Abstract
Two different disulfide (SS)-containing poly(amidoamine) (PAA) polymers were constructed using guanidino (Gua)-containing monomers (ie, arginine [Arg] and agmatine [Agm]) and N,N′-cystamine bisacrylamide (CBA) by Michael-addition polymerization. In order to characterize these two Gua-SS-PAA polymers and investigate their potentials as short hairpin RNA (shRNA)-delivery carriers, pSilencer 4.1-CMV FANCF shRNA was chosen as a model plasmid DNA to form complexes with these two polymers. The Gua-SS-PAAs and plasmid DNA complexes were determined with particle sizes less than 90 nm and positive ζ-potentials under 20 mV at nucleic acid:polymer weight ratios lower than 1:24. Bioresponsive release of plasmid DNA was observed from both newly constructed complexes. Significantly lower cytotoxicity was observed for both polymer complexes compared with polyethylenimine and Lipofectamine 2000, two widely used transfection reagents as reference carriers. Arg-CBA showed higher transfection efficiency and gene-silencing efficiency in MCF7 cells than Agm-CBA and the reference carriers. In addition, the cellular uptake of Arg-CBA in MCF7 cells was found to be higher and faster than Agm-CBA and the reference carriers. Similarly, plasmid DNA transport into the nucleus mediated by Arg-CBA was more than that by Agm-CBA and the reference carriers. The study suggested that guanidine and carboxyl introduced into Gua-SS-PAAs polymers resulted in a better nuclear localization effect, which played a key role in the observed enhancement of transfection efficiency and low cytotoxicity. Overall, two newly synthesized Gua-SS-PAAs polymers demonstrated great potential to be used as shRNA carriers for gene-therapy applications.
Keywords: short hairpin RNA, gene carrier, bioresponsive, guanidinylated disulfide-containing poly(amido amine), nuclear localization, transfection efficiency, cytotoxicity
Introduction
Over the past few decades, gene therapy has been developed as a promising strategy that has the potential to cure an innumerable range of diseases. A number of Phase I/II gene-therapy clinical trials have reported remarkable evidence of efficacy and safety for the treatment of various severe inherited diseases of the blood and immune and nervous systems, including primary immunodeficiencies, leukodystrophies, thalassemia, hemophilia, and retinal dystrophy, as well as such cancers as B-cell malignancies.1 RNA interference using small interfering RNAs (siRNAs) has emerged as a powerful tool for the knockdown of genes and holds great promise as a novel therapeutic strategy.2 However, since siRNAs are small molecules and highly negatively charged, they are not readily taken up by cells. Therefore, the development of efficient and safe delivery systems that have the capability to deliver siRNAs to action sites is essential for therapeutic activity.3 Currently, carriers for siRNA delivery include viral and nonviral carriers, such as peptides,4 cationic lipids,5 and cationic polymers, such as polyethylenimine (PEI),6 and poly-l-lysine.7 In addition, carriers based on inorganic nanoparticles, such as silica nanoparticles, are also members of the nonviral carriers.8 Viral-based delivery systems use natural viruses as delivery vectors and can provide high gene-transfection efficiency. Although applications of viral carriers have shown some success, there exist inherent disadvantages associated with viral carriers, such as inducing immune responses and mutagenicity. Nonviral gene carriers are cationic polymers or cationic lipids that can spontaneously attach to polynucleotides through electrostatic interactions to form polyplexes or lipoplexes, respectively.9 These nanoscale aggregates are rarely immunogenic and easy to scale up, which make them more promising as delivery carriers.10 In addition, the complex formation leads to improved protection of short hairpin RNA (shRNA) molecules from enzyme-mediated digestion and enhanced intracellular delivery.
Recently,11 a new class of biodegradable cationic polymers based on poly(amido amine) (PAA) with disulfide (SS) linkages in the backbone has been developed.12,13 In previous studies of SS-PAAs as gene-delivery carriers, it was shown that the presence of bioreducible SS linkages in these polymers resulted in significant increases in transfection efficiency, together with a reduction in cytotoxicity.14,15 These polymers can self-assemble with plasmid DNA (pDNA) into nanoscale complexes and display efficient gene-transfer properties. Due to the difference in redox status between oxidizing extracellular space and reducing intracellular space, SS bonds are stable outside the cell but are rapidly cleaved in the cytoplasm.16,17 This can result in rapid and favorable release of encapsulated therapeutic gene fragments in the intracellular environment. Cell selectivity of therapeutic siRNA delivery is critical to achieve its maximum therapeutic potential in many diseases. For cancer therapy, in order to reach distant tumors or metastases, systemic administration of siRNA polyplexes is inevitable.3 Upon intravenous injection, however, positively charged polyplexes might interact nonspecifically with serum proteins or erythrocytes and other blood cells, leading to the formation of aggregates, which causes rapid clearance by the reticuloendothelial system and sometimes significant toxicity.18 The biocompatibility of polyplexes can be enhanced by conjugation of polyethylene glycol (PEG) to the cationic polymer (PEGylation). In general, PEGylation of polyplexes results in a lower surface charge, reduced interaction with blood components, prolonged blood circulation, and lower cytotoxicity.19 On the other hand, steric shielding of polyplex particles also leads to reduced cellular association and uptake, diminished endosomal escape properties, and inefficient siRNA release.20 These contrasting effects associated with the use of PEG in oligonucleotide delivery are also referred to as the “PEG dilemma”.21
Guanidine is one of the strongest organic bases with some biological activities. Nitrogen and hydrogen atoms in the guanidino (Gua) group have a strong affinity to carbonic acid esters, phosphate esters, and peptides, leading to the formation of hydrogen bonds with some geometric configurations. The amino group of guanidine has the ability to react with various acids in the human body to form water-soluble salts. As a result, a drug-containing guanidine group can be easily transported and shows the selectivity of in vivo absorption and/or permeation.22 Kim et al23 further revealed that improved transfection efficiency of guanidinylated gene carriers could be attributed to two major reasons. Firstly, good cell-membrane permeability can be mediated by the charged nature of Gua groups. Secondly, peptidomimetic nuclear localization effects exist in guanidinylated polymeric gene carriers.
In order to accommodate better the needs in the research of nonviral gene carriers for therapeutic siRNA delivery, we designed and synthesized two different guanidinylated SS-PAA gene carriers. These two polymer carriers were loaded with a model pDNA, which could continuously express shRNA fragments. shRNA is a mimic for siRNA, which has the same ability to mediate gene silencing. The FANCF protein is encoded by the FANCF gene in humans. The FA–BRCA pathway participates in cell-cycle and apoptosis control and regulates detoxification, survival signal transduction, and DNA repair.24,25 Low expression and loss function of FANCF will interfere with the FA–BRCA pathway and result in tumorigenesis and development of drug resistance.26 Therefore, FANCF was chosen as a target gene, and the FANCF shRNA plasmid was constructed as a model pDNA. We expect a long-term RNA-interference effect for cancer therapy through intratumoral injection or other local administration of Gua-SS-PAA-based shRNA delivery. Overall, this study was designed to test the biological properties of two newly synthesized Gua-SS-PAA polymers and to investigate the relationships among the structure and biological properties of Gua-SS-PAAs as gene-delivery carriers.
Materials and methods
Materials
Branched PEI (water-free) with molecular weight of 25 kDa, ampicillin, reduced l-glutathione (GSH), MTT, tryptone, and yeast extracts were purchased from Sigma-Aldrich (St Louis, MO, USA). Lipofectamine 2000 (Lipo) reagent, Roswell Park Memorial Institute (RPMI) 1640 medium, fetal bovine serum (FBS), penicillin, streptomycin, DNase I, trypsin, and N,N′-cystamine bisacrylamide (CBA) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). A TianPure Midi Plasmid Kit was purchased from Tiangen Biotech Co Ltd (Beijing, China). Hoechst 33342 and DiO were purchased from Beyotime Institute of Biotechnology (Haimen, China). pSilencer 4.1-CMV FANCF shRNA was kindly provided by the Department of Pharmacology of China Medical University (Shenyang, China). A Lyso ID red detection kit and Total Nuclear ID green/red nucleolar/nuclear detection kit were purchased from Enzo Life Sciences (Farmingdale, NY, USA). FANCF antibody (D2) and goat antimouse IgG fluorescein isothiocyanate were purchased from Santa Cruz Biotechnology Inc (Dallas, TX, USA). Agmatine (Agm) sulfate and l-arginine (Arg) were purchased from Sinopharm Chemical Reagent (Shanghai, China). Acetone, N,N-dimethylformamide, and other common reagents were analytical grade.
Synthesis of guanidinylated SS-PAAs
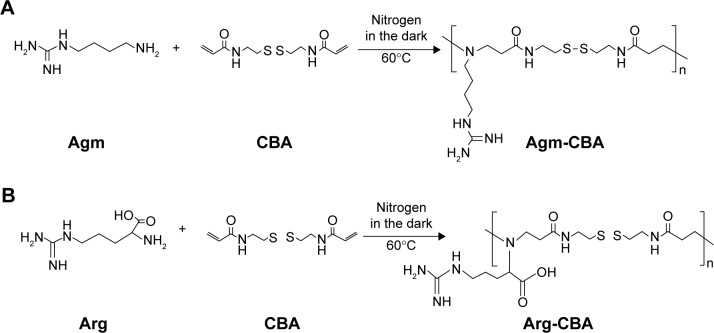
Two Gua-SS-PAA polymers (ie, Agm-CBA and Arg-CBA) were synthesized by Michael addition between CBA and Agm or Arg, in accordance with a previous report.14 Briefly, raw materials of Agm or Arg (0.38 mmol) and CBA (0.38 mmol) were added into a brown flask and dissolved with dimethylformamide (4 mL). Then, the reaction solution was heated in a water bath at 60°C under a nitrogen atmosphere and stirred continuously in the dark for at least 4 days. Subsequently, 10% of excess Agm or Arg was added to consume any unreacted acrylamide groups, and the reaction lasted for another 2 days. After that, the solution was diluted with water to 20 mL and then dialyzed against distilled water (molecular weight cutoff 1,000) for 2 days to remove the unreacted monomers. Lastly, the solution was lyophilized for 36 hours to obtain the final product. The synthesis route is shown in Scheme 1.
Scheme 1.
Schematic synthesis routes of Agm-CBA (A) and Arg-CBA (B).
Abbreviations: Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine.
Gua-SS-PAA polymer characterizations
The prepared Gua-SS-PAAs were dissolved in deuterated dimethyl sulfoxide, and their 1H nuclear magnetic resonance spectroscopy (NMR) spectra were recorded on a Varian Unity 600 MHz spectrometer (Bruker Corporation, Billerica, MA, USA).
Molecular weights of Gua-SS-PAAs were determined by gel-permeation chromatography using high-performance liquid chromatography (2690D equipped with Ultrahydrogel 120 and 250 columns; Waters Corporation, Milford, MA, USA). PEG was used as the molecular weight standard, and the molecular weight range for PEG was 1,500–21,600 Da. HAc-NaAc buffer solution (0.25 M, pH 4.5) was used as an eluent and run at a flow rate of 0.7 mL/min.
Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes
pDNA (pSilencer 4.1-CMV FANCF shRNA) was extracted from the pSilencer 4.1-CMV FANCF shRNA vector strain using the TianPure Midi plasmid-extraction kit, as previously described.25 The extracted pDNA was diluted to a final concentration of 10 μg/mL in HEPES buffer solution (HBS; 50 mM HEPES, 750 mM NaCl, pH 7.4). Then, the pDNA was complexed with Gua-SS-PAA polymers at different nucleic acid:polymer (pDNA:polymer) weight ratios from 1:0.5 to 1:96. The preparation process was as follows: a certain amount of polymer (Arg-CBA or Agm-CBA, 10 μg/μL in HBS) was added to 1 mL plasmid solution (10 μg/mL in HBS), followed by vortexing for 20 seconds, and then the solution was incubated for 30 minutes at room temperature. Lipo and PEI were complexed with pDNA as controls in this study. pDNA:Lipo and pDNA:PEI at concentrations (pDNA:Lipo 1:6 μg/μL weight:volume, pDNA:PEI 1:12 weight ratio) recommended by manufacturers were prepared by adding 60 μL Lipo or 12 μL PEI to 1 mL plasmid solution (10 μg/mL in HBS), followed by vortexing for 20 seconds, and the solution was incubated for 10 minutes at room temperature.
Determination of encapsulation efficiency
Polymer-encapsulation efficiency was determined by a Hoechst 33342 intercalation assay. The pDNA (0.4 μg, pSilencer) was mixed with 200 μL Hoechst 33342 (0.05 μg/mL) and then incubated with Gua-SS-PAAs (Agm-CBA and Arg-CBA) at different pDNA:polymer weight ratios from 1:0.5 to 1:96 for 30 minutes at room temperature. The fluorescence of all samples was measured by a microplate reader (SpectraMax M3; Molecular Devices LLC, Sunnyvale, CA, USA). Lipo and PEI were used as reference polymer carriers at concentrations (pDNA:Lipo 1:6 weight:volume, pDNA:PEI 1:12 weight ratio) recommended by manufacturers. The fluorescence of free DNA was used as a control. The encapsulation efficiency was determined from the following equation:27
| (1) |
where Flu represents fluorescence.
Measurements of particle size and ζ-potential
pDNA and Gua-SS-PAA polymer complexes were prepared as described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. Size and ζ-potential of the complexes were measured at 25°C on a Zetasizer Nano system (Malvern Instruments, Malvern, UK) using dynamic light scattering. PEI was used as a reference polymer carrier at a concentration (pDNA:PEI 1:12 weight ratio) recommended by the manufacturer. The particle-size and ζ-potential measurements were processed using Dispersion Technology version 7.01 (Malvern Instruments) software.
Atomic force microscopy analysis of complex particle formation
Samples were prepared on freshly cleaved mica using a mixed solution of pDNA (final concentration of 10 μg/mL) and Gua-SS-PAA polymers (final concentration of 0.4 mg/mL), resulting in complexes at a pDNA:polymer weight ratio of 1:48 in HBS. Lipo and PEI were used as reference polymer carriers at concentrations (pDNA:Lipo 1:6 weight:volume, pDNA:PEI 1:12 weight ratio) recommended by manufacturers. Complex formation was studied using a PicoForce Multimode NanoScope IIIa (Veeco Instruments, Oyster Bay, NY, USA) in tapping mode. Tapping-mode imaging was performed in situ in HBS, using a standard liquid cell and a standard Veeco NP cantilever. Similarly, in order to study different complexes in solid state, nitrogen was used to blow-dry the samples after they were prepared on the mica surface. The atomic force microscopy-system parameters were set the same as described earlier.
Agarose gel-electrophoresis study
Agarose gel-retardation assay
The polymer complexes pDNA/Lipo, pDNA/PEI, or pDNA/Gua-SS-PAAs at varying pDNA:polymer weight ratios ranging from 1:3 to 1:96 were prepared by the methods described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. After the addition of 2 μL of 6× loading buffer to 10 μL of plasmid solution, pDNA/Lipo, pDNA/PEI, pDNA/Agm-CBA, or pDNA/Arg-CBA, 12 μL of the mixture was loaded onto a 1% agarose gel containing 0.5 μg/mL Goodview, and run at 100 V for 30 minutes in TAE buffer (40 mM, Tris acetate, 1 mM ethylenediaminetetraacetic acid). DNA was then visualized with an ultraviolet lamp using BioSpectrum AC system water-cooled Gel Camera P:N 97-0154-05 (UVP LLC, Upland, CA, USA).
Reduction-sensitivity assay
Polymer complexes at varying pDNA:polymer weight ratios from 1:6 to 1:48 were prepared by the same method as described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. Then, GSH (final concentration of 20 mM) was added to 20 μL of pDNA and polymer-complex solutions, including pDNA/Lipo, pDNA/PEI, pDNA/Agm-CBA, or pDNA/Arg-CBA. The dispersions were then incubated for 30 minutes. The same complex solutions were also prepared without GSH as negative controls. After being mixed with 4 μL of 6× loading buffer, 24 μL of the mixture was loaded onto a 1% agarose gel containing 0.5 μg/mL Goodview and electrophoresed.28 The electrophoresis conditions were the same as described in the “Agarose gel-retardation assay” section.
Heparin-replacement assay
Polymer complexes at a pDNA:polymer weight ratio of 1:12 were prepared by the same method as described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. Heparin (10 U/μL) was diluted to different concentrations and added to 10 μL of pDNA and polymer-complex solutions (200 ng of pDNA), including pDNA/Lipo, pDNA/PEI, pDNA/Agm-CBA, or pDNA/Arg-CBA. The dispersions with different contents of heparin were then incubated for 30 minutes at 37°C. After the addition of 4 μL 6× loading buffer, 24 μL of the mixture was loaded onto a 1% agarose gel containing 0.5 μg/mL Goodview and electrophoresed.27 The electrophoresis conditions were the same as described in the “Agarose gel-retardation assay” section.
DNase I-degradation assay
Polymer complexes at different pDNA:polymer weight ratios (from 1:0.5 to 1:12) were prepared by the same method as described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section and incubated with 10 μL DNase I digestion system (DNase I 10 U, 50 mM KCl, 10 mM Tris–HCl, 10 mM MgCl2, 0.1% Triton X-100, pH 9) at 37°C for 30 minutes. The mixtures were then incubated with 5 μL termination solution (400 mM NaCl, 100 mM ethylenediaminetetraacetic acid, pH 8) for 10 minutes at room temperature.27 DNA integrity was assessed by agarose-gel electrophoresis after replacement with heparin solution (6 U/μL). The electrophoresis conditions were the same as described in the “Agarose gel-retardation assay” section.
Cell-viability measurement
MCF7 (human breast cancer) cell line was kindly provided by the Department of Pharmacology of China Medical University (Shenyang, China) and was Obtained originally from the ATCC (American type culture collection), were maintained in RPMI 1640 growth medium supplemented with 10% FBS and penicillin (100 U/mL)–streptomycin (100 μg/mL) at 37°C in a humidified atmosphere containing 5% CO2. MCF7 cells were seeded in 96-well plates (4×103 cells per well) and cultured overnight. Polymers and pDNA were dissolved in serum-containing cell-culture media. Complexes were prepared by adding 200 μL of polymers at concentrations of 60–960 μg/mL to 50 μL of pDNA at a concentration of 40 μg/mL, followed by vortexing for 10 seconds and incubation at room temperature for 30 minutes. MCF7 cells in each well were incubated with the desired amount of complexes for 24 hours, and then the complex solution in each well was removed. After that, 20 μL MTT (5 mg/mL) solution was added to each well and incubated for 4 hours. The MTT solution was then removed, and the purple crystals formed in each well were dissolved with 150 μL dimethyl sulfoxide. The absorbance (optical density [OD]) of samples was measured at 570 nm using a microplate reader (SpectraMax M3), and the absorbance of untreated cells was taken as 100% cell viability. Relative cell viability was calculated as:
| (2) |
where OD570 (control) was obtained with untreated cells, and OD570 (samples) was obtained in the presence of polymers.28
Determination of in vitro transfection efficiency
Transfection experiments were performed with MCF7 cells using the pSilencer plasmid. The pDNA and Gua-PP-SAA polymer complexes were prepared by the same methods as described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. MCF7 cells were seeded in six-well plates at a density of 2×105 cells per well and cultured in 1.5 mL RPMI 1640 complete culture medium until cell confluence reached about 60%. Cells were washed with fresh phosphate-buffered saline and incubated with 2 mL complete medium containing complexes with pDNA:polymer weight ratios from 1:6 to 1:96 for 4 hours at 37°C in a humidified 5% CO2-containing atmosphere. The complex containing medium was then replaced with 2 mL of fresh RPMI 1640 complete medium, and cells were incubated for another 44 hours. After that, cells in each well were incubated with FANCF antibody (D2) and then fluorescein isothiocyanate-labeled secondary antibody at 37°C for 1 hour and 30 minutes, respectively, following instructions from the antibody manufacturers. The treated MCF cells were then evaluated by flow cytometry and fluorescence microscopy. Briefly, one part of the MCF7 cells was observed under a TE2000 inverted fluorescence microscope (Nikon Corporation, Tokyo, Japan) with an excitation wavelength of 488 nm and a magnification of 200×. Another part of the MCF7 cells was collected for flow-cytometry measurement to determine transfection efficiency using a FACScan flow cytometer (BD, Franklin Lakes, NJ, USA). The flow cytometer was calibrated with untreated cells as a negative control and set at an excitation wavelength of 488 nm. The percentages of transfected cells were quantified from a gated viable population of 10,000 cells. Samples were treated following a routine flow-cytometry protocol.
Dynamic cellular uptake study
The dynamic uptake process of pDNA/Lipo, pDNA/PEI, pDNA/Agm-CBA, or pDNA/Arg-CBA in MCF7 cells was recorded at different time intervals by laser-scanning confocal microscopy. In order to display the intracellular localization of pDNA, the Total Nuclear ID green/red nucleolus/nuclear detection kit, Hoechst 33342, and DiO were used to stain the nucleolus, nucleus, pDNA, and cell membrane, respectively, according to the provided protocols from their respective manufacturers. In brief, pDNA (1.5 μg per well) and Hoechst 33342 (23 μg/mL) were incubated at 37°C for 1 hour. Then, the stained pDNA was precipitated with 70% ethanol and resuspended in phosphate-buffered saline. MCF7 cells were seeded at 1×105 cells per well (glass-bottom dish, diameter 35 mm) and cultured overnight in normal cell-culture conditions as described in the “Cell-viability measurement” section. After incubation, cells in each well were washed and stained with the Total Nuclear ID green/red nucleolar/nuclear detection kit and DiO sequentially. Simultaneously, the resuspended pDNA was incubated with polymers to prepare different complexes at a pDNA:polymer weight ratio of 1:48 following the method described in the “Preparation of Gua-SS-PAA pSilencer 4.1-CMV FANCF shRNA nanocomplexes” section. After the staining, cells were incubated in complete culture medium containing different complexes, and the localization of complexes was recorded at 5 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 4 hours using laser-scanning confocal microscopy (FV1000 SIM IX81; Olympus Corporation, Tokyo, Japan). The intracellular pDNA, nucleus, nucleolus, and cell membrane appeared in blue, red, green, and green, respectively.
Statistics
Statistical comparisons were performed using one-way analysis of variance, followed by Dunnett’s test. Pair-wise comparisons between treatments were achieved by using Student’s t-test. P-values less than 0.05 were considered statistically significant.
Results and discussion
Synthesis and characterization of Agm-CBA and Arg-CBA
In this study, two amino monomers – Agm and Arg – were used to introduce guanidine into SS-PAA gene-delivery carriers. The synthesis of Agm-CBA and Arg-CBA followed the route shown in Scheme 1 through one-step Michael-addition polymerization. The reaction occurs at the primary amine groups of Agm or Arg and the double-propenyl group of CBA. The 1H-NMR spectra of two polymerization products confirmed that Agm and Arg were successfully connected to CBA. Polymerization was further confirmed by the disappearance of a characteristic olefin peak of CBA at a chemical shift of 5.5–6.5 ppm, although tiny peaks at the same chemical shift were still visible, which indicated that unreacted CBA residual or end-propenyl groups of the bisacrylamide still existed (Figures S1–S5). However, this did not affect the measurement of molecular weight or polydispersity of either polymer, which had been confirmed by gel-permeation chromatography (Table 1). The average molecular weights of Agm-CBA and Arg-CBA were 8,289 and 7,975 g/mol, respectively, which were lower than that of PEI (12,894 g/mol). It appeared that there were no significant differences in absolute molecular weight between Arg-CBA and Agm-CBA, which suggested that Arg and Agm showed the same reactivity, and the carboxyl group in Arg did not affect the chain-extension process under the same polymerization conditions. Polydispersity indices of both polymers were found to be 1.51–1.83. The relatively low polydispersity indices showed the synthesized Gua-SS-PAAs had narrow molecular weight distribution.
Table 1.
Molecular weight and PD of Agm-CBA and Arg-CBA polymers measured by gel-permeation chromatography
| Polymer | Retention time (min) | Mn (g/mol) |
Mw (g/mol) |
Mp (g/mol) |
PD Mw/Mn |
|---|---|---|---|---|---|
| PEI | 18.292 | 12,894 | 23,610 | 17,658 | 1.83 |
| Agm-CBA | 17.819 | 8,289 | 13,614 | 9,543 | 1.64 |
| Arg-CBA | 17.871 | 7,975 | 12,033 | 9,162 | 1.51 |
Abbreviations: PD, polydispersity; Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; Mn, number-average molecular weight; Mw, weight-average molecular weight; Mp, peak molecular weight; PEI, polyethylenimine.
Encapsulation efficiency of polymeric carriers
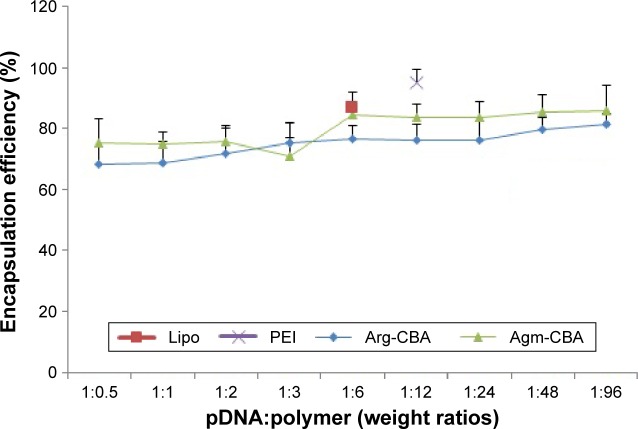
The DNA-encapsulation efficiency of Gua-SS-PAAs polymers at different pDNA:polymer weight ratios was determined with Hoechst 33342 intercalation. As shown in Figure 1, the encapsulation efficiencies of Agm-CBA and Arg-CBA were all above 65% at pDNA:polymer weight ratios ranging from 1:0.5 to 1:96. In contrast, the encapsulation efficiencies of PEI and Lipo were 95.12%±4.3% and 86.5%±5.61%, respectively, much higher. The encapsulation efficiency of Agm-CBA was higher than that of Arg-CBA, except at the pDNA:polymer weight ratio of 1:3. The results indicated that both polymers encapsulated pSilencer 4.1-CMV FANCF shRNA well and were suitable for gene delivery.29 Furthermore, the higher encapsulation efficiency of Agm-CBA compared to Arg-CBA might have been due to the carboxyl group in Arg, which influenced the charge density of Arg-CBA. From our results, it seemed that the positive-charge density of the polymer played an important role in encapsulation. For example, PEI had better encapsulation efficiency than Arg-CBA or Agm-CBA, while the ζ-potential of pDNA/PEI was found to be higher also.
Figure 1.
Encapsulation efficiency of Agm-CBA and Arg-CBA at weight ratios (pDNA:polymer) varying from 1:0.5 to 1:96.
Note: Results reported as mean ± standard deviation for three individual measurements.
Abbreviations: Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; pDNA, plasmid DNA; Lipo, Lipofectamine 2000; PEI, polyethylenimine.
Atomic force microscopy analysis of complex particle formation
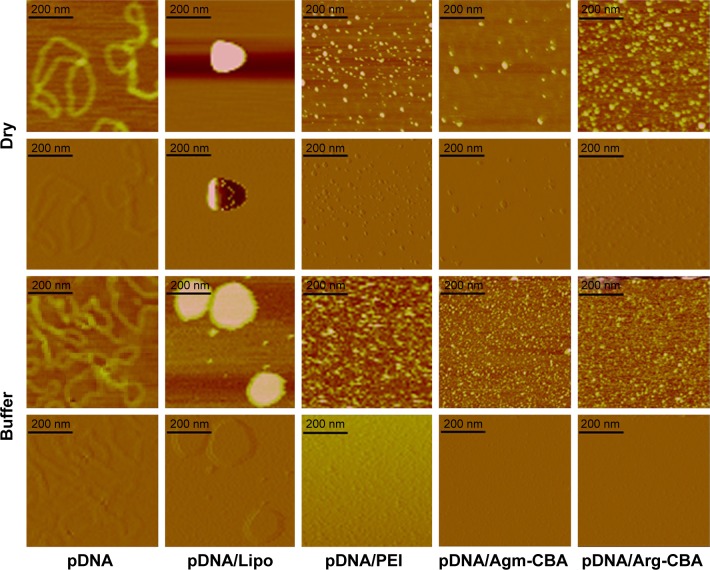
The condensation capability of Arg-CBA and Agm-CBA was investigated further by atomic force microscopy (Figure 2). The results showed that the polymers encapsulated pDNA after incubation with naked pDNA. The images on the top row in Figure 2 show typical plectonemic conformation of supercoiled plasmids and the conformation of different complexes in the absence of buffer on a freshly cleaved mica. We found that the sizes of pDNA/PEI, pDNA/Agm-CBA, and pDNA/Arg-CBA were all less than 40 nm, and pDNA/PEI was the smallest. The size of pDNA/Lipo was more than 200 nm, much larger than the other three complexes. Furthermore, the conformation of four complexes in the presence of buffer was also studied. There was a similar trend as that shown in the lower half of Figure 2, and the complexes were larger than those in the absence of buffer. In the buffer system, complexes exhibited higher particle density than in the system without the buffer. This might have been because the blow-dry process blew some particles away. However, the complexes in the buffer system were settling on the mica surface, which could make a difference in the images, since suspended complexes might show different morphological properties as well as particle size. In our study, complex particle sizes were further determined in a more quantitative manner, as shown in the next section.
Figure 2.
Atomic force microscopy images of Gua-SS-PAA complexes and naked pDNA in dry form or in buffer system.
Notes: Agm-CBA and Arg-CBA were complexed with pDNA at a weight ratio (pDNA:polymer) of 1:48; each image represents a 2×2 μm scan. Dry, complexes measured in a dry state; Buffer, complexes measured in HEPES buffer solution.
Abbreviations: Gua, guanidino; SS, disulfide; PAA, poly(amidoamine); pDNA, plasmid DNA; Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; Lipo, Lipofectamine 2000; PEI, polyethylenimine.
Particle-size and ζ-potential measurement
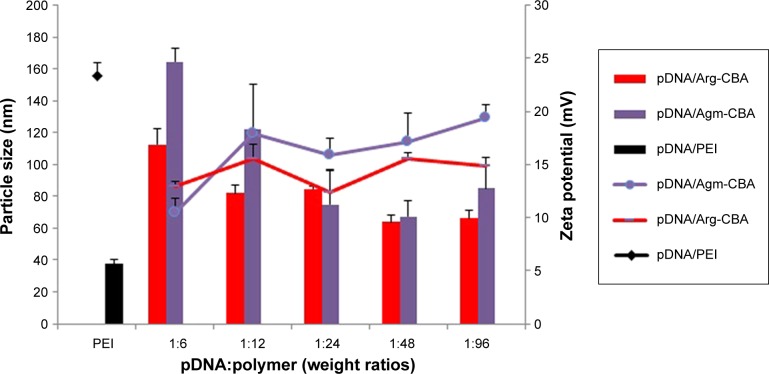
Dynamic light scattering was utilized to investigate the particle size and ζ-potential of complexes, including pDNA/PEI, pDNA/Agm-CBA, and pDNA/Arg-CBA. Figure 3 shows that particle sizes of pDNA/Agm-CBA and pDNA/Arg-CBA consistently declined as the pDNA:polymer weight ratios decreased from 1:6 to 1:96. The maximum sizes of pDNA/Agm-CBA and pDNA/Arg-CBA were 164.37±8.37 nm and 112.7±9.92 nm, respectively. When the pDNA:polymer weight ratio reduced to 1:96, the sizes decreased to 84.99±14.33 nm and 66.04±5.05 nm, respectively, indicating that the condensation capability of the two polymers increased along with the amount of polymers in complexes. There was no significant difference between pDNA/Agm-CBA and pDNA/Arg-CBA on particle condensation, except at the pDNA:polymer weight ratios of 1:6 and 1:12 (P<0.05), though pDNA/Arg-CBA exhibited stronger condensation ability than pDNA/Agm-CBA to a certain extent. In contrast, pDNA/PEI was much smaller (37.39±3.21 nm) than these two carriers when PEI was used at the recommended concentration (pDNA:polymer weight ratio 1:12), which suggested PEI had better condensation capability.
Figure 3.
Particle-size and ζ-potential measurements of Gua-SS-PAA complexes.
Notes: Agm-CBA and Arg-CBA were complexed with pDNA at weight ratios (pDNA:polymer) varying from 1:6 to 1:96. Results reported as mean ± standard deviation for three individual measurements.
Abbreviations: Gua, guanidino; SS, disulfide; PAA, poly(amidoamine); Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; pDNA, plasmid DNA.
The ζ-potentials of pDNA/Agm-CBA and pDNA/Arg-CBA increased along with the amount of polymers. When the particle sizes of complexes were stable, the ζ-potential did not change significantly. The ζ-potentials reached a maximum at the pDNA:polymer weight ratio of 1:12 for both complexes (15.5±1.41 mV and 17.9±0.4 mV for pDNA/Arg-CBA and pDNA/Agm-CBA, respectively). The ζ-potential of pDNA/PEI (23.4±1.2 mV) was higher than both Arg-CBA and Agm-CBA carriers (P<0.05). Interestingly, the ζ-potential of pDNA/Agm-CBA was higher than pDNA/Arg-CBA when the pDNA:polymer weight ratio was below 1:12. From these results, we concluded that ζ-potential was related to the conformation properties of complexes, and the differences on morphology might have been due to minor differences in the structure of polymers, such as the different side-chain groups.
In addition, the particle-size results were different in the atomic force microscopy and dynamic light-scattering studies, which might have been caused by differences in analytical techniques and treatment of samples.
Agarose gel-electrophoresis assay
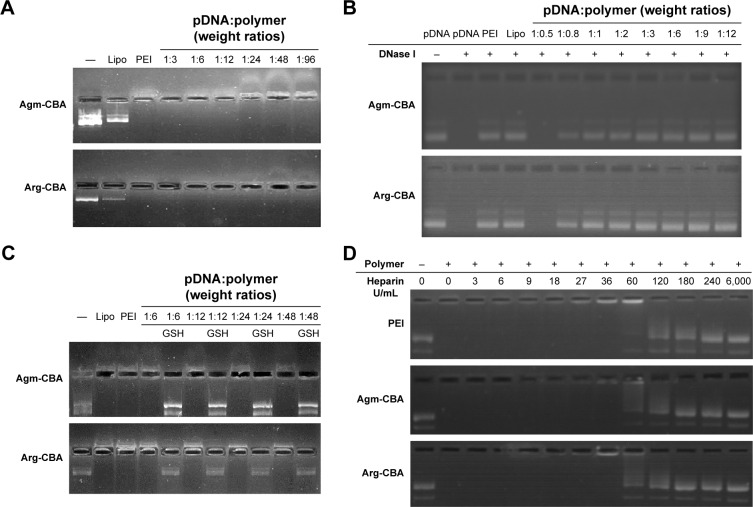
Agarose gel-electrophoresis assay was used to investigate the complexation ability and stability of different polymer carriers. As shown in Figure 4A, no retardation was found with pDNA alone and the retardation mediated by Lipo was incomplete since a band was still observed. pDNA/Agm-CBA and pDNA/Arg-CBA showed retardation at pDNA:polymer weight ratios from 1:6 to 1:96. pDNA/PEI at the recommended concentration also showed similar retardation, which indicated that these three complexes were stable and the complexation abilities of Agm-CBA and Arg-CBA were comparable to PEI. As shown in Figure 4B, DNase I assay indicated that similarly to Lipo and PEI, Agm-CBA and Arg-CBA protected the pDNA from degradation by DNase I. Furthermore, Arg-CBA showed a better protective effect than Agm-CBA, based on the observation that Arg-CBA at the pDNA:polymer weight ratio of 1:1 mediated the complete protection of pDNA, but Agm-CBA at the same ratio protected only part of the pDNA. This phenomenon was consistent with the results from particle-size and ζ-potential determination, which means that the complexation ability of Arg-CBA was better than Agm-CBA. As shown in Figure 4D, PEI, Agm-CBA, and Arg-CBA started to be replaced by heparin when the concentration of heparin reached 27 U/mL. At this concentration, a light band next to the sample-loading hole could be observed in the PEI lane, indicating that the resistance to heparin replacement of PEI was a little weaker than the other two carriers. Overall, we found that Agm-CBA and Arg-CBA possessed similar heparin-resistant ability as PEI. These two polymer carriers could have great potential for in vivo applications, since they might be resistant to other negatively charged proteins (such as heparin) in vivo.
Figure 4.
Stability and DNA-condensation ability of Gua-SS-PAA complexes determined by agarose-gel electrophoresis.
Notes: —, Naked pDNA. Agm-CBA and Arg-CBA were complexed with pDNA at weight ratios (pDNA:polymer) varying from 1:0.5 to 1:96. Complexes tested after being treated with nothing (A), DNase I (B), GSH (C), and heparin (D). (B) Heparin (6 U/μL) was used to replace pDNA fragments from complexes after treatment with DNase I.
Abbreviations: Gua, guanidino; SS, disulfide; PAA, poly(amidoamine); pDNA, plasmid DNA; Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; GSH, glutathione.
Reduction-sensitivity assay
Reduction-triggered disassembly of Agm-CBA and Arg-CBA was studied in vitro using agarose-gel electrophoresis. In our study, 20 mM of GSH was used to treat pDNA/polymer complexes, and the release of pDNA was determined by agarose-gel electrophoresis. As shown in Figure 4C, both Agm-CBA and Arg-CBA were sensitive to GSH at pDNA:polymer weight ratios ranging from 1:6 to 1:48. In addition, the released pDNA exhibited the same molecular length as naked pDNA, suggesting the existence of intact and functional pDNA.
Cell-viability measurement
Cytotoxicity is an important parameter in the development of safe nonviral carriers. High cytotoxicity of a carrier will negatively affect transfection efficiency and further influence the expression or silencing of the target gene. In order to investigate the cytotoxicity of Agm-CBA and Arg-CBA, MCF7 cells were incubated with different complexes prepared at pDNA:polymer weight ratios from 1:6 to 1:96 in the presence of 10% FBS, which was similar to the in vivo transfection environment. As shown in Figure 5, MTT results showed the cytotoxicity of Lipo or PEI was higher than Agm-CBA or Arg-DBA. The lower cytotoxicity of Agm-CBA and Arg-CBA carriers might be explained by the fact that both Agm and Arg were endogenous substances with limited toxic effects, and they rarely affected the normal activities of cells after release from polymer backbones. In addition, even the incubation time of complexes was extended to 24 hours, which was sixfold longer than the incubation time during the transfection-efficiency study. Furthermore, Martello et al30 reported that there were lot of factors associated with the cytotoxicity of carriers, such as molecular weight, charge density, monomer types, structure, intracellular degradability, and chain flexibility. Our data were consistent with Martello et al’s finding. Both carriers (Agm-CBA and Arg-CBA) showed similar molecular weight and particle size, but the ζ-potential of Agm-CBA was higher than that of Arg-CBA. The carboxyl structure in Arg-CBA might reduce its positive charge density on the surface of complexes, and so pDNA/Arg-CBA showed lower cytotoxicity than pDNA/Agm-CBA.
Figure 5.
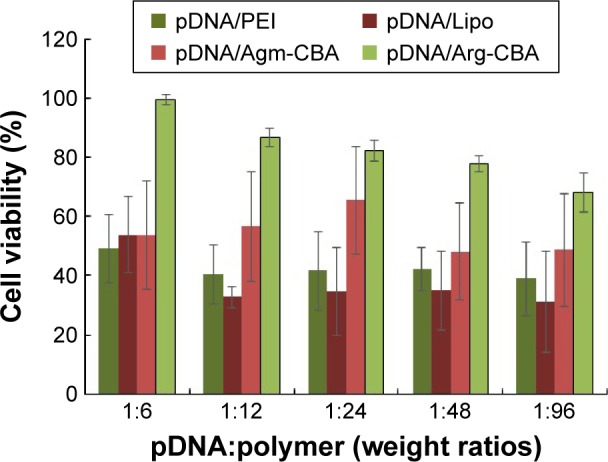
In vitro cytotoxicity of Agm-CBA and Arg-CBA complexed with pDNA at weight ratios (pDNA:polymer) varying from 1:6 to 1:96.
Notes: MCF7 cells were incubated with the desired amount of complexes for 24 hours. Results reported as mean ± standard deviation for three individual measurements.
Abbreviations: Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; pDNA, plasmid DNA; PEI, polyethylenimine; Lipo, Lipofectamine 2000.
Determination of in vitro transfection efficiency
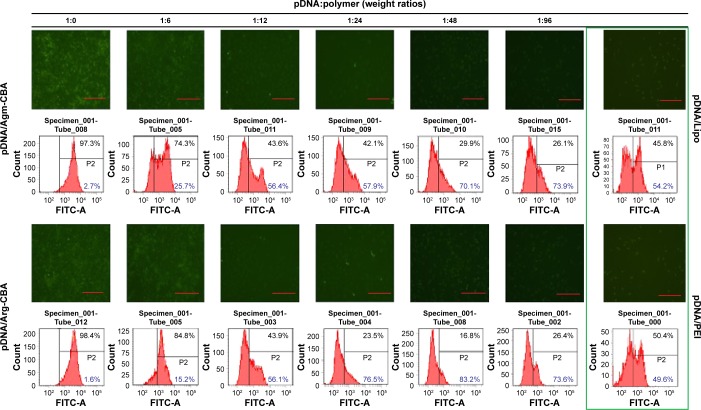
Transfection efficiencies of the complexes (pDNA/Agm-CBA and pDNA/Arg-CBA) at different pDNA:polymer weight ratios from 1:6 to 1:96 were investigated in MCF7 cells in the presence of serum. After transfection, immunofluorescence staining was performed following the standard protocol, and flow-cytometry and fluorescence-microscopy analysis were performed to evaluate the downregulation of FANCF expression. As shown in Figure 6, the silencing of FANCF mediated by pDNA/Agm-CBA or pDNA/Arg-CBA increased when the amount of polymer increased and reached a maximum of 73.9% and 83.2% at pDNA:polymer weight ratios of 1:96 and 1:48, respectively. Arg-CBA showed higher silencing efficiencies than Agm-CBA. At the higher pDNA:polymer weight ratios (1:24 and 1:48), the silencing efficiency of Arg-CBA (76.5% and 83.2%) was higher than Agm-CBA (57.9% and 70.1%). Furthermore, compared to Lipo or PEI at the recommended concentration, Arg-CBA and Agm-CBA at optimum conditions showed higher silencing efficiency, implying that the transfection efficiency of these two carriers meets the requirement of commercially available transfection reagents. In addition, pDNA/Arg-CBA showed lower efficiency at the pDNA:polymer weight ratio of 1:96 compared with 1:48. This phenomenon might be explained by 1) increased cytotoxicity of the complex at low pDNA:polymer weight ratio, which exerted a negative effect on transfection and 2) increased residual fragments after the degradation of the complex at low pDNA:polymer weight ratio, which hindered the release of pDNA.31
Figure 6.
In vitro transfection and expression results of Agm-CBA and Arg-CBA complexed with pDNA at weight ratios (pDNA:polymer) varying from 1:6 to 1:96.
Notes: The fluorescence intensity of FITC-labeled secondary antibodies connected to FANCF was observed by fluorescence microscopy (100×, scale bar 100 μm), and transfection efficiency was determined by flow cytometry 48 hours after gene transfection. The percentage marked as black represents the expression of FANCF, and the percentage marked as blue represents the silencing efficiency of FNACF (1 – expression of FANCF, transfection efficiency). Nonfluorescent MCF7 cell images were recorded under natural light as cell-density control.
Abbreviations: Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; pDNA, plasmid DNA; FITC, fluorescein isothiocyanate; Lipo, Lipofectamine 2000; PEI, polyethylenimine.
Cellular uptake study
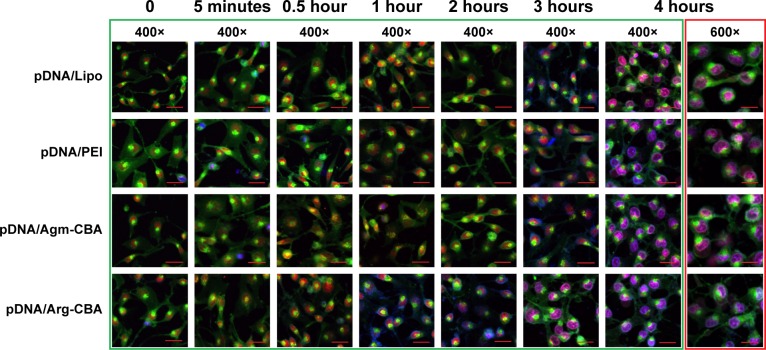
In order to understand the transfection efficiency and process of complexes, cell uptake and subcellular localization of different complexes were dynamically recorded by laser-scanning confocal microscopy. Lipo and PEI served as positive controls in this study. As shown in Figure 7, pDNA stained with blue could hardly be observed at 0–30 minutes in all groups. However, staining of the membrane (green), nucleus (red), and nucleolus (green) was clearly observed at these time points. At 1 or 2 hours after transfection, pDNA/Arg-CBA stained blue was observed in the cytoplasm and nucleus of MCF7 cells. In contrast, similar subcellular localizations of pDNA mediated by Agm-CBA, Lipo, or PEI occurred at 3 hours after transfection, and at this time point we found that the blue in the nucleus and cytoplasm was different among these three carriers. However, these three carriers exhibited the same color at the 4-hour time point. Staining in the cytoplasm and nucleus of each group at both 3- and 4-hour time points was in the following order (from purple to blue): Lipo, PEI, Agm-CBA, and Arg-CBA. The data suggested that Agm-CBA and Arg-CBA have better ability to enter the cell and localize in the nucleus compared with control groups (Lipo and PEI). There are two possible explanations for this. Firstly, after polymer degradation, residue fragment-containing guanidine was localized at the nucleus, leading to a charge change in this area and indirect attraction of pDNA to the nucleus. Secondly, the guanidine group in polymeric carriers enhanced the endosome-escape effect, leading to increased release and improved nucleus localization of free pDNA.32,33 Further studies will be needed to confirm these hypotheses. In addition, after MCF7 cells were treated with these carriers, no obvious change in the appearance of MCF7 cells was observed. This was consistent with cell-viability results, where MCF7 could recovered after 4-hour transfection.
Figure 7.
Intracellular trafficking of different complexes (pDNA/Agm-CBA and pDNA/Arg-CBA) at the weight ratio (pDNA:polymer) of 1:48.
Notes: Trafficking at 5-minute, 30-minute, 1-hour, 2-hour, 3-hour, and 4-hour time intervals. Scale bar 100 μm. The fluorescence signals were collected with three channels: pDNA stained with Hoechst 33342, blue; DiO-labeled cell membrane, green; nucleolus stained with the nucleolus tracker, green; nucleus stained with the nuclear tracker, red. Only merged images of three channels are shown.
Abbreviations: pDNA, plasmid DNA; Agm, agmatine; CBA, N,N′-cystamine bisacrylamide; Arg, arginine; FITC, fluorescein isothiocyanate; Lipo, Lipofectamine 2000; PEI, polyethylenimine.
Conclusion
In conclusion, two different Gua-SS-PAA carriers (Agm-CBA and Arg-CBA) were successfully constructed. Studies on their physicochemical properties, morphology, and biological properties demonstrated that these two reduction-sensitive carriers had the capability to encapsulate and deliver model pDNA by increasing membrane permeability and nucleus localization of pDNA. Both carriers showed suitable transfection efficiency and low cytotoxicity, suggesting that they have the potential for applications in in vivo gene therapy as delivery carriers. Further studies will be performed to evaluate the safety of these two carriers and to investigate the in vivo transfection efficiency and transport/degradation mechanisms.
Supplementary materials
1H nuclear magnetic resonance spectroscopy (NMR) of prepared Gua-SS-PAAs (AGM-CBA and ARG-CBA)
1H nuclear magnetic resonance spectroscopy of agmatine sulfate.
1H nuclear magnetic resonance spectroscopy of N,N′-cystamine bisacrylamide.
1H nuclear magnetic resonance spectroscopy (NMR) of Agm-CBA.
1H nuclear magnetic resonance spectroscopy (NMR) of L-arginine (Arg).
1H nuclear magnetic resonance spectroscopy (NMR) of Arg-CBA.
Acknowledgments
The study was financially supported by the Natural Science Foundation Committee of China (81373335) and supported in whole or in part by the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences, and School of Pharmacy, China Medical University.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526(7573):351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 2.Qiu K, Yu B, Huang H, et al. A dendritic nano-sized hexanuclear ruthenium(II) complex as a one- and two-photon luminescent tracking non-viral gene vector. Sci Rep. 2015;5:10707. doi: 10.1038/srep10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Aa LJ, Vader P, Storm G, Schiffelers RM, Engbersen JF. Optimization of poly(amido amine)s as vectors for siRNA delivery. J Control Release. 2011;150(2):177–186. doi: 10.1016/j.jconrel.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 4.Wong SP, Argyros O, Coutelle C, Harbottle RP. Non-viral S/MAR vectors replicate episomally in vivo when provided with a selective advantage. Gene Ther. 2011;18(1):82–87. doi: 10.1038/gt.2010.116. [DOI] [PubMed] [Google Scholar]
- 5.Maskell DP, Renault L, Serrao E, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523(7560):366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baliaka A, Zarogoulidis P, Domvri K, et al. Intratumoral gene therapy versus intravenous gene therapy for distant metastasis control with 2-diethylaminoethyl-dextran methyl methacrylate copolymer non-viral vector-p53. Gene Ther. 2014;21(2):158–167. doi: 10.1038/gt.2013.68. [DOI] [PubMed] [Google Scholar]
- 7.Lam AP, Dean DA. Progress and prospects: nuclear import of nonviral vectors. Gene Ther. 2010;17(4):439–447. doi: 10.1038/gt.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller JE, Zugates GT, Ferreira LS, et al. Intracellular delivery of core-shell fluorescent silica nanoparticles. Biomaterials. 2008;29(10):1526–1532. doi: 10.1016/j.biomaterials.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 10.Kim TI, Kim SW. Bioreducible polymers for gene delivery. React Funct Polym. 2011;71(3):344–349. doi: 10.1016/j.reactfunctpolym.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wu D, Ma Y, et al. Novel poly(amino ester)s obtained from Michael addition polymerizations of trifunctional amine monomers with diacrylates: safe and efficient DNA carriers. Chem Commun (Camb) 2003;(20):2630–2631. doi: 10.1039/b309487a. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Manickam DS, Chen J, Oupicky D. Effect of cell membrane thiols and reduction-triggered disassembly on transfection activity of bioreducible polyplexes. Eur J Pharm Sci. 2012;46(3):173–180. doi: 10.1016/j.ejps.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vader P, van der Aa LJ, Engbersen JF, Storm G, Schiffelers RM. Physicochemical and biological evaluation of siRNA polyplexes based on PEGylated poly(amido amine)s. Pharm Res. 2012;29(2):352–361. doi: 10.1007/s11095-011-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Blaauboer CJ, Timoneda MM, et al. Bioreducible poly(amido amine)s with oligoamine side chains: synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126(2):166–174. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Lin C, Zhong Z, Lok MC, et al. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug Chem. 2007;18(1):138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Qiu X, Ouyang J, Kong J, Zhong W, Xing MM. pH and reduction dual-sensitive copolymeric micelles for intracellular doxorubicin delivery. Biomacromolecules. 2011;12(10):3601–3611. doi: 10.1021/bm200804j. [DOI] [PubMed] [Google Scholar]
- 17.Xu YN, Liu L, Ji X, Huang SW, Zhuo RX. Bioreducible polycationic micelles for in vitro gene delivery. J Control Release. 2011;152(Suppl 1):e177–e179. doi: 10.1016/j.jconrel.2011.08.074. [DOI] [PubMed] [Google Scholar]
- 18.Piest M, Engbersen JF. Effects of charge density and hydrophobicity of poly(amido amine)s for non-viral gene delivery. J Control Release. 2010;148(1):83–90. doi: 10.1016/j.jconrel.2010.07.109. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, Engbersen JFJ. PEGylated bioreducible poly(amido amine)s for non-viral gene delivery. Mater Sci Eng C Mater Biol Appl. 2011;31(7):1330–1337. [Google Scholar]
- 20.Merdan T, Callahan J, Petersen H, et al. PEGylated polyethylenimine-Fab’ antibody fragment conjugates for targeted gene delivery to human ovarian carcinoma cells. Bioconjug Chem. 2003;14(5):989–996. doi: 10.1021/bc0340767. [DOI] [PubMed] [Google Scholar]
- 21.Sun YX, Zeng X, Meng QF, Zhang XZ, Cheng SX, Zhuo RX. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29(32):4356–4365. doi: 10.1016/j.biomaterials.2008.07.045. [DOI] [PubMed] [Google Scholar]
- 22.Wei CM, Jiang T, Li YZ, Zhu HJ, Wang JT. 5-(8-Hydroxyquinolyl) guanidinium dichloride. Acta Crystallogr E Struct Rep Online. 2004;60(3):o442–o443. [Google Scholar]
- 23.Kim TI, Lee M, Kim SW. A guanidinylated bioreducible polymer with high nuclear localization ability for gene delivery systems. Biomaterials. 2010;31(7):1798–1804. doi: 10.1016/j.biomaterials.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowal P, Gurtan AM, Stuckert P, D’Andrea AD, Ellenberger T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J Biol Chem. 2007;282(3):2047–2055. doi: 10.1074/jbc.M608356200. [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Zhao L, Li Y, et al. Silencing of Fanconi anemia complementation group F exhibits potent chemosensitization of mitomycin C activity in breast cancer cells. J Breast Cancer. 2013;16(3):291–299. doi: 10.4048/jbc.2013.16.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zhao L, Sun H, et al. Gene silencing of FANCF potentiates the sensitivity to mitoxantrone through activation of JNK and p38 signal pathways in breast cancer cells. PLoS One. 2012;7(8):e44254. doi: 10.1371/journal.pone.0044254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Hu H, Sun Y, et al. A pH-sensitive gene delivery system based on folic acid-PEG-chitosan: PAMAM-plasmid DNA complexes for cancer cell targeting. Biomaterials. 2013;34(38):10120–10132. doi: 10.1016/j.biomaterials.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Cohen S, Coué G, Beno D, Korenstein R, Engbersen JF. Bioreducible poly(amidoamine)s as carriers for intracellular protein delivery to intestinal cells. Biomaterials. 2012;33(2):614–623. doi: 10.1016/j.biomaterials.2011.09.085. [DOI] [PubMed] [Google Scholar]
- 29.Christensen LV, Chang CW, Yockman JW, et al. Reducible poly(amido ethylenediamine) for hypoxia-inducible VEGF delivery. J Control Release. 2007;118(2):254–261. doi: 10.1016/j.jconrel.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martello F, Piest M, Engbersen JF, Ferruti P. Effects of branched or linear architecture of bioreducible poly(amido amine)s on their in vitro gene delivery properties. J Control Release. 2012;164(3):372–379. doi: 10.1016/j.jconrel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Arote RB, Jiang HL, Kim YK, Cho MH, Choi YJ, Cho CS. Degradable poly(amido amine)s as gene delivery carriers. Expert Opin Drug Deliv. 2011;8(9):1237–1246. doi: 10.1517/17425247.2011.586333. [DOI] [PubMed] [Google Scholar]
- 32.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30(4):658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi JL, Tan JK, Sellers DL, Wei H, Horner PJ, Pun SH. Guanidinylated block copolymers for gene transfer: a comparison with amine-based materials for in vitro and in vivo gene transfer efficiency. Biomaterials. 2015;54:87–96. doi: 10.1016/j.biomaterials.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H nuclear magnetic resonance spectroscopy of agmatine sulfate.
1H nuclear magnetic resonance spectroscopy of N,N′-cystamine bisacrylamide.
1H nuclear magnetic resonance spectroscopy (NMR) of Agm-CBA.
1H nuclear magnetic resonance spectroscopy (NMR) of L-arginine (Arg).
1H nuclear magnetic resonance spectroscopy (NMR) of Arg-CBA.