Abstract
Long-lasting forms of synaptic plasticity and memory are dependent on new protein synthesis. Recent advances obtained from genetic, physiological, pharmacological, and biochemical studies provide strong evidence that translational control plays a key role in regulating long-term changes in neural circuits and thus long-term modifications in behavior. Translational control is important for regulating both general protein synthesis and synthesis of specific proteins in response to neuronal activity. In this review, we summarize and discuss recent progress in the field and highlight the prospects for better understanding of long-lasting changes in synaptic strength, learning, and memory and implications for neurological diseases.
Memories are usually divided into short-term memory, lasting 1–3 hr, and long-term memory, lasting for years or even a lifetime (Dudai, 2004; Kandel, 2001; McGaugh, 2000). In general, long-lasting memories require new protein synthesis, but there is no absolute time frame that differentiates protein-synthesis-dependent and -independent memories. For example, associative olfactory conditioning in Drosophila, known as anesthesia-resistant memory (ARM), can last over 24 hr in the absence of protein synthesis (Tully et al., 1994), while increases in postsynaptic responsiveness in Aplysia motor neurons depend on rapid protein synthesis after only 10 min (Villareal et al., 2007). The formation of new memories requires not just translation per se, but is dependent on regulation of specific mRNAs. Thus, a more thorough understanding of how these regulatory processes function in neurons should help to elucidate many important basic aspects of neuronal function. In this review, we focus on the regulation of translation during synaptic plasticity and memory formation, but it is noteworthy that translational control is important for additional neuronal functions, such as growth, axonal guidance, and other specialized neuronal functions.
Activity-dependent changes in the strength and/or number of synaptic connections are believed to underlie long-term changes in neural circuits and thus modulate behavior (Bliss and Collingridge, 1993; Malenka and Nicoll, 1999). To study memory at the “cellular” level, neuroscientists use very well defined models that measure changes in synaptic strength, termed long-term potentiation (LTP) and long-term depression (LTD) in vertebrates and long-term facilitation (LTF) in invertebrates (Kandel, 2001; Malenka and Bear, 2004). The idea of using LTP, which has received most of the attention as a cellular model for learning and memory, is supported by the evidence that LTP and memory share similar molecular and cellular mechanisms (Lynch, 2004; Neves et al., 2008). For instance, like memory, LTP occurs in two temporally distinct phases: early LTP (E-LTP) depends on modification of preexisting proteins, whereas late LTP (L-LTP) requires transcription and synthesis of new proteins. E-LTP is typically induced by a single train of high-frequency (tetanic) stimulation of an afferent pathway and lasts only 1–2 hr. In contrast, L-LTP is generally induced by several repetitions of such stimulations (typically four tetanic trains separated by 5–10 min) and persists for many hours (Costa-Mattioli and Sonenberg, 2008; Kandel, 2001; Kelleher et al., 2004b; Klann et al., 2004). In invertebrates, facilitation—an enhancement of synaptic strength induced by serotonin at sensory-motor synapses that is thought to underlie behavioral sensitization—also exhibits similar temporal phases, with short-term facilitation (STF) depending on modification of preexisting proteins and LTF being dependent on transcription and synthesis of new proteins (Kandel, 2001).
Mechanisms of Translation
The control of mRNA translation in eukaryotes is an important and frequent means to regulate gene expression. Initiation in eukaryotes is the rate-limiting step of translation under most circumstances and therefore serves as a major target for translational control. In eukaryotes, translation initiation is an exquisitely complex process catalyzed by at least 12 initiation factors (eIFs) and can be subdivided into three key events: (1) formation of the 43S ribosomal preinitiation complex, (2) binding of the mRNA to the 43S ribosomal complex, and (3) 80S ribosomal complex formation.
The binding of eIF2, which comprises three subunits (α, β, and γ), to GTP and Met-tRNAiMet to form a ternary complex is an early step in the initiation process. The ternary complex then associates with the small 40S ribosomal subunit, which is associated with other eIFs (see below) to form a 43S ribosomal preinitiation complex. Ribosome recruitment to the mRNA occurs by either (1) a cap-dependent process, in which ribosome binding is facilitated by the 5′-cap structure (m7GpppX, where X is any nucleotide and m is a methyl group) present on all nuclear-transcribed mRNAs (Shatkin, 1985), or (2) a less frequently used cap-independent mechanism that involves recruitment of the ribosome to an internal sequence in the mRNA 5′ untranslated region (UTR), termed internal ribosome entry site (IRES) (Doudna and Sarnow, 2007; Elroy-Stein and Merrick, 2007). A critical factor involved in cap-dependent translation is eIF4F, which consists of three subunits: (1) eIF4E, the cap-binding protein; (2) eIF4A, a bidirectional ATP-dependent RNA helicase that is thought to unwind the secondary structure of the 5′ UTR of the mRNA; and (3) eIF4GI or eIF4GII, two large scaffolding proteins that bridge the mRNA to the 43S preinitiation complex through interactions with eIF3 (which is bound to the 40S ribosomal subunit) (Gingras et al., 1999b). Once bound to the 5′ end of the mRNA, the 43S ribosomal complex is thought to traverse the 5′ UTR in a 5′-3′ direction, until it encounters the initiation codon (AUG or a cognate thereof). Initiation codon selection is effected by several factors, including eIF1, eIF2, and eIF3 (Hinnebusch et al., 2007; Pestova et al., 2007). Because these eIFs bind to the surfaces of 40S, which are critical for 40S-60S intersubunit interactions, they preclude 60S ribosomal joining. To release eIF2-bound GTP from the ribosome, GTP is hydrolyzed by eIF5, a GTPase-activating protein (GAP). eIF5 together with a distinct GTPase eIF5B are thought to displace eIF2 and other factors, thus enabling 60S ribosome subunit joining (Pestova et al., 2007). After initiation is completed, elongation factors are recruited to carry out the elongation of the polypeptide chain. eEF1A is a GTPase required for the entry of the tRNA onto the ribosome, and eEF1B (consisting of three subunits (α, β, and γ) is the guanine nucleotide exchange factor (GEF) for eEF1A. eEF2 catalyzes the translocation of ribosome on the mRNA after peptide bond formation. Upon recognition of a stop codon, termination factors promote the release of the polypeptide chain from the mRNA and ribosome.
Regulation of Translation by eIF2α Phosphorylation
eIF2 associates with the small 40S ribosomal subunit in its GTP-bound form. GTP is hydrolyzed by eIF5 upon 60S joining to release eIF2 from the ribosome in a GDP-bound state. To reconstitute a functional eIF2•GTP•Met-tRNAiMet ternary complex for a new round of translation initiation, the GEF, eIF2B, catalyzes the exchange of GDP for GTP on eIF2 (Hinnebusch et al., 2007).
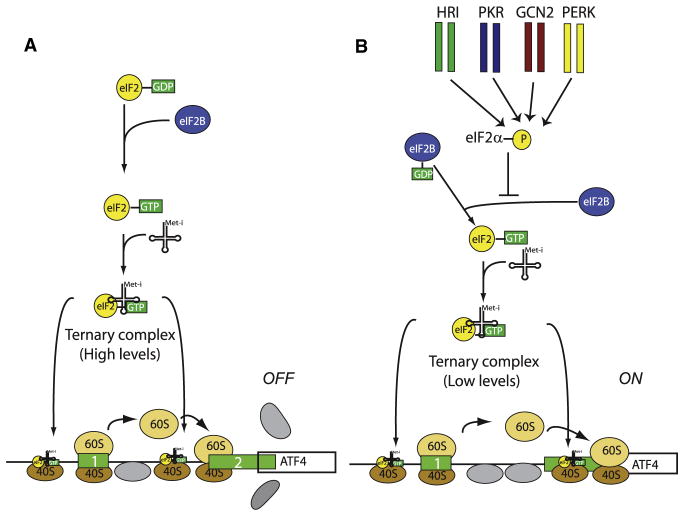
Phosphorylation of Ser51 in the α subunit of eIF2 converts eIF2α into a dominant inhibitor of the GEF activity of eIF2B and therefore causes a decrease in general translation initiation (Dever, 2002; Hinnebusch et al., 2007). Because most cells contain more eIF2 than eIF2B, phosphorylation of only a fraction of eIF2α is sufficient to inhibit eIF2B’s function and thus decrease translation rates. In higher eukaryotes, the phosphorylation of eIF2α is controlled by four protein kinases, for which eIF2α is most likely their only known substrate (Figure 1) (Dever et al., 2007). The kinases are the double-stranded (ds) RNA-activated protein kinase (PKR), the hemin-regulated inhibitor kinase (HRI), the pancreatic eIF2α or PKR-endoplasmic reticulum (ER)-related kinase (PEK/PERK), and the general control nonderepressible-2 (GCN2) kinase, each of which is activated by distinct stresses that decrease protein synthesis by an appropriate response (PKR by double-stranded RNA [dsRNA], HRI by heme deficiency, PEK/PERK by misfolded proteins in the ER, and GCN2 by amino acid deprivation and UV irradiation) (Figure 1 and Table 1). GCN2 is the only eIF2α kinase that is evolutionarily conserved from yeast to mammals (Costa-Mattioli et al., 2007b; Hinnebusch, 2000) and is enriched in the brain of flies (Santoyo et al., 1997) and mammals (Berlanga et al., 1999; Sood et al., 2000), especially in the hippocampus (Costa-Mattioli et al., 2005).
Figure 1. Molecular Model for ATF4 mRNA Translational Control.
The ATF4 mRNA (straight line), harbors uORFs 1 and 2 (green boxes). Upon translation of uORF1, the 80S ribosome dissociates and the 40S ribosomal subunit remains attached to the mRNA and resumes scanning in a 5′ to 3′ direction.
(A) Under normal conditions, there is a sufficient supply of ternary complex (eIF2-Met-tRNAiMet-GTP), which rapidly associate with the 40S ribosomal subunit. This enables the 40S subunit to reinitiate translation at the AUG of uORF2. The 80S ribosome dissociates after terminating at uORF2. As the 40S ribosomal subunit cannot scan backward, the AUG codon of ATF4 cannot be translated.
(B) Under conditions in which eIF2α is phosphorylated, the amount of ternary complex is reduced. Thus, a significant portion of scanning 40S ribosomal subunits scan pass the AUG of uORF2 and initiate at the AUG of ATF4.
Table 1.
Translation Initiation/Elongation Factors Regulated via Phosphorylation
| Translation Initiation/Elongation Factor | Kinase | Effect on Translation |
|---|---|---|
| eIF2α | GCN2, PKR, HRI, PERK | decrease |
| 4E-BP1 (the best-characterized 4E-BP) | mTORC1, Erk1/2 | increase |
| eIF4E | MNK1/2 | increase? (see text) |
| eEF2 | eEF2Kinase | decrease |
| eIF4B | S6K1/2, Rsk | increase |
| eIF4G | mTORC1 | increase? |
Although phosphorylation of eIF2α impairs general translation, it also paradoxically results in translational upregulation of a subset of mRNAs that contain upstream open reading frames (uORFs) (Hinnebusch et al., 2007; Ron and Harding, 2007) (Figure 1). The molecular mechanism underlying this selective translational upregulation was extensively studied and elucidated for the general amino acid control response in the yeast S. cerevisiae (Hinnebusch et al., 2007) and is conserved throughout evolution. In yeast, when amino acids are scarce, translation of the transcriptional activator GCN4 mRNA, which contains four uORFs, is stimulated by eIF2α phosphorylation (Hinnebusch et al., 2007). In mammalian cells, the translation of the GCN4 metazoan counterpart, the transcriptional modulator ATF4, which contains two uORFs, is enhanced in response to eIF2α phosphorylation (Harding et al., 2000; Vattem and Wek, 2004) (Figure 1). The first ATF4 mRNA uORF encodes a 3 amino acid peptide. The second uORF which encodes a 59 amino acid peptide, overlaps with the first 83 nt of the ATF4-coding region. When levels of the ternary complex are high, scanning ribosomes translate the first ORF, and a large fraction of them reinitiate on the second ORF, thus terminating downstream of the ATF4 major initiation codon. Because the ribosome cannot scan backward, translation from the major initiation codon is low. In contrast, when eIF2α is phosphorylated, the levels of ternary complex are reduced, thus a significant fraction of the scanning 40S subunits cannot reinitiate on the second ORF, thus bypassing it and continue scanning to allow initiation at the major ATF4 start codon (Figure 1). Therefore, as in yeast, eIF2α phosphorylation regulates both general and gene-specific translation in mammalian cells (Figure 1).
Importantly, ATF4 and its homologs are repressors of cAMP response element binding protein (CREB)-mediated gene expression, which is widely considered to be required for long-lasting changes in synaptic plasticity and memory in diverse phyla (Bartsch et al., 1995; Chen et al., 2003). Thus, eIF2α phosphorylation regulates two fundamental processes that are crucial for the storage of new memories: new protein synthesis and CREB-mediated gene expression via translational control of ATF4 mRNA.
eIF2α Phosphorylation Is Critical for the Induction of Long-Lasting Changes in Synaptic Strength and Memory
Stimuli that generate sustained, gene expression-dependent increases in synaptic strength, such as tetanic stimulation, treatment with brain-derived neurotrophic factor (BDNF), or the cAMP activator forskolin, decrease the phosphorylation of eIF2α (Costa-Mattioli et al., 2005; Takei et al., 2001). Moreover, eIF2α phosphorylation is also reduced in rats trained in a Pavlovian (associative) fear conditioning task that induces gene expression-dependent long-term memory (Costa-Mattioli et al., 2007a). Strikingly, genetic reduction of eIF2α phosphorylation in hippocampal slices from mice, either lacking GCN2 (the major eIF2α kinase in the mammalian brain) or heterozygous for an eIF2α mutation (that converts the phosphorylation site serine 51 to alanine), reduced the threshold for the induction of both L-LTP and learning in several behavioral tasks, such as the Morris water maze, associative fear conditioning, and conditioned taste aversion (Costa-Mattioli et al., 2005, 2007a). In agreement with these data, preventing eIF2α dephosphorylation with Sal003, a pharmacological inhibitor of eIF2α phoshphatases, blocks both L-LTP and long-term memory formation. Furthermore, the impairment of L-LTP by Sal003 is mediated by ATF4, as late LTP induced in hippocampal slices from ATF4 knockout mice is resistant to Sal003 (Costa-Mattioli et al., 2007a).
One question that remains to be addressed is the molecular mechanisms by which GCN2 is modulated in response to neuronal activity. It is possible that activity-dependent changes in synaptic strength increase amino acid import into the neurons and thereby inactivate GCN2 through the conventional mechanism, which depends on the extent of tRNA charging with amino acids (Hinnebusch et al., 2007). Indeed, in the anterior piriform cortex, like in yeast, GCN2-mediated eIF2α phosphorylation is regulated by the levels of uncharged tRNAs (Hao et al., 2005; Maurin et al., 2005). Alternatively, it is possible that L-LTP inducing protocols boost the levels of IMPACT, a GCN2 inhibitor, which is enriched in the mammalian brain (Pereira et al., 2005).
The data summarized above strongly support the idea that eIF2α phosphorylation plays a key role in the expression of genes required for long-lasting synaptic plasticity and memory consolidation. Thus, translational regulation is a key regulator of the transcriptional activation program that is required to form long-term memories.
Regulation of mTOR and Its Downstream Effectors: 4E-BP, S6K1, S6K2, and TOP mRNAs
Although eIF4F complex assembly is regulated via several modes, the best-characterized mechanism is through interaction with members of a family of small molecular weight proteins, the eIF4E-binding proteins (4E-BPs). 4E-BP1 and 4E-BP2, which share 56% identity, were first identified as eIF4E-binding proteins by Far-Western interactions (Pause et al., 1994). The third member of the family, 4E-BP3, was discovered later (Poulin et al., 1998). The 4E-BPs specifically inhibit cap-dependent translation initiation in vitro and in vivo by preventing the assembly of the eIF4F complex and, consequently, ribosome recruitment to the mRNA (Haghighat et al., 1995; Pause et al., 1994). 4E-BPs and eIF4G share a canonical eIF4E-binding site (YXXXXLΦ, where X is any amino acid and Φ is a hydrophobic amino acid), through which they compete for binding to the convex dorsal surface of eIF4E (Mader et al., 1995; Marcotrigiano et al., 1999). 4E-BP1 (the best-characterized 4E-BP) binding to eIF4E is regulated by phosphorylation on serine and threonine residues (Lin et al., 1994; Pause et al., 1994). Hypophosphorylated 4E-BPs bind with high affinity to eIF4E and preferentially inhibit translation of a subset of mRNAs, many of which contain G + C rich and highly structured 5′ UTRs (Gingras et al., 1999b).
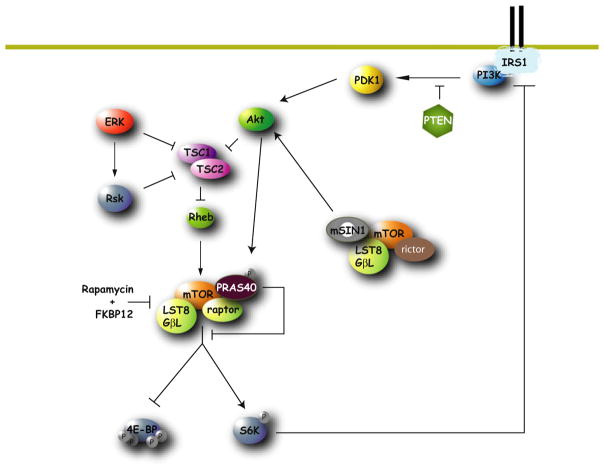
In contrast, hyperphosphorylated 4E-BPs bind with low affinity to eIF4E and thus can no longer inhibit translation (Beretta et al., 1996; Pause et al., 1994). Many kinds of extracellular stimuli, such as serum, growth factors, or hormones (e.g., insulin), promote the phosphorylation of 4E-BPs (Gingras et al., 1999b). The major protein kinase that phosphorylates 4E-BPs is the mammalian target of rapamycin (mTOR) (Figure 2) (Hay and Sonenberg, 2004). mTOR is a critical downstream target of the phosphatidylinositol-3 kinase (PI3K) signaling pathway. PI3K phosphorylates the membrane-bound phospholipid, phosphatidylinositol-4,5-bisphosphate (PIP2), converting it to phosphatidylinositol-3,4,5-trisphosphate (PIP3), which then recruits Akt to the membrane where it is phosphorylated and activated by PI3K-dependent kinase 1 (PDK1) and mTORC2 (see below) (Sabatini, 2006) (Figure 2; see below). Akt activates mTOR via phosphorylation and inhibition of the TSC2 subunit of the tuberous sclerosis complex (TSC). TSC is a heterodimer consisting of TSC1 (hamartin) and TSC2 (tuberin). TSC2 is a GTPase-activating protein (GAP), which hydrolyzes the GTP bound to the small G protein Ras-homolog enriched in brain (Rheb). When TSC2 is phosphorylated, its GAP activity is decreased, resulting in Rheb and mTOR activation (Hay and Sonenberg, 2004) (Figure 2). In Drosophila, AKT may also activate TOR by additional pathways, as nonphosphorylatable TSC can rescue the growth phenotype seen in TSC mutants (Dong and Pan, 2004).
Figure 2. mTOR Complex 1 and mTOR Complex 2 Signaling Network.
The mTOR kinase is a component of two distinct multiprotein complexes called mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). In addition to mTOR, mTORC1 contains RAPTOR, mLST8, and PRAS40. mTORC1’s activity is modulated by the PI3K/Akt signaling pathway. Akt phosphorylates and inhibits TSC2, which leads to activation of Rheb, which in turns activates mTORC1. Akt also phosphorylates PRAS40. mTORC1 activation releases mTORC1 from PRAS40 repression and leads to phosphorylation of 4E-BPs and S6K. mTORC2 contains mLST8, RICTOR, mSIN1. The best-characterized substrate of mTORC2 is the AKT kinase.
In addition to the dominant PI3K signaling pathway described above, the extracellular signal-regulated kinase (ERK) can activate mTOR under certain conditions (Figure 2). ERK phosphorylates and activates p90S6K (RSK), which in turn can phosphorylate and activate PDK1 (Frodin et al., 2000). ERK can also impact mTOR function further downstream by phosphosphorylating TSC2 either directly or indirectly via RSK (Ma et al., 2005; Roux and Blenis, 2004). The ERK signaling pathway to mTOR appears to be particularly important in the hippocampus, as activation of downstream mTORC effectors by different stimulation paradigms (forskolin, high-frequency stimulation, mGluR agonists, etc.) are partially or completely blocked by both PI3K and ERK inhibitors (Banko et al., 2004; Kelleher et al., 2004b; Tsokas et al., 2005).
mTOR can form two distinct complexes. One complex termed mTORC1 contains Raptor and LST8/GβL and is sensitive to the drug rapamycin (a macrolide that binds mTORC1 as a complex with the immunophilin FKBP12) (Figure 2). mTORC1 phosphorylates its target proteins through their recruitment by the adaptor protein Raptor. These substrates possess a conserved TOR signaling motif (TOS) that mediates binding to Raptor. Rapamycin decreases mTOR-Raptor interactions, thereby preventing mTORC1 from phosphorylating its targets proteins (Kim et al., 2002; Oshiro et al., 2004) (Figure 2). In addition to 4E-BPs, other substrates of mTORC1, such as S6K1, S6K2, and PRAS40, have been documented (Sabatini, 2006; Sancak et al., 2007; Vander Haar et al., 2007) (Table 1).
mTORC1 specifically regulates the translation of a subset of mRNAs that contain extensive secondary structure at their 5′ UTR or an oligopyrimidine tract in their 5′ end (TOP mRNAs) (Ruvinsky and Meyuhas,2006). The latter mRNAs largely encode components of the translational machinery itself, including ribosomal proteins and elongation factors. The TOP sequence represses translation, and this repression is removed in an mTORC1-dependent manner via a mechanism that has yet to be identified.
The second mTOR complex (mTORC2), which contains LST8/ GβL and Rictor instead of Raptor, is typically rapamycin insensitive and phosphorylates Akt/PKB and several PKC isoforms (Hara et al., 2002; Jacinto et al., 2004; Kim et al., 2002; Loewith et al., 2002; Sarbassov et al., 2004) (Figure 2). Importantly, while rapamycin specifically inhibits mTORC1 after a short treatment, prolonged treatment with rapamycin also blocks mTORC2 activity (Sarbassov et al., 2006).
The mTOR Signaling Pathway Is Important for Synaptic Plasticity
Both L-LTP and mGluR-LTD-inducing stimulation triggers the phosphorylation of mTOR downstream targets, 4E-BPs and S6K1/2, which is correlated with an increase in the translation of a number of TOP mRNAs (Hou and Klann, 2004; Kelleher et al., 2004a; Tsokas et al., 2005; Antion et al., 2008a). The involvement of mTORC1 in neuronal plasticity is evolutionarily conserved, as induction of long-term facilitation in the invertebrate Aplysia also triggers the phosphorylation of S6 kinase and 4E-BP as well as the translation of TOP mRNAs (Carroll et al., 2004, 2006; Khan et al., 2001). Immunocytochemical studies have shown that all of these changes also occur locally in dendrites (Cammalleri et al., 2003; Carroll et al., 2004; Gobert et al., 2008; Tsokas et al., 2005).
Rapamycin blocks both long-lasting synaptic changes and memory consolidation in mammals in a number of behavioral tasks (Dash et al., 2006; Tang et al., 2002; Tischmeyer et al., 2003), as well as long-term changes in plasticity in Aplysia (Casadio et al., 1999; Hu et al., 2006). What downstream targets of mTORC1 are important for plasticity? 4E-BP2 is the major 4E-BP isoform present in the adult brain (Banko et al., 2005). 4E-BP2 knockout mice display impaired L-LTP and spatial learning in both Morris water maze and contextual fear conditioning (Banko et al., 2005). Other types of memory impacted by the deletion of 4E-BP2 include motor memory, working memory, and associative memory for aversive taste (Banko et al., 2007). However, although these studies demonstrate the importance of 4E-BP2 in these processes, they do not show that the requirement for mTORC1 is mediated by 4E-BP2 regulation.
mTOR downstream targets S6K1 and S6K2 are also involved in memory formation. Although L-LTP is normal in hippocampal slices from either S6K1- or S6K2-deficient mice, they exhibit an early-onset contextual fear memory deficit within 1 hr of training, a deficit in conditioned taste aversion (CTA), and impaired spatial learning and memory in the Morris water maze (Antion et al., 2008b). S6K2-deficient mice exhibit decreased contextual fear memory 7 days after training, a reduction in latent inhibition of CTA, but normal spatial learning in the Morris water maze. Because S6K1 and S6K2 can functionally compensate for one other, it would be pertinent to study mice with a deletion in both S6K1 and S6K2 to obtain a better understanding of the role of S6Ks in long-lasting synaptic plasticity and long-term memory.
Upstream effectors of mTORC1 are also implicated in long-lasting forms of synaptic plasticity and memory. Recent evidence supports the notion that regulation of mTORC1 activity is critical for activity-dependent changes in synaptic strength and memory. TSC2+/− heterozygous mice, in which the mTORC1 signaling pathways is constitutively active, exhibit a lowered threshold for the induction of L-LTP and impaired hippocampus-dependent memory similar to the GCN2 and 4E-BP2 knockout mice (Banko et al., 2005; Costa-Mattioli et al., 2005; Ehninger et al., 2008). Remarkably, rapamcyin treatment reversed the facilitated L-LTP and rescued the memory impairment in TSC2+/− heterozygous mice (Ehninger et al., 2008). Consistent with these findings, TSC1+/− mice also exhibit hippocampus-dependent learning impairments and abnormal social behavior (Goorden et al., 2007). It would be important to determine whether, as was the case for TSC2+/− mice, rapamcyin rescues the learning and social deficit in TSC1+/− mice. These data conform to the idea that proper control of mTORC1 signaling is crucial for long-lasting synaptic plasticity and memory consolidation because either inhibiting (rapamycin) or activating (TSC2+/− and TSC1+/− mice) mTOR leads to memory impairments. Since loss of TSC1 and TSC2 leads to an activation of the unfolded protein response (Ozcan et al., 2008), an attractive, but merely speculative hypothesis would be that the memory impairment in TSC1+/− and TSC2+/− mice is due to abnormally elevated eIF2α phosphorylation levels.
Translational Control by eIF4E Phosphorylation
eIF4E is phosphorylated at a single site, Ser209 in mammals (Joshi et al., 1995; Whalen et al., 1996), by MAPK signal-integrating kinase/MAPK-interacting kinase 1 and 2 (Mnk1/2) (Fukunaga and Hunter, 1997; Pyronnet et al., 1999; Waskiewicz et al., 1997) (Table 1). Mnk-dependent phosphorylation decreases the affinity of eIF4E for the cap structure by ~4-fold (Scheper et al., 2002; Zuberek et al., 2003). Mnk1/2 binds to the C-terminal region of eIF4GI and eIF4GII, which serves as a docking site, to phosphorylate eIF4E. In general, the phosphorylation of eIF4E is tightly correlated with the rate of translation, which may be due to the fact that eIF4E association with eIF4G (and thus with eIF4G bound Mnk) is also correlated with the rate of translation (Table 1). However, the molecular mechanism by which eIF4E phosphorylation affects translation remains controversial (Scheper and Proud, 2002).
When eIF4E phosphorylation is increased in response to overexpression of Mnks in Aplysia, there is a reduction in cap-dependent translation (Ross et al., 2006). Also, increased dosage of Mnk (LK6) in Drosophila caused a reduction of growth (Reiling et al., 2005). Blocking Mnk activity either pharmacologically or genetically did not affect overall protein synthesis (Ueda et al., 2004; Morley and Naegele, 2002).
Given that eIF4E phosphorylation is evolutionarily conserved, it is noteworthy that a reduction of eIF4E phosphorylation does not impact general translation rates. However, regulation of eIF4E phosphorylation is important under certain conditions. For instance, flies lacking Mnk grew poorly under starvation conditions (Arquier et al., 2005) and constitutively activated MNK1 expression in a lymphoma model promoted tumorigenesis, and a dominant-negative MNK exhibited an opposite effect (Wendel et al., 2007). The finding that eIF4E phosphorylation is required under certain conditions could be explained by the hypothesis that translation of some specific mRNAs are more dependent on eIF4E phosphorylation than the translation of bulk mRNAs. However, the identity of the mRNAs that are translationally controlled by eIF4E phosphorylation remains to be determined.
Induction of both LTP and mGluR-LTD leads to increased eIF4E phosphorylation via ERK-dependent activation of Mnk I (Banko et al., 2006; Kelleher et al., 2004a), consistent with the requirement of ERK for translational-dependent forms of plasticity and learning (Kelleher et al., 2004a). However, the evidence that eIF4E phosphorylation is the critical step that is dependent on ERK is only correlative. Moreover, in the nervous system, ERK appears to be required not only for eIF4E phosphorylation but also for activation of mTOR signaling (Kelleher et al., 2004a).
Translational Control by IRES
In general, ribosome recruitment to the mRNA is facilitated by the 5′-cap structure that is present on all nuclear transcribed eukaryotic mRNAs (Shatkin, 1985). The cap structure is separated from the initiation codon, which is usually the first AUG (or its cognate) triplet downstream of the 5′ end, by 50–100 nucleotides (Gingras et al., 1999a; Pestova et al., 2001).
In an alternative and less frequent process, the recruitment of the ribosome to the mRNA is independent of the cap structure, but it is instead mediated by an IRES. IRES-mediated translation was first discovered in picornaviruses (Doudna and Sarnow, 2007), where the viral RNA is uncapped and contains long 5′ UTRs (>400 nucleotides), which are highly structured and act as barriers for scanning ribosomes. Thus, the ribosome is directly recruited to the 5′ UTR of mRNA via direct interaction or the IRES provides sites for interaction with specific eIFs, such as the eIF4 group of initiation factors that recruit the ribosome.
IRES-mediated translation is not restricted to viral RNAs since some cellular mRNAs also contain IRESes (Elroy-Stein and Merrick, 2007). Cellular IRESes were identified in the 5′ UTR of mRNAs encoding proteins which need to be synthesized under stress condition such as apoptosis, virus infection, hypoxia, nutrient deprivation, etc., where cap-dependent translation is impaired (Holcik and Sonenberg, 2005).
Since IRES-driven translation allows for the control of translation of specifics mRNAs, it is possible that neurons could use this mechanism to upregulate the translation of a subset of mRNAs during a learning experience. Consistent with this idea, some mRNAs that are transported to dendrites appear to exhibit IRES activity (Pinkstaff et al., 2001). In Aplysia bag cell neurons, a decrease in eIF4E phosphorylation is associated with an increase in translation of a neuropeptide mRNA that has an IRES in its 5′ UTR (Dyer et al., 2003). Notably, during facilitation induced by continuous, as opposed to spaced, application of serotonin, a decrease in eIF4E phosphorylation correlated with a decrease in cap-dependent translation (Dyer and Sossin, 2000; Yanow et al., 1998). However, no mRNAs have been identified that are specifically upregulated through an IRES-dependent mechanism at this time.
Translational Regulation by Elongation Factors
Although initiation is usually the rate-limiting step in translation, elongation can also be regulated (Herbert and Proud, 2007). The major mechanism for regulating elongation is phosphorylation of eEF2 by the eEF2 kinase (Table 1). eEF2 kinase is regulated by calcium/calmodulin and thus by neuronal activity (Herbert and Proud, 2007). Indeed, miniature excitatory postsynaptic potentials (mini EPSPS), caused by the spontaneous release of neurotransmitter, have been demonstrated to inhibit local translation through activation of eEF2 kinase (Sutton et al., 2007). In turn, eEF2 kinase can be inhibited through phosphorylation by S6 kinase (Herbert and Proud, 2007). These findings serve as the basis for an attractive model for establishing synapse specificity of translation, because calcium entry would inactivate translation at all synapses but could be reversed by mTORC1 activation at specific synapses. mTOR- and calcium-dependent regulation of eEF2 is conserved in Aplysia (Carroll et al., 2004).
While phosphorylation of eEF2 causes a decrease in general translation, it appears that it is also able to promote the translation of some mRNAs (Chotiner et al., 2003; Park et al., 2008). The exact molecular mechanism underlying this regulation is not clear. One idea is that the block of elongation frees up a rate-limiting initiation factor (such as eIF4E) that then allows initiation on poorly translated mRNAs (Brendler et al., 1981; Godefroy-Colburn and Thach, 1981; Scheetz et al., 2000; Walden et al., 1981). Indeed, blocking elongation by another mechanism, using low concentrations of cycloheximide (an inhibitor of peptide bond formation), also appears to cause the upregulation of some neuronal mRNAs (Scheetz et al., 2000; Park et al., 2008). Translational control by eEF2 phosphorylation appears to be particularly important in mGluR-LTD, where two mRNAs encoding the proteins Arc/Arg3.1 and MAP-1B are translationally activated in an eEF2 kinase-dependent manner (Davidkova and Carroll, 2007; Park et al., 2008). Interestingly, in both eEF2 kinase and Arc/Arg3.1 knockout mice, mGluR-LTD is impaired (Park et al., 2008).
A number of studies have documented an elevation in the levels of the elongation factors eEF1A and eEF2 after synaptic activity in both mammals and Aplysia that likely occurs via translation of mRNAs possessing a TOP sequence at the 5′ end (Antion et al., 2008a; Giustetto et al., 2003; Huang et al., 2005; Tsokas et al., 2005). However, it is not clear whether the levels of eEF1A are rate limiting for elongation (Condeelis, 1995). An increase in eEF1A may be important due to an additional role outside of translation. Indeed, eEF1A is a major regulator of the actin cytoskeleton independent of its role in translational control (Gross and Kinzy, 2005).
Local Protein Synthesis and Synaptic Plasticity
There is general agreement that the late phase of LTP, which is induced with electrical or chemical stimulation, is dependent on new gene expression, as it is blocked by both transcription and translation inhibitors (for reviews see Costa-Mattioli and Sonenberg, 2008; Kandel, 2001; Kelleher et al., 2004b; Klann and Dever, 2004; Sutton and Schuman, 2006). Because major components of the protein synthesis machinery, including ribosomes, translation factors, and mRNAs, are present in dendrites and dendritic spines (Steward and Schuman, 2001; Sutton and Schuman, 2006), it was also posited that long-lasting plasticity could be engendered through the activation of local protein synthesis, i.e., protein synthesis in dendrites without the requirement for transcription in the neuronal soma. Indeed, there are hundreds of distinct mRNAs present in isolated hippocampal dendrites (Eberwine et al., 2001; Miyashiro et al., 1994; Poon et al., 2006). Evidence that local protein synthesis is required for long-lasting synaptic plasticity was first provided by Kang and Schuman (Kang and Schuman, 1996). They demonstrated that BDNF could induce LTP that was blocked by protein synthesis inhibitors even when pre- and postsynaptic pyramidal neurons were severed from their somas (Kang and Schuman, 1996). Recently, it was shown that other forms of synaptic plasticity require dendritic translation: mGluR-LTD and L-LTP induced by pairing E-LTP-inducing stimulation (normally protein synthesis independent) with activation of β-adrenergic receptors are dependent on local dendritic protein synthesis, but not on new gene expression (Gelinas and Nguyen, 2005; Huber et al., 2000).
Moreover, there are several examples where dendritic, protein synthesis-dependent gene expression-independent LTP can be induced with patterns of electrical stimulation that differ from those conventionally used to induce L-LTP (Cracco et al., 2005; Vickers et al., 2005). Even under standard L-LTP stimulation paradigms, pharmacological and genetic evidence indicates that translation inhibitors impact L-LTP at an earlier temporal window than transcription inhibitors (Banko et al., 2005; Kelleher et al., 2004a). These data suggest that L-LTP has an early component that is transcription independent but likely dendritic and translation dependent and a late phase that is somatic, requiring both transcription and translation. Even the gene expression-dependent phase may require local translation of the newly synthesized mRNAs since L-LTP was reduced with local, dendritic application of a protein synthesis inhibitor (Bradshaw et al., 2003).
In mollusks, there is also a well-characterized translation-dependent, transcription-independent form of memory (Sangha et al., 2003; Sutton et al., 2001). This form of memory is often termed intermediate-term memory (ITM), although it is noteworthy that not all forms of ITM require translation (Sutton et al., 2001; Yin et al., 1994). Translation-dependent forms of memory are regulated locally and often do not require the cell soma (Liu et al., 2003; Martin et al., 1997; Sherff and Carew, 2004).
Local translation is also required to stabilize new synapses in Aplysia that are formed after learning (Casadio et al., 1999; Martin et al., 1997). Rapamycin-sensitive activation of local translation is stimulated by a single 5 min application of serotonin, but the stabilization occurs between 24 and 72 hr after this application, suggesting a sustained upregulation of translation after a transient signal. Indeed, inhibition of local protein synthesis starting 24 hr after induction selectively removes the new synapses, leaving previously formed synapses intact (Miniaci et al., 2008). A proposed mechanism for this prolonged plasticity is the translational upregulation of cytoplasmic polyadenylation element binding protein (CPEB), which in a prion-like manner remains activated for a prolonged period of time, upregulating translation locally in dendrites (Si et al., 2003). However, this interpretation is not entirely consistent with the requirement for continued local synthesis of CPEB even 48 hr after induction (Miniaci et al., 2008). Instead, it appears that CPEB is part of a positive-feedback mechanism required for the increased translation needed to stabilize the new synapses. Another possibility is that the increase in translation of TOP mRNAs, an abundant fraction of the mRNAs present at Aplysia processes, could contribute to the persistent long-term increase in translation required to stabilize new synapses (Khan et al., 2001; Moccia et al., 2003).
Translation of Transported mRNAs
Local translation depends on transport of mRNAs from the cell body to distal sites. In neurons, transport of mRNAs to dendrites has a major impact on their translational regulation. Below we discuss the contribution of RNA-binding proteins (RBPs) to translation repression and mRNA transport. During mRNA transport to dendrites, translation is repressed by RBPs. For translation to be activated at local sites, such a repression needs to be removed, raising the possibility that this could be a rate-limiting step in translational control at local sites. A number of translational inhibitors have been implicated in this step, including fragile X mental retardation protein (FMRP), CPEB, Zip code binding protein/insulin-like growth factor binding protein 1 (ZBP/IMP1), hnRNPA2 and RNA Granule 105 (see below). A well-documented study of translational repression during mRNA transport, which serves as an excellent paradigm, is that of β-actin mRNA translation. In mammalian neurons, ZBP1 inhibits translation of β-actin mRNA during sorting to growing neurites. Phosphorylation of ZBP1/IMP1 by Src reduces its RNA-binding affinity and relieves translational repression (Huttelmaier et al., 2005).
It should also be noted that recent reports have demonstrated splicing to occur in dendrites (Glanzer et al., 2005). Thus, transport of an intron-containing mRNA followed by dendritic splicing is another example of local translational control.
FMRP
An important translational repressor in the brain is FMRP. Mutations causing a loss of FMRP result in fragile X mental retardation, which is the most common inherited disease causing mental retardation (Turner et al., 1996). This disorder is in most cases caused by the expansion of the trinucleotide sequence CGG in the 5′ UTR of the FMR1 gene on the X chromosome, which leads to hypermethylation and silencing of the FMR1 gene. FMRP binds to a subset of mRNAs encoding proteins involved in synaptic plasticity and development and inhibits their translation (Darnell et al., 2001).
The strongest evidence for a role of FMRP in mRNA repression is based on genetic models (flies and mice lacking FMRP) (Bolduc et al., 2008; Dolen et al., 2007). In these animals, the translation of mRNAs to which FMRP binds is thought to be derepressed because protein levels encoded by the FMRP-binding mRNAs are increased (Brown et al., 2001; Garber et al., 2006). Indeed, in fragile X mutant flies, an excess in protein synthesis causes an impairment in memory (Bolduc et al., 2008). Many mRNA targets of FMRP are transported normally in FMRP knockout mice (Steward et al., 1998a); however, the number of mRNA granules is reduced (Aschrafi et al., 2005). This may be due to the precocious disruption of the granules and translational activation of their mRNAs in the absence of FMRP.
The precise mechanism by which FMRP represses translation of target mRNAs remains unresolved (Bagni, 2008; Iacoangeli et al., 2008a, 2008b). One model posits that FMRP regulates translation of its target mRNAs via interaction with the noncoding neuronal RNA BC1 (Zalfa et al., 2003). The binding to BC1 may enhance FMRP functions in a number of ways; such as increasing binding to target mRNAs or inhibiting eIF4A, and thus represses translation initiation of mRNAs harboring a structured 5′ UTR (Lin et al., 2008; Wang et al., 2002). Recently, Napoli et al. provided evidence that FMRP represses translation initiation via interaction with CYFIP1/Sra-1, a newly identified neuronal eIF4E-binding protein. CYFIP1/Sra-1 competes with eIF4G for binding to eIF4E in a manner similar to 4E-BPs, although not through a canonical eIF4E-binding site (Napoli et al., 2008).
In contrast to a block of translation initiation through BC1 or CYFIP1/Sra-1, an alternative model posits that FMRP acts at a postinitiation step because it associates with functional polyribosomes (Stefani et al., 2004). FMRP phosphorylation is reported to affect its translational activity. Phosphorylated FMRP cosediments with stalled ribosomes, whereas nonphosphorylated FMRP associates with actively translating ribosomes (Ceman et al., 2003). S6K1 phosphorylates FMRP on a conserved serine residue required for mRNA binding, and FMRP phosphorylation was abolished at this site in S6K1 knockout mice or following rapamycin treatment (Narayanan et al., 2008). In both S6K1 and FMRP knockout mice, the levels of SAPAP3, a FMRP target mRNA, were increased. Thus, S6K1 activation, normally thought to be an activator of translation, appears to be required to repress translation of some FMRP target mRNAs.
A number of the FMRP-regulated target mRNAs encode proteins that play a role in a form of LTD induced by activation of group I mGluR receptors (Volk et al., 2007). As mentioned earlier, this form of LTD requires local protein synthesis (Huber et al., 2000). Interestingly, in FMRP knockout mice, mGluR-LTD is enhanced and no longer requires protein synthesis (Hou et al., 2006; Huber et al., 2002; Nosyreva and Huber, 2006). Thus, the proteins that are required for enhanced mGluR-LTD are presumably present before stimulation due to the loss of repression by FMRP. However, production of these proteins may still require other aspects of mGluR signaling, as many phenotypes of FMRP knockout mice are rescued when mGluR5 activity is reduced either genetically or with pharmacological inhibitors (Dolen et al., 2007; Yan et al., 2005). Strikingly, a similar rescue has been observed in Drosophila, suggesting a strong evolutionary link between mGluR5 receptors and FMRP signaling (Pan et al., 2008).
In agreement with the notion that S6K-dependent phosphorylation of FMRP is required for translation repression, mGluR LTD is enhanced in S6K2 and S6K1/2 double-knockout mice, although mGluR-LTD appears to be normal in S6K1 knockout mice (Antion et al., 2008a). Similar to FMRP knockout mice, enhanced mGluR-LTD in S6K2 knockout mice is resistant to protein synthesis inhibition (Antion et al., 2008a).
If the major mechanism by which FMRP inhibits translation of its targets is through BC1 RNA, then one would expect that BC1 knockout mice would also exhibit protein synthesis-independent enhanced mGluR LTD. This question remains to be examined.
Although protein synthesis is required for mGluR-LTD-induced plasticity, it is not sufficient, because the elicitation of LTD is dependent on mGluR receptor stimulation even when the proteins (such as Arc and MAP1B) are already present. Also, it suggests that local protein synthesis is important for regulating protein levels, and these levels are only rate limiting under certain conditions. For example, manipulations of the proteasome system can lead to a switch from protein synthesis-dependent plasticity to protein synthesis-independent plasticity by regulating the basal level of important proteins normally produced by local synthesis (Fonseca et al., 2006; Karpova et al., 2006).
CPEB
CPEB is an established translational repressor in Xenopus oocytes where translation of many mRNAs is dormant due to the lack of a polyA tail. CPEB binds to well-defined sites, often located close to the poly-A addition site (Richter, 2007). Significantly, the number and position of the CPE and the Pumilio-binding elements on the mRNA determine whether an mRNA is translationally repressed by CPEB (Pique et al., 2008). In neurons, CPEB not only represses translation but also contributes to the transport of mRNAs containing a CPEB-binding site (Huang et al., 2003). At synapses, neuronal activity can activate CPEB phosphorylation leading to an increase in the length of the polyA tail and mRNA translation (Huang et al., 2002; Wu et al., 1998). In addition, expression of a dominant-negative of CPEB that lacks the phosphorylation sites blocks a protein synthesis-dependent form of LTD in the cerebellum (McEvoy et al., 2007).
Whereas in late-stage oocytes, translational repression by CPEB is due to its binding to the eIF4E-binding protein Maskin, the eIF4E-binding region of Maskin is not present in mammals. However, other proteins can simultaneously bind to both CPEB and eIF4E, such as neuroguidin, that may act in a manner similar to Maskin (Jung et al., 2006). In Aplysia, translation of CPEB at synapses is required for activation of local translation and engendering a long-term form of synaptic plasticity (Si et al., 2003). CPEB is also required for long-term memory formation in Drosophila (Keleman et al., 2007).
Additional RBPs and Modes of RNA Transport Are Candidates for Regulation of Plasticity
Other RBPs have been implicated in the repression of translation of transported mRNAs. RNA granule 105 is a translational repressor found in RNA granules, and its removal is coupled to translational activation (Shiina et al., 2005). hnRNP-A2 binds to many of the important transported mRNAs, including Arc/ Arg3.1 and αCaMKII, and regulates their transport (Gao et al., 2008; Shan et al., 2003).
While the repression by mRNA-binding proteins is essential for the regulation of translation of transported mRNAs, the type of transport structure may also be critical. There are a number of distinct types of transport complexes used in neurons, and the mechanisms of repression in the different complexes are probably distinct (Sossin and DesGroseillers, 2006). Thus, if mRNAs are transported in mRNA granules containing stalled polysomes, translation would be blocked at the elongation step. It should be stressed that at the present time the evidence that mRNA granules contain stalled polysomes, as opposed to coaggregations of mRNAs and ribosomes, is solely based on the molecular composition of these structures (Elvira et al., 2006; Krichevsky and Kosik, 2001). In contrast, if mRNAs are transported in RNA particles, without polysomes, translation would be blocked at the initiation step. It is not clear whether the repression of mRNA-binding protein determines the type of transport mechanism or whether there are other factors involved in this process. Many RBPs, such as FMRP, are found in both RNA granules and in RNA particles and thus probably additional proteins regulate the type of structure FMRP-bound mRNAs are present on (Zalfa et al., 2006).
A significant RBP implicated in transport, as opposed to translational repression, is Staufen, first identified as being required for RNA localizaton in Drosophila oocytes (St Johnston et al., 1991). Staufen has been particularly implicated in neuronal mRNA transport (Krichevsky and Kosik, 2001). A dominant-negative form of Staufen reduced overall transport of mRNAs to dendrites (Tang et al., 2001). In addition, treatment of hippocampal neurons with siRNA against Staufen1, one of two isoforms of Staufen in mammals, blocks L-LTP without affecting E-LTP (Lebeau et al., 2008). The role of Staufen in synaptic plasticity appears to be evolutionarily conserved, as reducing levels of Staufen in Aplysia also blocks LTF (Liu et al., 2006). In addition, a Staufen mutant exhibited impaired memory in Drosophila (Dubnau et al., 2003). Whether the requirement of Staufen for L-LTP is due to the loss of mRNA transport or to other actions of Staufen in translational regulation remains to be determined (Sossin and DesGroseillers, 2006).
Translational Regulation by miRNAs
The discovery of microRNAs (miRNAs) has revolutionized the biology field. miRNAs are a family of small RNAs (~21 nucleotides long) that regulate as much as 50% of all gene expression post-transcriptionally, in different phyla (Bushati and Cohen, 2007). Mammalian miRNAs are encoded by monocystronic and polycistronic gene clusters sometimes found within intronic regions of noncoding genes. RNA polymerase II transcribes long-miRNA precursors (pri-miRNA). pri-miRNAs are first processed in the nuclei by the RNase III enzyme Drosha: cleavage by Drosha results in a 70 nucleotide long RNA stem loop, termed pre-miRNA. Following export to the cytoplasm, pre-miRNAs are further processed by Dicer into miRNA duplexes ~21 nucleotides long. Following cleavage by Dicer, one strand, termed the guide strand, is incorporated into an RNA-induced silencing complex (RISC), a complex that consists of proteins crucial for silencing the target mRNA. The molecular mechanism by which miRNAs silence the expression of their target mRNAs remains unclear. Evidence to support several disparate mechanisms has been reported: (1) inhibition of translation elongation, (2) inhibition of translation initiation, (3) ribosome drop-off, (4) cotranslational protein degradation, and (5) mRNA degradation (Eulalio et al., 2007; Filipowicz et al., 2008; Jackson and Standart, 2007). In addition, the interaction of the p-body marker protein GW182 with Argonaute appears to be essential for both miRNA-mediated translational repression and mRNA decay in Drosophila cells (Eulalio et al., 2008).
miRNA-Mediated Control of Synaptic Plasticity
The brain contains many miRNAs (Kosik, 2006). In mammals, conditional ablation of essential components of the miRNA machinery appears to be critical for brain function. For example, inactivation of Dicer in forebrain, Purkinje, and dopaminergic neurons results in microcephaly, progressive neurodegeneration, and increased apoptosis and neurodegeneration, respectively (Davis et al., 2008; Hebert and De Strooper, 2007; Kim et al., 2007; Schaefer et al., 2007).
Because miRNAs regulate translation of its target mRNAs, it is highly likely that this tool is used for gene-specific regulation of translation in dendrites (Kosik, 2006). It has been recently suggested that even processing of miRNAs can occur in dendrites (Lugli et al., 2005, 2008). In Drosophila, during olfactory conditioning, local translational upregulation of CAMKII is due to a decrease in miRNA-mediated repression (Ashraf et al., 2006). Proteolysis of a component of the RISC complex appears to explain the removal of repression. This miRNA-binding site is conserved in vertebrates and thus may represent a conserved mechanism (Ashraf et al., 2006). In mammalian hippocampal neurons, the brain enriched miRNA 134 inhibits translation of Limk1 (a protein involved in spine development), thus blocking excitatory synaptic transmission and the size of dendritic spines. Neuronal activity such as BDNF treatment derepressed the miR-134-mediated translation inhibition of Limk1 mRNA (Schratt et al., 2006).
Plasticity-Induced Proteins
A question that has been raised persistently but not answered satisfactorily is the identity of the proteins that are synthesized following the induction of LTP and LTD that are important for plasticity. Two prominent proteins that are thought to be upregulated locally are αCa2+/calmodulin-dependent protein kinase II (αCaMKII) (Ouyang et al., 1999) and PKMζ (Osten et al., 1996). What is particularly interesting about these two proteins is that they both have the potential to stabilize L-LTP via positive, feed-forward mechanisms. For example, L-LTP inducing stimulation increases the phosphorylation of CPEB in a CaMKII-dependent manner, triggering CPE-mediated protein synthesis (Atkins et al., 2004). Because phosphorylation of CPEB increases polyadenylation of αCaMKII mRNA and synthesis of αCaMKII in neurons in response to NMDA receptor stimulation (Wells et al., 2001; Wu and Bag, 1998), it is possible that a persistent increase in the levels of αCaMKII could be maintained following induction of L-LTP. Similarly, there is a requirement for protein kinase Mζ (PKMζ) activity for the synthesis of new PKMζ during LTP (Kelly et al., 2007). Remarkably, application of a peptide that blocks PKMζ inhibits both L-LTP maintenance and memory storage (Pastalkova et al., 2006; Shema et al., 2007). The details of the positive PKMζ feedforward loop and whether genetic deletion of PKMζ leads to impaired L-LTP maintenance and memory storage remain to be established.
Probably, one of the best-characterized LTP-induced proteins is Arc/Arg3.1, which is quickly synthesized after learning, and its mRNA travels down dendrites and is localized to active synapses, as is the translated protein (Steward et al., 1998b). L-LTP is impaired in Arc/Arg3.1 knockout mice (Plath et al., 2006), and Arc/Arg3.1 may play a role in cytoskeletal rearrangements underlying L-LTP (Messaoudi et al., 2007). Another major role for Arc/ Arg3.1 is in the endocytosis of AMPA receptors associated with LTD and the homeostatic response of neurons (Chowdhury et al., 2006; Rial Verde et al., 2006). Indeed, reducing Arc/Arg3.1 mRNA levels blocks mGluR-LTD (Park et al., 2008; Waung et al., 2008). The gene encoding Arc/Arg3.1 has several introns following the exon, which contains the natural stop codon (Giorgi et al., 2007). This configuration is rare, as it constitutes a signal for triggering nonsense-mediated decay (NMD) response, a quality-control process in which the mRNA is destabilized by the presence of a premature termination codon (Amrani et al., 2006). Notably, eliminating this pathway in neurons leads to increased synaptic strength, although overexpressing Arc/Arg3.1 decreases synaptic strength, suggesting thatNMD isused in neurons to regulate levels of additional synaptic proteins as well (Giorgi et al., 2007).
Another mRNA whose translation is linked to mGluR-LTD is the cytoskeletal MAP1B mRNA (Hou et al., 2006; Davidkova and Carroll, 2007). Notably, MAP1B is an FMRP target mRNA (Darnell et al., 2001), mGluR-LTD triggers protein synthesis-dependent increases in MAP1B (Hou et al., 2006), and basal levels of MAP1B are increased in FMRP knockout mice (Lu et al., 2004; Hou et al., 2006). Taken together, these data suggest that MAP1B and Arc/Arg3.1 are both attractive candidates for the critical proteins synthesized during mGluR-LTD (Davidkova and Carroll, 2007; Muddashetty et al., 2007; Weiler et al., 1997).
LTP and mGluR-LTD and long-term facilitation in Aplysia increase the translation of 5′ TOP mRNAs (Antion et al., 2008a; Carroll et al., 2004; Huang et al., 2005; Khan et al., 2001; Ronesi and Huber, 2008; Tsokas et al., 2005, 2007). The increase in translation of 5′ TOP mRNAs has been demonstrated directly using reporters in dendrites and can be attributed to TOR-dependent removal of TOP repression (Gobert et al., 2008). Strikingly 5′ TOP mRNAs are enriched in dendrites of hippocampal neurons (Poon et al., 2006) and neurites of Aplysia neurons (Moccia et al., 2003). The increased levels of proteins encoded by 5′ TOP mRNAs were demonstrated in dendrites as well as the neuronal soma (Antion et al., 2008a; Tsokas et al., 2005, 2007). Consistent with these data, proteomic studies examining mTORC1-dependent increases in BDNF-induced translation also identified 5′ TOP mRNA encoded proteins as a major component of the translational response (Liao et al., 2007). As mentioned earlier, 5′ TOP mRNAs encode translation factors and ribosomal proteins and therefore could promote local protein synthesis-dependent synaptic plasticity in neurons by facilitating translation in response to specific patterns of activity and receptor activation at synapses. There is not yet a pharmacological or genetic mechanism to selectivity block mTORC1-mediated upregulation of 5′ TOP mRNAs. A better understanding of the mechanism regulating 5′ TOP mRNA translation will help to answer the question whether the translation of 5′ TOP mRNAs is important for synaptic plasticity.
In Aplysia, a protein whose local translation appears to be important for plasticity is the neuropeptide sensorin (Hu et al., 2006). Sensorin is important for activation of ERK and consequently LTF (Hu et al., 2004). Translation of sensorin may also play a role in the stabilization of new synapses, since local synthesis of sensorin plays a role in stabilization of initial synapse formation (Hu et al., 2007; Lyles et al., 2006). Secreted factors are also attractive products of local translation in vertebrates. BDNF is important for late-LTP under some stimulation paradigms, and it has been reported that LTP becomes protein synthesis independent if BDNF is added exogenously (Pang et al., 2004). Moreover, exogenous BDNF can also rescue some memories from protein synthesis inhibition (Bekinschtein et al., 2007). However, mutations that abrogate transport of BDNF mRNA to dendrites cause a deficit in pruning and enlargement of dendritic spines, suggesting that the specific role of BDNF in dendrites is to regulate spine density and the ability to induce protein synthesis-independent forms of LTP (An et al., 2008). In contrast, the BDNF, which is important for late LTP, may be released from the presynaptic neuron, presumably from a regulated secretory vesicle (Zakharenko et al., 2003).
Conclusions and Future Questions
Translational control has an enormous impact on synaptic plasticity and memory via the regulation of the synthesis of proteins that are critical for many aspects of these processes. Precise control of translation is important, because either enhancing or reducing activity of specific signaling pathways, such as mTORC1, causes memory deficits. One significant issue that has not been adequately addressed is whether translational control is more important for regulating translation of a subset of mRNAs or general translation. Several studies suggest that translational control is critical for specific transcripts. For example, in mammalian neurons, changes in eIF2α phosphorylation specifically regulate translation of ATF4 mRNA (Costa-Mattioli et al., 2005, 2007b). This process is conserved since in yeast levels of eIF2α phosphorylation that do not cause general inhibition of translation are sufficient for stimulation of Gcn4p synthesis (Hinnebusch et al., 2004; Hinnebusch and Natarajan, 2002). Thus, a mild (physiological) change in eIF2α, which does not significantly alter translation rates of abundant mRNAs, is sufficient to affect ATF4 mRNA translation. It is noteworthy that in some studies extreme conditions were employed to strongly activate eIF2α kinases, resulting in a severe block to general translation initiation. Neurons would probably never experience such insults, except under pathological conditions such as hypoxia and stroke (DeGracia et al., 1997), but weak levels of kinase activation are likely to be the rule (Dever, 2002). Similar to translational control by eIF2α, phosphorylation of eEF2 inhibits general translation but paradoxically stimulates the translation of specific transcripts that appear to be important for mGluR-LTD (Park et al., 2008; Scheetz et al., 2000).
Along similar lines, specific mRNAs regulated by 4E-BPs have not yet been identified in the nervous system. However, in mouse embryonic fibroblasts, 4E-BPs regulate translation of a subset of mRNAs, encoding protein involved in type-I interferon response (Colina et al., 2008). Therefore, it is expected that in neurons 4E-BPs will regulate translation of specific mRNAs rather than general translation. Consistent with these data, rapamycin preferentially represses translation of a subset of mRNAs rather than general translation (Grolleau et al., 2002). Thus, it is possible that the requirement for mTORC1 in plasticity entails translational control of specific mRNAs. Finally, miR-134 is thought to specifically regulate LimK1 mRNA translation and thus control synaptic development (Schratt et al., 2006). In contrast, it was recently suggested that general translation may also be important for long-lasting processes. There is substantial evidence that LTP and LTD-inducing protocols activate the signaling pathways that control general translation (Hou and Klann, 2004; Kelleher et al., 2004b; Tsokas et al., 2005; Banko et al., 2006). However, whether learning leads to large increases in general translation remains to be determined. It is thought that an increase in 5′ TOP mRNAs would lead to enhanced general translation.
Another unresolved issue is related to the time window in which protein synthesis is needed for memory consolidation. In general, the application of protein synthesis inhibitors around the time of training blocks long-term memory consolidation but does not alter already established memories. However, recent evidence suggests that there is more than one wave of protein synthesis during the memory consolidation process (Bekinschtein et al., 2007; Bourtchouladze et al., 1998). In contrast, the stabilization of new synapses in Aplysia appears to require long-lived upregulation of protein synthesis for a prolonged period of time (Miniaci et al., 2008). Therefore, three models could contribute to the role of protein synthesis in long-term memories: (1) a transient burst of protein synthesis probably in response to environmental cues or experiences, (2) multiple bursts of protein synthesis required for distinct windows after training, or (3) a persistent prolonged local increase in protein synthesis to support new growth.
In conclusion, we have described a relatively large number of specific mechanisms of translational control that are operating in neurons. In the coming years, it will be important to match specific translational control mechanisms with specific transcripts and with distinct memory traces. Understanding the complexities of translational control will allow for the molecular dissection of long-lasting plasticity mechanisms and eventual mnemonic processes.
Acknowledgments
We are grateful to Michael Sutton for comments on this manuscript, and we apologize to those whose data were not discussed due to space limitations. This work was supported by grants from Canadian Institute of Health (CIHR) to N.S., W.S.S., and M.C.-M. and National Institute of Health (NIH) to E.K. N.S. is a Howard Hughes Medical Institute (HHMI) International scholar.
References
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion MD, Hou L, Wong H, Hoeffer CA, Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008a;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion MD, Merhav M, Hoeffer CA, Reis G, Kozma SC, Thomas G, Schuman EM, Rosenblum K, Klann E. Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn Mem. 2008b;15:29–38. doi: 10.1101/lm.661908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquier N, Bourouis M, Colombani J, Leopold P. Drosophila Lk6 kinase controls phosphorylation of eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr Biol. 2005;15:19–23. doi: 10.1016/j.cub.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Cunningham BA, Edelman GM, Vanderklish PW. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc Natl Acad Sci USA. 2005;102:2180–2185. doi: 10.1073/pnas.0409803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008;105:E19. doi: 10.1073/pnas.0801034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Klann E. NMDA receptor activation results in PKA- and ERK-dependent Mnk1 activation and increased eIF4E phosphorylation in hippocampal area CA1. J Neurochem. 2004;91:462–470. doi: 10.1111/j.1471-4159.2004.02734.x. [DOI] [PubMed] [Google Scholar]
- Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25:9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Merhav M, Stern E, Sonenberg N, Rosenblum K, Klann E. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiol Learn Mem. 2007;87:248–256. doi: 10.1016/j.nlm.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Bradshaw KD, Emptage NJ, Bliss TV. A role for dendritic protein synthesis in hippocampal late LTP. Eur J Neurosci. 2003;18:3150–3152. doi: 10.1111/j.1460-9568.2003.03054.x. [DOI] [PubMed] [Google Scholar]
- Brendler T, Godefroy-Colburn T, Yu S, Thach RE. The role of mRNA competition in regulating translation. III Comparison of in vitro and in vivo results. J Biol Chem. 1981;256:11755–11761. [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA. 2003;100:14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M, Warren O, Fan X, Sossin WS. 5-HT stimulates eEF2 dephosphorylation in a rapamycin-sensitive manner in Aplysia neurites. J Neurochem. 2004;90:1464–1476. doi: 10.1111/j.1471-4159.2004.02634.x. [DOI] [PubMed] [Google Scholar]
- Carroll M, Dyer J, Sossin WS. Serotonin increases phosphorylation of synaptic 4EBP through TOR, but eukaryotic initiation factor 4E levels do not limit somatic cap-dependent translation in aplysia neurons. Mol Cell Biol. 2006;26:8586–8598. doi: 10.1128/MCB.00955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey CH, Kandel ER. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–237. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- Chotiner JK, Khorasani H, Nairn AC, O’Dell TJ, Watson JB. Adenylyl cyclase-dependent form of chemical long-term potentiation triggers translational regulation at the elongation step. Neuroscience. 2003;116:743–752. doi: 10.1016/s0306-4522(02)00797-2. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N. Chapter 5 Translational control of gene expression: A molecular switch for memory storage. Prog Brain Res. 2008;169:81–95. doi: 10.1016/S0079-6123(07)00005-2. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007a;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N, Klann E. Translational control mechanism of synaptic plasticity and memory. In: Byrne JH, editor. Learning and Memory: A Comprehensive Reference. Elsevier; 2007b. pp. 675–694. [Google Scholar]
- Cracco JB, Serrano P, Moskowitz SI, Bergold PJ, Sacktor TC. Protein synthesis-dependent LTP in isolated dendrites of CA1 pyramidal cells. Hippocampus. 2005;15:551–556. doi: 10.1002/hipo.20078. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Dash PK, Orsi SA, Moore AN. Spatial memory formation and memory-enhancing effect of glucose involves activation of the tuberous sclerosis complex-Mammalian target of rapamycin pathway. J Neurosci. 2006;26:8048–8056. doi: 10.1523/JNEUROSCI.0671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27:13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGracia DJ, Sullivan JM, Neumar RW, Alousi SS, Hikade KR, Pittman JE, White BC, Rafols JA, Krause GS. Effect of brain ischemia and reperfusion on the localization of phosphorylated eukaryotic initiation factor 2 alpha. J Cereb Blood Flow Metab. 1997;17:1291–1302. doi: 10.1097/00004647-199712000-00004. [DOI] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Dever TE, Dar AC, Sicheri F. The eIF2alpha kinases. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 319–345. [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Pan D. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 2004;18:2479–2484. doi: 10.1101/gad.1240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Sarnow P. Translation initiation by viral Internal Ribosome Entry Sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 129–154. [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Sossin WS. Regulation of eukaryotic initiation factor 4E phosphorylation in the nervous system of Aplysia californica. J Neurochem. 2000;75:872–881. doi: 10.1046/j.1471-4159.2000.0750872.x. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Michel S, Lee W, Castellucci VF, Wayne NL, Sossin WS. An activity-dependent switch to cap-independent translation triggered by eIF4E dephoshphorylation. Nat Neurosci. 2003;6:219–220. doi: 10.1038/nn1018. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Miyashiro K, Kacharmina JE, Job C. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc Natl Acad Sci USA. 2001;98:7080–7085. doi: 10.1073/pnas.121146698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2(+/−) mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 155–172. [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Frodin M, Jensen CJ, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of {alpha} calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K, Smith KT, Reines D, Warren ST. Transcription, translation and fragile X syndrome. Curr Opin Genet Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Gelinas JN, Nguyen PV. Beta-adrenergic receptor activation facilitates induction of a protein synthesis-dependent late phase of long-term potentiation. J Neurosci. 2005;25:3294–3303. doi: 10.1523/JNEUROSCI.4175-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999a;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999b;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci USA. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert D, Topolnik L, Azzi M, Huang L, Badeaux F, Desgroseillers L, Sossin WS, Lacaille JC. Forskolin induction of late-LTP and up-regulation of 5′ TOP mRNAs translation via mTOR, ERK, and PI3K in hippocampal pyramidal cells. J Neurochem. 2008;106:1160–1174. doi: 10.1111/j.1471-4159.2008.05470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godefroy-Colburn T, Thach RE. The role of mRNA competition in regulating translation. IV Kinetic model. J Biol Chem. 1981;256:11762–11773. [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- Grolleau A, Bowman J, Pradet-Balade B, Puravs E, Hanash S, Garcia-Sanz JA, Beretta L. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J Biol Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, Gietzen DW. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Molecular biology. miRNAs in neuro-degeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- Herbert TP, Proud CG. Regulation of translation elongation and the cotranslationa protein target pathway. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 601–624. [Google Scholar]
- Hinnebusch AG. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey JW, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. pp. 185–244. [Google Scholar]
- Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Asano K, Olsen DS, Phan L, Nielsen KH, Valasek L. Study of translational control of eukaryotic gene expression using yeast. Ann N Y Acad Sci. 2004;1038:60–74. doi: 10.1196/annals.1315.012. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Dever TE, Asano K. Mechanism of translation initiation in yeast Saccharomycies cerevisiae. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 225–268. [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Hu JY, Glickman L, Wu F, Schacher S. Serotonin regulates the secretion and autocrine action of a neuropeptide to activate MAPK required for long-term facilitation in Aplysia. Neuron. 2004;43:373–385. doi: 10.1016/j.neuron.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Hu JY, Wu F, Schacher S. Two signaling pathways regulate the expression and secretion of a neuropeptide required for long-term facilitation in Aplysia. J Neurosci. 2006;26:1026–1035. doi: 10.1523/JNEUROSCI.4258-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Chen Y, Schacher S. Protein kinase C regulates local synthesis and secretion of a neuropeptide required for activity-dependent long-term synaptic plasticity. J Neurosci. 2007;27:8927–8939. doi: 10.1523/JNEUROSCI.2322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. The mRNA for elongation factor 1alpha is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci. 2005;25:7199–7209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008a;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. Reply to Bagni: On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008b;105:E29. doi: 10.1073/pnas.0803737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- Jung MY, Lorenz L, Richter JD. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol. 2006;26:4277–4287. doi: 10.1128/MCB.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Karpova A, Mikhaylova M, Thomas U, Knopfel T, Behnisch T. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. J Neurosci. 2006;26:4949–4955. doi: 10.1523/JNEUROSCI.4573-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004a;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004b;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kelly MT, Crary JF, Sacktor TC. Regulation of protein kinase Mzeta synthesis by multiple kinases in long-term potentiation. J Neurosci. 2007;27:3439–3444. doi: 10.1523/JNEUROSCI.5612-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Pepio AM, Sossin WS. Serotonin activates S6 kinase in a rapamycin-sensitive manner in Aplysia synaptosomes. J Neurosci. 2001;21:382–391. doi: 10.1523/JNEUROSCI.21-02-00382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Klann E, Antion MD, Banko JL, Hou L. Synaptic plasticity and translation initiation. Learn Mem. 2004;11:365–372. doi: 10.1101/lm.79004. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille JC. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol. 2008;28:2896–2907. doi: 10.1128/MCB.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Pilotte J, Xu T, Wong CC, Edelman GM, Vanderklish P, Yates JR., 3rd BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: an analysis using high-throughput proteomics. J Proteome Res. 2007;6:1059–1071. doi: 10.1021/pr060358f. [DOI] [PubMed] [Google Scholar]
- Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Hu JY, Wang D, Schacher S. Protein synthesis at synapse versus cell body: enhanced but transient expression of long-term facilitation at isolated synapses. J Neurobiol. 2003;56:275–286. doi: 10.1002/neu.10242. [DOI] [PubMed] [Google Scholar]
- Liu J, Hu JY, Wu F, Schwartz JH, Schacher S. Two mRNA-binding proteins regulate the distribution of syntaxin mRNA in Aplysia sensory neurons. J Neurosci. 2006;26:5204–5214. doi: 10.1523/JNEUROSCI.4917-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Nat Acad Sci. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA. Long-term potentiation—a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, Ron D, Fafournoux P. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005;1:273–277. doi: 10.1016/j.cmet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- McEvoy M, Cao G, Llopis PM, Kundel M, Jones K, Hofler C, Shin C, Wells DG. Cytoplasmic polyadenylation element binding protein 1-mediated mRNA translation in Purkinje neurons is required for cerebellar long-term depression and motor coordination. J Neurosci. 2007;27:6400–6411. doi: 10.1523/JNEUROSCI.5211-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. Memory—a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER, Bailey CH. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E, EY, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley SJ, Naegele S. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J Biol Chem. 2002;277:32855–32859. doi: 10.1074/jbc.C200376200. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates FMRP with the neuronal protein synthesis-dependent mTOR signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- Oshiro N, Yoshino K, Hidayat S, Tokunaga C, Hara K, Eguchi S, Avruch J, Yonezawa K. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- Osten P, Valsamis L, Harris A, Sacktor TC. Protein synthesis-dependent formation of protein kinase Mzeta in long-term potentiation. J Neurosci. 1996;16:2444–2451. doi: 10.1523/JNEUROSCI.16-08-02444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Rosenstein A, Kreiman G, Schuman EM, Kennedy MB. Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J Neurosci. 1999;19:7823–7833. doi: 10.1523/JNEUROSCI.19-18-07823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Woodruff E, 3rd, Liang P, Broadie K. Mechanistic relationships between Drosophila fragile X mental retardation protein and metabotropic glutamate receptor A signaling. Mol Cell Neurosci. 2008;37:747–760. doi: 10.1016/j.mcn.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, Lu B. Cleavage of proBDNF by tPA/ plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006;313:1141–1144. doi: 10.1126/science.1128657. [DOI] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pereira CM, Sattlegger E, Jiang HY, Longo BM, Jaqueta CB, Hinnebusch AG, Wek RC, Mello LE, Castilho BA. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J Biol Chem. 2005;280:28316–28323. doi: 10.1074/jbc.M408571200. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Doepfner KT, Hafen E, Stocker H. Diet-dependent effects of the Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr Biol. 2005;15:24–30. doi: 10.1016/j.cub.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Ron D, Harding HP. eIF2alpha phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 345–368. [Google Scholar]
- Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross G, Dyer JR, Castellucci VF, Sossin WS. Mnk is a negative regulator of cap-dependent translation in Aplysia neurons. J Neurochem. 2006;97:79–91. doi: 10.1111/j.1471-4159.2006.03704.x. [DOI] [PubMed] [Google Scholar]
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sangha S, Scheibenstock A, McComb C, Lukowiak K. Intermediate and long-term memories of associative learning are differentially affected by transcription versus translation blockers in Lymnaea. J Exp Biol. 2003;206:1605–1613. doi: 10.1242/jeb.00301. [DOI] [PubMed] [Google Scholar]
- Santoyo J, Alcalde J, Mendez R, Pulido D, de Haro C. Cloning and characterization of a cDNA encoding a protein synthesis initiation factor-2alpha (eIF-2alpha) kinase from Drosophila melanogaster. Homology to yeast GCN2 protein kinase. J Biol Chem. 1997;272:12544–12550. doi: 10.1074/jbc.272.19.12544. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of micro-RNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem. 2002;277:3303–3309. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- Shema R, Sacktor TC, Dudai Y. Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKM zeta. Science. 2007;317:951–953. doi: 10.1126/science.1144334. [DOI] [PubMed] [Google Scholar]
- Sherff CM, Carew TJ. Parallel somatic and synaptic processing in the induction of intermediate-term and long-term synaptic facilitation in Aplysia. Proc Natl Acad Sci USA. 2004;101:7463–7468. doi: 10.1073/pnas.0402163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Shinkura K, Tokunaga M. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J Neurosci. 2005;25:4420–4434. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Steward O, Bakker CE, Willems PJ, Oostra BA. No evidence for disruption of normal patterns of mRNA localization in dendrites or dendritic transport of recently synthesized mRNA in FMR1 knockout mice, a model for human fragile-X mental retardation syndrome. Neuroreport. 1998a;9:477–481. doi: 10.1097/00001756-199802160-00022. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998b;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Masters SE, Bagnall MW, Carew TJ. Molecular mechanisms underlying a unique intermediate phase of memory in aplysia. Neuron. 2001;31:143–154. doi: 10.1016/s0896-6273(01)00342-7. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J Biol Chem. 2001;276:42818–42825. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischmeyer W, Schicknick H, Kraus M, Seidenbecher CI, Staak S, Scheich H, Gundelfinger ED. Rapamycin-sensitive signalling in long-term consolidation of auditory cortex-dependent memory. Eur J Neurosci. 2003;18:942–950. doi: 10.1046/j.1460-9568.2003.02820.x. [DOI] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Ma T, Iyengar R, Landau EM, Blitzer RD. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J Neurosci. 2007;27:5885–5894. doi: 10.1523/JNEUROSCI.4548-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Turner G, Webb T, Wake S, Robinson H. Prevalence of fragile X syndrome. Am J Med Genet. 1996;64:196–197. doi: 10.1002/(SICI)1096-8628(19960712)64:1<196::AID-AJMG35>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers CA, Dickson KS, Wyllie DJ. Induction and maintenance of late-phase long-term potentiation in isolated dendrites of rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;568:803–813. doi: 10.1113/jphysiol.2005.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal G, Li Q, Cai D, Glanzman DL. The role of rapid, local, postsynaptic protein synthesis in learning-related synaptic facilitation in aplysia. Curr Biol. 2007;17:2073–2080. doi: 10.1016/j.cub.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk LJ, Pfeiffer BE, Gibson JR, Huber KM. Multiple Gq-coupled receptors converge on a common protein synthesis-dependent long-term depression that is affected in fragile X syndrome mental retardation. J Neurosci. 2007;27:11624–11634. doi: 10.1523/JNEUROSCI.2266-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Godefroy-Colburn T, Thach RE. The role of mRNA competition in regulating translation. I Demonstration of competition in vivo. J Biol Chem. 1981;256:11739–11746. [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen SG, Gingras AC, Amankwa L, Mader S, Branton PE, Aebersold R, Sonenberg N. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J Biol Chem. 1996;271:11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- Wu J, Bag J. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J Biol Chem. 1998;273:34535–34542. doi: 10.1074/jbc.273.51.34535. [DOI] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Yan QJ, Rammal M, Tranfaglia M, Bauchwitz RP. Suppression of two major Fragile X Syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology. 2005;49:1053–1066. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Yanow SK, Manseau F, Hislop J, Castellucci VF, Sossin WS. Biochemical pathways by which serotonin regulates translation in the nervous system of Aplysia. J Neurochem. 1998;70:572–583. doi: 10.1046/j.1471-4159.1998.70020572.x. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- Zakharenko SS, Patterson SL, Dragatsis I, Zeitlin SO, Siegelbaum SA, Kandel ER, Morozov A. Presynaptic BDNF required for a presynaptic but not postsynaptic component of LTP at hippocampal CA1–CA3 synapses. Neuron. 2003;39:975–990. doi: 10.1016/s0896-6273(03)00543-9. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Zuberek J, Wyslouch-Cieszynska A, Niedzwiecka A, Dadlez M, Stepinski J, Augustyniak W, Gingras AC, Zhang Z, Burley SK, Sonenberg N, et al. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003;9:52–61. doi: 10.1261/rna.2133403. [DOI] [PMC free article] [PubMed] [Google Scholar]