Abstract
Rational design of organic 2D (O2D) materials has made some progress, but is still in its initial stages. Here, we report on a class of self-assembling small molecules that form nano/microscale supramolecular 2D materials in aqueous media. We show that a judicial combination of four different intermolecular interactions forms the basis for the robust formation of these ultra-thin assemblies. We also show that these assemblies can be programmed to disassemble in response to a specific protein and release its non-covalently bound guest molecules.
Keywords: Organic 2D Materials, Self-assembly, Stimuli Responsive, Controlled Release, Ultra-thin assemblies
Graphical abstract
Two-dimensional (2D) materials have attracted great attention due to their potential in many applications ranging from semiconductors to to biomedicine.[1] In contrast to the more prosperous inorganic 2D materials, study of organic 2D (O2D) materials is still in its launching phase, but is believed to be promising owing to their soft characteristics and thus suitability in a variety of biomedical applications.[2] Among the O2D materials, significant efforts have been directed towards graphene analogs.[1a] Although few successful attempts in making polymeric[3] and supramolecular[4] O2D materials have been reported, examples for the effective formation of free-standing ultra-thin O2D materials are still quite limited. Also, current mainstream, top-down preparation strategies contain additional exfoliation steps, while the traditional bottom-up methods such as self-assembled monolayer formation rely on pre-organization of molecules at a liquid-air interface or on a substrate surface.[5] Thus, the development of a facile molecular design strategy for the preparation of ultra-thin O2D materials addresses an urgent need with both fundamental and practical implications.
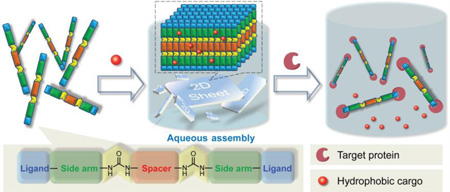
Moreover, though research on making organic-inorganic hybrid “smart” 2D materials[6] have become a hotspot in recent years, studies on pure organic smart 2D materials still quite rare.[7] Thus, designing organic “smart” 2D crystalline materials with stimuli-sensitive characteristics, especially biomolecule-sensitivity, remains as a realm in mist. Herein, we report a simple bottom-up strategy, which uses multiple supramolecular interactions in a concerted fashion, to prepare smart O2D materials by non-interfacial self-assembly of organic small molecules in aqueous media at room temperature (Scheme 1). We show that these autonomously-formed mono/few-layer thick O2D sheets require no extra exfoliation. We also show that these O2D sheets can be programmed to disassemble in in response to a specific protein. Since small hydrophobic guest molecules can be loaded into these sheets, this binding-induced disassembly process can be conveniently visualized.
Scheme 1.
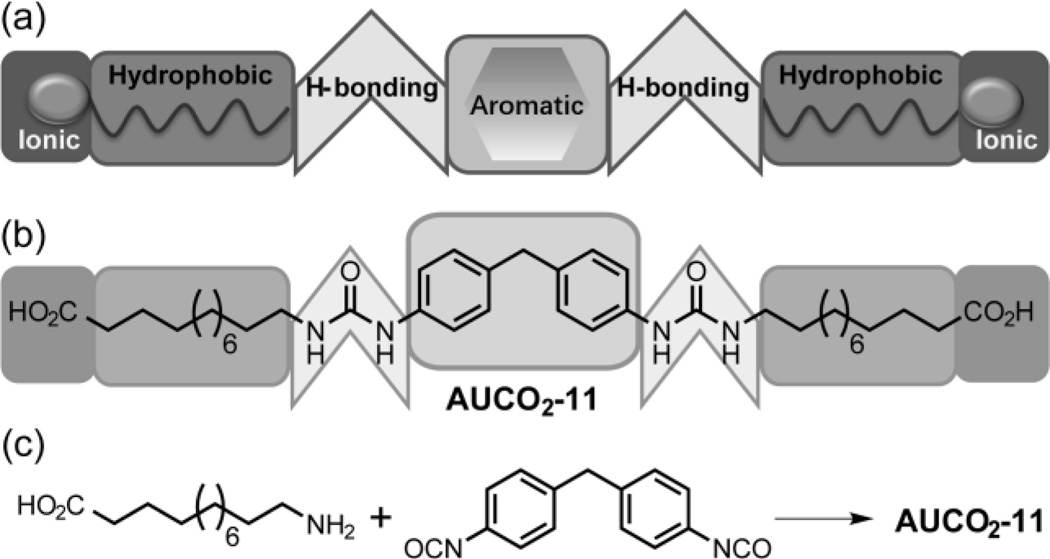
(a) Illustration of the molecular design containing four complementary intermolecular interactions. (b) Example of a moelcule for the formation of the 2D assemblies in the aqueous phase. (c) Example of the simplicity in the synthetic scheme to achieve the targeted materials.
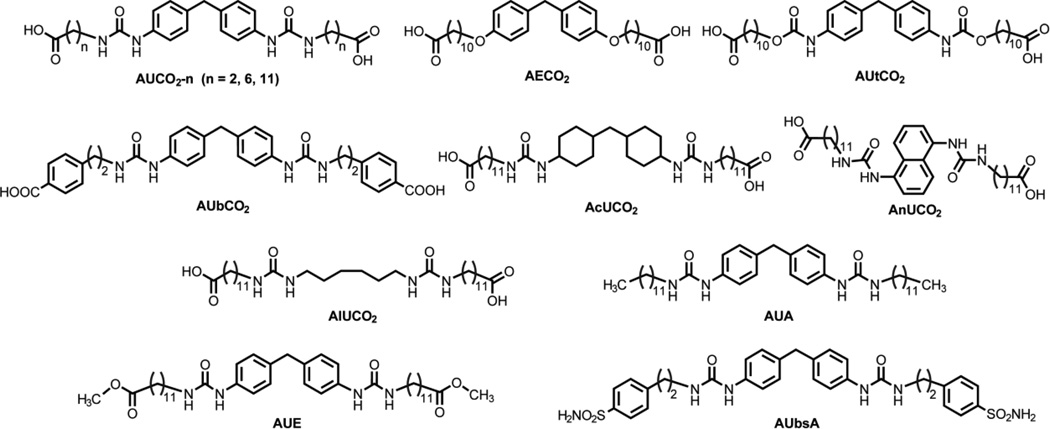
To fundamentally unravel the factors that influence the formation of the O2D structures, a concerted series of twelve molecules has been designed (Scheme 2) and synthesized using a simple approach (Scheme 1c). The basic molecular design strategy here involves (a) the utility of hydrogen bonding interactions that are proximally located with aromatic rings to have correlated hydrogen-bonding and π−π interactions; In fact, molecules with similar structure have been reported as small molecule gelators, owing to the self-assembly ability provided by strong interactions.[8a] (b) an alkyl chain linking the urea-based aromatic units with an ionic head group, where the alkyl chain provides the hydrophobic interactions to reinforce the assembly in the aqueous phase and finally (c) the ionic head groups on either side of the molecules to introduce repulsive forces that presumably mitigate inter-layer interactions and thus avoid the need for exfoliation steps to achieve ultrathin O2D sheets.
Scheme 2.
Structure of the aromatic urea (AU) based amphiphiles and the appropriate controls to test the design principles.
The O2D sheets were prepared by simply injecting a DMSO-based solution of the small molecules into deionized water. The resultant suspensions were allowed to stand still for ~12 hours to let the automated self-assembly finish. These samples were then analyzed by transmission electron microscopy (TEM), High-resolution TEM (HRTEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM). AUCO2-11 was investigated first, since the relatively long alkyl chains on both sides may provide for the targeted self-assembly due to strong hydrophobic effect and van der Waals interactions.
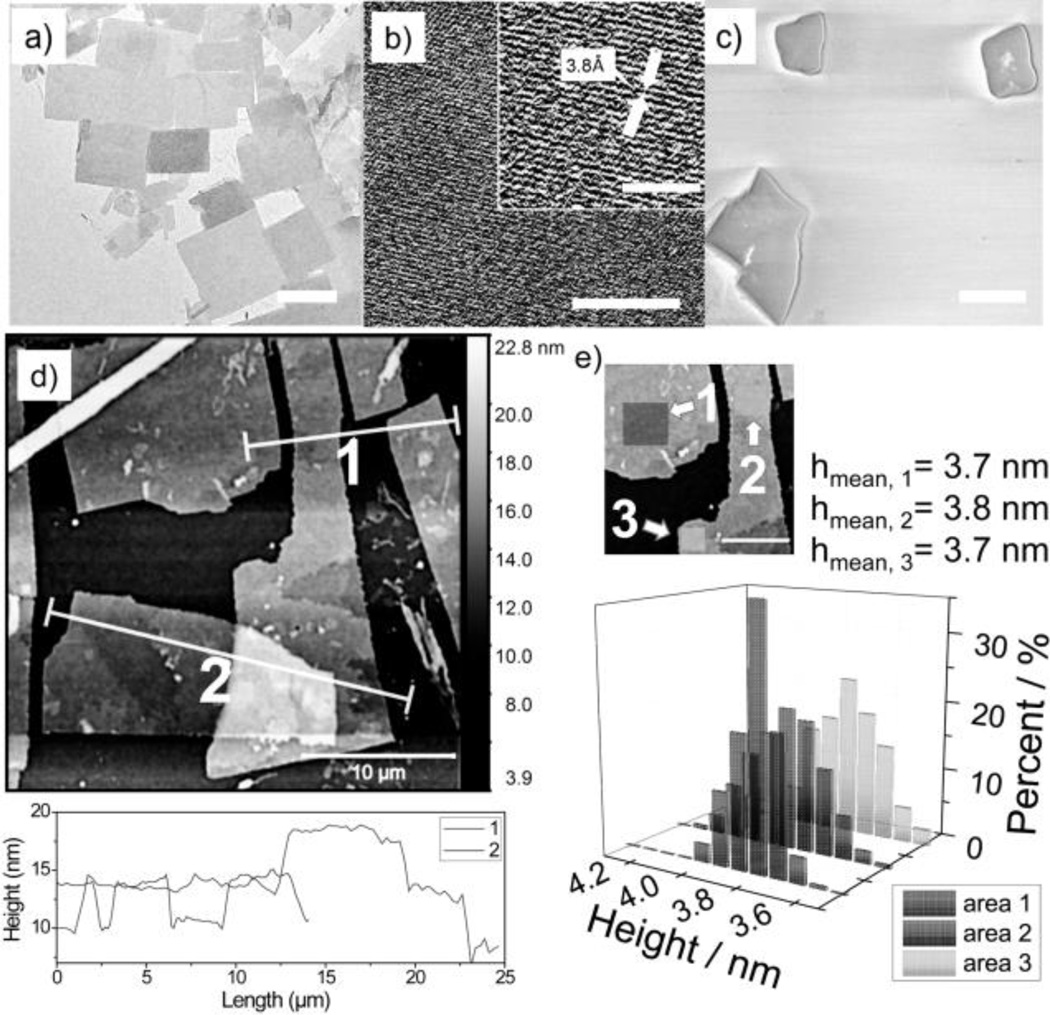
TEM and (HRTEM) images shows that AUCO2-11 form 2D sheets and that the structures are highly ordered (Figure 1a and 1b). The distance of the visible lattice fringes roughly corresponds to ~0.376 nm, in agreement with the interspacing of (101̄0) planes (0.38 nm). The corresponding selected area electron diffraction (SAED) pattern (Figure S1d) displays clear phases, suggesting the high crystal quality within the 2D sheet. SEM images further support the formation of the O2D structure (Figure 1c).
Figure 1.
Microscopy analyses of AUCO2-11 a) TEM image of the 2D sheets in water. b) HRTEM image of the 2D sheets in water with a zoomed in image on upper-right corner. c) SEM images of 2D sheets (Au-coated). d) AFM image with height profiles, e) Histogram of height distribution. Scale bar in (a)=1 µm, (b)=8 nm, (b, zoomed-in image)=4 nm, (c)=1 µm, (d)=10 µm, (e)=5 µm.
The fact that these O2D sheets were stably formed with or without the gold-coating in SEM suggests that the O2D assembly is robust (Figure S1a). AFM studies also suggest that the O2D sheets are relatively smooth with lateral dimension at micrometer scale (Figure 1d). Average thickness of these sheets, based on AFM height profiles, reveals their relatively uniform thickness of (3.7 ± 0.1) nm. A statistical study of the monolayer thickness suggests a mean average height of ~3.7 nm (Figure 1e). XRD investigation also suggests a highly ordered structure with a d-spacing of 3.7 nm (Figure S1b) which matches the results of small angle X-ray scattering (SAXS) of the 2D sheets in powder form (Figure S1e). Note that shape and size control is not a current target for this O2D self-assembly and will be a focus in the future.
As an initial test to identify whether this is a hydrophobicity-induced self-assembly, formation of 2D assemblies in pure DMSO was tested and found that these do not provide any significant assembly (Figure S1c). To further understand the design rules in aqueous media, compared to those in organic solvents,[8] we systematically varied the structure of the molecule.
First, the effect of the hydrophobic linker was studied by comparing AUCO2-2 and AUCO2-6 with AUCO2-11. The self-assembly was performed at three different pH values and the longer linkers favored the 2D structure in all cases (Table 1). This is attributed to the increasing hydrophobic effect and van der Waals interactions, akin to those seen in surfactant self-assembly. Interestingly AUbCO2, containing a benzoic acid as head group in addition to only a two carbon linker, also exhibits a strong propensity to form 2D assemblies. This may be due to the additional reinforcement provided by π−π interactions. The self-assembly variations observed at different pH values is likely due to the optimal balance between the head-group repulsions that could prevent the 2D self-assembly and that are necessary to prevent interlayer interactions to provide the targeted ultrathin structures. In fact, these molecules were found to form highly aggregated 2D structures at pH<4.
Table 1.
Comparison of AUCO2-1 and AUbCO2 in their assembly behavior in aqueous solution
| Name | pH = 4 | pH = 7 | pH = 12 |
|---|---|---|---|
| AUCO2-2 | Nano sheets or defective sheets |
No film or defective film |
No film |
| AUCO2-6 | µm sheets | µm sheets or defective film |
No film or defective film |
| AUCO2-11 | µm sheets | µm sheets | µm ribbon or defective film |
| AUbCO2 | µm sheets | µm sheets | µm ribbon or defective film |
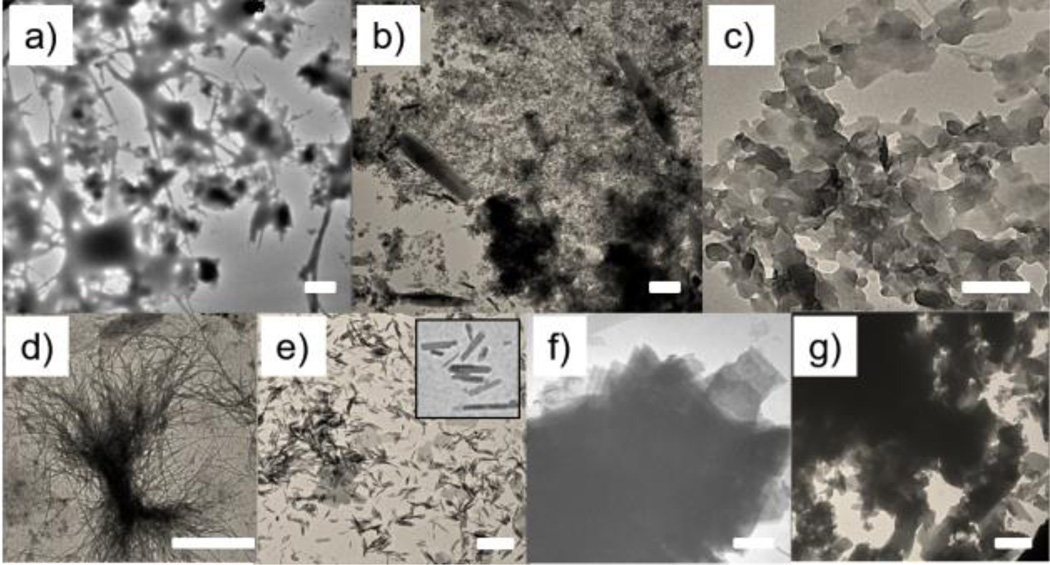
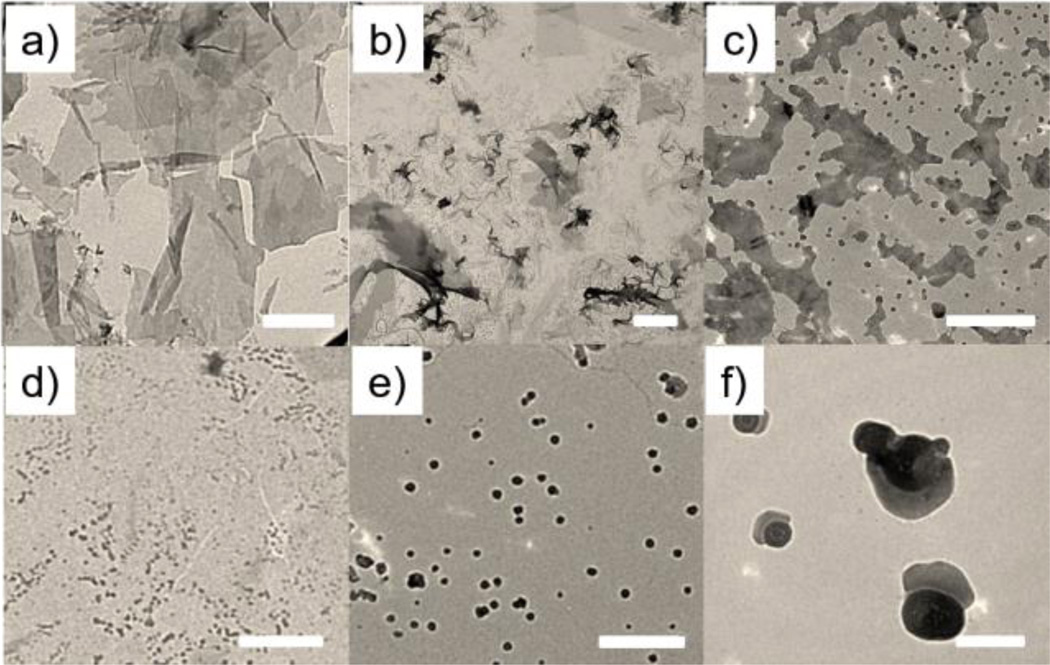
The coordinated use of multiple interactions to drive the formation of the O2D assemblies is the basis for our molecular design. To test this, we examined the importance of the intermolecular hydrogen-bonding due to the urea functionalities. Specifically, we replaced the urea moieties with ether (AECO2) or urethane (AUtCO2) functionalities, where the hydrogen-bonding interaction is completely obviated or partially weakened respectively. Indeed, the assemblies were irregular aggregates in both cases (Figure 2a, 2b). These results suggest that the urea units, with their hallmark hydrogen-bonding self-complementarity, are critical for the observed 2D assemblies.
Figure 2.
SEM images of assemblies of a) AECO2, b) AUtCO2, c) AcUCO2, d) AlUCO2, e) AnUCO2 with a zoomed in image on upper-right corner, f) AU and g) AUE in water. Scale bar (a–g)=1 µm.
Next, we investigated the importance of the rigid aromatic units at the core. AcUCO2, where the two phenyl rings were replaced by cyclohexyl units, afforded assemblies with significant crosslinking (Figure 2c). Though the remnant rigidity in the structure allowed 2D growth, the loss of the assortative π−π interactions seems to have increased the interlayer interactions, presumably due to mismatched hydrogen-bonding interactions, to cause crosslinking among the layered structures. On the other hand, if a completely flexible alkyl spacer is used (as in AlUCO2), needle-like structures were observed indicative of a linear propagation of the assemblies (Figure 2d). These assemblies were also bundled or aggregated, presumably due to mismatched interactions among the linear assemblies. Interestingly, there seems to be a need for a small amount of flexibility between the two urea units. If there’s no flexibility in the spacer, as with AnUCO2, small ribbon-like assemblies were formed (Figure 2e). This is attributed to the possibility that the naphthyl units provide a too-rigid of a linker, where the hydrogen-bonding interaction and the aromatic π−π interaction directions are not complementarily placed.
We hypothesized that an optimal head-group repulsion is needed to balance the 2D assembly formation and the prevention of interlayer interactions. To test this hypothesis, we replaced the carboxylate moiety with an alkyl group or masked it as an ester in the form AUA and AUE respectively. Consistent with our design hypothesis, both molecules exhibited a strong tendency to form 2D structures, but a very limited amount of free-standing mono/few-layer can be observed. Majority of the 2D sheets were stacked together and formed bulk aggregates (Figures 2f and 2g). These results suggest that the hydrophilic head groups are necessary to avoid the extra exfoliation steps to achieve ultra-thin 2D assemblies.
As a preliminary demonstration of the utility of these O2D materials, we tested these molecules for binding-induced disassembly in response to specific proteins. Binding-induced disassembly has been previously demonstrated with micelle-like amphiphilic aggregates.[9] We conceived that such a demonstration with O2D assemblies will greatly expand the repertoires of both the 2D scaffold and the protein-triggered disassembly process. We employed a benzenesulfonamide moiety, a well-known ligand for carbonic anhydrases,[10] as the surface exposed functionality in the self-assembling small molecule (AUbsA). We first ascertained that this molecule indeed forms a 2D assembly based on TEM, SEM, AFM and XRD (Figure 3a and Figure S2a). The O2D sheets were found to be ~2.4 nm thick, close to the single molecule dimension.
Figure 3.
TEM image of AUbsA 2D sheet a) in water, and after incubation of the assembly with the target protein, BCA, for b) 2 hours, c) 4 hours. d) 2 days. TEM images of e) 7 days after addition of control protein, f) a zoom-in image of Figure 1e. Scale bar (a–e)=1 µm, (f)=0.1 µm.
When bovine carbonic anhydrase (BCA) was added to an aqueous suspension containing the 2D sheets (CAUbsA=1.5 µM), the morphology of the 2D assembly seems to be compromised over time (Figure 3b–d). TEM studies confirmed that majority of these 2D assemblies were completely dissolved at longer time scales. However, the morphological change was also observed for proteins that are non-complementary to the sulfonamides, viz. avidin and hyaluronidase. Interestingly, this non-specific interaction produced a consistently spherical nanoparticle morphology that persisted even after two weeks (Figure 3e–f).
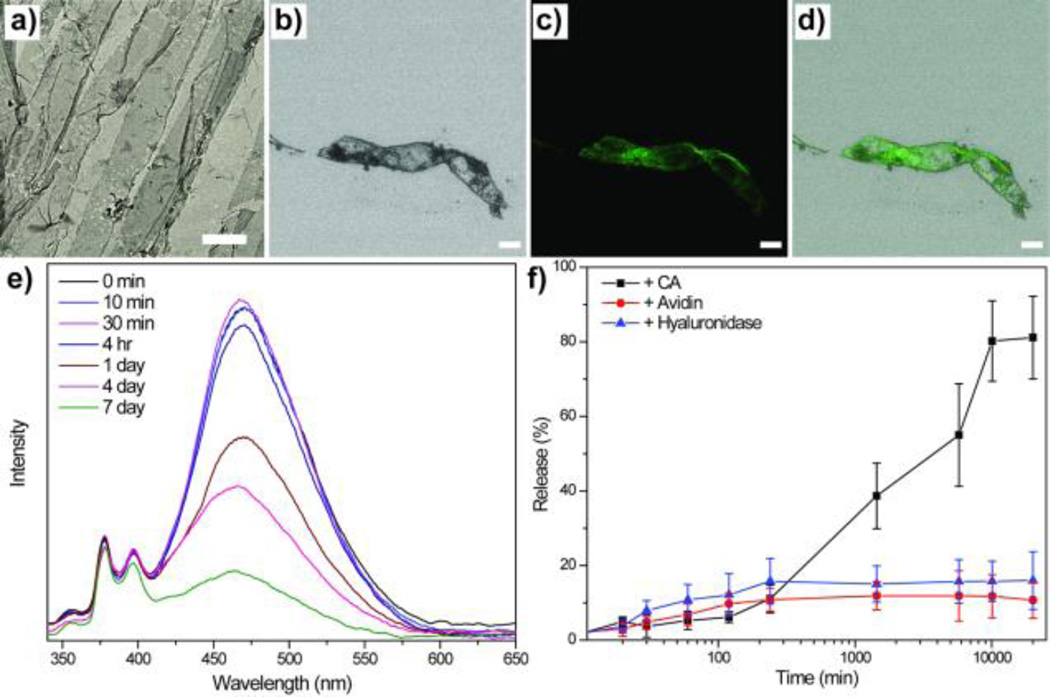
Considering the difference in the persistence of the protein-induced morphologies, we tested the possibility of releasing hydrophobic guest molecules within these assemblies in the presence of specific proteins. Accordingly, we loaded pyrene within 2D sheets, where the overall 2D structure was maintained after guest loading (Figure 4a). Confocal microscopy also shows that the fluorescent dyes were successfully loaded to the sheets (Figure 4b–4d). When the suspension of 2D sheet was exposed to BCA, the possible release of pyrene was monitored by absorption spectroscopy. Since the hydrophobic guest is insoluble in water, it would simply precipitate out of solution. Indeed, the absorbance of pyrene decreased over time in the presence of BCA (~80% release over a few days, Figure 4e and 4f). No such release was observed in the absence of BCA or in the presence of non-complementary proteins. These observations suggest that a sustained, specific protein-responsive release of guest molecules is indeed possible.
Figure 4.
TEM images of AUbsA a) after loading of the pyrene. Confocal images of the 2D sheet, b) bright field, c) pyrene, d) merged image of (b) and (c). e) Pyrene release from 2D sheet in the presence of BCA. f) Plot of % release with time and comparison with controls. Scale bar (a)=5 µm, (b–d)=10 µm.
In conclusion, we have designed and synthesized a new series of self-assembling molecules that readily afford molecular-thin O2D materials in aqueous media. We show here that aromatic π−π interactions, hydrogen-bonding interactions, hydrophobic interactions, and electrostatic repulsions all play a key role in the formation of these assemblies. Finally, to the best of our knowledge, there has been no report on ‘smart’ O2D materials before. Here, by employing a protein-specific ligand as the hydrophilic head group, we have shown that these 2D sheets can undergo a binding–induced disassembly in response to a specific protein. This feature, the simplicity of the synthesis/preparation method, and our understanding of the factors that underlie the assembly formation provide a foundation for pursuing O2D assemblies for future applications in smart materials-based systems.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM-065255) and the MURI program of the Army Research Office (W911NF-15-1-0568) for support. We also thank Prof. Dhandapani Venkataraman for his kind help in analyzing the SAED data.
Contributor Information
Dr. Wei Bai, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA 01003 (USA)
Ziwen Jiang, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA 01003 (USA).
Dr. Alexander E. Ribbe, Department of Polymer Science and Engineering, University of Massachusetts Amherst, Amherst, MA 01003 (USA)
Prof. S. Thayumanavan, Email: thai@chem.umass.edu, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA 01003 (USA).
References
- 1.a) Zhuang X, Mai Y, Wu D, Zhang F, Feng X. Adv. Mater. 2015;27:403–427. doi: 10.1002/adma.201401857. [DOI] [PubMed] [Google Scholar]; b) Chimene D, Alge DL, Gaharwar A. Adv. Mater. 2015;27:7261–7284. doi: 10.1002/adma.201502422. [DOI] [PubMed] [Google Scholar]; c) Butler S, Hollen S, Cao L, et al. ACS Nano. 2013;7:2898–2926. doi: 10.1021/nn400280c. [DOI] [PubMed] [Google Scholar]; d) Miró P, Audiffreda M, Heine T. Chem. Soc. Rev. 2014;43:6537–6554. doi: 10.1039/c4cs00102h. [DOI] [PubMed] [Google Scholar]; e) Guptaa A, Sakthivela T, Seal S. Progess in Materials Science. 2015;73:44–126. [Google Scholar]
- 2.a) Boott CE, Nazemi A, Manners I. Angew. Chem. Int. Ed. 2015;54:13876–13894. doi: 10.1002/anie.201502009. [DOI] [PubMed] [Google Scholar]; b) Xiang Z, Cao D. Polym. Chem. 2015;6:1896–1911. [Google Scholar]; c) Wang Z, Zhu W, Qiu Y, Yi X, Bussche A, Kane A, Gao H, Koski K, Hurt R. Chem. Soc. Rev. 2016 doi: 10.1039/c5cs00914f. Advance Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Keller A. Phil. Mag. 1957;2:1171–1175. [Google Scholar]; b) Nam K, Shelby S, Choi P, Marciel A, Chen R, Tan L, Chu T, Mesch R, Lee B, Connolly M, Kisielowski C, Zuckermann R. Nat. Mater. 2010;9:454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]; c) Zhou H, Liu J, Du S, Zhang L, Li G, Zhang Y, Tang B, Gao H. J Am. Chem. Soc. 2014;136:5567–5570. doi: 10.1021/ja501308s. [DOI] [PubMed] [Google Scholar]; d) Kory M, Wörle M, Weber T, Payamyar P, Poll S, Dshemuchadse J, Trapp N, Schlüter AD. Nat. Chem. 2014;6:779–784. doi: 10.1038/nchem.2007. [DOI] [PubMed] [Google Scholar]; e) Pfeffermann M, Dong R, Graf R, Zajaczkowski W, Gorelik T, Pisula W, Narita A, Müllen K, Feng X. J Am. Chem. Soc. 2015;137:14525–14532. doi: 10.1021/jacs.5b09638. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Zhang X, Hsu C, Ren X, Gu Y, Song B, Sun H, Yang S, Chen E, Tu Y, Li X, Yang X, Li Y, Zhu X. Angew. Chem. Int. Ed. 2015;54:114–117. doi: 10.1002/anie.201408438. [DOI] [PubMed] [Google Scholar]
- 4.a) Wu D, Liu R, Pisula W, Feng X, Müllen K. Angew. Chem. Int. Ed. 2011;50:2791–2794. doi: 10.1002/anie.201004245. [DOI] [PubMed] [Google Scholar]; b) Bauer T, Zheng Z, Renn A, Enning R, Stemmer A, Sakamoto J, Schlüter AD. Angew. Chem. Int. Ed. 2011;50:7879–7884. doi: 10.1002/anie.201100669. [DOI] [PubMed] [Google Scholar]; c) Zhang K, Tian J, Hanifi D, Zhang Y, Sue AC, Zhou T, Zhang L, Zhao X, Liu Y, Li Z. J Am. Chem. Soc. 2013;135:17913–17918. doi: 10.1021/ja4086935. [DOI] [PubMed] [Google Scholar]; d) Payamyar P, Kaja K, Ruiz-Vargas C, et al. Adv. Mater. 2014;26:2052–2058. doi: 10.1002/adma.201304705. [DOI] [PubMed] [Google Scholar]; e) Wu G, Verwilst P, Liu K, Smet M, Faulc C, Zhang X. Chem. Sci. 2013;4:4486–4493. [Google Scholar]; f) Song B, Wang Z, Chen S, Zhang X, Fu Y, Smet M, Dehaen W. Angew. Chem. Int. Ed. 2005;44:4731–4735. doi: 10.1002/anie.200500980. [DOI] [PubMed] [Google Scholar]
- 5.a) Kim J, Kwon S, Cho D, et al. Nat. Comm. 2015;6:8294. doi: 10.1038/ncomms9294. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang N, Xu Q, Xu S, Qi Y, Chen M, Li H, Han B. Sci. Rep. 2015;5:16764. doi: 10.1038/srep16764. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shen J, He Y, Wu J, Gao C, Keyshar K, Zhang X, Yang Y, Ye M, Vajtai R, Lou J, Ajayan P. Nano Lett. 2015;15:5449–5454. doi: 10.1021/acs.nanolett.5b01842. [DOI] [PubMed] [Google Scholar]; d) Mitra S, Kandambeth S, Biswal B, et al. J Am. Chem. Soc. 2016;138:2823–2828. doi: 10.1021/jacs.5b13533. [DOI] [PubMed] [Google Scholar]
- 6.a) Yu B, Jiang X, Yin J. Chem. Commun. 2013;49:603–605. doi: 10.1039/c2cc37645h. [DOI] [PubMed] [Google Scholar]; b) Kim J, Bohra M, Singh V, Cassidy C, Sowwan M. ACS Appl. Mater. Interfaces. 2014;6:13339–13343. doi: 10.1021/am5041708. [DOI] [PubMed] [Google Scholar]; c) Mo R, Jiang T, Sun W, Gu Z. Biomaterials. 2015;50:67–74. doi: 10.1016/j.biomaterials.2015.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Cao Z, Wang G, Chen Y, Liang F, Yang Z. Macromolecules. 2015;48:7256–7261. [Google Scholar]; e) Hao Y, Wang L, Zhang B. Nanotechnology. 2016;27:025101. doi: 10.1088/0957-4484/27/2/025101. [DOI] [PubMed] [Google Scholar]
- 7.a) Shin S, Lim S, Kim Y. J Am. Chem. Soc. 2013;135:2156–2159. doi: 10.1021/ja400160j. [DOI] [PubMed] [Google Scholar]; b) Zheng Y, Zhou H, Liu D. Angew. Chem. Int. Ed. 2013;52:4845–4848. doi: 10.1002/anie.201210090. [DOI] [PubMed] [Google Scholar]; c) Betthausen E, Dulle M, Hanske C. Macromolecules. 2014;47:6289–6301. [Google Scholar]; d) Lay C, Lee M, Lee H, Phang I, Ling X. ACS Nano. 2015;9:9708–9717. doi: 10.1021/acsnano.5b04300. [DOI] [PubMed] [Google Scholar]; e) Vybornyi M, Rudnev A, Häner R. Chem. Mater. 2015;27:1426–1431. [Google Scholar]
- 8.a) Zhang C, Wang J, Wang J, Li M, Yang X, Xu H. Chem. Eur. J. 2012;18:14954–14956. doi: 10.1002/chem.201202721. [DOI] [PubMed] [Google Scholar]; b) Davis R, Berger R, Zentel R. Adv. Mater. 2007;19:3878–3881. [Google Scholar]; c) Kim J, Davis R, Zentel R. J Colloid Interf. Sci. 2011;359:428–435. doi: 10.1016/j.jcis.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 9.a) Guo J, Zhuang J, Wang F, Raghupathi K, Thayumanavan S. J Am. Chem. Soc. 2014;136:2220–2223. doi: 10.1021/ja4108676. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Torres D, Garzoni M, Subrahmanyam A, Pavan G, Thayumanavan S. J Am. Chem. Soc. 2014;136:5385–5399. doi: 10.1021/ja500634u. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Molla M, Prasad P, Thayumanavan S. J Am. Chem. Soc. 2015;137:7286–7289. doi: 10.1021/jacs.5b04285. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang H, Zhuang J, Raghupathi K, Thayumanavan S. Chem. Commun. 2015;51:17265–17268. doi: 10.1039/c5cc07408h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Raghupathi K, Guo J, Munkhbat O, Rangadurai P, Thayumanavan S. Acc. Chem. Res. 2014;47:2200–2211. doi: 10.1021/ar500143u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem. Rev. 2008;108:946. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Alterio V, Di Fiore A, D’Ambrosio K, Supuran CT, De Simone G. Chem. Rev. 2012;112:4421. doi: 10.1021/cr200176r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.