Abstract
Isolated in vitro more than half a century ago, the H37Rv strain of Mycobacterium tuberculosis still remains the strain of choice for the majority of laboratories conducting in vivo studies of TB pathogenesis. In this report we reveal that H37Rv is highly prone to losing the ability to synthesize the cell wall lipid phthiocerol dimycocerosate (PDIM) during extended periods of in vitro culture. In addition, H37Rv stocks that have been held in vitro for even a short length of time should be thought of as a heterogeneous population of PDIM-positive and PDIM-negative cell types. We demonstrate that after weekly subculture of PDIM-positive isolates over a period of 20 weeks, the proportion of PDIM-negative cells rises above 30 %. That PDIM biosynthesis is negatively selected in vitro is evident from the broad range of mutation types we observe within cultures originating from a single PDIM-positive parental clone. Moreover, the appearance of these multiple mutation types coupled with an enhanced growth rate of PDIM-negative bacteria ensures that ‘PDIM-less’ clones rapidly dominate in vitro cultures. It has been known for almost a decade that strains of M. tuberculosis that lack PDIM are severely attenuated during in vivo infection. Therefore, the loss of PDIM raises a very serious issue in regard to the interpretation of putative virulence factors where heterogeneous parental cultures are potentially being compared in vivo to recombinant clones isolated within a PDIM-negative background. It is essential that researchers undertaking in vivo virulence studies confirm the presence of PDIM within all recombinant clones and the parental strains they are derived from.
INTRODUCTION
Despite the availability of effective chemotherapy and a partially effective vaccine for more than half a century, there are still around two million human deaths each year due to infection with Mycobacterium tuberculosis, the bacterial agent responsible for tuberculosis (TB) (World Health Organization, 2006). The effects of this devastating disease are exacerbated in many regions of the world as a result of malnutrition, HIV co-infection and multiple drug resistance (Dye, 2006). Encouragingly, there have been some tremendous advances in our knowledge of the genetics underlying M. tuberculosis metabolic pathways and the biosynthesis of potential virulence-associated molecules in the decade since the publication of the complete genome sequence of M. tuberculosis (Cole et al., 1998). The challenge now is to try to translate these ‘post-genomic’ findings into measurable improvements in terms of diagnosis, treatment and prevention of TB disease.
To date, the overwhelming majority of studies aimed at deciphering the molecular and biochemical basis of M. tuberculosis pathogenesis at both the in vitro and in vivo (macrophage or animal model) level have involved the ubiquitous H37Rv strain. The progenitor of H37Rv (known as H37) was isolated in 1905 from the sputum of a patient with chronic pulmonary TB and following an extended period of laboratory selection was ‘dissociated’ into avirulent (H37Ra) and virulent (H37Rv) forms (Steenken et al., 1934; Steenken, 1950). Due to its widespread distribution in laboratories around the world and a long history of use in a variety of biochemical and virulence assays, the H37Rv strain was the obvious choice for the original TB genome-sequencing project (Cole et al., 1998). To date, it has also been the strain of choice for the majority of targeted mutagenesis studies, which have involved both plasmid and phage-based technologies. H37Rv was also employed in the in vitro and in vivo gene essentiality screens based on the TraSH (transposon site hybridization) technique (Rengarajan et al., 2005; Sassetti et al., 2003; Sassetti & Rubin, 2003). The results of the latter are frequently consulted in the context of identifying potential drug targets for future TB drug development programmes (Murphy & Brown, 2007).
One of the first examples of the application of a ‘post-genomic’ approach to investigating M. tuberculosis pathogenesis was the demonstration that the wax-like compound phthiocerol dimycocerosate (PDIM) was necessary for full virulence in the mouse model of tuberculosis (Camacho et al., 1999; Cox et al., 1999). This cell-wall-associated lipid is composed of a long-chain β-diol (phthiocerol) esterified to two multi-methyl branched fatty acids (mycocerosic acids). At least three independent studies have confirmed that M. tuberculosis mutants deficient in PDIM synthesis or translocation are severely attenuated in vivo (Camacho et al., 1999; Cox et al., 1999; Domenech et al., 2005). None of these studies have reported an in vitro defect associated with the loss of PDIM. The relative importance of this lipid to the biology of M. tuberculosis infection can also be gleaned from the fact that approximately 50 kb (>1 %) of the genome is dedicated to the biosynthesis and transport of this complex molecule (Cole et al., 1998; Onwueme et al., 2005). Although there are publications pointing to a role for PDIM in TB pathogenesis that go back to the early 1970s (Goren et al., 1974), its precise mode of action is still not entirely clear. Recent articles have suggested multiple roles ranging from host plasma membrane modification (Astarie-Dequeker et al., 2009) to innate immune modulation (Rousseau et al., 2004) and protection of the bacteria from host-derived reactive nitrogen intermediates (Rousseau et al., 2004).
In spite of the obvious advantages to using H37Rv for laboratory-based virulence assays (sequence availability, drug sensitivity, amenability to genetic manipulation, etc.), during our previous work on polyketide synthase (PKS) biosynthetic pathways and putative lipid-transport proteins in M. tuberculosis (Domenech et al., 2004, 2005; Reed et al., 2004) we observed that a disproportionately high number of gene-replacement mutants that we had generated in the H37Rv background were completely devoid of PDIM. This was despite the fact that we were targeting genes in pathways that were not predicted to have an impact on PDIM biosynthesis. Moreover, these same mutants constructed in strains other than H37Rv retained the ability to synthesize PDIM (authors’ unpublished observations). Indeed, there are even a few recorded instances in the literature where independent laboratories have published discordant results with respect to the proposed involvement of particular genes in PDIM biosynthesis (Matsunaga et al., 2004; Saxena et al., 2003; Sirakova et al., 2003; Waddell et al., 2005). In hindsight, it seems likely that the loss of PDIM in these instances was unrelated to the pathways being studied. Mayer Goren and colleagues first noted that H37Rv had a propensity for losing the ability to synthesize PDIM during in vitro culture back in 1974 (Goren et al., 1974). That this phenomenon occurs relatively frequently is evident from three recent publications that document the loss of PDIM from H37Rv (Andreu & Gibert, 2008; Kana et al., 2008; Manjunatha et al., 2006).
If we accept that (a) PDIM appears to be lost during in vitro culture with some regularity, (b) the loss of PDIM is associated with attenuation in vivo, and (3) the majority of researchers carrying out in vivo pathogenesis studies of mutants derived from H37Rv (or other M. tuberculosis strains) do not routinely screen for the presence of PDIM, then there is every likelihood that at least a portion of virulence-attenuating mutations described thus far should have been attributed to the loss of PDIM rather than to the particular genes/pathways under investigation. This possibility prompted us to carry out the present study aimed at identifying the mechanisms underlying the loss of PDIM synthesis, and the frequency with which this occurs for two H37Rv lines (ATCC and Pasteur) and two recent clinical isolates. We also wanted to investigate whether transformation and/or drug selection procedures might enhance the selection of PDIM-deficient mutants in the laboratory. Our data indicate that the loss of PDIM occurs spontaneously within liquid culture and can involve multiple mechanisms, including gene deletion and regulatory mutations. Shortly thereafter, these PDIM-negative clones are able to dominate within a mixed cell population due to a slight enhancement in their in vitro growth rate as compared to PDIM-positive clones. The potential implications of these findings for researchers carrying out in vivo virulence studies employing H37Rv are discussed.
METHODS
Bacterial strains and in vitro culture
Two independent stocks of M. tuberculosis H37Rv were originally obtained from the American type Culture Collection (ATCC 27294) and the Institut Pasteur (Paris, France) and are referred to herein as H37Rv-ATCC and H37Rv-Pasteur, respectively. H37Rv-Pasteur was the strain used in the M. tuberculosis genome-sequencing project of Cole et al. (1998). In addition, two recent clinical isolates belonging to the East-Asian, or W/Beijing, lineage were included in this study (Gagneux & Small, 2007). One, referred to here as B-4, was kindly provided by Dr Sebastien Gagneux (NIMR, Mill Hill, UK). The other, HN878, was originally obtained from Dr James Musser (Methodist Hospital Research Institute, Houston, TX, USA) (Sreevatsan et al., 1997). All strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 10 % ADC [NaCl, 8.1 g l−1; BSA Fraction V (Calbiochem), 50 g l−1; glucose, 20 g l−1], 0.2 % glycerol and 0.05 % Tween 80, or on Middlebrook 7H11 agar (Difco) supplemented with 10 % OADC enrichment (as ADC plus 0.6 ml oleic acid l−1 and 3.6 mM NaOH). Kanamycin (25 μg l−1) was included where indicated. All chemicals were supplied by Sigma-Aldrich unless otherwise noted. For the in vitro growth curves, bacterial cultures were adjusted to an OD600 of 0.1, diluted 1 : 100 in 7H9 and allowed to grow for 12 days at 37 °C with constant rolling. On days 9 and 12, cultures were diluted 1 : 3 prior to measuring the OD600. The OD600 values of the undiluted cultures were obtained by multiplying these values by a factor of 3. To confirm that the loss of PDIM does not lead to a significant alteration in OD600, c.f.u. were determined for each culture on day 0.
Lipid analysis
Apolar lipid extraction and analysis was carried out essentially as described by Slayden & Barry (2001). Individual colonies were grown at 37 °C in 1.5 ml 7H9/ADC (in 5 ml tubes with a 2-position ventilation cap; Sarstedt) with occasional shaking for 3–4 weeks. Then 100 μl aliquots of these cultures were added to 1 ml 7H9/ADC (in 4 ml cryovials; Simport) containing 0.1 μCi (3.7 kBq) ml−1 [1-14C]propionic acid (3.7×103 Bq ml−1; American Radiolabelled Chemicals) and incubated at 37 °C with continuous rolling for 7 days prior to lipid extraction. Following centrifugation, the bacterial pellets were resuspended in 600 μl methanol/0.3 % aq. NaCl (10 : 1, v/v) to which 300 μl petroleum ether was added. After 10 min of vigorous mixing, the samples were centrifuged and the upper (petroleum ether) phase containing the apolar lipid fraction was removed and dried. Lipid samples were resuspended in 25 μl petroleum ether and 5 μl aliquots were spotted onto TLC plates (250 μm silica gel 60 plates; EM Science). Plates were run three times in petroleum ether/ethyl acetate (98 : 2, v/v) for the detection of PDIM, or once in chloroform/methanol/water (100 : 14 : 0.8, by vol.) for sulfolipid-1 detection. TLC plates were visualized using a Storm 840 PhosphorImager (GE Healthcare).
General nucleic acid techniques
Electrocompetent M. tuberculosis were prepared as described by Snapper et al. (1990) and transformed with the pMV361 integrative vector containing a kanamycin-resistance marker (Stover et al., 1991). Mycobacterial DNA was isolated according to Pelicic et al. (1997) and total RNA was purified from cultures grown to OD600 0.2 using the method of Sherman et al. (2001). Residual genomic DNA was removed from RNA samples via treatment with RNase-free DNase I (Fermentas). cDNA for qRT-PCR analysis was generated using RevertAid M-MuLV Reverse Transcriptase (Fermentas) together with random hexamer oligonu-cleotide primers (Invitrogen).
Microarray analysis
Whole-genome microarray analysis was carried out for both genomic DNA and RNA according to established protocols (Mostowy et al., 2004) with some minor modifications. A 2 μg sample of purified genomic DNA was directly labelled with Cy3 or Cy5-dCTP (GE Healthcare) using DNA polymerase I Klenow fragment and a mixture of 6-mer and 9-mer random primers (New England Biolabs). Four-microgram samples of RNA were used in the preparation of cDNA labelled with either Cy3 or Cy5-NHS esters (GE Healthcare) via the aminoallyl indirect labelling method (Yu et al., 2002) in the presence of RevertAid M-MuLV Reverse Transcriptase (Fermentas). Labelled genomic DNA and cDNA was subsequently purified using the QIAquick PCR Purification kit (Qiagen), and samples to be co-hybridized on the same array were mixed and concentrated to 6 μl in a Savant SpeedVac Concentrator. The M. tuberculosis whole-genome microarrays used in these experiments were kindly provided by Dr M. Behr (McGill University). These arrays are composed of 70 bp oligonucleotides (TB Array-Ready Oligo Set, Operon) printed in duplicate as sixteen 20 (rows)×21 (columns) grids on Sigmascreen microarray slides (Sigma). A Virtek Chipwriter (model SDDC2; Virtek) was used for printing the arrays. Microarray hybridization was carried out in GE Healthcare hybridization buffer in the presence of yeast tRNA (0.2 μg ml−1) at 42 °C for 16 h. Following hybridization, microarray slides were washed three times (15 min per wash) with 0.5× SSC+0.01 % SDS, 0.06× SSC+0.01 % SDS and 0.06× SSC. The first and second washes were performed at 37 °C, while the last was at room temperature.
Microarrays were scanned using a confocal laser scanner (ScanArray Lite; Packard) and fluorescence intensities were quantified with ScanArray Express software (Packard). Mean fluorescence values for each spot, minus the surrounding background, were used in the analysis. Spots flagged as misrepresentative or those with a negative value following background correction were excluded from further analysis. The fluorescence intensity ratios (Cy3/Cy5 or Cy5/Cy3) were calculated for each spot, log10 transformed and then normalized with respect to the mean value of all fluorescence ratios. A Z-score [Z=(log10 fluorescence ratio−mean)/standard deviation] was calculated for each spot and the Z-scores for each gene were averaged across all replicates. For genomic DNA comparisons, two independent arrays were analysed for each pair of samples (Cy3/Cy5 and Cy5/ Cy3). In the same manner, two independent biological replicates were analysed for each M. tuberculosis strain for the RNA expression analysis (a total of 2–4 arrays for each experiment). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession numbers GSE17232 and GSE17234 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17232; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17234).
Competition experiments
In vitro growth competition experiments were carried out for H37Rv-ATCC isolate 8 (PDIM+) and isolate 3 (PDIM−). Cultures (OD650 0.4) of both isolates were adjusted to OD600 0.1 and mixed in a 1 : 1 ratio. This ‘starter’ culture was diluted 1 : 100 and incubated at 37 °C in a rolling incubator. At days 7 and 14 the culture was diluted 1/1000 and incubated at 37 °C for an additional week. Quantification of both isolates within the mixed culture was performed by qRT-PCR. At days 7, 14 and 21, 10 ml of the culture was used in the purification of genomic DNA. At day 0, 10 ml of the starter culture was used to quantify the initial starting ratio of both isolates. Two independent biological replicates were included in these competition studies.
Quantitative real-time PCR (qRT-PCR)
Determination of the relative proportion of each strain within the competition experiments was accomplished by qRT-PCR using two sets of primers. Primers specific for the hspX gene (hspX-F, 5′-GAATTCGCGTACGG-TTCCTTC-3′, and hspX-R, 5′-TGTCGTCCTCGTCAGCACC-3′) were used for the relative quantification of the total bacterial population within the growing culture. Primers specific for ppsD (ppsD-A, 5′-GCGGATCGCGTATCTGTTG-3′, and ppsD-B, 5′-TCTGACACGCCAAGTGAATTG-3′) allowed us to quantify the proportion of bacteria that were PDIM positive due to the fact that ppsD is deleted in H37Rv-ATCC isolate 3 (PDIM−). Primers specific for sigA (sigA1-F, 5′-TCGCGCCTAC CTCAAACAG-3′, and sigA1-R, 5′-CGTACAGGCCAGCCTCGAT-3′) were used as an endogenous control to normalize the amount of DNA template added to each qRT-PCR sample. qRT-PCRs were carried out in triplicate using a 7300 Real-Time PCR System (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems). The relative standard curve method was used for quantification. Standard deviations (SD) were calculated according to the formula SD=(cv)(X), as specified by Applied Biosystems (Applied Biosystems, 2005). X is the ratio of the mean values of the nominator (n) and denominator (d). cv is the coefficient of variation (standard deviation normalized to the mean values of each gene) calculated from the formula cv= √[(cvn)2+(cvd)2].
Transcriptional profile differences identified via microarray were confirmed by qRT-PCR of cDNA samples using primers specific for the ppsA gene (ppsA-A, 5′-CCAAATCAGCACTTCGAAACC-3′, and ppsA-B, 5′-GGCCATTCAGTTTGTGTGTCA-3′).
RESULTS
PDIM-deficient mutants arise spontaneously in vitro in the absence of selective pressure
Prior to the current study, two publications in the last 30 years had reported that M. tuberculosis H37Rv could spontaneously lose the ability to synthesize PDIM during in vitro culture (Goren et al., 1974; Manjunatha et al., 2006). The relative frequency with which we were observing PDIM-negative transformants for H37Rv (unpublished observations) had also led us to speculate that routine techniques such as electro-transformation (electroporation) and drug selection may have been selecting for PDIM-deficient clones, in a manner analogous to the highly transformable mc2155 variant of Mycobacterium smegmatis (Etienne et al., 2005; Snapper et al., 1990). Due to the tremendous implications that the loss of PDIM has for M. tuberculosis virulence and pathogenesis studies, we felt it was necessary to understand the range of conditions and the frequency with which the loss of PDIM occurs.
We began by analysing the PDIM content of individual clones derived from two distinct H37Rv strains (H37Rv-ATCC, H37Rv-Pasteur) as well as a more recent clinical isolate, HN878, grown under four different experimental conditions. Both the ATCC and Pasteur lines of H37Rv were included in the study because of genetic evidence indicating that, at least to some degree, they should be thought of as distinct strains (Dubey et al., 2002). Standard in vitro exponential-phase cultures were plated at a range of dilutions onto either 7H11 plates (‘no treatment’) or 7H11 plates containing kanamycin (Kan) for the selection of spontaneous KanR mutants. The same cultures were used in the preparation of electrocompetent cells for transformation with the pMV361 (KanR) integrative vector and plated onto 7H11/Kan (‘pMV361’). Lastly, the H37Rv cultures were also electroporated in the absence of any plasmid DNA and plated onto 7H11 (‘no plasmid’) or 7H11/Kan. With these four sets of conditions we evaluated the impact of antibiotic selection, electroporation and a combination of the two (antibiotic selection following plasmid transformation) on the isolation of PDIM-deficient mutants. Individual colonies from each treatment group (30–68 clones per group) were subcultured in 7H9 broth and metabolically labelled with [14C]propionic acid. The apolar lipid fraction was then extracted and assayed for PDIM via TLC. As indicated in Table 1, PDIM-deficient mutants were detected for all three M. tuberculosis strains under each of the conditions tested. Relative to the untreated controls, pairwise comparisons (Fisher’s exact test, GraphPad Prism v4.0) revealed no statistically significant effect of any of the treatments on the appearance of PDIM-negative clones. As such, we conclude that the loss of PDIM occurs spontaneously as a consequence of in vitro culture, rather than due to any selective process encountered during plasmid transformation and drug selection.
Table 1.
Effect of antibiotic selection and electro-transformation on PDIM content in vitro
| M. tuberculosis strain | No treatment**
|
Spontaneous KanR mutants**
|
||||||
|---|---|---|---|---|---|---|---|---|
| PDIM-positive | PDIM-negative | Total | % Negative | PDIM-positive | PDIM-negative | Total | % Negative | |
| H37Rv-ATCC (27294) | 14 | 54 | 68 | 79 | 13 | 34 | 47 | 72 |
| H37Rv-Pasteur | 47 | 1 | 48 | 2 | 47 | 1 | 48 | 2 |
| HN878 | 31 | 4 | 35 | 11 | 46 | 1 | 47 | 2 |
|
| ||||||||
| pMV361**
|
No plasmid*
|
|||||||
| PDIM-positive | PDIM-negative | Total | % Negative | PDIM-positive | PDIM-negative | Total | % Negative | |
|
| ||||||||
| H37Rv-ATCC (27294) | 14 | 22 | 36 | 61 | 5 | 27 | 32 | 84 |
| H37Rv-Pasteur | 28 | 4 | 32 | 12 | 22 | 2 | 24 | 8 |
| HN878 | 26 | 4 | 30 | 13 | ND | ND | ND | ND |
Results are from 1 (*) or 2 (**) independent experiments for each strain; ND, not determined.
Whilst we can only assume that our bacterial stocks are representative of those held in most other laboratories, our results suggest that any in vitro M. tuberculosis culture could potentially consist of a heterogenous mix of PDIM-positive and PDIM-negative clones. The exact ratio of the two is likely to change according to variations in culture conditions, length of time in culture, number of serial passages and the M. tuberculosis strain being used. Perhaps the most striking observation is that on average, 74 % of all H37Rv-ATCC colonies analysed in these experiments (n=183) were PDIM negative. It is not surprising, therefore, that we previously had some difficulty in isolating recombinants in this strain background that had retained PDIM. It should be noted that all efforts were made in the preceding experiments to use bacterial stocks that had been subject to the least amount of in vitro passage since obtaining them from their original source.
Loss of PDIM is the consequence of mutations accumulated during in vitro passage
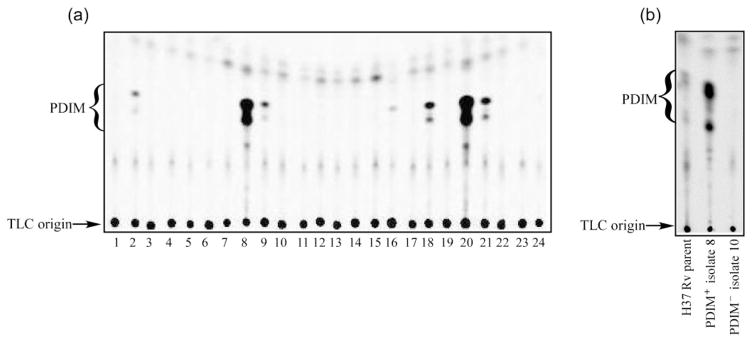
Fig. 1(a) shows a representative TLC of 24 colonies isolated from the H37Rv-ATCC strain (Fig. 1b) after plating directly from an exponential-phase culture (‘no treatment’). Aside from the large percentage of colonies that lack detectable PDIM, there also appears to be variation amongst the PDIM-positive clones with respect to the amount of PDIM that is produced. From Fig. 1(a), isolates 8 and 20 show the greatest PDIM production whereas isolates 2, 9, 16, 18 and 21 are much weaker producers. We have also observed this variable PDIM production for the H37Rv-Pasteur and HN878 strains (data not shown). Although we continue to refer to these isolates in this study as being ‘weakly PDIM positive’ or ‘PDIM+/−’, it is conceivable that this phenotype could result from PDIM-positive and PDIM-negative cells forming microscopic aggregates (‘clumps’) in liquid culture that are not dispersed upon plating. In turn, the amount of PDIM extracted from subcultures derived from these mixed colonies would ultimately reflect the proportion of PDIM-positive and -negative cell types present.
Fig. 1.
PDIM-deficient mutants arise spontaneously in vitro in the absence of additional selection pressure. (a) TLC of apolar lipid extracts prepared for 24 individual clones derived from an untreated M. tuberculosis H37Rv (ATCC 27294) liquid culture. Cultures were radiolabelled with [1-14C]propionic acid prior to extraction and the TLC was developed three times in petroleum ether/ethyl acetate (98 : 2, v/v). (b) TLC of apolar lipid extracts prepared from the parental H37Rv (ATCC 27294) stock and representative PDIM-positive (no. 8) and PDIM-negative (no. 10) subclones identified in (a).
To ascertain whether the variation in PDIM synthesis is a stable phenotype or is subject to some form of phase variation, three of the clones identified in Fig. 1(a) (two PDIM-positive and one PDIM-negative clone) were grown in 7H9 broth for 3 weeks and plated once more on 7H11. In each case, 24 daughter colonies were picked into 7H9, metabolically labelled and assayed for PDIM as described above. All subclones derived from the PDIM-positive clones retained the ability to synthesize PDIM (data not shown). None of the colonies derived from the PDIM-negative parent had reverted to become PDIM positive. Once the ability to produce PDIM is lost, strains appear unable to recover. The identification of distinct mechanisms leading to the loss of PDIM in H37Rv-ATCC is described below.
Rapid loss of PDIM synthesis during serial passage in vitro
In order to estimate the rate at which PDIM-negative clones are generated during standard in vitro culture, we selected two strongly PDIM-positive clones from the ‘no treatment’ groups (see above) of H37Rv-ATCC (clones 8 and 20; Fig. 1a) and H37Rv-Pasteur. For each of the four isolates, two separate 7H9 broth cultures were inoculated and grown at 37 °C with constant rolling over a period of 20 weeks. Each week, one set of the duplicated cultures was diluted 1 : 25 in fresh 7H9 broth (‘passage’ group), while the other set of cultures was left rolling without additional fresh medium or passage for the entire duration of the experiment (‘no passage’ group). After 20 weeks, serial dilutions of the former were plated onto 7H11 for the isolation of single colonies prior to PDIM analysis. The second group was subcultured once in fresh 7H9 prior to plating so as to disperse the bacterial clumps that had formed during the 20-week incubation period. At the end of the 20 weeks, the frequency of PDIM-negative colonies was significantly higher in the ‘passage’ group (31 %) for H37Rv-ATCC than in the cultures that were left undisturbed (11 %; Table 2). This difference is likely to reflect the fact that the cultures being continuously passaged had undergone many more cycles of DNA replication and bacterial division. Once again we observed that the Pasteur variant of H37Rv appeared much less likely to generate PDIM-negative offspring in vitro than the ATCC-derived strain (Table 2). Only one isolate within the group that was continuously passaged showed a defect in PDIM biosynthesis in this case (Table 2).
Table 2.
Effect of serial passage on the spontaneous loss of PDIM from H37Rv in vitro
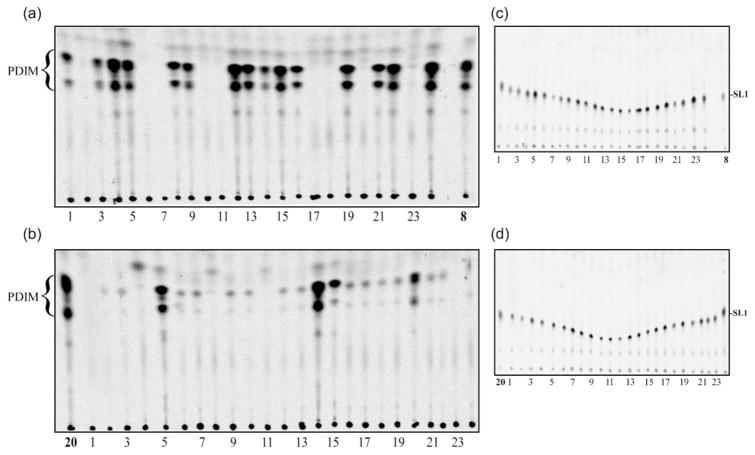
Fig. 2 shows the TLC profile of the isolates collected from the two H37Rv-ATCC cultures that were subject to weekly subculture over a 20-week period. The PDIM content of the parental isolates (8 and 20) and the 24 subclones derived from each are shown in panels (a) and (b), respectively. The isolates derived from clone 8 appear to fall into two categories: those that completely lack PDIM (e.g. isolates 2, 6, 7 and 10) and those that produce PDIM in amounts similar to the parent. As described above, a third category appears to exist amongst the clone 20 derivatives – those that are weakly PDIM positive (e.g. isolates 2, 3, 6 and 7). In order to rule out the possibility that the observed differences were due to variation in the amount of lipid being loaded onto the TLC plates, we ran the same plates in a more polar solvent system that allows visualization of sulfolipid-1 (SL-1) (Fig. 2c, d). As the amount of SL-1 appears to be relatively constant, we conclude that the differences in PDIM content are not attributable to sample-to-sample variation in lipid extraction or TLC loading technique, nor are they due to a more general cellular defect in lipid biosynthesis.
Fig. 2.
Rapid loss of PDIM during serial passage of H37Rv in vitro. After weekly subculture over a period of 20 weeks, apolar lipid extracts were prepared from 24 subclones derived from two of the strongly PDIM-positive cultures identified in Fig. 1(a). Liquid cultures were radiolabelled with [1-14C]propionic acid prior to extraction and the TLCs for PDIM detection were developed three times in petroleum ether/ethyl acetate (98 : 2, v/v) (panels a, b). Subclones derived from isolates 8 (panels a, c) and 20 (panels b, d) are shown and the parental isolates are indicated. The same TLC plates were also analysed for sulfolipid-1 (SL-1) (panels c, d). The solvent system in this case was chloroform/methanol/water (100 : 14 : 0.8, by vol.).
A range of mutations is responsible for the loss of PDIM in vitro
To investigate the range of mechanisms involved in PDIM loss in vitro, we performed whole-genome DNA microarray comparisons for six PDIM-negative isolates (Fig. 1a, nos 3–6, 10 and 16) and a weakly PDIM-positive isolate (no. 9) from the no-treatment group of strain H37Rv-ATCC (Table 1; Fig. 1a). The two PDIM-positive isolates used as the comparators in these experiments were clones 8 and 20 (Fig. 1a). For two of the PDIM-negative isolates (nos 3 and 4) we identified a deletion of approximately 29 kb that encompasses genes Rv2931 (ppsA) to Rv2938 (drrC) (Table 3). The ppsA–E genes encode a PKS involved in phthiocerol synthesis (Trivedi et al., 2005), and Mycobacterium bovis mutants disrupted in ppsC and ppsD have been shown to lack PDIM (Azad et al., 1997). Similarly, M. tuberculosis mutants bearing inactivated drrB and drrC genes are defective in PDIM secretion, indicating that these two proteins transport PDIM across the mycobacterial cell wall (Camacho et al., 1999, 2001; Waddell et al., 2005). The deletion of both the pps and drr operons is undoubtedly responsible for the absence of PDIM from these two strains. Isolate 3 was also found to contain several other small deletions (e.g. iniA, purA and fadD14; Table 3) that most likely have arisen independently of the loss of PDIM. For the remaining PDIM-negative isolates, there were no obvious genetic lesions that could account for their deficiency in PDIM synthesis.
Table 3.
Whole-genome comparisons of PDIM-positive and PDIM-negative isolates
Data represent the mean of two independent DNA arrays for each strain.
| Locus ID | Gene or product | Z-score* | Function |
|---|---|---|---|
| Isolate 3 (PDIM−)/isolate 20 (PDIM+) | |||
| Rv0342 | iniA | −2.49 | Isoniazid-inducible unknown protein |
| Rv0357c | purA | −2.46 | Probable adenylosuccinate synthase |
| Rv0531 | CHP | −2.91 | Conserved hypothetical protein |
| Rv0541c | CHP | −2.18 | Probable conserved integral membrane protein |
| Rv0607 | HP | −2.48 | Hypothetical protein |
| Rv1058 | fadD14 | −2.50 | Probable medium-chain fatty-acyl-CoA synthetase |
| Rv2931 | ppsA | −13.13 | PGL and PDIM biosynthesis |
| Rv2932 | ppsB | −5.10 | PGL and PDIM biosynthesis |
| Rv2933 | ppsC | −14.17 | PGL and PDIM biosynthesis |
| Rv2934 | ppsD | −2.80 | PGL and PDIM biosynthesis |
| Rv2935 | ppsE | −4.69 | PGL and PDIM biosynthesis |
| Rv2936 | drrA | −2.94 | Probable daunorubicin resistance and PDIM transporter |
| Rv2937 | drrB | −7.84 | Probable daunorubicin resistance and PDIM transporter |
| Rv2938 | drrC | −3.84 | Probable daunorubicin resistance and PDIM transporter |
| Isolate 4 (PDIM−)/isolate 8 (PDIM+) | |||
| Rv2931 | ppsA | −9.19 | PGL and PDIM biosynthesis |
| Rv2932 | ppsB | −9.84 | PGL and PDIM biosynthesis |
| Rv2933 | ppsC | −11.46 | PGL and PDIM biosynthesis |
| Rv2934 | ppsD | −0.94 | PGL and PDIM biosynthesis |
| Rv2935 | ppsE | −2.96 | PGL and PDIM biosynthesis |
| Rv2936 | drrA | −9.45 | Probable daunorubicin resistance and PDIM transporter |
| Rv2937 | drrB | −7.21 | Probable daunorubicin resistance and PDIM transporter |
| Rv2938 | drrC | −9.72 | Probable daunorubicin resistance and PDIM transporter |
| Rv3169 | CHP | −2.40 | Conserved hypothetical protein |
Negative Z-score values indicate the absence of a particular gene within PDIM-negative isolates.
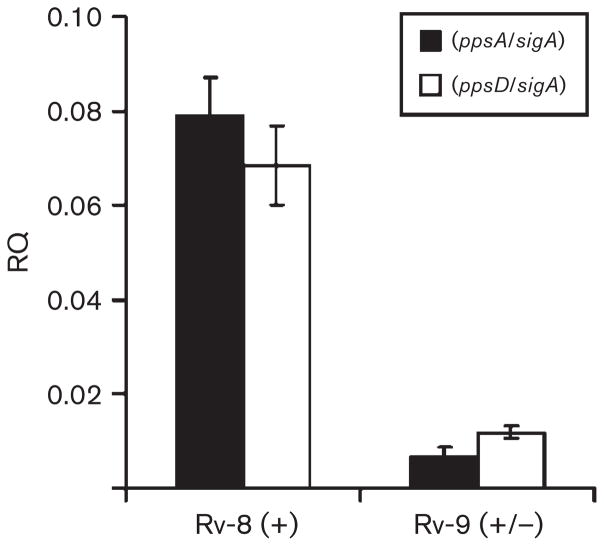
In an attempt to identify regulatory or transcriptional defects that may be associated with the loss of PDIM, we carried out whole-genome transcriptional analysis for the PDIM-negative strains that were not deleted in either the pps or drr loci. Among the five isolates studied, only isolate 9 showed a decrease in the expression of genes known to be involved in PDIM synthesis. In this particular isolate, the fadD26, ppsA, ppsB and ppsC (Rv2930–Rv2933) genes were all found to be downregulated relative to the PDIM-positive isolate 8 (Table 4). This result was subsequently confirmed via qRT-PCR analysis of ppsA gene expression within an independently obtained cDNA sample (Fig. 3). fadD26 encodes a fatty-acyl-AMP ligase that is involved in PDIM biosynthesis by converting long-chain fatty acids into acyl-adenylates, which then become substrates for the pps PKS system (Trivedi et al., 2005). M. tuberculosis strains in which fadD26 has been inactivated are unable to synthesize PDIM and are attenuated in the mouse model of TB infection (Camacho et al., 1999, 2001). As stated above, the genes that are known to be involved in the biosynthesis of PDIM are organized into a single large (~50 kb) cluster (Cole et al., 1998). Within this cluster, fadD26, ppsA–E, drrA–C and papA5 [an acyl transferase that catalyses the esterification of phthiocerol and mycocerosic acid to form PDIM (Onwueme et al., 2005)] are also reported to form a single transcriptional unit (Camacho et al., 2001). Although the ppsD–papA5 genes did not appear to be downregulated when this clone was analysed by micorarray, qRT-PCR carried out using cDNA prepared from an independent RNA sample confirmed the downregulation of ppsD (Fig. 3).
Table 4.
Transcriptional profile comparing PDIM-positive and PDIM-negative isolates
Data represent the mean of two independent biological replicates and four cDNA arrays per strain.
| Locus ID | Gene or product | Z-score* | Function |
|---|---|---|---|
| Isolate 9 (PDIM+/−)/isolate 8 (PDIM+) | |||
| Rv2930 | fadD26 | −5.41 | Fatty-acyl-AMP ligase |
| Rv2931 | ppsA | −4.83 | PGL and PDIM biosynthesis |
| Rv2932 | ppsB | −5.96 | PGL and PDIM biosynthesis |
| Rv2933 | ppsC | −4.25 | PGL and PDIM biosynthesis |
| Isolate 10 (PDIM−)/isolate 8 (PDIM+) | |||
| Rv0335c | PE6 | +6.5 | PE6 |
| Rv0899 | ompA | +4.3 | Porin of low specific activity |
| Rv0914c | Rv0914c | +9.4 | Possible lipid carrier protein or ketoacyl-CoA thiolase |
| Rv0942 | CHP | +7.6 | Conserved hypothetical protein |
| Rv0973c | accA2 | +7.3 | Probable acetyl/propionyl-CoA carboxylase |
| Rv2107 | PE22 | +6.5 | PE22 |
| MT1321 | HP | +6.8 | Hypothetical protein |
Negative Z-scores indicate downregulation of a particular gene within PDIM-negative isolates. Positive Z-scores indicate upregulation within the PDIM-negative isolates.
Fig. 3.
Repression of genes involved in phthiocerol biosynthesis is associated with a defect in PDIM production in vitro. qRT-PCR analysis was carried out on cDNA samples prepared from the PDIM-positive isolate 8 (Rv-8, PDIM+) and the weakly positive isolate 9 (Rv-9, PDIM+/−). Relative expression data (RQ, or relative quantity) were obtained using primers specific for the ppsA and ppsD genes normalized to the sigA housekeeping gene. The cDNA used in these experiments was prepared from RNA samples obtained independently of the samples analysed via microarray (Table 4) and serves to confirm the array data. Each cDNA sample was assayed twice (in triplicate) and standard deviations are indicated.
When analysed by microarray, none of the other PDIM-negative isolates (nos 5, 6, 10 and 16) revealed any consistent transcriptional differences across independent replicates that could account for the deficiency in PDIM. Interestingly, one isolate (no. 10) showed an increase in the expression of a small number of genes, two of which are associated with lipid metabolism (Table 4). The Rv0914c gene encodes a putative lipid carrier protein or keto acyl-CoA thiolase thought to be involved in fatty acid β-oxidation, whilst accA2 encodes one of the subunits of an acetyl/propionyl-CoA carboxylase. The latter is involved in malonyl-CoA or methylmalonyl-CoA formation, the essential first step in fatty acid biosynthesis. Although not tested as part of this study, it is possible that the upregulation of these two genes is a direct consequence of this isolate’s inability to synthesize PDIM.
Loss of PDIM confers a growth advantage in vitro
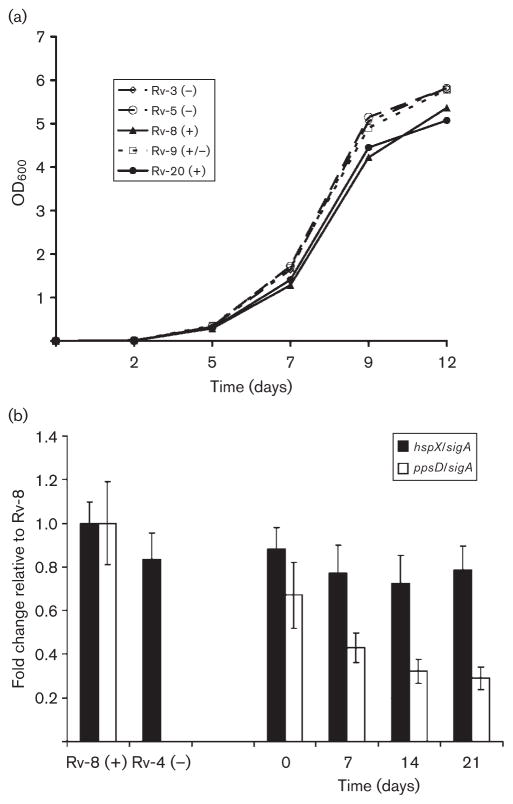
Despite the fact that we had detected a range of mutation types associated with PDIM loss in vitro, it would seem highly unlikely that we would observe such a high proportion of PDIM-negative clones over such a short time-frame (see Table 2) unless there was at least some growth advantage attributable to the loss of PDIM. To test this hypothesis, we initially carried out a series of standard in vitro growth curves for H37Rv-ATCC isolates 8 and 20 (PDIM+), 3 and 5 (PDIM−) and the weakly positive isolate, 9 (PDIM+/−). Based on OD600 measurements, we were able to demonstrate a slight, yet consistent, growth increase for each of the isolates defective in PDIM synthesis relative to the two PDIM-positive controls (Fig. 4a).
Fig. 4.
Loss of PDIM confers a growth advantage in vitro. (a) In vitro growth curves for two PDIM-positive isolates (8 and 20; solid lines), a weakly positive isolate (9) and two PDIM-negative isolates (3 and 5). Data obtained from one of two independent biological replicates are presented. (b, left) qRT-PCR analysis of genomic DNA prepared from PDIM-positive isolate 8 (Rv-8) and PDIM-negative isolate 4 (Rv-4). Data obtained with primers specific for the hspX and ppsD genes were normalized to sigA. The normalized values are plotted relative to the wild-type PDIM-positive strain (no. 8). These data confirm that ppsD is absent from the PDIM-negative strain. (b, right) The proportion of isolate 8 (PDIM+) and isolate 4 (PDIM−) when grown in mixed culture was assayed by qRT-PCR over a period of 21 days. Primers specific for hspX were used for quantification of PDIM+ and PDIM− isolates in the mixture, whilst ppsD primers were specific for the PDIM+ isolate. Data are normalized to sigA and the results are expressed relative to Rv-8. Standard deviations are indicated in (b).
Measuring growth within independent liquid cultures does not address the question of relative growth dynamics for cultures that are a heterogeneous mixture of PDIM-positive and -negative clones. Therefore, we also conducted an in vitro competition experiment for mixed cultures of isolates 8 (PDIM+) and 4 (PDIM−). Isolate 4 had previously been shown to have a large genomic deletion spanning the ppsA–drrC region (Table 3). The two cultures were initially adjusted to the same cell density, mixed in a 1 : 1 ratio and incubated at 37 °C with rolling for a total of 21 days. As detailed in Methods, the mixture was subcultured on days 7 and 14. Relative quantification of each isolate within the mixed population was accomplished by qRT-PCR of genomic DNA purified from culture aliquots harvested on days 0, 7, 14 and 21. Primers specific for the ppsD gene, absent from isolate 4 (PDIM−), were used to estimate the proportion of the mixed culture that was PDIM+. Primers specific for hspX (present in both isolates) were used as a reference to estimate the total cell population. Fig. 4(b, left) shows the results of the qRT-PCR using DNA purified from the two starter cultures prior to mixing. Both the ppsD and hspX genes are detected in the PDIM-positive isolate 8 (Rv-8), whereas ppsD is clearly absent from the PDIM-negative isolate 4 (Rv-4). This latter result confirms the results obtained for the microarray comparisons (Table 3). At the initial time point of the competition experiment (day 0), 67 % of bacteria in the mixed culture were determined to be ppsD+ (Fig. 4b, right). By day 21, this percentage had dropped to 29 %, indicating that the relative proportion of the PDIM-positive clone had decreased approximately twofold. By subtraction, the proportion of the PDIM-negative clone had risen from 33 to 71 % over the course of 3 weeks. These same trends were observed for a competition experiment carried out with isolates 3 and 8 (data not presented). Like isolate 4, isolate 3 also bears a large deletion in the ppsA–drrC region (Table 3). These data confirm that the inability to synthesize PDIM can, at least in some instances, confer an enhancement of growth rate in vitro that contributes to these strains’ ability to dominate over time within a heterogeneous cell population.
DISCUSSION
To date, the overwhelming majority of laboratory studies aimed at deciphering the mechanistic basis for M. tuberculosis virulence have employed the ubiquitous H37Rv strain. Derived in vitro from the H37 pulmonary isolate by researchers at the Trudeau Institute in the mid-1940s (Steenken, 1950), it has served as the primary reference strain for M. tuberculosis research across the globe ever since. To a large extent, the widespread acceptance of H37Rv has recently been driven by the availability of the complete genome sequence for this strain (Cole et al., 1998). The availability of sequence information has also contributed to the situation where the vast majority of recombinant M. tuberculosis clones, including those used for in vivo pathogenesis and vaccination experiments, have now been generated in the H37Rv background. Unfortunately, our data presented herein add to a mounting body of recent evidence (Andreu & Gibert, 2008; Kana et al., 2008; Manjunatha et al., 2006) indicating that a substantial proportion of H37Rv stock cultures held around the world contain a high percentage of cells lacking PDIM – a cell-wall-associated wax that is essential for full virulence in vivo.
A direct link between PDIM loss and an attenuation defect in vivo was confirmed at the genetic level for two independent strains of M. tuberculosis back in 1999 (Camacho et al., 1999; Cox et al., 1999). In each of these two studies, the level of in vivo attenuation afforded through the loss of PDIM was approximately 50-fold in mouse lungs by just 3 weeks post-infection. We have subsequently described the same level of attenuation for an MmpL7 mutant generated in H37Rv (Domenech et al., 2005). MmpL7 is the transmembrane transport protein necessary for PDIM export. To the best of our knowledge, the association between the spontaneous loss of PDIM from H37Rv in vitro and attenuation in vivo was first reported in 1974 (Goren et al., 1974). Prior to initiating the present study, we were aware of only one other publication that had commented on the isolation of PDIM-negative clones from H37Rv-ATCC cultivated in vitro (Manjunatha et al., 2006). In that particular study, mutants selected for resistance to the nitroimidazole compound PA-824 were found to have lost the ability to synthesize PDIM independently of the drug selection process. Sixty per cent of PA-824R clones that were subject to comparative genome sequencing in that study were found to contain genomic deletions or point mutations within the PDIM locus. While our study was under way, two independent publications have reported similar findings with respect to PDIM loss in H37Rv-ATCC. One reported the complete absence of PDIM from the parental H37Rv culture (ATCC 25618) that was used in the construction of a series of resuscitation-promoting factor (rpf)-like gene mutants (Kana et al., 2008). Interestingly, the ATCC stock in that case is apparently distinct from the ATCC 27294 stock used in our work and the work of Andreu & Gibert (2008). Similar to the findings reported herein, these two authors have recently reported on a PDIM-positive stock culture that had become 75 % PDIM negative after routine subculture on solid media (7H10 agar) every 4–6 weeks over a period of 2 years (≈20 passages). We observed loss of PDIM from approximately 30 % of clones after the same number of serial passages carried out in liquid culture (Table 2).
Two factors seem to contribute to the rapid emergence of PDIM-negative clones in vitro. Firstly, multiple independent mutations appear to be occurring simultaneously that result in the loss of PDIM. Secondly, the fact that these mutants are no longer synthesizing PDIM affords them a competitive growth advantage within a heterogeneous population (Fig. 4a). We describe two distinct mutation types in this study: genomic deletion and regulatory mutations that affect the expression of genes within the PDIM biosynthetic pathway (Tables 3 and 4). For two of our PDIM-less isolates (nos 3 and 4; Fig. 1a), we have uncovered a 29 kb deletion encompassing ppsA to drrC (Table 3). Although not confirmed by sequencing, we assume these two clones are both derived from the same progenitor, given that they appear to contain the same deletion. For isolate 9, either a mutation affecting promoter activity or some other type of regulatory mutation has taken place that impacts expression of genes within the fadD26–papA5 transcriptional unit. It is most likely that one or more unspecified point mutations affecting translation or protein function have occurred within the other PDIM-deficient strains. As mentioned above, genomic deletions and point mutations resulting in the loss of PDIM synthesis have been described by Manjunatha et al. (2006), whilst Andreu & Gibert (2008) have also reported regulatory changes leading to the downregulation of the mas gene. The mas gene encodes the PKS that synthesizes the multi-methyl branched mycocerosic acids that form part of the complex PDIM structure (Azad et al., 1996).
Although throughout this study we have referred to the loss of PDIM from in vitro cultures as being a ‘spontaneous’ process, a more accurate description would probably be that PDIM synthesis in vitro is being strongly selected against. This is perhaps not surprising, given the high energetic cost of replicating, transcribing and translating a ≈50 kb DNA cluster consisting of 15 separate open reading frames encoding proteins involved in at least 35 catalytic steps (Trivedi et al., 2005). On top of this is the additional energy necessary for the synthesis and transport of an abundant >90-carbon wax-like compound. The extreme length of time that H37Rv has been adapted to in vitro culture (the H37 progenitor was first isolated in 1905) is presumably the reason why the proportion of PDIM-negative clones in stocks of this strain is so high. However, it is still not apparent to us why H37Rv-ATCC would be more likely to lose the ability to synthesize PDIM in vitro than an alternative line of H37Rv we included in this study (H37Rv-Pasteur). It is possible that mutations that have already been described in other lipid biosynthetic pathways (e.g. pks3/pks4)) in H37Rv-Pasteur are responsible for this distinction (Dubey et al., 2002).
M. tuberculosis stocks (H37Rv or otherwise) that are a heterogeneous mixture of PDIM-positive and PDIM-negative cell types clearly present a major issue with regard to the identification and interpretation of putative virulence factors. If researchers are unaware of this situation, it is not difficult to imagine a scenario where PDIM-negative recombinants are isolated and compared in vivo alongside mixed parental cultures that contain a proportion of cells producing PDIM and that are fully virulent. A role in virulence could easily be misassigned in this case. Conversely, it is also possible to envisage a situation where particular molecules that are essential for full virulence in wild-type bacteria may be obscured from detection in the complete absence of PDIM. Likewise, unless extreme care is taken to start out with a culture that is 100 % PDIM-positive (e.g. derived from a single PDIM+ clone or from a stock culture passaged through mice to enrich for virulence) when carrying out whole-genome in vivo screens involving, for example, TraSH (Rengarajan et al., 2005; Sassetti et al., 2003; Sassetti & Rubin, 2003) or signature-tagged mutagenesis (Mazurkiewicz et al., 2006), then the potential for false-positive ‘hits’ with these types of techniques is very high. Indeed, the two original M. tuberculosis signature-tagged mutagenesis screens and an in vivo TraSH screen were very efficient at identifying clones that had specific insertions that inactivated PDIM biosynthetic pathways (Camacho et al., 1999; Cox et al., 1999; Sassetti & Rubin, 2003). As a matter of course, we now screen all our recombinants for PDIM production and our parental strains are frozen down from cultures inoculated with isolated PDIM-positive clones (e.g. clones 8 and 20, Fig. 1a). The findings described herein should also serve to further emphasize the importance of including the relevant complementation strains when carrying out phenotypic analyses that involve recombinant strains.
Although we have focused much of our attention in this study on H37Rv, the problems associated with PDIM loss in vitro are not unique to this strain. For instance, we have also identified a small proportion of PDIM-negative subclones within an in vitro liquid culture of HN878 (Table 1). This W/Beijing isolate was first routinely cultured in laboratories around 1999 (Manca et al., 1999). Other researchers have noted that clinical isolate MT103 became defective in PDIM synthesis after being held in culture continuously for 18 months (Cardona et al., 2006). Finally, we have also screened a more recent W/Beijing clinical isolate (‘B-4’) that has spent only a minimal amount of time in in vitro culture. Out of 44 individual colonies that were analysed for this particular isolate, 100 % were PDIM positive (data not shown). These results tend to support the idea that length of time in culture is a predisposing factor that contributes to the loss of PDIM in vitro.
In summary, routine in vitro culture of M. tuberculosis appears to select for the loss of PDIM via a variety of mechanisms. All strains, and in particular H37Rv, that have been repeatedly subcultured should be suspected of containing a heterogeneous population of PDIM-positive and -negative cell types. In the future, it is incumbent upon all researchers undertaking comparative virulence studies to confirm the presence of PDIM within both recombinant clones and the parental strains they are derived from.
Acknowledgments
The authors are indebted to Fiona McIntosh for her assistance in conducting the microarray experiments. This work was funded by Canadian Institutes of Health Research grant MOP82931 and Natural Sciences and Engineering Research Council of Canada grant RGPIN342048-07. M. R. is supported by a New Investigator Salary Award from the Canadian Institutes of Health Research. P. D. is supported by the Montreal General Hospital 175th Anniversary Fellowship.
Abbreviations
- PDIM
phthiocerol dimycocerosate
- PKS
polyketide synthase
- qRT-PCR
quantitative real-time-PCR
- TB
tuberculosis
Footnotes
References
- Andreu N, Gibert I. Cell population heterogeneity in Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2008;88:553–559. doi: 10.1016/j.tube.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Applied Biosystems. Real-Time PCR Systems: Chemistry Guide. Foster City, CA: Applied Biosystems; 2005. Part Number 4348358 Rev. E edn. [Google Scholar]
- Astarie-Dequeker C, Le Guyader L, Malaga W, Seaphanh FK, Chalut C, Lopez A, Guilhot C. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009;5:e1000289. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AK, Sirakova TD, Rogers LM, Kolattukudy PE. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc Natl Acad Sci U S A. 1996;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Constant P, Raynaud C, Laneelle MA, Triccas JA, Gicquel B, Daffé M, Guilhot C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- Cardona PJ, Soto CY, Martin C, Giquel B, Agusti G, Andreu N, Guirado E, Sirakova T, Kolattukudy P, et al. Neutral-red reaction is related to virulence and cell wall methyl-branched lipids in Mycobacterium tuberculosis. Microbes Infect. 2006;8:183–190. doi: 10.1016/j.micinf.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE., III The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Barry CE., III Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey VS, Sirakova TD, Kolattukudy PE. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol. 2002;45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne G, Laval F, Villeneuve C, Dinadayala P, Abouwarda A, Zerbib D, Galamba A, Daffé M. The cell envelope structure and properties of Mycobacterium smegmatis mc2155: is there a clue for the unique transformability of the strain? Microbiology. 2005;151:2075–2086. doi: 10.1099/mic.0.27869-0. [DOI] [PubMed] [Google Scholar]
- Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: phthiocerol dimy-cocerosate and the attenuation indicator lipid. Infect Immun. 1974;9:150–158. doi: 10.1128/iai.9.1.150-158.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Barry CE, III, Bergtold A, Freeman S, Haslett PA, Musser JM, Freedman VH, Kaplan G. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol. 1999;162:6740–6746. [PubMed] [Google Scholar]
- Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Cleto C, Sherman DR, Behr MA. The Mycobacterium tuberculosis complex transcriptome of attenuation. Tuberculosis (Edinb) 2004;84:197–204. doi: 10.1016/j.tube.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Brown JR. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis. 2007;7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwueme KC, Vos CJ, Zurita J, Ferreras JA, Quadri LE. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog Lipid Res. 2005;44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., III A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Winter N, Pivert E, Bordat Y, Neyrolles O, Ave P, Huerre M, Gicquel B, Jackson M. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell Microbiol. 2004;6:277–287. doi: 10.1046/j.1462-5822.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Saxena P, Yadav G, Mohanty D, Gokhale RS. A new family of type III polyketide synthases in Mycobacterium tuberculosis. J Biol Chem. 2003;278:44780–44790. doi: 10.1074/jbc.M306714200. [DOI] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirakova TD, Dubey VS, Kim HJ, Cynamon MH, Kolattukudy PE. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect Immun. 2003;71:3794–3801. doi: 10.1128/IAI.71.7.3794-3801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Barry CE., III . Analysis of the lipids of Mycobacterium tuberculosis. In: Parish T, Stoker NG, editors. Mycobacterium Tuberculosis Protocols. Totowa, NJ: Humana Press; 2001. pp. 229–245. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken W., Jr Dissociation of the tubercle bacillus; a review. Am Rev Tuberc. 1950;62:22–33. doi: 10.1164/art.1950.62.1-2.22. [DOI] [PubMed] [Google Scholar]
- Steenken W, Jr, Oatway WH, Jr, Petroff SA. Biological studies of the tubercle bacillus: iii. Dissociation and pathogenicity of the r and s variants of the human tubercle bacillus (H37) J Exp Med. 1934;60:515–540. doi: 10.1084/jem.60.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Waddell SJ, Chung GA, Gibson KJ, Everett MJ, Minnikin DE, Besra GS, Butcher PD. Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Lett Appl Microbiol. 2005;40:201–206. doi: 10.1111/j.1472-765X.2005.01659.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO Report 2006. Geneva, Switzerland: World Health Organization; 2006. Global Tuberculosis Control–Surveillance, Planning, Financing. [Google Scholar]
- Yu J, Othman MI, Farjo R, Zareparsi S, MacNee SP, Yoshida S, Swaroop A. Evaluation and optimization of procedures for target labeling and hybridization of cDNA microarrays. Mol Vis. 2002;8:130–137. [PubMed] [Google Scholar]