Abstract
Many leukemias are characterized by well-known mutations that drive oncogenesis. Mice engineered with these mutations provide a foundation for understanding leukemogenesis and identifying therapies. However, data from whole genome studies provide evidence that malignancies are characterized by multiple genetic alterations that vary between patients, as well as inherited genetic variation that can also contribute to oncogenesis. Improved outcomes will require precision medicine approaches–targeted therapies tailored to malignancies in each patient. Preclinical models that reflect the range of mutations and the genetic background present in patient populations are required to develop and test the combinations of therapies that will be used to provide precision medicine therapeutic strategies. Patient-derived xenografts (PDX) produced by transplanting leukemia cells from patients into immune deficient mice provide preclinical models where disease mechanisms and therapeutic efficacy can be studied in vivo in context of the genetic variability present in patient tumors. PDX models are possible because many elements in the bone marrow microenvironment show cross-species activity between mice and humans. However, several cytokines likely to impact leukemia cells are species-specific with limited activity on transplanted human leukemia cells. In this review we discuss the importance of PDX models for developing precision medicine approaches to leukemia treatment. We illustrate how PDX models can be optimized to overcome a lack of cross-species cytokine activity by reviewing a recent strategy developed for use with a high-risk form of B-cell acute lymphoblastic leukemia (B-ALL) that is characterized by overexpression of CRLF2, a receptor component for the cytokine, TSLP.
INTRODUCTION
Leukemia, like other malignancies, is a disease of cells with selective survival and proliferation advantages due to multiple diverse genetic alterations. These include acquired genetic alterations layered on inherited genetic variation to produce a genetic landscape that is highly variable from patient to patient. Mutant mouse and engineered cell models provide valuable tools for defining the roles of particular pathways in cancer cells. However, the patient’s cells remain the best model for studying and identifying effective therapies to target the complex and diverse interactions between acquired mutations and background genetic landscape that are unique to the development and progression of cancer in each patient. Successful precision medicine approaches will require preclinical models, like patient-derived xenografts (PDX), that allow investigators to evaluate therapies and model the emergence of chemoresistant clones using patient cells in an in vivo environment optimized to mimic conditions present in the patient. Here we describe a high-risk B-cell acute lymphoblastic leukemia (B-ALL) characterized by overexpression of a cytokine receptor (CRLF2) that lacks cross-species cytokine activation. We review the strategy used to develop a PDX model that produces physiological levels of the human cytokine, thymic stromal lymphopoietin (TSLP), for activating this receptor.
LEUKEMOGENESIS AND CHEMORESISTANCE ARE THE OUTCOME OF SELECTIVE PROCESSES DEPENDENT ON THE GENETIC LANDSCAPE
Leukemia is caused by genetic alterations that allow cells to proliferate more and survive better than normal cells. Normal blood cells differentiate continuously from rapidly dividing progenitors produced by hematopoietic stem cells in the bone marrow. Somatic mutations due to intrinsic causes (errors in DNA repair, replication) and external causes (mutagens, cytotoxic therapies) occur over the life of an individual. These mutations, are passed from parent to daughter cells, and accumulate, along with epigenetic changes, over time.1 Leukemia cells compete with normal cells and with each other for nutrients and for space in their microenviroment. Cells with detrimental mutations are selected against and those with survival advantages are selected for.
A range of genetic features comprise the genetic landscape that is acted on by selective pressures. Mutations that give cells a fitness advantage contribute to oncogenesis and are referred to as driver mutations. Mutations that co-occur with driver mutations but do not contribute to oncogenesis are described as passenger mutations.2 Inherited genetic variability is an additional factor that can impact the development of leukemia as well as treatment outcomes.3 In addition to rare germline mutations, common genetic variants, identified based on their overrepresentation in patients with leukemia, have the potential to contribute to genetic susceptibility to this disease. Whole genome studies also provide increasing evidence that inherited genetic variations impact relapse through multiple mechanisms, including altering the response of cancer cells to cytotoxic agents.3
The number of known driver mutations, as well as passenger mutations, has soared with input from Genome Wide Association Studies (GWAS). Solid tumors average 33–66 mutations per patient that affect protein structure.4 Distinguishing driver mutations from passenger mutations can be a challenge. A number of statistical and theoretical methods for evaluating GWAS data have been developed to predict which mutations are drivers and passengers.4 Engineered animal and cell model systems are likely to provide biological information important for determining the driver versus passenger mutation status and for identifying the oncogenic mechanisms of individual driver mutations. However, passenger status can be context dependent —with time passenger mutations may modify the disease, conferring survival advantages in context of additional mutations or under the selective pressure of chemotherapy.5 Leukemias exhibit fewer genetic alterations (~9 per patient) than most other malignancies, nevertheless the variation in genetic factors from patient to patient and at different points in the course of leukemia is substantial.4
Thus, leukemogenesis and chemoresistance are selective processes that act on the unique combinations of driver and passenger mutations present in context of inherited genetic variation. Effective precision medicine approaches will depend on identifying combinations of drugs that target the multiple driver mutations in context of a range of unique passenger mutations and inherited genetic variation with minimal adverse effects. Experimental systems that can model the diverse genetic landscapes present in patient populations, as well as the dynamics of disease progression under the selective pressure of chemotherapy will be essential to this process.
LEUKEMIA MODELS AND THEIR LIMITATION
Mouse models of leukemia
The earliest mouse models of malignancy were generated by exposure to carcinogens. These models were important tools for evaluating cytotoxic agents and understanding the process of oncogenesis. They provided support for the use of cytarabine, which continues to be a widely used therapy for leukemia. A major drawback to these models is that few cases of human leukemia arise from exposure to carcinogens, and the molecular basis of the malignancies are not known.6 These gave way to transgenic mice that expressed fusion proteins associated with human cancers. Mice transgenic for the BCR-ABL fusion protein that arises from the Philadelphia positive chromosome translocation produces murine B-ALL that mirrors the human disease. This model has been used to test targeted therapies and obesity effects on the progression of ALL.6 Knockout mice with deletions of the IKZF1 (Ikaros) provided the first evidence that this DNA binding protein was a tumor suppressor.7 Subsequent studies showed that that deletions/mutations in IKZF1 are strongly associated with poor prognosis in B-ALL.8 Similarly, mosaic models created by engineering fusion proteins into hematopoietic stem cells and subsequent transplantation back into mice have provided important models for understanding disease mechanisms of particular genetic alterations and for identifying therapies to effectively target these mutations. The identification of combination therapies that effectively target multiple combinations of driver mutations in mouse models becomes complex and costly. Evaluating drug efficacy in context of inherited genetic variability across the human genome is not possible using mouse models. Complementary approaches are needed.
Cell lines and primary patient samples
Human cell lines cloned from patient samples provide a renewable source of human cells but with limitations. Once large scale molecular analyses established the heterogeneity of tumors, there was a move toward the use of panels of cell lines that could better reflect the heterogeneity across the population. Platforms of up to 1200 cell lines from multiple cancer types are now available.9 The ability to engineer combinations of mutations by genome editing using the clustered regularly-interspaced short palindromic repeats (CRISPR) technology offers new opportunities for developing cell lines with unique combinations of driver mutations.10 However, cell lines are derived from rare cells selected for their ability to grow well in vitro over time. After multiple passages cell lines can undergo extensive modifications.9 Despite this, cell lines can provide the reproducible results necessary for dissecting signaling pathways that contribute to leukemogenesis in humans.
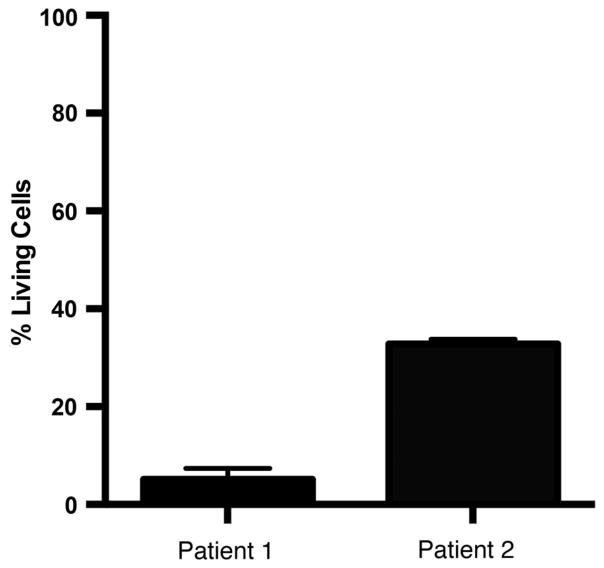
It is important that data obtained from cell lines is validated in primary patient samples, although primary samples have their own limitations. Primary leukemia samples are typically available in small quantities, limiting the information that can be obtained from them. Primary cells often die quickly in culture outside of the in vivo environment. For example, it is not unusual to find that 90% of primary leukemia cells are dead in untreated cultures within 3 days. The health of the remaining cells is unlikely to be optimal, making it difficult to evaluate drug efficacy. In vitro co-cultures that mimic the bone marrow microenvironment by providing bone marrow stromal cells11 and/or exogenous cytokines can improve survival. However, as shown in figure 1, the viability of primary samples in co-culture with stromal cells and cytokines is variable, and can still be as low as 10% after 3 days. Although primary human cells, and to a lesser extent, human cell lines, can model diverse combinations of driver mutations in context of inherited genetic variability, the data in figure 1 serve to illustrate that they do not model critical in vivo factors.
Figure 1.
Primary leukemia samples show poor in vitro survival. Primary leukemia cells were obtained as waste samples from two patients with high-risk B-ALL and viably frozen and stored in liquid nitrogen. Cells were thawed and placed immediately in co-culture with a mixture of mouse (MS-5 cell line) and human (HS27 +T cell line) bone marrow stromal cells that provide a source of interleukin-7 (IL-7) and TSLP.11,24 Following 72 h in culture cells were harvested and stained for flow cytometry to detect living (7-AAD–) leukemia cells. Graphed are the percent of living cells among total leukemia cells following 3 days of culture. B-ALL, B-cell acute lymphoblastic leukemia; TSLP, thymic stromal lymphopoietin.
PDX models
PDX models produced by transplanting primary patient samples into immune deficient mice offer several advantages not present in mouse models. They provide an in vivo model of disease heterogeneity across the human population and also model the polyclonal nature of leukemia. PDX models also have the potential of modeling the range of driver mutation combinations present in the patient population and in context of existing inherited genetic variability. However, PDX have a number of limitations, foremost among these are (1) the absence of normal immune components and (2) absent or reduced ability of murine cytokines and chemokines to replace their human counterparts in stimulating human leukemia cells. Experiments to optimize PDX models by circumventing these limitations are ongoing and summarized in a recent review.12 Here we will focus on one example.
CRLF2 B-ALL: A PERFECT LEUKEMIA STORM WHERE PDX MODELS COULD BE MOST EFFECTIVE
Children with ALL who are at high-risk for relapse can be identified based on clinical features at diagnosis.13 GWAS studies have identified specific genetic lesions and their associated signaling pathways as driving factors in chemoresistant B-ALLs.14,15 One example is the genetic alteration leading to overexpression of the CRLF2 cyto-kine receptor component that results in CRLF2 B-ALL.16–18
CRLF2 B-ALL is a high-risk ALL with multiple associated driver mutations and inherited germline variations that are also likely to contribute to disease. CRLF2 is characterized by deletions/mutations in the IKZF1 (Ikaros) tumor suppressor gene (80% of cases) as well as mutations in JAK kinases (~50% of cases).19 In addition, CRLF2 B-ALL frequently includes deletions and/or mutations in EBF and PAX5, two genes essential for B cell differentiation.20 CRLF2 B-ALL is five times more common in Hispanic children than others.19,21 Further, children with Down syndrome have a 10-fold to 20-fold increased risk of developing ALL, and half of these cases are CRLF2 B-ALL.16,17,22 This increased susceptibility suggests that inherited genetic variations are a likely contributor to leukemogenesis and/or disease progression. Hispanic children who develop B-ALL also experience a higher mortality rate than others, raising the possibility that inherited genetic variations could also contribute to chemoreistance and relapse in CRLF2 B-ALL. Thus, in CRLF2 B-ALL, diverse combinations of driver mutations and inherited genetic variations come together to create a perfect leukemia storm where PDX models could be most helpful in defining disease mechanisms and identifying effective therapeutic strategies.
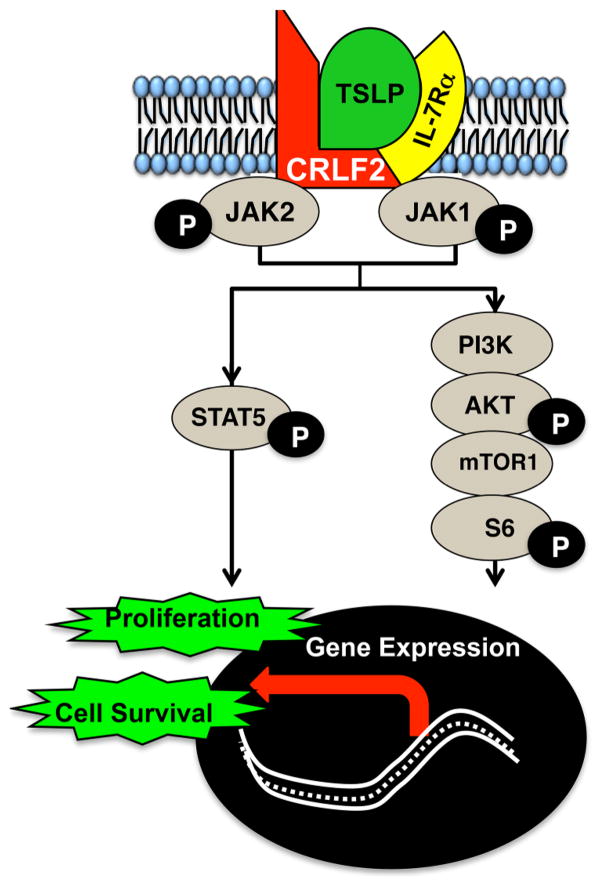
Biologically, CRLF2 is part of the receptor complex for the cytokine, TSLP. CRLF2 and the IL-7 receptor α chain (IL-7Rα) come together to form a cytokine receptor signaling complex that is activated by TSLP. Binding of TSLP induces downstream activation of the JAK2-STAT5 (Janus Kinase 2-Signal Transducer and Activator of Transcription 5) (figure 2) and PI3K/AKT/mTOR (Phosphoinositide3-kinase/protein kinase B/mechanistic target of rapomycin) pathways in CRLF2 B-ALL cells.23,24 Activation of these pathways induces changes in gene expression and cellular functions that are associated with survival and proliferation of leukemia cells.25–27 CRLF2-mediated signals are therefore likely to contribute to leukemogenesis, disease progression and chemoresistance. However, mouse TSLP is species-specific and does not activate human CRLF2.24 Thus, classic PDX mice do not model the TSLP-induced CRLF2 signaling present in vivo in patients.
Figure 2.
Pathways activated following binding of the TSLP cytokine to its receptor complex. CRLF2 together with the IL-7Rα chain form a cytokine receptor signaling complex activated by TSLP. Binding of TSLP stimulates downstream JAK-STAT5 and PI3K/AKT/mTOR pathway activation and subsequent expression of genes that promote cell survival and proliferation. JAK2-STAT5, Janus Kinase 2-Signal Transducer and Activator of Transcription 5; PI3K/AKT/mTOR, phosphoinositide3-kinase/protein kinase B/mechanistic target of rapomycin; TSLP, thymic stromal lymphopoietin.
A PRECLINICAL PDX MODEL THAT PROVIDES PHYSIOLOGICAL LEVELS OF HUMAN TSLP FOR STUDIES OF CRLF2 B-ALL
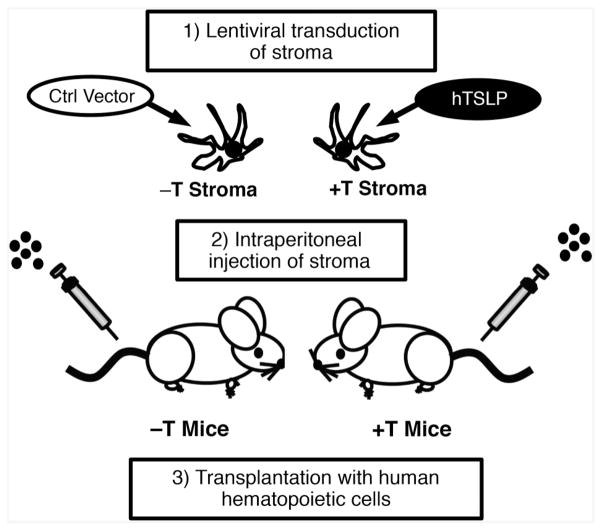
To overcome the lack of CRLF2 stimulation in classic PDX, we developed and implemented a strategy (figure 3) for engineering PDX mice to express physiological levels of human TSLP (hTSLP).24 We first transduced a human stromal cell line to express high levels of hTSLP and then injected the stromal cells intraperitoneally into immune deficient mice. Weekly injections produced sustained physiological levels of hTSLP (12–32 pg/mL). Levels of TSLP correlated with numbers of stromal cells injected. Thus by varying the number of injected stroma we could manipulate circulating hTSLP levels. In addition to mice that expressed hTSLP (+T mice), we also generated control mice (−T mice) using stroma transduced with empty vector. hTSLP was not detectable in the sera of −T mice.24
Figure 3.
Strategy for engineering PDX mice to express hTSLP. Human stromal cells were transduced to express high levels of hTSLP (+T stroma) or with control (ctrl) vector (−T stroma). Immune deficient mice were engineered to express physiological levels of hTSLP by weekly intraperitoneal injection of +T stroma (+T mice). hTSLP was not detectable in −T mice that were produced by weekly injections with −T stroma. Together +T and −T mice produce a model system to study the role of TSLP in CRLF2 B-ALL. +T mice provide a PDX model for identifying combinations of therapies that can be used to effectively treat CRLF2 B-ALL arising from diverse genetic landscapes and in context of physiological levels of hTSLP. B-ALL, B-cell acute lymphoblastic leukemia; hTSLP, human TSLP; TSLP, thymic stromal lymphopoietin.
We validated our model by evaluating its in vivo functional effects on normal B cell precursors and on primary CRLF2 B-ALL cells. TSLP has been shown to induce in vitro proliferation of normal human B cell precursors.28 To test our model we transplanted +T and −T mice with normal human CD34+ hematopoietic stem cells and compared the production of B cell precursors. The production of normal B cell precursors was increased threefold to sixfold in +T as compared to −T mice, providing one validation of the +T PDX model.24 To provide additional validation, we transplanted +T and −T mice with primary CRLF2 B-ALL cells. Leukemia cells were allowed to expand for several weeks and then harvested for comparisons of whole genome expression. Gene set enrichment analysis (GSEA) showed that genes downstream of mTOR were upregulated in leukemia cells from +T mice as compared to −T mice,24 thus providing a second validation of +T PDX mice in CRLF2 B-ALL cells.
The true test of the model is whether +T PDX provides an in vivo environment that is more like the patient than −T PDX. To test this, we compared whole genome expression in CRLF2 B-ALL cells harvested from +T and −T mice to the original patient sample used to generate them. We found that gene expression in in +T PDX was significantly closer than −T PDX to the original patient sample.24 Taken together our studies showed that PDX mice can be engineered to express physiological levels of hTSLP that induces gene expression profiles that are more similar to those in patients than classic PDX.
The +T PDX model and others like are likely to be important tools for identifying combinations of targeted therapies that can be used to effectively treat leukemia with unique combinations of driver mutations in context of inherited genetic variations that can impact disease progression as well as chemoresistance. Such models are likely to be a critical bridge between the laboratory and the use of precision medicine approaches in the clinic.
Acknowledgments
This work was supported by NIH R21CA162259, NIH P20 MD006988, NIH 2R25 GM060507, a St. Baldrick’s Research Grant and a Hyundai Hope on Wheels Scholar Hope Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors have no conflicts of interest to disclose.
Funding Hyundai Hope On Wheels; National Institute on Minority Health and Health Disparities (grant number P20 MD006988); National Cancer Institute (R21CA162259); National Institute of General Medical Sciences (2 R25 GM060507); St. Baldrick’s Foundation.
Footnotes
Contributors OLF, CB, TMM, and KJP analyzed the data discussed in this work and participated in the conceptual design of the manuscript. KJP wrote the manuscript. OLF, CB and TMM participated in critically revising the manuscript for intellectual content.
Competing interests None declared.
Ethics approval Loma Linda University Institutional Review Board.
Provenance and peer review Commissioned; externally peer reviewed.
References
- 1.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriyama T, Relling MV, Yang JJ. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood. 2015;125:3988–95. doi: 10.1182/blood-2014-12-580001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech. 2014;7:941–51. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook GJ, Pardee TS. Animal models of leukemia: any closer to the real thing? Cancer Metastasis Rev. 2013;32:63–76. doi: 10.1007/s10555-012-9405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–99. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 8.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–8. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrish YK, Baez I, Milford TA, et al. IL-7 Dependence in human B lymphopoiesis increases during progression of ontogeny from cord blood to bone marrow. J Immunol. 2009;182:4255–66. doi: 10.4049/jimmunol.0800489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theocharides AP, Rongvaux A, Fritsch K, et al. Humanized hemato-lymphoid system mice. Haematologica. 2016;101:5–19. doi: 10.3324/haematol.2014.115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowda C, Francis OL, Ding Y, et al. Chapter 1: Pediatric High Risk Leukemia — Molecular Insights. In: Guenovam M, Balatzenko G, editors. Leukemias—Updates and New Insights. 2015. pp. 1–25. [Google Scholar]
- 14.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–84. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunger SP, Raetz EA, Loh ML, et al. Improving outcomes for high-risk ALL: translating new discoveries into clinical care. Pediatr Blood Cancer. 2011;56:984–93. doi: 10.1002/pbc.22996. [DOI] [PubMed] [Google Scholar]
- 16.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–6. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–98. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 18.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115:5393–7. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 19.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–21. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ensor HM, Schwab C, Russell LJ, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 2011;117:2129–36. doi: 10.1182/blood-2010-07-297135. [DOI] [PubMed] [Google Scholar]
- 22.Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–17. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 23.Tasian SK, Doral MY, Borowitz MJ, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–42. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis OL, Milford TM, Martinez SR, et al. A novel xenograft model to study the role of TSLP-induced CRLF2 signals in normal and malignant human B lymphopoiesis. Haematologica. 2015 doi: 10.3324/haematol.2015.125336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie S, Wang Y, Liu J, et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 2001;20:6188–95. doi: 10.1038/sj.onc.1204834. [DOI] [PubMed] [Google Scholar]
- 26.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–54. [PubMed] [Google Scholar]
- 27.Fransecky L, Mochmann LH, Baldus CD. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell Ther. 2015;3:2. doi: 10.1186/s40591-015-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheeren FA, van Lent AU, Nagasawa M, et al. Thymic stromal lymphopoietin induces early human B-cell proliferation and differentiation. Eur J Immunol. 2010;40:955–65. doi: 10.1002/eji.200939419. [DOI] [PubMed] [Google Scholar]