteins to DNA in cells treated with 6-thioguanine and UVA. Nucleic Acids Res. 2011, 39, 5057–5066. 10.1093/nar/gkr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R.; Karran P. Oxidation-mediated DNA cross-linking contributes to the toxicity of 6-thioguanine in human cells. Cancer Res. 2012, 72, 4787–4795. 10.1158/0008-5472.CAN-12-1278. [DOI] [PubMed] [Google Scholar]
- Misdaq M.; Ziegler S.; von Ahsen N.; Oellerich M.; Asif A. R. Thiopurines induce oxidative stress in T-lymphocytes: a proteomic approach. Mediators Inflammation 2015, 2015, 1. 10.1155/2015/434825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett C. M.; Walker S. L.; O’Donovan P.; Warwick J.; Harwood C. A.; Karran P.; McGregor J. Azathioprine treatment sensitizes human skin to ultraviolet A radiation. Br. J. Dermatol. 2008, 159, 198–204. 10.1111/j.1365-2133.2008.08610.x. [DOI] [PubMed] [Google Scholar]
- Euvrard S.; Kanitakis J.; Claudy A. Skin cancers after organ transplantation. N. Engl. J. Med. 2003, 348, 1681–1691. 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- Peyrin-Biroulet L.; Khosrotehrani K.; Carrat F.; Bouvier A.-M.; Chevaux J.-B.; Simon T.; Carbonnel F.; Colombel J.-F.; Dupas J.-L.; Godeberge P.; Hugot J.-P.; Lemann M.; Nahon S.; Sabate J.-M.; Tucat G.; Beaugerie L. for the Cesame Study Group. Increased risk for nonmelanoma skin cancers in patients who receive thiopurines for inflammatory bowel disease. Gastroenterology 2011, 141, 1621–1628. 10.1053/j.gastro.2011.06.050. [DOI] [PubMed] [Google Scholar]
- Attard N. R.; Karran P. UVA photosensitization of thiopurines and skin cancer in organ transplant recipients. Photochem. Photobiol. Sci. 2012, 11, 62–68. 10.1039/C1PP05194F. [DOI] [PubMed] [Google Scholar]
- Ren X.; Li F.; Jeffs G.; Zhang X.; Xu Y.-Z.; Karran P. Guanine sulphinate is a major stable product of photochemical oxidation of DNA 6-thioguanine by UVA irradiation. Nucleic Acids Res. 2010, 38, 1832–1840. 10.1093/nar/gkp1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M. S.; Duarte T. L.; Cooper D.; Chen J.; Nandagopal S.; Evans M. D. Combination of azathioprine and UVA irradiation is a major source of cellular 8-oxo-7,8-dihydro-2′-deoxyguanosine. DNA Repair 2008, 7, 1982–1989. 10.1016/j.dnarep.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Gueranger Q.; Li F.; Peacock M.; Larnicol-Fery A.; Brem R.; Macpherson P.; Egly J.-M.; Karran P. Protein oxidation and DNA repair inhibition by 6-thioguanine and UVA radiation. J. Invest. Dermatol. 2014, 134, 1408–1417. 10.1038/jid.2013.509. [DOI] [PubMed] [Google Scholar]
- Montaner B.; O’Donovan P.; Reelfs O.; Perrett C. M.; Zhang X.; Xu Y.-Z.; Ren X.; Macpherson P.; Frith D.; Karran P. Reactive oxygen-mediated damage to a human DNA replication and repair protein. EMBO Rep. 2007, 8, 1074–1079. 10.1038/sj.embor.7401084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven M.; Brem R.; Macpherson P.; Peacock M.; Karran P. Oxidative damage to RPA limits the nucleotide excision repair capacity of human cells. J. Invest. Dermatol. 2015, 135, 2834–2841. 10.1038/jid.2015.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam E.; Brem R.; Karran P. Oxidative stress-induced protein damage inhibits DNA repair and determines mutation risk and anticancer drug effectiveness. Mol. Cancer Res. 2016, 14, 612–622. 10.1158/1541-7786.MCR-16-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhiaubet-Vallet V.; Bosca F.; Miranda M. A. Photosensitized DNA damage: The case of fluoroquinolones. Photochem. Photobiol. 2009, 85, 861–868. 10.1111/j.1751-1097.2009.00548.x. [DOI] [PubMed] [Google Scholar]
- Peacock M.; Brem R.; Macpherson P.; Karran P. DNA repair inhibition by UVA photoactivated fluoroquinolones and vemurafenib. Nucleic Acids Res. 2014, 42, 13714–13722. 10.1093/nar/gku1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Jeffs G.; Ren X.; O’Donovan P.; Montaner B.; Perrett C. M.; Karran P.; Xu Y.-Z. Novel DNA lesions generated by the interaction between therapeutic thiopurines and UVA light. DNA Repair 2007, 6, 344–354. 10.1016/j.dnarep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Reelfs O.; Macpherson P.; Ren X.; Xu Y.-Z.; Karran P.; Young A. Identification of potentially cytotoxic lesions induced by UVA photoactivation of DNA 4-thiothymidine in human cells. Nucleic Acids Res. 2011, 39, 9620–9632. 10.1093/nar/gkr674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C. A.; Nuhlen S.; Judith D.; Frith D.; Snijders A. P.; Behrends C.; Tooze S. A. TBC1D14 regulated autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016, 35, 281–301. 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1378. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Wickham H.ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, 2009. [Google Scholar]
- LePage G. A. Incorporation of 6-thioguanine into nucleic acids. Cancer Res. 1960, 20, 403–408. [PubMed] [Google Scholar]
- Costas C.; Yuriev E.; Meyer K. L.; Guion T. S.; Hanna M. M. RNA-protein crosslinking to AMP residues at internal positions in RNA with a new photocrosslinking ATP analog. Nucleic Acids Res. 2000, 28, 1849–1858. 10.1093/nar/28.9.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G.; Wills K. L. H.; Furlan C.; Rappsilber J. Chromatin enrichment for proteomics. Nat. Protoc. 2014, 9, 2090–2099. 10.1038/nprot.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G.; Butter F.; Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2008, 19, 284–293. 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S. D.; Raman A.; Taverna S. D.; Tackett A. J. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012, 2, 198–205. 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliszczak A. E.; Rainey M. D.; Harhen B.; Boisvert F. M.; Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci. Rep. 2011, 1, 95. 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H.; Taylor C.; Brown G. D.; Papachristou E. K.; Carroll J. S.; D’Santos C. S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016, 11, 316–326. 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- Soldi M.; Bonaldi T.. The ChroP approach combines ChIP and mass spectrometry to dissect locus-specific proteomic landscapes of chromatin. J. Visualized Exp. 2014, 11, ( (86), ). DOI: 10.3791/51220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan I. D.; Drevensek P.; Turel I.; Poklar Ulrih N. Characterization of ciprofloxacin binding to the linear single- and double-stranded DNA. Biochim. Biophys. Acta, Gene Struct. Expression 2003, 1628, 111–122. 10.1016/S0167-4781(03)00135-0. [DOI] [PubMed] [Google Scholar]
- Cahill M. A.; Nordheim A.; Xu Y.-Z. Crosslinking of SRF to the c-fos SRE CArG box guanines using photo-active thioguanine oligonucleotides. Biochem. Biophys. Res. Commun. 1996, 229, 170–175. 10.1006/bbrc.1996.1775. [DOI] [PubMed] [Google Scholar]
- Favre A.; Saintomé C.; Fourrey J.-L.; Clivio P.; Laugaa P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J. Photochem. Photobiol., B 1998, 42, 109–124. 10.1016/S1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- Pelin M.; De Iudicibus S.; Fusco L.; Taboga E.; Pellizzari G.; Lagatolla C.; Martelossi S.; Ventura A.; Decorti G.; Stocco G. Role of oxidative stress mediated by glutathione-S-transferase in thiopurines’ toxic effects. Chem. Res. Toxicol. 2015, 28, 1186–1195. 10.1021/acs.chemrestox.5b00019. [DOI] [PubMed] [Google Scholar]
- Altman S. A.; Zastawny T. H.; Randers-Eichhorn L.; Cacciuttolo M. A.; Akman S. A.; Dizdaroglu M.; Rao G. Formation of DNA-protein cross-links in cultured mammalian cells upon treatment with iron ions. Free Radical Biol. Med. 1995, 19, 897–902. 10.1016/0891-5849(95)00095-F. [DOI] [PubMed] [Google Scholar]
- Johansen M. E.; Muller J. G.; Xu X.; Burrows C. J. Oxidatively Induced DNA–Protein Cross-Linking between Single-Stranded Binding Protein and Oligodeoxynucleotides Containing 8-Oxo-7,8-dihydro-2′-deoxyguanosine. Biochemistry 2005, 44, 5660–5671. 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- Perrier S.; Hau J.; Gasparutto D.; Cadet J.; Favier A.; Ravanat J.-L. Characterization of lysine-guanine cross-links upon one electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J. Am. Chem. Soc. 2006, 128, 5703–5710. 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- Xie M.-Z.; Shoulkamy M. I.; Salem A. M. H.; Oba S.; Goda M.; Nakano T.; Ide H. Aldehydes with high and low toxicities inactivate cells by damaging distinct cellular targets. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2016, 786, 41–51. 10.1016/j.mrfmmm.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Baker D. J.; Wuenschell G.; Xia L.; Termini J.; Bates S. E.; Riggs A. D.; O’Connor T. R. Nucleotide excision repair eliminates unique DNA-protein crosslinks from mammalian cells. J. Biol. Chem. 2007, 282, 22592–22604. 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- Ridpath J. R.; Nakamura A.; Tano K.; Luke A. M.; Sonoda E.; Arakawa H.; Buerstedde J.-M.; Gillespie D. A. F.; Sale J. E.; Yamazoe M.; Bishop D. K.; Takata M.; Takeda S.; Watanabe M.; Swenberg J. A.; Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007, 67, 11117–11122. 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- Quievryn G.; Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 2000, 21, 1573–1580. 10.1093/carcin/21.8.1573. [DOI] [PubMed] [Google Scholar]
- Ide H.; Shoulkamy M. I.; Nakano T.; Miyamoto-Matsubara M.; Salem A. M. H. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2011, 711, 113–122. 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Langevin F.; Crossan G. P.; Rosado I. V.; Arends M. J.; Patel K. J. FancD2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 2011, 475, 53–58. 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Rosado I. V.; Langevin F.; Crossan G. P.; Takata M.; Patel K. J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct. Mol. Biol. 2011, 18, 1432–1434. 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Stingele J.; Habermann B.; Jentsch S. DNA-protein crosslink repair:proteases as DNA repair enzymes. Trends Biochem. Sci. 2015, 40, 67–71. 10.1016/j.tibs.2014.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.