Abstract
T cells have dramatic functional and proliferative shifts in the course of maintaining immune protection from pathogens and cancer. To support these changes, T cells undergo metabolic reprogramming upon stimulation and again after antigen clearance. Depending on the extrinsic cell signals, T cells can differentiate into functionally distinct subsets that utilize and require diverse metabolic programs. Effector T cells (Teff) enhance glucose and glutamine uptake, whereas regulatory T cells (Treg) do not rely on significant rates of glycolysis. The dependence of these subsets on specific metabolic programs makes T cells reliant on these signaling pathways and nutrients. Metabolic pathways, such as those regulated by mTOR and Myc, augment T cell glycolysis and glutaminolysis programs to promote T cell activity. These pathways respond to signals and control metabolism through both transcriptional or post-transcriptional mechanisms. Epigenetic modifications also play an important role by stabilizing the transcription factors that define subset specific reprogramming. In addition, circadian rhythm cycling may also influence energy use, immune surveillance, and function of T cells. In this review, we focus on the metabolic and nutrient requirements of T cells, and how canonical pathways of growth and metabolism regulate nutrients that are essential for T cell function.
Keywords: T cell metabolism, mTOR, Glut1, glutamine, epigenetics, circadian rhythms
1.1 INTRODUCTION
Human inflammatory diseases and immunological clearance depend on efficient and appropriate T cell activation, balance, and subsequent inactivation. Deficits in these processes are a growing concern in medical care and it is estimated that 5–7% of individuals experience an autoimmune and inflammatory disorder[1]. Maintaining proper T cell activation and function is a complicated process that requires signaling pathway integration, initiation of metabolic reprogramming, and effector cell proliferation and cytokine production[2, 3]. Activated T cells switch from oxidative to glycolytic metabolism. This shift is somewhat counterintuitive, as glycolysis is less efficient than oxidative phosphorylation when considered as a source of ATP. Known as the Warburg Effect or aerobic glycolysis, ATP is generated primarily from glycolysis even in the presence of oxygen. This metabolic program was famously discovered in cancer cells[4], but it has been known for decades that T lymphocytes also induce aerobic glycolysis during effector responses[5]. Aerobic glycolysis can be highly efficient at promoting biosynthesis essential for effector function and rapid proliferation, but also relies on high levels of nutrient uptake, which may change with tissue location, inflammation, or even time of day.
Metabolic flexibility is critical to allow cells to rapidly adjust to changing signals and environments to support cell survival, signaling, biosynthesis, and growth. The interplay between cell extrinsic and intrinsic signals is tightly connected, and cytokines, growth factors, and receptor signaling are all integrated by well-characterized pathways, including JAK/STAT, mTOR/AMPK, and T cell receptor (TCR) signaling, among many others. These signaling pathways are controlled at both the transcriptional level, such as circadian cycling of protein expression, and post-transcriptional, as in the case of mTOR. Nutrient access also regulates signaling and availability of essential amino acids which is crucial to promote mTOR signaling[6]. The activity of T cells and their function is also modified by circadian rhythm. Circulating lymphocyte number can vary dramatically depending on the time of day, likely due to expression of homing molecules on the cell surface[7]. Studies in mice with disrupted circadian rhythm show increased incidences of obesity and metabolic syndrome, and in humans, increased cholesterol levels and obesity[8, 9]. Though the role of canonical intrinsic circadian rhythm cycling in T cells is not firmly established, altered circadian rhythms may modify circulating nutrients[10] and hormones[11] available in the environment that influence T cell responses.
1.1 Basics of T cell metabolism
The primary duty of naïve T cells is immune surveillance. T cells stay in close proximity to B cells and antigen presenting cells (APCs) in secondary lymphoid tissues and are poised to respond to presentation of specific antigen[12, 13]. Upon stimulation, T cells undergo a dramatic shift in metabolism that is marked by increased nutrient uptake and glycolysis. Mitochondrial oxidative phosphorylation (oxphos) also increases, but to a lesser extent[14]. This leads to a general shift in the metabolic flux such that activated T cells are considered predominantly glycolytic, with increased glycolysis and lactate production, and large changes in uptake of anabolic precursors such as glucose and amino acids[15–17]. Metabolic switching is likely due to increased metabolic demand for both energy, reducing equivalents, and precursors for cell components[2]. Cells that fail to meet this metabolic demand undergo programmed cell death[18]. Carbon tracing for glucose and glutamine has recently shown that a majority of carbon cell mass in rapidly proliferating cells, including T cells, is derived from amino acids, and not glucose[19]. However, a high flux of both glucose and glutamine is required for effector T cell (Teff) function[16]. After successful T cell proliferation and immunological clearance of pathogens, Teff responses are diminished and memory T cells emerge with naïve-like oxidative phosphorylation metabolism[20]. This metabolic reprogramming event is paramount to transition of effector cells to memory, as memory T cells require oxidative metabolism. Indeed, inhibition of glycolytic pathways enhances T cell memory while inhibition of mitochondrial respiration prevents memory cell formation[21].
1.2 T cell subsets and metabolic specification
Although the balance of inflammatory Teff and suppressive regulatory T cells (Treg) is tightly regulated during immune responses, dysregulation occurs as nutrients change in obesity, chronic inflammation, and cancer. These observations supported the potential for distinct metabolic programs for different T cell subsets. Indeed, direct measurement of the metabolism of CD4 subsets or CD8 effector or memory populations showed the use of clearly distinct metabolic programs[15, 22]. Teff activate and generally utilize aerobic glycolysis reminiscent of cancer cells. In contrast, Treg were shown to predominantly rely on mitochondrial oxidative pathways. Similarly, activated CD8 T cells utilize glycolysis while memory CD8 T cells utilize lipid oxidation. Altered nutrients in obesity, hyperglycemia, and hyperlipidemia thus can lead to shifts in immune cell subsets that promote insulin resistance and lead to an increased occurrence of autoimmune and other immune disorders[23]. While direct causes remain uncertain, histological analysis of visceral adipose tissue in obese subjects shows changes in T cell subsets including increased effector Th1 cells and macrophage infiltration but decreased Treg[24]. Hyperglycemia itself has been shown to enhance cell survival in general by increasing glycolysis and inhibiting caspases via NF-kB expression[25]. These shifts in T cell subsets may have significant implications for immunity. For example, increased Th17 cells can cross the blood brain barrier and induce inflammation in Experimental Autoimmune Encephalomyelitis (EAE) that induces neuronal cell death. Depletion of Treg exacerbates[26] while addition of induced Treg protect[27] from this excessive inflammation and tissue damage. As Treg are required for peripheral tolerance and prevention of inflammatory responses, it is not surprising that System Lupus Erythematosus (SLE) patients have low Treg numbers circulating in peripheral blood when compared to normal patients[28]. Th17 cell numbers increase in SLE flares, with concomitant rise in IL-17 production[29]. It should be noted, however, that circulating populations could be very different from those at sites of inflammation. Conversely, inhibition of Treg suppression could hamper treatment to induce anti-tumor immunity. Tumor elimination depends upon robust Teff activity and depletion of Treg during tumor growth enhances systemic tumor responses[30]. Clearly, affecting the balance of Teff and Treg clinically will require a solid understanding of the underlying disease and how T cell subsets function.
It is also important to consider that T cell subsets are not fixed and instead have a great deal of plasticity. Rather than T cell differentiation advancing from a naïve, quiescent state to stable effector or regulatory lineages, with little to no change in cell identity after differentiation, it is now clear that CD4 subsets can switch from one functional subset to another. We now know that T cells can also display characteristics of multiple subsets. For example, in vivo co-expression of Tbet and GATA3 has been shown, including co-production of IL-4 and IFNy cytokines[31, 32]. Likewise, Th17 cells can become Th1 cells, and regulatory T cells can become effector cells[33, 34] based on microenvironmental cues and epigenetic programing. The flexibility of T cells to reprogram themselves as “Th1-like” or “Th17-like” is becoming an accepted characteristic of T cell biology. However, the function and metabolic programs of these heterogeneous T cell populations have not been well explored, nor is it understood to what extent metabolic and nutrient shifts may impact T cell plasticity. Delineating the differences between naïve cells and dual-type or phenotypically plastic T cells may show how altered nutrient-limited conditions such as sites of inflammation or tumor microenvironment impact T cell populations and inform future therapeutics.
2. METABOLIC REGULATORS OF T CELLS
2.1 mTOR and AMPK
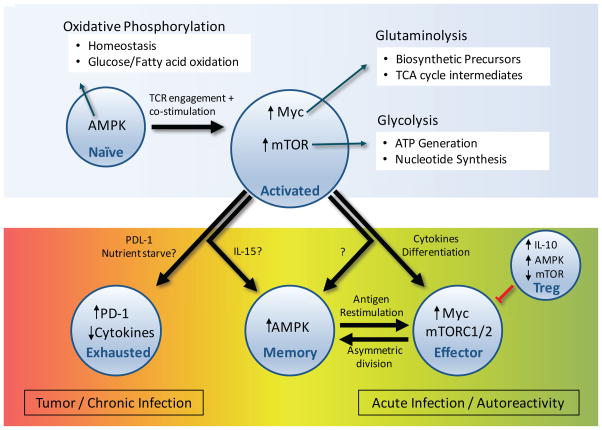
There are several canonical regulators of cell metabolism that are now know to play key roles in the metabolism of T cells[35–37]. The mTOR and AMPK pathways have significant and opposing roles in T cell metabolic programming and differentiation. AMPK is an AMP-sensitive signaling kinase – in times of stress, when nutrient availability is low or ATP synthesis decreased, AMPK activates cellular machinery to boost catabolic metabolism and inhibit anabolic growth[38, 39]. AMPK can also phosphorylate TSC2 and RAPTOR to inhibit mTOR signaling[40]. When biosynthetic nutrients are abundant, mTOR integrates extracellular signaling and sensing of essential amino acids. This promotes anabolic metabolism through transcriptional activity and direct action to promote glycolysis and lipogenesis[41]. The mTOR and AMPK pathways can be generalized as a rheostat, where AMPK induces catabolic metabolism and oxidative phosphorylation that slow proliferation, whereas the mTOR pathway promotes anabolic growth pathways to drive proliferation during activation (Figure 1). However, the role of mTOR in T cell metabolism is quite complicated and differential mTOR signaling can modify the balance of effector or regulatory T cell lineages. When mTOR is entirely absent, Teff cannot be generated, and T cells are pushed into the Treg fate[42]. mTOR kinase exists in two independent complexes: mTOR complex 1 or 2 (mTORC1 or mTORC2), which have distinct effects on T cell subsets. Inhibition of mTORC1 signaling with intact mTORC2 signaling[43] or overactive mTORC1 signaling[44] can lead to accumulation of non-functional Treg and onset of autoimmunity. Teff are also highly sensitive to mTORC complex regulation and loss of mTORC1 through deletion of the GTPase Rheb specifically eliminates the formation of Th1 and Th17, but not Th2 cells[45].
Figure 1.
Naïve T cells are predominantly oxidative and constantly monitor their environment for immune insult. Upon T cell receptor stimulation and co-stimulation by antigen presenting cells, T cells upregulate Myc expression and mTOR signaling, enhancing glycolysis and glutaminolysis (above). In tumor environment, some T cells exhibit high PD-1 expression and reduced activation. During the immune response, some cells will become memory T cells and return to naïve-like oxidative metabolism, whereas large populations of clonally selected T cells remain effector cells with high Myc and mTOR pathway signaling to respond to acute infection. In autoimmune settings, effector T cells react against self-antigens erroneously, enhancing effector responses that escape Treg suppression. Induced Treg (iTreg) are important in suppressing this peripheral autoreactivity.
Selective regulation of mTORC1 or mTORC2 pathways is also important during T cell activation, as TCR-stimulation can lead to asymmetric division of mTORC1 activity in proximal mother and distal daughter cells. It was recently shown that the APC proximal cell maintains strong effector programming and mTORC1 signaling, whereas the distal cell has reduced levels of nutrient transporters and mTORC1 signaling[42, 46], potentially leading to generation of effector versus memory cell populations. In Treg, the mTOR contribution to cell proliferation is even more complicated. Inhibiting mTOR with rapamycin before TCR stimulation counterintuitively induces proliferation in normally-anergic Treg in vitro[47]. Interestingly, PTEN- or TSC2-deficient Treg that have high mTORC1 activity not only lose suppressive function, but also have decreased lineage stability and may gain inflammatory activity as ‘ex-Treg’[48, 49].
2.2 Myc
The transcription factor Myc is also an important modulator of T cell metabolism. Myc is growth factor-induced and commonly mutated or overexpressed in cancer cells to promote glycolytic programming and glutamine uptake and catabolism[50]. Myc interacts with a variety of signaling and transcriptional networks [51] that regulate cell growth, cell death, and metabolism[52]. Indeed, there are few anabolic pathways untouched by Myc. T cell activation leads to Myc upregulation and Myc is essential for appropriate Teff activation[2]. Consistent with the role of Myc in cancer to promote aerobic glycolysis, Myc-deficient T cells fail to upregulate glycolysis and glutaminolysis following activation [53]. The asymmetric division of mTORC1 activity upon T cell activation has profound implications on Myc in proximal and distal daughter cells. Myc protein has a short half-life and mTORC1-dependent translation is critical to maintain high Myc levels. Higher levels of mTORC1 signaling in proximal cell lead to differential partitioning of Myc protein with lower Myc levels and reduced activation, mTOR signaling, and glycolytic metabolism in the distal daughter cell. This may be part of the asymmetric division of cell fates, where Myc-high cells become effectors and Myc-low daughter cells become memory T cells[46] with reduced glycolysis and mTOR signaling, as described above.
2.3 HIF1 α
Hypoxia Inducible Factor 1α (HIF1α) is an important transcriptional regulator of T cell metabolism in response to environmental stress. HIF1α can strongly promote glycolysis in conditions of hypoxia and is regulated by oxygen sensing mechanisms and mTORC1 signaling, which promotes HIF1α protein translation[54]. While mTORC1-mediated regulation provides a link to other nutrient sensing mechanisms, HIF1α was initially discovered because it is upregulated in low-oxygen environments[55]. When lymphocytes move into environments with low oxygen tension, HIF1α becomes important to promote adaptation. B and T cells appear to have a differential response to HIF1α, as HIF1αdeficient B cells develop poorly and generate autoantibodies that drive autoimmunity[56] while HIF1α-deficient T cells can still activate normally in vitro[2]. Upon T cell activation, however, HIF1α is selectively required for generation of Th17 cells[57]. This may act through interaction with the Th17 transcription factor RORγT. It may also specifically regulate the degradation of Foxp3, thus preventing Treg generation[58]. While the role of HIF1α in T cell metabolism remains poorly understood, HIF1α is known to directly target checkpoint receptor ligand interactions through regulation of PDL-1, which may have implications in the tumor microenvironment[56].
2.4 Metabolites as signaling molecules
In addition to signaling pathways that directly regulate T cell metabolism, it is also now apparent that metabolites and metabolic pathways in turn regulate signaling and differentiation events. This bidirectional connection ensures that cells only proceed to activate or differentiate if nutrient conditions are appropriate. In response to limiting glucose, it was recently shown that reduced glycolysis allows GAPDH, the enzyme responsible for catalyzing glyceraldehyde-3-phosphate to glycerate 1,3-bisphosphate in the glycolysis pathway, to translocate to the nucleus and modify IFNγ translation in T cells[14]. While a role for GAPDH in translation has been previously shown[59], the modification of transcript for IFNy specifically correlated with induction of glycolytic metabolism to provide a new link between this metabolic enzyme and inflammation[14]. Similarly, the glycolytic metabolite phosphoenolpyruvate may directly modify calcium signaling and T cell activation, to link glucose availability and T cell receptor induced signaling[60]. It is likely that other metabolic enzymes and metabolites will have additional moonlighting jobs but have yet to be discovered. Our incomplete understanding of the metabolic regulation of T cells is slowly being filled in, opening new targets of potential treatment for diseases of autoimmunity, immune-deficiency, and cancer.
3. SUBSTRATE AVAILABILITY
3.1 Glucose uptake
Beyond the changes in signaling pathways that directly modify metabolic pathways intrinsic to T cells, access to extracellular nutrients and the regulation of nutrient uptake are also essential for T cell function. In some cases, nutrient levels are altered or may not be present, such as in tumor environment. Even if adequate nutrients are available, however, appropriate transporters must be present on the cell surface. Indeed, T cell activation leads to a sharp upregulation of nutrient transporter expression and cell surface trafficking to increase metabolic substrate availability and remove waste products. The glucose transporter Glut1 and the amino acid transporters SLC1a5 and SLC7a5 are upregulated and traffic to the cell surface in response to PI3K/Akt/mTORC1 signaling to allow T cells to uptake glucose and amino acids, including large neutral amino acids and glutamine[17, 61, 62]. Likewise, Mct1 and Mct4 transport lactate out of cells to maintain glycolysis and redox balance and are also upregulated upon activation [63]. These changes can have significant impact on T cell activation and fate. T cells rely strongly on high rates of glucose uptake[64] and elevated Glut1 expression can enhance activation and glycolysis of effector T cells, leading to a Systemic Lupus Erythematosus (SLE) like-disease[65]. Conversely, Glut1-deficient T cells have decreased glucose uptake and reduced Teff proliferation, activation, and function. Mice grafted with Glut1-deficient T cells are protected from Graft-vs-Host Disease (GvHD) and Inflammatory Bowel Disease (IBD), showing that glucose transporter alterations affect in vivo function. Treg, which rely on oxidative metabolism more so than glycolysis, were unaffected by Glut1 deficiency[61]. However, Glut1 protein expression is upregulated in hypoxic conditions and Treg may instead rely more on glycolysis in low oxygen environments[66].
3.2 Glutamine and amino acids
In addition to glucose, availability and metabolism of amino acids has significant impacts on T cell metabolism. Glutamine in particular is an important substrate for highly proliferative cells[22]. Glutamine is the most abundant amino acid in circulation[67] and is a critical fuel for anabolic metabolism. As proliferative cells remove citrate from the TCA cycle to generate fatty acids for various structures such as cell membrane, glutamine is internalized and converted to glutamate, then the TCA intermediate, α-ketoglutarate (αKG). In addition, glutamine is essential for nucleotide synthesis, generation of glutathione, and provides glutamate that is used to facilitate transport of additional amino acids[68].
Drugs such as BPTES slow the growth of tumor cells in vitro by inhibiting the first enzymatic step in glutaminolysis, Glutaminase. Knockout of the Glutaminase enzyme inhibits tumor growth[69, 70], supporting Glutaminase an anti-cancer target. Because proliferative T cells closely resemble the metabolic state of cancer cells, glutamine metabolism also likely contributes to T cell metabolism and function. Indeed, removal of glutamine from cell culture media completely abrogates T cell activation and IL-2 production[16]. Multiple cell surface transporters have been shown to uptake glutamine[71], but a specific role for Slc1A5 (ASCT2) has been demonstrated in T cells. In a mouse model deficient of Slc1A5 deficiency, Th1 and Th17 effector T cells were reduced while Treg numbers were unaffected, showing differential effects on T cell subsets that correlate with dependence of these subsets on mTORC1. Strangely, proliferation of these effector cells was not affected, as would be expected if alterations in amino acid levels were reduced. It was further shown that ASCT2 knockout mice were less susceptible to Experiment Autoimmune Encephalomyelitis (EAE), a model of multiple sclerosis, which was attributed to reduced Th17 cells. Though Treg suppressive capacity was not directly assessed, the idea that Teff responses could be selectively modified by targeting glutamine availability is enticing[62].
In addition to providing metabolic substrates, amino acids have direct activating effects on mTORC1. Low amino acid levels inhibit mTOR association with lysosome, and thus prevent downstream signaling for cell growth and proliferation[46]. How the mTOR complexes and its related proteins identify amino acid levels remains uncertain, though SES2[72] or CASTOR proteins[73] are responsible for some amino acid sensing. While the essential amino acid leucine is likely the key substrate for mTORC1 amino acid sensing, T cells deficient in the large neutral amino acid transporter, SLC7a5, also fail to maintain mTORC1 activity and support anabolic metabolism[74]. In this case, however, it is unclear if mTORC1 responds directly these amino acids or if other metabolic deficiencies caused by lack of sufficient neutral amino acids prevent mTORC1 activation. Regardless of the precise mechanism, Slc7a5 deficient T cells failed to properly activate and maintain mTORC1 signaling and, as a consequence, failed to upregulate Glut1 and induce glycolysis necessary for effector T cell activation.
4. METABOLISM AND EPIGENETIC MODIFICATIONS IN T CELLS
Another mechanism by which nutrient status and metabolic pathways may influence T cell signaling and differentiation is through epigenetic modification of DNA and histones. The two primary epigenetic modifications, methylation and acetylation, each use metabolites as substrates. DNA methylation is controlled by DNA methyltransferases[75], and require methyl groups donated by S-adenosylmethionine derived from the one carbon metabolism pathway. This pathway integrates nutrients from glycolysis, amino acids, and choline or folate[76]. Associated with reduced gene transcription, methylation of DNA is heritable and stable [77], but responds to dietary influences. Indeed, reduced dietary methionine was found to lower DNA methylation[78]. Conversely, the dioxygenase enzymes that de-methylate DNA, such as TET2, require αKG as a substrate[79]. Thus both methylation and demethylation events are metabolically sensitive.
Acetylation and deacetylation of histones and other proteins also play significant roles in gene transcription and cell function, affecting growth and proliferation. In rapidly dividing cells, for example, Akt-induced glycolytic flux has a direct impact on acetyl-CoA levels, which is the required coenzyme substrate for acetyl-transferase activity. Increased availability of acetyl-CoA enhanced global acetylation of histones, even when nutrients were limited[80]. Lysine acetylation can also affect enzymatic activity and protein interactions[81]. The Treg-associated transcription factor, Foxp3, for example, is stabilized by hyperacetylation, leading to increased Foxp3 expression and higher Treg numbers [82]. It was also recently shown that acetyl-CoA availability enhances CD8+ effector function, as increased acetate uptake is required for effector CD8+ T cell responses to L. monocytogenes. This increased acetyl-CoA allowed for the acetylation of GAPDH, enhancing glycolysis and boosting CD8+ function[83]. Because T cells rely so strongly on glycolytic reprogramming, epigenetic and protein modifications could be a significant contributor to T cell function and inflammatory responses by linking substrate availability, diet, and metabolism.
Modification of epigenetic marks can have major functional consequences. Myc expression is required for T cell activation but methylation of the enhancer regions usually bound by Myc prevents Myc association and reduces gene expression[84]. A study of CD8+ T lymphocytes from geriatric and young patients showed methylation status of T cells was significantly more variable in geriatric CD8+ T cells and this correlated with decreased immune function. Several genes important in CD8+ T cell differentiation and immune response, such as IFNy and CCL5, had decreased methylation which correlated with higher transcription levels[85]. These studies were confounded, however, by decreased naïve T cells and increased memory T cells in the older population, which could skew the methylation status data. Further, during differentiation of Th1 and Th2 human T cells, the IFNy locus itself is differentially methylated. The locus becomes hypomethylated in Th1 cells while hypermethlyated in Th2 cells[86, 87]. In naïve T cells deficient for the methyltransferase Dnmt1, activation led to reduced proliferation, concomitant loss of methylation on DNA globally, and increased IFNy production[88].
In addition to cytokines, the Foxp3 locus is tightly regulated by methylation to control Treg stability. Stable nTreg have fully demethylated DNA upstream of exon-1 of Foxp3, while iTreg and less stable Treg generally show only partial demethylation. Interestingly, it seems that full demethylation is required for stable Foxp3 expression, as iTreg can lose Foxp3 levels when TGF-β is removed from culture[89]. Stability of Foxp3 expression modified by methylation status may be a reoccurring theme, as the ‘why’ of methylation is under investigation. Moreover, from a purely clinical standpoint, methylation status of Foxp3+ cells could be used to ensure stable Treg lineages are identified correctly in graft transplantation. Additional studies on T cell-specific transcription factors shows epigenetic regulation for Tbet, GATA3, and RORγt, and these are affected both as epigenetic modifiers and as agents of chromatin remodeling[90, 91]. How substrate availability and metabolic pathways impact methylation status of lymphocyte-regulatory loci remains an important question.
5. Tumor Infiltrating Lymphocytes and Metabolic Adaptation
Tumor cells can be highly glycolytic and poorly vascularized, causing the tumor microenvironment to have reduced glucose and amino acid levels[92], while simultaneously accumulating lactate, increased acidity, and other waste products[93]. Tumor infiltrating lymphocytes (TILs) thus face the challenge of engaging in active cell metabolism necessary for effector function when substrates are limited by nutrient availability. High lactate levels inhibited proliferation and cytokine production in human cytotoxic lymphocytes, which was attributed to disruption of a required lactate gradient, prevention of lactate export, and metabolic perturbation[94]. Additionally, in squamous tumors, highly glycolytic tumors had reduced CD8+ T cell infiltration[95].
TILs isolated from tumors exhibit characteristics of stereotypically lowered activity called exhaustion[96]. This exhaustion is due in part to chronic stimulation, but T cell activation in low nutrient conditions is also sufficient to induce an exhausted-like state of anergy[97]. Though the regulation of exhaustion remains complex, upregulation of the death receptor and ligand PD-1 and PDL-1, respectively, correlates with reduced activation and proliferation in CD4+ T cells[98] and tolerance in CD8+ T cells[99]. It is interesting that blockade of the immune checkpoint PD-1 can improve T cell function even in this metabolically challenging microenvironment[100]. Blockade of PD-1 in T cells enhances activation and proliferation[101]. Perhaps improved T cell function despite low nutrient levels is due to an increase in the ability of T cells to uptake and metabolize those nutrients. Indeed, PD-1 was shown to directly impair Akt and mTORC1 signaling and T cell glucose metabolism[102]. Blocking PD-1 interaction with its ligand, therefore, leads to increased T cell glycolysis that may improve the metabolic competitiveness of T cells in the tumor setting[103].
6. CIRCADIAN RHYTHM REGULATION OF CELL METABOLISM
It has been appreciated for some time that immune responses are influenced by time of day and circadian rhythms that are closely linked to metabolic status[104]. Entrainment of the circadian rhythm is generally provided by sunlight, sleep cycles, and feeding[10]. The daily cycles of feeding and sleeping connect energy homeostasis with metabolic substrate availability, as feeding schedules correlate with availability of glucose and amino acids in the blood[105], and loss of the normal circadian cycle can effect bodyweight and metabolic disorder[106–108]. A key function, therefore, of circadian rhythms is to coordinate metabolism with nutrient availability. The transcription factors Clock and BMAL1 constitute the original basis for circadian cycling and these form dimers to induce transcription of oscillatory inhibitory proteins PER (PER1-3), Cry (Cry1 and 2), and REV-ERB[109]. The rise and fall of these transcription factors Clock and BMAL and their downstream protein targets establish the cell-intrinsic circadian rhythm. Lymphocytes show the same 24-hour circadian cycling as most other mammalian cells[110]. Indeed, circadian cycling could affect autoimmune disease pathogenicity as well, as loss-of-function Clock mutant mice had reduced IL-23 levels and improved dermatitis, a psoriasis-like model. It was shown that Clock actually bound the promoter region of IL-23 receptor, which exhibited circadian behavior. Further, a loss-of-function mutation in the inhibitory protein Per1 exacerbated the dermatitis, pointing to the circadian pathway as altering autoimmune conditions[111]. Because the expression of Clock and BMAL1 is under control of retinoic acid receptor-related orphan nuclear receptors (RORs)[112], it is possible that transcriptional regulation of Clock genes are partially under control of RORyt, the regulator of Th17 development. However, the connection between T cell differentiation and Clock:BMAL1 expression remains unexplored.
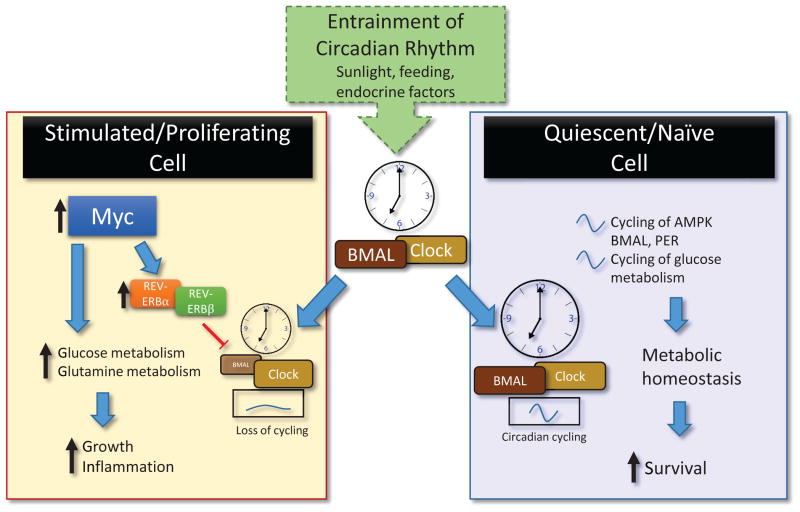
T cells and cancer cells are both strongly driven by Myc which may circumvent the normal process of growth cycles limited by circadian rhythm. In cultured U2OS cells, overexpression of Myc enhanced expression of REV-ERBα and REV-ERBβ, which are responsible for a feedback inhibition loop of circadian-controlled BMAL protein expression[113], abolishing circadian cycling. This also eliminated a previously unknown cycling of glycolytic metabolism, which varied glucose consumption based on the time of day[114]. Combined with increased glutamine metabolism, Myc expression can thus push circadian rhythm-limited cells beyond their regular programming (Figure 2). Remarkably, despite cell-intrinsic circadian cycling of proteins and metabolism, activated T lymphocytes also appear unaffected by elimination of the circadian clock, at least in terms of differentiation and viral clearance. Deletion of Bmal1 in T cells did not prevent the generation of Th17 cells, though there were slight reductions in IL-2 production[115], which is important in immune regulation and activation. This points to potential other extrinsic regulators of the circadian cycles in lymphocytes. As circadian rhythms in T cells have not been fully explored, the role of circadian cycling on T cell metabolism and function remains an open question.
Figure 2.
The circadian clock exists in most somatic cells and is entrained by the Suprachiasmatic Nucleus in the brain via sunlight. This affects feeding behavior, sleep/wake cycles, circulating T cell counts, and more. In quiescent cells (right), cycling of metabolic sensor AMPK is coordinated with Clock:BMAL and PER1/PER2 expression, with concomitant cycling of glucose metabolism and potentially important in T cell maintenance and surveillance. Overexpression of Myc (left), increases REV-ERBα and REV-ERBβ expression to inhibit BMAL1 transcription and prevent circadian cycling. Myc expression induces glucose and glutamine metabolism and is required for T cell effector function.
7. OPEN QUESTIONS
The interplay of signaling proteins, metabolic status, and transcription factor regulation is in constant flux to control T cell fate. While it is clear that metabolism must be tightly linked with cell differentiation and function, it remains an open question to what extent signaling pathways drive metabolism or vice versa. Though T cells need transcription factors to enact the metabolic programs required for function, we only poorly understand how the metabolic status of a cell in turn affects T cell specification and function. Perhaps there are more enzymes-turned-promotors lurking in the labyrinth of metabolic regulation, like GAPDH. What is becoming increasingly evident is that the surrounding environment and nutrient accessibility can strongly affect T cell function. In tumor infiltrating lymphocytes, effector function and tumor growth are in constant battle, with tumor cells out-competing T cells during tumor progression. How do we use our knowledge of metabolic vulnerabilities to tip the balance toward favoring T cell function in tumor? Perhaps metabolic inhibitors or activators can tweak the availability of substrate that reduces the specific activity of inflammatory Teff during autoimmunity, modifies tumor use of Teff nutrients, or enhances cytotoxic lymphocyte activity to prevent tumor growth. These dual and multi-function T cells may be an interesting new area for research, where a cell’s metabolic plasticity allow for survival and function in distinct nutrient environments. As we unravel the key metabolic characteristics of these different classes of T cells, it will be important to understand the role they play in immune modulation and tumor infiltration. These strategies may provide new and significant benefit to patients without the unfortunate side effect profiles of the old and new treatment regimens.
Highlights.
T cells require glucose, glutamine, and associated transporters to activate
Myc, AMPK, and mTOR are key molecular switches of T cell metabolism
Activation of T cells induce Myc that may circumvent normal circadian rhythm
Epigenetic marks respond to metabolic status and control T cell fate
T cells under metabolic stress in inflammation or tumors decrease function
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health (R01 DK105550 and R21 CA194829).
We would like to thank members of the Rathmell lab for intellectual support.
Footnotes
CONFLICT OF INTEREST
The authors report no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.EL-GABALAWY H, GUENTHER LC, BERNSTEIN CN. Epidemiology of Immune-Mediated Inflammatory Diseases: Incidence, Prevalence, Natural History, and Comorbidities. The Journal of Rheumatology. 2010;85:2–10. doi: 10.3899/jrheum.091461. [DOI] [PubMed] [Google Scholar]
- 2.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. Journal of Biological Chemistry. 1994;269(50):31484–90. [PubMed] [Google Scholar]
- 4.House SW, Warburg O, Burk D, Schade AL. On Respiratory Impairment in Cancer Cells. Science. 1956;124(3215):267–272. [PubMed] [Google Scholar]
- 5.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261(5562):702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 6.Wise DR, Thompson CB. Glutamine Addiction: A New Therapeutic Target in Cancer. Trends in biochemical sciences. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besedovsky L, Born J, Lange T. Endogenous glucocorticoid receptor signaling drives rhythmic changes in human T-cell subset numbers and the expression of the chemokine receptor CXCR4. The FASEB Journal. 2014;28(1):67–75. doi: 10.1096/fj.13-237958. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson BH, Knutsson AK, Lindahl BO, Alfredsson LS. Metabolic disturbances in male workers with rotating three-shift work. Results of the WOLF study. International Archives of Occupational and Environmental Health. 2003;76(6):424–430. doi: 10.1007/s00420-003-0440-y. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27 485 people. Occupational and Environmental Medicine. 2001;58(11):747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froy O. The relationship between nutrition and circadian rhythms in mammals. Frontiers in Neuroendocrinology. 2007;28(2–3):61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrov S, Benedict C, Heutling D, Westermann J, Born J, Lange T. Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood. 2009;113(21):5134–5143. doi: 10.1182/blood-2008-11-190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential Localization of Effector Memory Cells in Nonlymphoid Tissue. Science. 2001;291(5512):2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 13.von Andrian UH, Mackay CR. T-Cell Function and Migration — Two Sides of the Same Coin. New England Journal of Medicine. 2000;343(14):1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 14.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–51. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. The Journal of Experimental Medicine. 2015;212(9):1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185(2):1037–44. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14(5):500–8. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alves NL, Derks IAM, Berk E, Spijker R, van Lier RAW, Eldering E. The Noxa/Mcl-1 Axis Regulates Susceptibility to Apoptosis under Glucose Limitation in Dividing T Cells. Immunity. 24(6):703–716. doi: 10.1016/j.immuni.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, Manalis SR, Vander Heiden MG. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Developmental cell. 2016;36(5):540–9. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. The Journal of Clinical Investigation. 123(10):4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Windt Gerritje JW, Everts B, Chang C-H, Curtis Jonathan D, Freitas Tori C, Amiel E, Pearce Edward J, Pearce Erika L. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–83. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukens J, Dixit VD, Kanneganti T-D. Inflammasome activation in obesity-related inflammatory diseases and autoimmunity. Discovery medicine. 2011;12(62):65–74. [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Youm Y-H, Vandanmagsar B, Ravussin A, Gimble JM, Greenway F, Stephens JM, Mynatt RL, Dixit VD. Obesity Increases the Production of Proinflammatory Mediators from Adipose Tissue T Cells and Compromises TCR Repertoire Diversity: Implications for Systemic Inflammation and Insulin Resistance. Journal of immunology (Baltimore, Md: 1950) 2010;185(3):1836–1845. doi: 10.4049/jimmunol.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan P, Kahn DA, Baltimore D. Anti-apoptotic effect of hyperglycemia can allow survival of potentially autoreactive T cells. Cell Death and Differentiation. 2011;18(4):690–699. doi: 10.1038/cdd.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeachy MJ, Stephens LA, Anderton SM. Natural Recovery and Protection from Autoimmune Encephalomyelitis: Contribution of CD4+CD25+ Regulatory Cells within the Central Nervous System. The Journal of Immunology. 2005;175(5):3025–3032. doi: 10.4049/jimmunol.175.5.3025. [DOI] [PubMed] [Google Scholar]
- 27.Selvaraj RK, Geiger TL. Mitigation of Experimental Allergic Encephalomyelitis by TGF-β Induced Foxp3+ Regulatory T Lymphocytes through the Induction of Anergy and Infectious Tolerance. The Journal of Immunology. 2008;180(5):2830–2838. doi: 10.4049/jimmunol.180.5.2830. [DOI] [PubMed] [Google Scholar]
- 28.Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, Nochy D, Debré P, Piette J-C, Gorochov G. Global Natural Regulatory T Cell Depletion in Active Systemic Lupus Erythematosus. The Journal of Immunology. 2005;175(12):8392–8400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Chu Y, Yang X, Gao D, Zhu L, Yang X, Wan L, Li M. Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis & Rheumatism. 2009;60(5):1472–1483. doi: 10.1002/art.24499. [DOI] [PubMed] [Google Scholar]
- 30.Marabelle A, Kohrt H, Sagiv-Barfi I, Ajami B, Axtell RC, Zhou G, Rajapaksa R, Green MR, Torchia J, Brody J, Luong R, Rosenblum MD, Steinman L, Levitsky HI, Tse V, Levy R. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. The Journal of Clinical Investigation. 123(6):2447–2463. doi: 10.1172/JCI64859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegazy AN, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Löhning M. Interferons Direct Th2 Cell Reprogramming to Generate a Stable GATA-3+T-bet+ Cell Subset with Combined Th2 and Th1 Cell Functions. Immunity. 2010;32(1):116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Shi G, Cox CA, Vistica BP, Tan C, Wawrousek EF, Gery I. Phenotype Switching by Inflammation-Inducing Polarized Th17 Cells, but not by Th1 Cells. Journal of immunology (Baltimore, Md: 1950) 2008;181(10):7205–7213. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou L, Chong MMW, Littman DR. Plasticity of CD4+ T Cell Lineage Differentiation. Immunity. 2009;30(5):646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Koenen HJPM, Smeets RL, Vink PM, van Rijssen E, Boots AMH, Joosten I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17–producing cells. Blood. 2008;112(6):2340–2352. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 35.Spiegelman BM, Flier JS. Obesity and the Regulation of Energy Balance. Cell. 104(4):531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 36.Dang CV, Kim J-w, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8(1):51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 37.Laplante M, Sabatini David M. mTOR Signaling in Growth Control and Disease. Cell. 149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill HM, Holloway GP, Steinberg GR. AMPK regulation of fatty acid metabolism and mitochondrial biogenesis: Implications for obesity. Molecular and Cellular Endocrinology. 2013;366(2):135–151. doi: 10.1016/j.mce.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase – development of the energy sensor concept. The Journal of Physiology. 2006;574(Pt 1):7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoki K, Zhu T, Guan K-L. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell. 2003;115(5):577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 41.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung Y-L, Schulze A. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metabolism. 2008;8(3-3):224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832–44. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499(7459):485–90. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, Bluestone JA, Turka LA. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16(2):188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verbist KC, Guy CS, Milasta S, Liedmann S, Kamiński MM, Wang R, Green DR. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. 2016;532(7599):389–393. doi: 10.1038/nature17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Procaccini C, De Rosa V, Galgani M, Abanni L, Calì G, Porcellini A, Carbone F, Fontana S, Horvath TL, La Cava A, Matarese G. An Oscillatory Switch in mTOR Kinase Activity Sets Regulatory T Cell Responsiveness. Immunity. 33(6):929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, Bluestone JA, Turka LA. The phosphatase PTEN-mediated control of PI-3 kinase in T(regs) cells maintains homeostasis and lineage stability. Nature immunology. 2015;16(2):188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Regulatory T cells require the phosphatase PTEN to restrain type 1 and follicular helper T-cell responses. Nature immunology. 2015;16(2):178–187. doi: 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao P, Tchernyshyov I, Chang T-C, Lee Y-S, Kita K. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eilers M, Eisenman RN. Myc’s broad reach. Genes & Development. 2008;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A Large Scale Genetic Analysis of c-Myc-regulated Gene Expression Patterns. Journal of Biological Chemistry. 2003;278(14):12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- 53.Wang R, Green DR. Metabolic reprogramming and metabolic dependency in T cells. Immunol Rev. 2012;249(1):14–26. doi: 10.1111/j.1600-065X.2012.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of Hypoxia-Inducible Factor 1α Expression and Function by the Mammalian Target of Rapamycin. Molecular and CellularBiology. 2002;22(20):7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Transcription Factor HIF-1 Is a Necessary Mediator of the Pasteur Effect in Mammalian Cells. Molecular and Cellular Biology. 2001;21(10):3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojima H, Gu H, Nomura S, Caldwell CC, Kobata T, Carmeliet P, Semenza GL, Sitkovsky MV. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1α-deficient chimeric mice. Proceedings of the National Academy of Sciences. 2002;99(4):2170–2174. doi: 10.1073/pnas.052706699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of T(H)17 and T(reg) cells. The Journal of Experimental Medicine. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang Eric V, Barbi J, Yang H-Y, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen H-R, Luo W, Zeller K, Shimoda L, Topalian Suzanne L, Semenza Gregg L, Dang Chi V, Pardoll Drew M, Pan F. Control of TH17/Treg Balance by Hypoxia-Inducible Factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonafé N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-Phosphate Dehydrogenase Binds to the AU-Rich 3′ Untranslated Region of Colony-Stimulating Factor–1 (CSF-1) Messenger RNA in Human Ovarian Cancer Cells: Possible Role in CSF-1 Posttranscriptional Regulation and Tumor Phenotype. Cancer Research. 2005;65(9):3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 60.Ho P-C, Bihuniak Jessica D, Macintyre Andrew N, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales Jose C, Kleinstein Steven H, Abel ED, Insogna Karl L, Feske S, Locasale Jason W, Bosenberg Marcus W, Rathmell Jeffrey C, Kaech Susan M. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, Sun SC. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40(5):692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray CM, Hutchinson R, Bantick JR, Belfield GP, Benjamin AD, Brazma D, Bundick RV, Cook ID, Craggs RI, Edwards S, Evans LR, Harrison R, Holness E, Jackson AP, Jackson CG, Kingston LP, Perry MWD, Ross ARJ, Rugman PA, Sidhu SS, Sullivan M, Taylor-Fishwick DA, Walker PC, Whitehead YM, Wilkinson DJ, Wright A, Donald DK. Monocarboxylate transporter MCT1 is a target for immunosuppression. Nat Chem Biol. 2005;1(7):371–376. doi: 10.1038/nchembio744. [DOI] [PubMed] [Google Scholar]
- 64.Jacobs SR, Herman CE, MacIver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways. Journal of immunology (Baltimore, Md: 1950) 2008;180(7):4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neildez-Nguyen TMA, Bigot J, Da Rocha S, Corre G, Boisgerault F, Paldi A, Galy A. Hypoxic culture conditions enhance the generation of regulatory T cells. Immunology. 2015;144(3):431–443. doi: 10.1111/imm.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newsholme P, Procopio J, Lima MMR, Pithon-Curi TC, Curi R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochemistry and Function. 2003;21(1):1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 68.Cheng RJD. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2009;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Matés JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–9. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, Rutter J, Merritt ME, DeBerardinis RJ. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56(3):414–24. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pochini L, Scalise M, Galluccio M, Indiveri C. Membrane transporters for the special amino acid glutamine: structure/function relationships and relevance to human health. Frontiers in Chemistry. 2014;2:61. doi: 10.3389/fchem.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng M, Yin N, Li Ming O. Sestrins Function as Guanine Nucleotide Dissociation Inhibitors for Rag GTPases to Control mTORC1 Signaling. Cell. 2014;159(1):122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chantranupong L, Scaria Sonia M, Saxton Robert A, Gygi Melanie P, Shen K, Wyant Gregory A, Wang T, Harper JW, Gygi Steven P, Sabatini David M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165(1):153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poncet N, Mitchell FE, Ibrahim AFM, McGuire VA, English G, Arthur JSC, Shi Y-B, Taylor PM. The Catalytic Subunit of the System L1 Amino Acid Transporter (Slc7a5) Facilitates Nutrient Signalling in Mouse Skeletal Muscle. PLoS ONE. 2014;9(2):e89547. doi: 10.1371/journal.pone.0089547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones PA, Takai D. The Role of DNA Methylation in Mammalian Epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 76.Niculescu MD, Zeisel SH. Diet, Methyl Donors and DNA Methylation: Interactions between Dietary Folate, Methionine and Choline. The Journal of Nutrition. 2002;132(8):2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 77.Trerotola M, Relli V, Simeone P, Alberti S. Epigenetic inheritance and the missing heritability. Human Genomics. 2015;9(1):17. doi: 10.1186/s40246-015-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mentch Samantha J, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman Marcas M, Thalacker-Mercer Anna E, Nichenametla Sailendra N, Locasale Jason W. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metabolism. 22(5):861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakajima H, Kunimoto H. TET2 as an epigenetic master regulator for normal and malignant hematopoiesis. Cancer Science. 2014;105(9):1093–1099. doi: 10.1111/cas.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee Joyce V, Carrer A, Shah S, Snyder Nathaniel W, Wei S, Venneti S, Worth Andrew J, Yuan Z-F, Lim H-W, Liu S, Jackson E, Aiello Nicole M, Haas Naomi B, Rebbeck Timothy R, Judkins A, Won K-J, Chodosh Lewis A, Garcia Benjamin A, Stanger Ben Z, Feldman Michael D, Blair Ian A, Wellen Kathryn E. Akt-Dependent Metabolic Reprogramming Regulates Tumor Cell Histone Acetylation. Cell Metabolism. 20(2):306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 82.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YYJ, Beekman JM, van Beekum O, Brenkman AB, Hijnen D-J, Mutis T, Kalkhoven E, Prakken BJ, Coffer PJ. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115(5):965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 83.Balmer Maria L, Ma Eric H, Bantug Glenn R, Grählert J, Pfister S, Glatter T, Jauch A, Dimeloe S, Slack E, Dehio P, Krzyzaniak Magdalena A, King Carolyn G, Burgener A-V, Fischer M, Develioglu L, Belle R, Recher M, Bonilla Weldy V, Macpherson Andrew J, Hapfelmeier S, Jones Russell G, Hess C. Memory CD8+ T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity. doi: 10.1016/j.immuni.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 84.Prendergast G, Ziff E. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 85.Tserel L, Kolde R, Limbach M, Tretyakov K, Kasela S, Kisand K, Saare M, Vilo J, Metspalu A, Milani L, Peterson P. Age-related profiling of DNA methylation in CD8+ T cells reveals changes in immune response and transcriptional regulator genes. Scientific Reports. 2015;5:13107. doi: 10.1038/srep13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones B, Chen J. Inhibition of IFN-γ transcription by site-specific methylation during T helper cell development. The EMBO Journal. 2006;25(11):2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. Effect of Promoter Methylation on the Regulation of IFN-γ Gene During In Vitro Differentiation of Human Peripheral Blood T Cells into a Th2 Population. The Journal of Immunology. 2003;171(5):2510–2516. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- 88.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A Critical Role for Dnmt1 and DNA Methylation in T Cell Development, Function, and Survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 89.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang H-D, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic Control of the foxp3 Locus in Regulatory T Cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gray SM, Kaech SM, Staron MM. The interface between transcriptional and epigenetic control of effector and memory CD8(+) T-cell differentiation. Immunological reviews. 2014;261(1):157–168. doi: 10.1111/imr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang S, Collins PL, Aune TM. T-Bet Dependent Removal of Sin3A-Histone Deacetylase Complexes at the Ifng Locus Drives Th1 Differentiation. Journal of immunology (Baltimore, Md: 1950) 2008;181(12):8372–8381. doi: 10.4049/jimmunol.181.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. The EMBO Journal. 2010;29(12):2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Romero-Garcia S, Moreno-Altamirano MMB, Prado-Garcia H, Sánchez-García FJ. Lactate Contribution to the Tumor Microenvironment: Mechanisms, Effects on Immune Cells and Therapeutic Relevance. Frontiers in Immunology. 2016;7:52. doi: 10.3389/fimmu.2016.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, Renner K, Timischl B, Mackensen A, Kunz-Schughart L, Andreesen R, Krause SW, Kreutz M. Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood. 2007;109(9):3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 95.Ottensmeier CH, Perry KL, Harden EL, Stasakova J, Jenei V, Fleming J, Wood O, Woo J, Woelk CH, Thomas GJ, Thirdborough SM. Upregulated glucose metabolism correlates inversely with CD8+ T cell infiltration and survival in squamous cell carcinoma. Cancer Research. 2016 doi: 10.1158/0008-5472.CAN-15-3121. [DOI] [PubMed] [Google Scholar]
- 96.Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T Cells Are Metabolically Anergic. Journal of immunology (Baltimore, Md: 1950) 2009;183(10):6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. CTLA-4 and PD-1 Receptors Inhibit T-Cell Activation by Distinct Mechanisms. Molecular and Cellular Biology. 2005;25(21):9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Probst HC, McCoy K, Okazaki T, Honjo T, van den Broek M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat Immunol. 2005;6(3):280–286. doi: 10.1038/ni1165. [DOI] [PubMed] [Google Scholar]
- 100.Chang C-H, Qiu J, O’Sullivan D, Buck Michael D, Noguchi T, Curtis Jonathan D, Chen Q, Gindin M, Gubin Matthew M, van der Windt Gerritje JW, Tonc E, Schreiber Robert D, Pearce Edward J, Pearce Erika L. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proceedings of the National Academy of Sciences. 2008;105(39):15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun. 2015;6 doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siska PJ, Rathmell JC. T cell metabolic fitness in antitumor immunity. Trends in Immunology. 36(4):257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nature reviews. Immunology. 2013;13(3):190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qian J, Scheer FAJL. Circadian System and Glucose Metabolism: Implications for Physiology and Disease. Trends in Endocrinology & Metabolism. 2016;27(5):282–293. doi: 10.1016/j.tem.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takahashi JS, Hong H-K, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paschos GK, Ibrahim S, Song W-L, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, FitzGerald GA. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18(12):1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dibner C, Schibler U, Albrecht U. The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annual Review of Physiology. 2010;72(1):517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 110.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102(12):4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 111.Ando N, Nakamura Y, Aoki R, Ishimaru K, Ogawa H, Okumura K, Shibata S, Shimada S, Nakao A. Circadian Gene Clock Regulates Psoriasis-Like Skin Inflammation in Mice. The Journal of Investigative Dermatology. 2015;135(12):3001–3008. doi: 10.1038/jid.2015.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marciano David P, Chang Mi R, Corzo Cesar A, Goswami D, Lam Vinh Q, Pascal Bruce D, Griffin Patrick R. The Therapeutic Potential of Nuclear Receptor Modulators for Treatment of Metabolic Disorders: PPARγ, RORs, and Reverbs. Cell Metabolism. 2014;19(2):193–208. doi: 10.1016/j.cmet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 113.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes & Development. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Altman Brian J, Hsieh Annie L, Sengupta A, Krishnanaiah Saikumari Y, Stine Zachary E, Walton Zandra E, Gouw Arvin M, Venkataraman A, Li B, Goraksha-Hicks P, Diskin Sharon J, Bellovin David I, Simon MC, Rathmell Jeffrey C, Lazar Mitchell A, Maris John M, Felsher Dean W, Hogenesch John B, Weljie Aalim M, Dang Chi V. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metabolism. 2015;22(6):1009–1019. doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hemmers S, Rudensky Alexander Y. The Cell-Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell Reports. 11(9):1339–1349. doi: 10.1016/j.celrep.2015.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]