Abstract
Affinity-based chromatography assays encompass the use of solid supports containing immobilized biological targets to monitor binding events in the isolation , identification and/or characterization of bioactive compounds. This powerful bioanalytical technique allows the screening of potential binders through fast analyses that can be directly performed using isolated substances or complex matrices. An overview of the recent researches in frontal and zonal affinity-based chromatography screening assays, which has been used as a tool in the identification and characterization of new anti-cancer agents, is discussed. In addition, a critical evaluation of the recently emerged ligands fishing assays in complex mixtures is also discussed.
Keywords: Anti-cancer active compounds, screening method, affinity-based chromatography assays
1. INTRODUCTION
Currently, the identification of novel anti-cancer compounds is predominantly carried out with functional cell assays using established cell lines to measure the cytotoxic effects. Of these, tetrazolium salt-based assays including MTT, MTS, XTT or WST are the most widely used to assess cell proliferation, cell viability and drug cytotoxicity [1]. The screening of these compounds has to be carried out on both tumoral as well as normal cell lines, to identify compounds that are selective for the tumoral cell line. While these methods have been used successfully as evidenced by their widespread use, the entire process of screening new anti-cancer active compounds is time-consuming and typically is low throughput.
More recently, bioaffinity chromatography has been used as a novel approach to identify potential new anti-cancer active compounds [2]. Using this method, a specific target, typically a protein that could target cell growth and/or cell survival of tumoral cells [3], is immobilized and screened against a complex mixture or synthetic combinatorial library for the identification of novel active compounds for cancer treatment. The screening of natural products is of paramount importance, as the majority of anti-cancer compounds are nature-based or derived from a nature-based product. As a result, the chromatography-based bioassays have been exploited as a promising approach for the identification of novel active compounds [4–7]. Once a compound is identified with this approach, the compound can be fully characterized including its affinity for the targeted protein [8, 9]. Several recent reviews [2, 4, 9] have discussed in great detail the variety of supports and method of immobilization. In addition, the recent review carried out by Hage et al. [9] provides a good overview of the various classifications, including high performance affinity chromatography (HPAC), high performance liquid affinity chromatography (HPLAC) and affinity monolith chromatography (AMC). Herein, we are going to use the term bioaffinity chromatography as a general term describing the aforementioned classifications.
Bioaffinity chromatography is typically characterized using a variety of elution modes, including zonal (linear and non-linear) chromatography and frontal affinity chromatography. These methods can be used to calculate equilibrium and kinetics constants [4, 10]. Zonal chromatography has been successfully used for fragment-based drug discovery/design (FBDD) for the determination of retention factors [11]. In this case, fragments were analyzed by mass spectrometry (MS/MS) and the specific retention times were obtained by comparing the retentions between the activated and non-activated bioaffinity column [12]. The bioaffinity columns for these assays are usually prepared by in-situ immobilization in capillary columns (100 µm × 0.5 mm) packed with porous spherical silica [12, 13]. Zonal chromatography has also been used in screening inhibitors for immobilized enzyme reactors (IMERs) [4, 8, 14–17]. In these studies, inhibition of enzymatic activity can be studied by measuring changes in product formation. A limitation of this method is the necessity for the enzyme to have a relatively high turnover numbers due to the limited contact time between the enzyme and the binder. Using zonal chromatography, non-linear conditions have also been used for assessing binding affinities by monitoring the signals out-put and the deviation from the Gaussian band profile. The association/dissociation constants (kon and koff) and equilibrium dissociation constant (Kd) can be calculated by the use of the nonlinear isotherms [18, 19].
Frontal affinity chromatography unlike zonal affinity chromatography is carried out under dynamic equilibrium conditions. It is frequently used in association with mass spectrometry and has several advantages including the capability of determining equilibrium dissociation constants (Kd) and the number of active binding sites (Bt) which can be calculated from the breakthrough curves [12, 20]. In addition, it was recently demonstrated that the binders can be ranked based on their affinity towards the column [4, 14]. The main drawback of this approach is the large concentration of binders required as they are continuously infused over the entire run and thus result in the use of larger amounts of the binder mixtures. The presence of a displacer ligand in the mobile phase results in another frequently used method termed frontal and/or zonal displacement chromatography. In this case, a set concentration of a known binder (marker) is placed in the mobile phase with increasing concentrations of a displacer. Based on the change in retention volume, binding affinity of the displacer can be calculated. The experiments can be used to qualitatively rank compounds according to their EC50 values [9, 21, 22].
More recently, the versatility of these approaches has been demonstrated in bioconjugation experiments, where proteins were immobilized onto the surface of magnetic particles, to ‘fish’ binders out of a complex mixture. The experiments are usually associated with bioaffinity chromatography for evaluating the equilibrium dissociation constant of the identified binders [4, 18].
In this review, the use of bioaffinity chromatography for probing ligand-protein and protein–protein interactions will be explored with respect to targeting anti-cancer active compounds. All the reported studies in each section used bioaffinity chromatography-based methodologies to prospect new cancer treatments.
2. CHROMATOGRAPHY-BASED ASSAYS
2.1. Frontal Affinity Chromatography
Frontal chromatography is a widely used affinity-based chromatographic approach used for screening active compounds through the frontal elution. It is a quantitative method firstly described by Kasay et al in 1975 [23] and carried out under dynamic equilibrium conditions, where the sample is continuously infused in the chromatographic column and each constituent of the sample emerges (breaks through) at a different time depending on its concentration and affinity for the stationary phase, forming a stepped chromatogram. The term frontal affinity chromatography (FAC) is used when the chromatographic column contains an immobilized biological target, like a protein, and the infused sample contains potential binders [4].
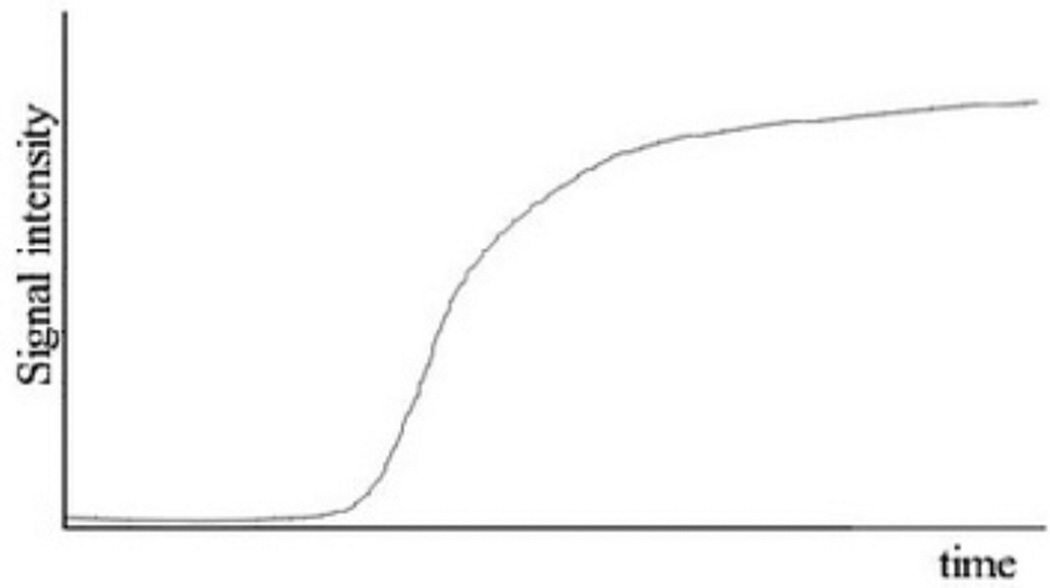
In FAC assays, usually the potential binders are added to the mobile phase and the sample is continuously delivered onto the column. Each sample constituent migrates through the column at different rates, depending on its affinity for the immobilized biological target, and breaks through as a series of fronts. The continuous infusion of the binders results in a titration of the binding sites of the immobilized target: in the beginning, the binder concentration eluting from the column is low, since the number of available binding sites on the surface of the immobilized target is large. When the binding sites become saturated, the binder concentration eluting from the column increases gradually, producing a vertical rise in the chromatographic trace, often referred as breakthrough curve. At the end of this curve, the infused and eluted binder concentration will be identical, forming a plateau, as illustrated in the Figure 1 [4].
Fig. 1.
Typical breakthrough curve obtained in a FAC assay.
The required chromatographic system to carry out frontal affinity analysis consists of a liquid chromatography pump, an injection system, a chromatographic column containing the immobilized biological target, and a detector. The employed detector depends on the complexity and concentration of the infused sample and could include a radioflow, UV or fluorescence detector, and a mass spectrometer. The detection of the analyte requires either a selective labeling of a binder and the use of a corresponding detector,for example radiolabeled marker with a radioflow detector or a fluorescently labeled binder with a fluorescent detector; or a detector capable of discriminating between coeluting compounds, for example a mass spectrometer (MS), as long as each compound has a unique m/z or a characteristic transition (MS/MS) for tandem systems, thus allowing a label-free assay, making it the most flexible and generalized strategy [24–26].
FAC can be applied to the investigation of ligand-protein and protein-protein interactions and in this review we will highlight recent applications of FAC in the investigation of binders-protein interactions for targeting anti-cancer active compounds.
Generally, FAC assays can be classified in two different groups: direct and indirect assays (Figs. 2 and 3). In the direct assays, the evaluated compound (analyte) is directly monitored by the detector of the chromatographic system and its retention time is directly associated with its concentration and affinity for the immobilized biomolecule. While, indirect assays (displacement chromatography) include the use of a known ligand as a marker and the interaction of the analyte with the immobilized biomolecule is indirectly observed through a displacement of the marker breakthrough curve. Both approaches can be used for binders screening and characterization purposes.
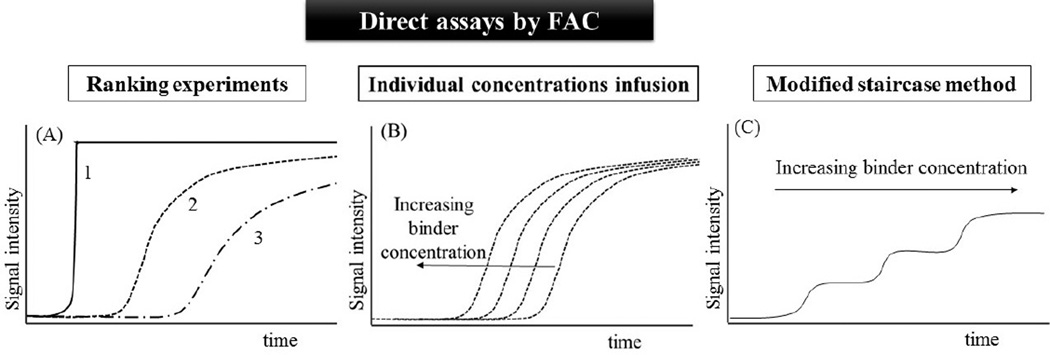
Fig. 2.
Representative illustrations of FAC direct assays for binders screening and characterization. In ranking experiments (A), a compounds mixture is continuously infused at a known concentration onto the bioaffinity column: the breakthrough curve 1 represents the elution of a non-affinity compound; the breakthrough curve 2 represents the elution of a compound with moderate affinity to the immobilized target; the breakthrough curve 3 represents the elution of the most potent binder presents in the evaluated mixture. The dissociation constant (Kd) can be accurately obtained by the infusion of individual concentrations of the binder (B), or in a single assay with the sequential infusion of the binder at increasing concentrations by the modified staircase method (C).
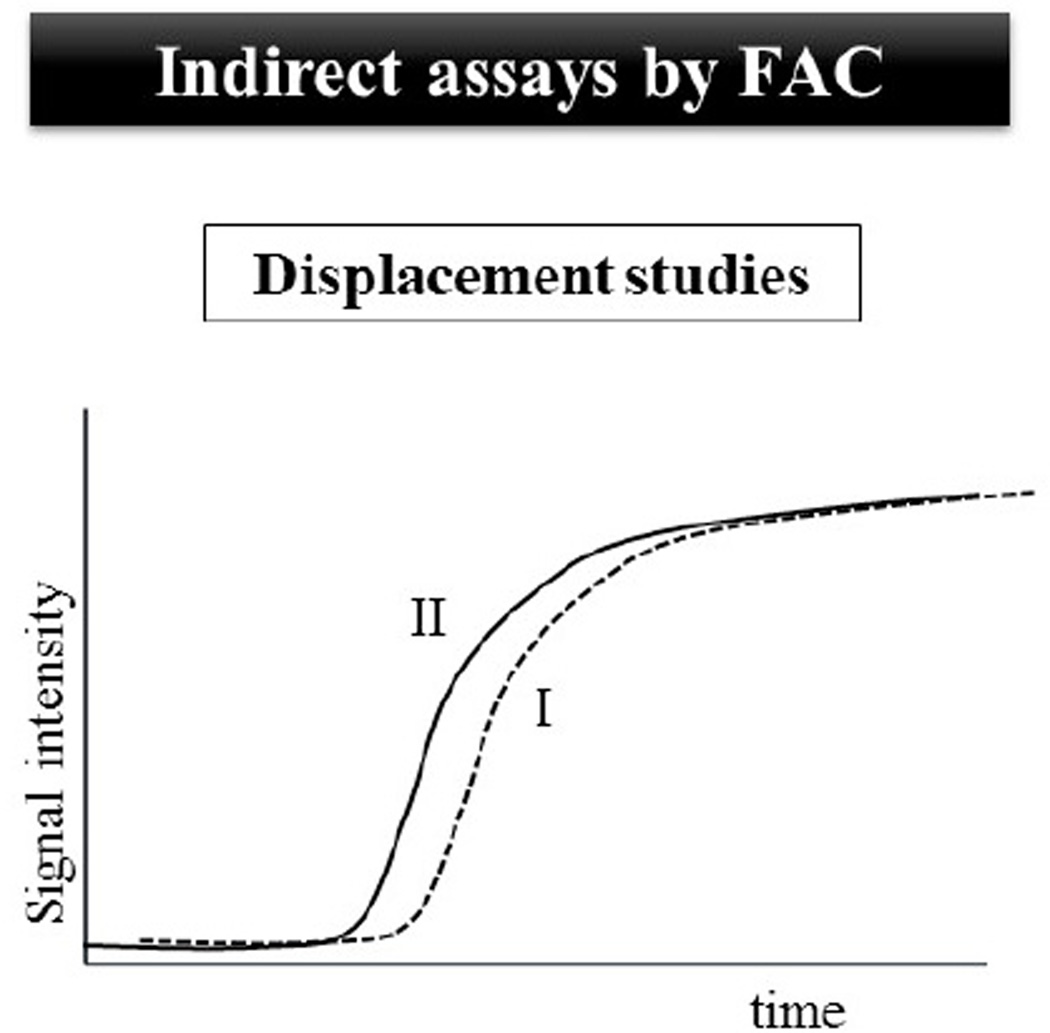
Fig. 3.
In displacement studies, the breakthrough curve of a marker (I) is monitored. When the analyzed solution contains the marker and an evaluated compound (or a compounds mixture), the breakthrough curve of the marker could be displaced (II), suggesting a direct competition between the binder and the evaluated compound (or at least one compound in a mixture) competes directly for a specific binding site.
2.1.1. Direct assays by FAC
FAC direct assays are a valuable tool to screen and characterize ligands. The mean position of the ligand breakthrough curve (breakthrough time or volume) depends on the ligand concentration [A], the number of available active binding sites (Bt, in mol) and the dissociation constant of the binder-target interaction (Kd). The basic FAC equation (Eq.1) encompasses all these parameters, where V is the retention volume of the ligand obtained from the midpoint of the breakthrough curve and V0 is the retention volume in the absence of the binding event. V0 can be calculated from the retention volume of a compound with no affinity for the immobilized target, from the retention volume of the binders using a similar chromatographic column without the immobilized target or the retention volume of a saturating concentration of the ligand. The chromatographic profile can be analyzed with a polynomial equation to derive the inflection point corresponding to the breakthrough volume (V) [4, 25, 26].
| Equation 1 |
One of the most important FAC direct assays is the ranking experiment, which is carried out by ranking the affinity order on the basis of breakthrough volume. The most potent binders from the mixture will elute last due to the higher affinity for the immobilized target (Fig. 2A). To ensure that the elution order is specifically related to the binder-immobilized biomolecule interaction, it is crucial to investigate nonspecific interactions. Thus, a column without the targeted protein is frequently used, either by using an irreversible inhibitor [12] or by preparing an identical column without the immobilized target [4, 20]. Furthermore, the used of a void marker (a non affinity compound) is critical to verify the fastest elution time, and rank the constituents from the compounds mixture.
Ranking experiments by FAC were used to screen inhibitors of epidermal growth factor receptor (EGFR) in Caragana jubata crude extract [27]. A polyclonal antibody raised against piceatannol (a known anti-EGFR inhibitor) was coupled with bovine serum albumin for mimicking the receptor and used as stationary phase. The FAC assay was performed by infusion of 100 µg.mL−1 of the crude extract from which the six most abundant ions were monitored. The elution order and the breakthrough volumes showed the efficiency of the method of recognizing and ranking different anti-EGFR inhibitors.
Two synthetized β-D-Galp-(1–3)-β-D-GlcpN (lacto-N-biose) disaccharide libraries were screened for galectin-3 binding on a recombinant human galectin-3 column through ranking assays [28]. A known non-binder trisaccharide was used as the void volume marker. The 17-synthetized disaccharides were split into four mixtures containing an equimolar ratio of each compound. To ensure that the results were not affected by non-specific interaction, a control experiment was performed with a blank column. The relative retention times of the library components were used to classify the most potent binders.
Direct assays by FAC can be also employed in the characterization of binders through the determination of the dissociation constant of the binder-target interaction (Kd), as illustrated in Fig. 2B and 2C. This approach is useful for the characterization of the immobilized protein, by determining the number of active binding sites on the column (Bt) and the binding affinity (Kd) of the tested ligand. These parameters are obtained using a series of concentrations of a ligand and determining the breakthrough volume at each concentration. By analyzing changes in (V-V0)−1 versus [A] using a Lineweaver-Burk type double reciprocal plot, one can determine Kd and Bt from the intercepts on the ordinate and from the slope, respectively [4, 25, 29]. Several other standard non-linear regression analyses can also be employed.
Human purine nucleoside phosphorylase (HsPNP) bioaffinity-based capillary columns were prepared for affinity screening and characterization studies [20]. The dissociation constant for a fourth-generation immucillin derivative, an HsPNP inhibitor, and the number of available active binding sites in the immobilized enzyme were assessed by FAC experiments. To this end, increasing concentrations of this inhibitor were continuously infused until a typical sigmoidal profile was obtained. The injection of increasing inhibitor concentration resulted in traces with reduced breakthrough volumes. This approach allowed the determination of Kd for the binder characterization, and the characterization of the bioaffinity columns through the Bt investigation.
A second approach to assess the Kd and Bt is a frontal binding assay called modified staircase method (Fig. 2C) [4]. In this approach, the binder is sequentially infused at increasing concentrations until saturation, starting from the lowest concentration, with the simultaneously infusion of a void marker, at a fixed concentration. Equation 2 is used to calculate Kd and Bt.
| Equation 2 |
Where V – V0 is the corrected breakthrough volume for the binder, Bt is the number of available active binding sites in mol, A0 refers to the binder infusion concentration and the summed concentrations ([A]0+y) refer to the initial concentration of the binder for the first step of the staircase; while for the subsequent steps will be the sum of that step and all its predecessors [4, 26]. The slope of a plot of [A]0+y versus reciprocal (V-V0) provides the column capacity Bt and the negative intercept provides Kd. The ability to obtain Bt and Kd for the binder-biomolecule binding event from a single course of experiments is a unique feature of FAC [24, 30].
A modified staircase method was used for Kd determination of human recombinant protein kinase (PKCα) and chelerythrine chloride, a PKCα substrate site competitive inhibitor [31]. The determined value (698 nM) is comparable to the literature IC50 value of 660 nM, and demonstrates that in a simple assay this approach can be used to accurately assess the Kd value.
The performance of the modified staircase method and the individual concentrations infusion for determining Kd value was compared by Temporini et al [32]. A human recombinant A2A adenosine receptor [33] bioaffinity column was employed to assess Kd and Bt using ANR 152, a known human recombinant A2A receptor. Individual concentrations of the binder (2.5–12.5 nM) were infused, with a 12h washing procedure between each analysis for column regeneration. By this approach, the Kd and Bt determination experiments were conducted in 4–5 days. Using the modified staircase method, Bt and Kd were assessed in 4–5h of experiments.
2.1.2. Indirect Assays by FAC (Frontal Displacement Chromatography)
Screening and Kd determination of high- and low-affinity protein interactions can be assessed by the use of a marker ligand, a compound with high affinity for the target protein. The infusion of a solution containing the marker and the evaluated compound results in shorter breakthrough volumes for the marker if the analyte competes for the same binding site of the marker ligand (Figure 3). In this case, the analyte can be called displacer. Therefore, these assays are less susceptible to the interference of non-specific interactions. The larger the percentage shift, the higher is the degree of competition between the marker and the displacer for the specific binding site. High productivity can be obtained with this assay for identifying active compounds in complex mixtures. This approach is also referred as frontal displacement chromatography. The percentage shift can be calculated by equation 3 [25, 34].
| Equation 3 |
Where tM is the marker breakthrough time in the absence of a displacer, t is the corrected marker breakthrough time in the presence of a displacer (the difference between the breakthrough time of the marker and the breakthrough time of the void marker), tNSB is the nonspecific binding breakthrough time difference in the absence of the immobilized target [25, 34].
The breast cancer resistance protein (BCRP) is expressed in the nuclear membranes of human-derived glioblastoma and astrocytoma cell lines [35]. To prepare a bioaffinity column containing immobilized BCRP, a nuclear membrane affinity column was produced by the immobilization of nuclear membrane fragments from the LN-229 astrocytoma cell line onto an immobilized artificial membrane stationary phase (IAM) [36]. [3H]-Etoposide, a BCRP substrate, was used as a marker ligand to confirm the presence of functional BCRP using frontal displacement chromatography. Increasing concentrations of unlabeled etoposide resulted in a decrease in the breakthrough volume of the marker, representing specific binding to the bioaffinity column. The Kd for the displacer could be determined following the equation 4:
| Equation 4 |
where [D] is the displacer concentration, V is the breakthrough volume of the marker, V0 is the retention volume in the absence of the binding event, P is the number of available active binding sites in mol (Bt) multiplied by the ratio of the Kd of displacer over the Kd of the marker, and Kd is the dissociation constant for the displacer. A non-linear regression plot of [D] (V-V0) versus [D] furnishes the Kd value for the displacer [36]. The calculated Kd for the displacer etoposide on the bioaffinity column was consistent with those one obtained from another approaches.
Table 1 summarizes the application of affinity-based chromatography assays by frontal elution in the search and characterization of bioactive anti-cancer compounds. Evaluating the papers depicted at Table 1, it is clear that an increase in this assay approach may result in faster lead times for hit identification.
Table 1.
Studies described in the literature using FAC assays in the identification and/or characterization of new anti-cancer agents.
| Immobilized biomolecule | Chromatography assay | Binders/Analytes | Ref |
|---|---|---|---|
| Angiogenesis inhibitor Kingle 5 |
Infusion of individual concentrations of each binder for Kd and Bt determination |
L-lysine, epsilon-aminocaproic acid, 7-aminohepatanoic acid,trans-4-(aminomethyl)cyclohexane carboxylic acid and benzylamine |
[37] |
| Carbohydrates (α-man, β- gal and β-glc) |
Infusion of individual concentrations of each binder (lectins) for Kd and Bt determination |
Lectins (Con A, LCA, PNA) | [38, 39] |
| Lectins (Aleuria aurantia and Aspergillus oryzae lectins) |
Infusion of individual concentrations of each binder for Kd and Bt determination |
p-nitrophenyl α-L-fucopyranoside and 113 pyridylaminated oligosaccharides |
[40] |
| Dihydrofolate reductase | Infusion of individual concentrations of each binder for Kd and Bt determination and ranking experiments |
Folic acid, pyrimethamine and trimethoprim | [41] |
| Sirtuin-6 (SIRT6 protein) | Displacement studies for ranking studies and characterization of the binding site |
Quercetin and others structurally related flavonoids | [42, 43] |
| Human galectin-1 and other mammalian galectins |
Infusion of individual concentrations of each binder for Kd and Bt determination; displacement studies |
Pyridylaminated sugars | [44] |
| P-glycoprotein | Displacement studies for Kd and Bt determination |
Vinblastine, verapamil, doxorubicin and cyclosporine A | [45] |
| G protein-coupled receptor- 17 |
Infusion of individual concentrations of each binder for Kd and Bt determination and ranking experiments |
Cangrelor, MRS2179 and uridine diphosphate | [46] |
| Heat shock protein 90 | Infusion of individual concentrations of each binder for Kd and Bt determination |
Coumermycin A, novobiocin, geldanamycin, 17-AAG and radicicol |
[47] |
| Erythropoietin-producing hepatocellular B2 |
Displacement studies for screening of 468 compounds (52 mixtures of 9 compounds) |
11 kinase inhibitors | [34, 48] |
2.2. Zonal Affinity Chromatography
Antitumor drug discovery has become one of the most challenging and researched fields in cancer therapy, and numerous screening techniques, based on target receptors or enzymes, and virtual screening systems, using computer-aided drug design, have been employed for investigating lead compounds or drugs [49]. Here we focus specifically on small molecule interactions with an immobilized anticancer target, with an emphasis on enzymes, membrane-bound receptors and a multitarget lipid-raft-coated silica beads by zonal elution analysis.
2.2.1. Principles of Zonal Elution
Zonal elution generally involves the injection of a small amount of binder through a column under linear elution conditions; an online detector monitors the elution time or volume of the binder. In zonal bioaffinity chromatography (ZBC), the chromatographic process includes binding/affinity interactions between the bio-molecule and the binder. It is possible to obtain information on the equilibrium constants describing this interaction [50–52]. Zonal elution can measure the degree of affinity of a binder–protein binding by either varying the mobile-phase composition and/or temperature or studying the alterations in binder and/or protein structures. Furthermore, the shape of binder’s elution furnishes information on the rates of these binding processes. Zonal elution has also been used to investigate the activity of enzymes during ligand screening [4].
The retention factor (k) of injected solute generally characterizes the chromatographic data obtained from zonal elution. The k value measures how strongly a compound interacts with the bioaffinity column. By comparing the k value of different substances, one can determine their relative affinity for the immobilized protein [53]. ZBC is most commonly applied in competition and displacement studies. It allows one to study the ligand–protein interactions occurring on a single binding site. To this end, the known ligand is injected into the system while a fixed concentration of a trial competing agent is eluted through the column [54].
Ideal zonal chromatography should afford a peak with Gaussian shape; however, asymmetric peaks usually arise during this process. The asymmetry is usually due to heterogeneous mass transfer, heterogeneity of the stationary phase and, extra column effects, which is a problem in analytical separations, but an excellent tool to characterize the separation process by non-linear chromatography (NLC). NLC peak tailing is concentration-dependent, and deviation from the Gaussian distribution varies with analyte concentration [55].
NLC measures the kinetic parameters involved in the formation and dissociation of the solute-stationary phase complex – the association (kon) and dissociation (koff) rate constants, as well as the equilibrium (Ka) and affinity constants (Kd) [4, 56].
Advantages of chromatographic assays include versatility, as many different protein classes can be studied, and the ability to screen binders from mixtures of compounds and extracts, as well as interface with detectors such as tandem mass spectrometers to allow screening and deconvolution of mass-encoded libraries [14].
2.2.2. Multidimensional (2D LC) Approaches
In zonal chromatography, the retention factor of a compound is comparable with its affinity for the target. An important factor is to identify false positive results related to nonspecific binding of the tested compound to the chromatographic support. For complex samples the lower chromatographic efficiency of used bioaffinity columns preclude adequate resolution of a mixture of compounds on the basis of their affinities. Multidimensional chromatographic system (Figure 4) has been used to improve analyses and information about binders in complex samples (Table 2). Jia et al. [57] described a method based on an online comprehensive two dimensional HepG2/CMC/enrich columns/HPLC/time-of-flight mass spectrometry system to screen potential anti-hepatoma components from drug-containing serum of rats after oral administration of Radix scutellariae. By this system the screening and identification of active parent components and metabolites binding to HepG2 cell membrane receptors was realized.
Fig. 4.
Block diagram of 2DLC system. (A) Position 1 and (B) position 2.
Table 2.
Target-based screening for antitumoral agents in various mixtures [74].
| Molecular target | Screening model | Active compounds | Mixtures | Reference |
|---|---|---|---|---|
| HepG2 | HepG2/CMC | baicalein, wogonin, chrysin, oroxylin A, neobaicalein and rivularin |
Radix scutellariae | [57] |
| HepG2 | HepG2/CMC | Oxymatrine and matrine Berberine and tetrahydropalmatine |
Radix sophorae flavescentis Cortex phellodendri amurensis, |
[83] |
| EGFR | HEK293/CMC | Asarinin | Radix et rhizoma asari | [84] |
| EGFR | HEK293/CMC | Vauquline, strychnine | Semen strychni | [76] |
| EGFR | HEK293/CMC | Resveratrol | Rhizoma polygoni cuspidati | [85] |
| EGFR | A431/CMC (2D–LC-MS) | Taspine, caulophine | Radix caulophylli | [86] |
| EGFR | A431/CMC | Oxymatrine, matrine | Radix sophorae flavescentis | [87] |
| TrkA receptor | Lipid raft | NA | Albizziae cortex, Galla chinensis | [75, 88] |
| β2-Adrenoceptor | Purified receptor | Amygdalin | Semen armeniacae amarum | [89] |
| VEGFR-2 | HEK293/CMC | Mesaconitine, aconitine, hypaconitine | Aconitum carmichaeli debx | [77] |
| alpha(1A)- adrenoceptor |
(α1AAR/CMC) | Methoxiamine, magnoflorine and caulophine | Radix Caulophylli | [90] |
Abbreviations: HepG2 carcinoma cell line, A431, epidermoid carcinoma cell line; 17-AAG, 17-allylaminogeldanamycin; CMC, cell membrane chromatography; DNA, deoxyribo-nucleic acid; EGFR, epidermal growth factor receptor; HEK293, human embryonic kidney 293 cells; Hsp90, heat shock protein 90; NA, non applicable; TrkA, tropomyosin-related tyrosine kinase A; VEGFR-2, vascular endothelial growth factor receptor-2.
Membrane and transmembrane receptor proteins are the targets of almost 75% of current pharmaceuticals [58]. In fact, membrane-bound receptors serve in transduction and amplification of signals across the cell membrane and allow cells to signal growth or apoptosis [59], or release chemicals in response to a physical or chemical stimulus from extracellular signaling [60]. Kinases are a major therapeutic as they are involved in signaling pathways and regulate process such as gene transcription, cell cycle, apoptosis, and differentiation through phosphorylation of various substrates [61, 62]. Membrane-bound proteins are the most challenging targets for development of small molecules screening assay. The development and use of cellular membrane affinity chromatography (CMAC) columns have been extensively demonstrated and different strategies have been described to immobilize membrane proteins [63, 64]. CMAC has been used to screen active components from complex samples such as herbal medicines (HMs) [65, 66] as demonstrated in Table 2, and combined with LC-MS has been used for identifying leading anticancer compounds [67–70].
CMAC models have resulted in successful isolation of numerous bioactive components from complex samples that interact with membrane receptors. The dynamic simulation of the action of drug in vivo by the CMAC presents a direct screening technique for active compounds [68, 70, 71]. However, CMAC showed some drawbacks regarding its selectivity, specificity, stability and service life span. These are due to the use of homogenized cell membrane containing multiple receptors at relatively low densities [72]. More recently, a shift from a single target to a multiple target approach has been sought as several effective drugs have been demonstrate to produce their action via interaction with multiple targets [73, 74]. In this context, Xu and coauthors [74, 75] reported a novel and promising technology with inherent high selectivity and specificity. The biomaterial prepared from the TrkA (tropomyosin-related tyrosine kinase) receptor-rich lipid raft for identifying antitumor agents and online application. The overexpressed TrkA receptors extracted lipid raft was immobilized on activated silica beads to form lipid raft coated silica beads and the biomaterial was packed into a column to serve as a stationary phase for online analysis of potential antitumor components. The bioactive components, gefitinib and lestaurtinib (standard anticancer drugs), exhibited longer retention time as compared to non-target (gemcitabine).
In spite of comprehensive studies on well-known cancer target receptors such as epidermal growth factor receptor (EGFR), vascular epidermal growth factor receptor (VEGFR) and Fast receptor (FasR), there is still very limited report regarding their application for bioscreening antitumor agents [75, 76]. So far the best known studies have been limited to cell membrane chromatography with highly expressed receptors like EGFR and VEGFR. The medical benefits of employing these receptors for affinity screening cannot be overemphasized in the quest for effective chemotherapeutic drugs [74].
An online analytical method based on VEGFR-2 cell membrane chromatography (VEGFR-CMC) and mass spectrometry was described for screening and identification of active component from Aconitum carmichaeli Debx. Fractions separated by VEGFR-CMC column (first dimension) were transferred and adsorbed on an enrichment column. The system was hyphenated through a 10-port column switcher. Enrichment fractions were sent into LC-MS system (second dimension) for separation and preliminary identification, respectively. Sunitinib malate was used as positive control. To confirm that sunitinib and compound(s) screened from Aconitum carmichaeli Debx. were both active on the same site of VEGFR-2, competitive displacement test were performed. From this extract mesaconitine (MSC), aconitine (AC), and hypaconitine (HPC) were identified as the active constituents acting on VEGFR-2. To confirm the usefulness of the method, the in vitro inhibition activity of MSC, AC, and HPC on vascular endothelial growth factor (VEGF) secretion of HEK293/VEGFR cell was tested by VEGF-ELISA assay [77].
Based on the same strategies, high expression EGFR/CMAC-online-LC-MS was used for screening Semen Strychni components and investigating their biological effects [76]. EGFR-mediated signaling can induce cells into a continuous and uncontrolled dividing state, which leads to increased malignant cell production and augmented tumors [78, 79] and it is an important target for screening anti-tumor inhibitors [80–82]. Sun et al [76] described a method based on a comprehensive two-dimensional EGFR/CMC-online-LC-MS. The EGFR cell membrane column was prepared “in situ” by the adsorption of cell membrane suspension on activated silica. The EGFR/CMC column was used in the first dimension and any fraction retained on the EGFR/CMC was enriched using an enrichment column and eluted into the second dimension for separation. The active compounds vauquline and strychnine were simultaneously detected in Semen Strychni extract and this model can be usefully for screening binders from other extracts. According to the cell proliferation assay results vauquline and strychnine inhibited cell proliferation of HEK293/EGFR and inhibited Erk phosphorylation, which indicated that could effectively reduce expression of downstream signaling molecules. Some target screening models for antitumor agents are presented in Table 2.
Fragment-based drug discovery (FBDD) has become a new strategy for drug discovery [11, 12, 91]. One of the distinctive features of fragment-based discovery is the need for suitable screening methods to reliably detect the low affinity hits, typically binding with a Kd in the high µM to mM range. These low affinity hits still represent good starting points for hits-to-leads chemistry due to the often high ligand efficiency [13]. This initial low affinity has led to the development and refinement of a wide variety of biophysical methods to detect such binding [92]. Kinases are involved in a range of different biological processes such as signaling, proliferation apoptosis, and differentiation [93, 94], and many different types of pathological states, such as cardiovascular diseases, cancer [64, 95] and neurodegenerative diseases [96].
Meiby et al. have demonstrated the potential of using bioaffinity zonal chromatography in combination with MS detection for fragment screening of cyclin G-associated kinase (GAK). After a virtual screening, a fragments library was selected and assayed using a capillary GAK protein column. Results indicated the possibility to identify compounds with higher affinity (Kd ≤ 200 M) by comparison with the reference column (with inhibited enzyme). Other interesting observation was the ability of bioaffinity zonal chromatography to perform a chiral separation and hence determine affinity constants for individual enantiomers. This gives a significant advantage as compared with other technologies for fragment screening such as surface plasmon resonance SPR. A single run is required to give an estimate of the fragment affinity and theoretically of its kinetics. The drawback was that required relatively long elution times and each sample was eluted for 140 min with a total of 33 h of analysis [11].
2.3. Ligands Fishing
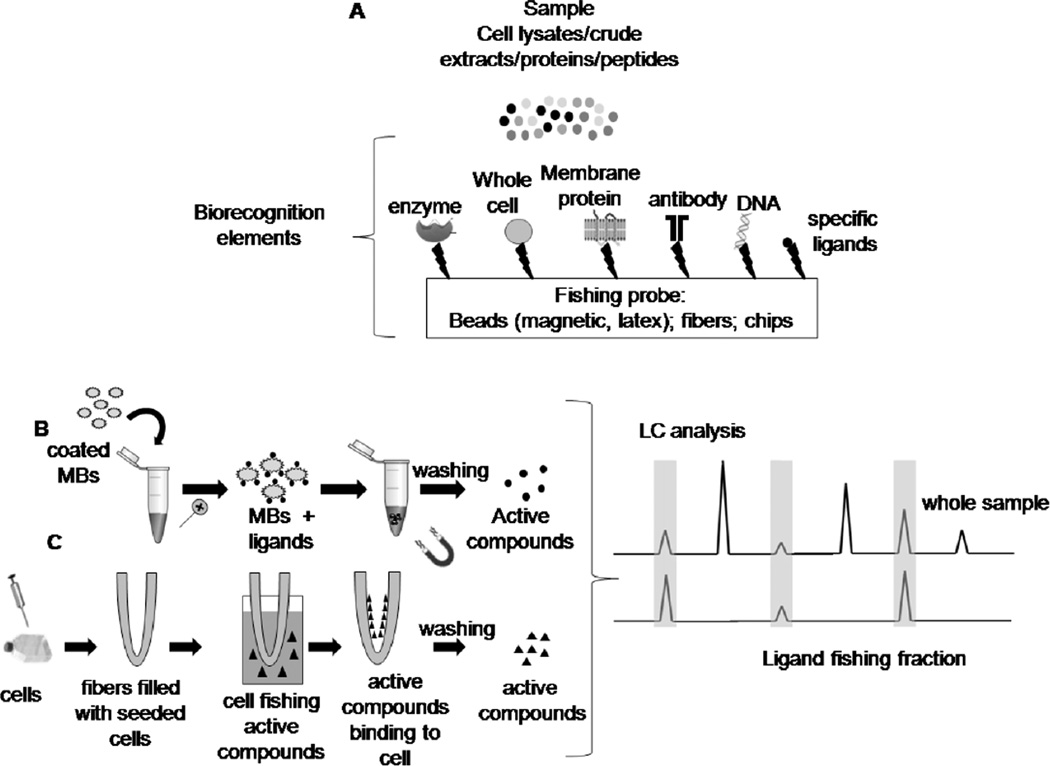
Ligands fishing assay is another widely used process in the screening and isolation of active compounds in complex mixtures, as cell lysates [29] and natural product extracts [4, 6, 18, 97]. Most common ligands fishing assays methods for targeting anticancer active compounds involves the immobilization of whole cells or membrane proteins onto different matrices such as biosensors, fibers, micro- and nano-sized beads to haul out proteins from complex matrices (Fig. 5A). Nowadays, a great effort is being made towards the development of new methods capable of simulating the actual conditions of interactions between active compounds and cells or even more specifically, isolate binders.
Fig. 5.
(A) Schematic workflow for some general ligand fishing assays. (B) Scheme for the ligand fishing assay with magnetic beads and (C) with hollow fibers.
Hollow fibers filled with living cells or seeded cells have been used to screen active compounds from traditional Chinese medicine (TCM) (Fig. 5C). This method consists of a hollow fiber internal lumen surface filled with a certain amount of living cell suspension or seeded cell. The fiber is then bent into a U-shape and inserted into the sample solution to fish binders out. Hollow cell fiber cell fishing (HFCF) prepared with HCT116 colorectal cancer cells was used to screen anthraquinones active compounds from extracts of Polygonum cuspidatum, Cecropia obtusifolia L. and Polygoni multiflori radix praeparata [98]. MCF7 human breast cancer cells, MADB106 mouse breast cancer cells, and SGC7901 gastric cancer cells were seeded on the internal surface of hollow fibers that were used to screen an antitumor-active protoberberine alkaloid group from a Coptis chinensis decoction [99]. HFCF using three types of tumor cells (MCF-7, SGC7901, and MADB-106) was used to screen flavonoid and anthraquinone active compound groups simultaneously from TCMs [100].
Variables such as the surface properties of the hollow fibers, the non-specific binding between active centers in the fiber and the binders, the cell survival rate under different conditions before and after screening, the repeatability and recovery of HFCF-LC method can be investigated in detail. The structures screened from TCMs were identified by comparing to the retention time of the reference substances and confirmed by mass spectrometry [98].
Nonetheless, this method has some disadvantages as the resulting activity may be due to non-specific interactions between hollow fiber activity centers and binders. Moreover the mechanism of action and pharmacologic effects of the binders fished by HFCF require further research through laborious bioassays and the novel structurally binders require subsequent spectroscopic and spectrometric analysis to identify their possible structure [99].
The use of magnetic beads (MBs) for binders and proteins fishing have gained a significant amount of interest specially for screening complex matrices due to the ease in isolating binders without additional purification procedures (Figure 5B) [97]. Magnetic particles are preferred carriers for biomolecules such as cells, nucleic acids and proteins. They provide an excellent support for the immobilization of proteins since protein–protein complexes are maintained intact on the surface of the protein-coated magnetic beads [47, 101]. Moreover, MBs can be tailored to specifically bind the biomarkers and concentrate them from the complex specimen under magnetic actuation, avoiding interference before testing [102]. Therefore, their interaction with a magnetic force enables separation of MBs from a given aqueous matrix or a biological environment without filtration or centrifugation step allowing the identification of a compound(s) that is not concentration dependent but rather affinity dependent [97, 103].
A series of magnetic beads already functionalized is commercially available (Adembeads1, Dynabeads1, BioMag1, SiMAG1, MACS1MPs, BioCLon). They are synthesized containing a magnetic element in their core such as iron, nickel, neodymium or magnetite and they can be modified with derivatives such as tosyl, amine, carboxyl or epoxy groups, for the immobilization of whole organisms, proteins and peptides, enzymes, antibodies, DNA, among others [102, 104]. Magnetic separation techniques advantageously replaced classical separation techniques in order to eliminate the disadvantages such as decomposition, inactivation or deformation of the biomolecules [98].
Heat shock protein 90α (Hsp90α) is a molecular chaperone that has been targeted for the development of new anticancer therapies. It has been successfully immobilized on a silica-based stationary phase through either the amino- or carboxy-terminus of the protein to produce Hsp90-NT (immobilization via N-terminus) and Hsp90α -(CT) (immobilization via C-terminus) columns and that the resulting column was used in liquid chromatography experiments to identify small molecule Hsp90 binders [47]. However, a limitation of this approach was the screening of complex matrices for protein-protein interactions, could not be carried out on-line. As a result, Hsp90α was immobilized onto the surface of silica-based magnetic beads. Apart from the isolation of known Hsp90α ligands from a mixture containing binders and non-binders it allowed the isolation of proteins from a mixture of proteins, as well as a cellular extract. Therefore, these magnetic beads coated with Hsp90α were used for the first time to “fish out” new lead drug candidates and client proteins from complex chemical and biological mixtures [101].
Prostate specific antigen (PSA) is an extracellular serine protease belonging to the kallikrein family and has been used as a screening tool for the diagnosis and prognosis of prostate cancer. The lectine concanavalin A (Con A) was covalently immobilized directly and through a spacer arm (1,6-diaminohexane-HDMA) on magnetic poly(glycidyl methacrylate) (mPGMA) beads. Total PSA (tPSA) and free PSA (fPSA) binding capacities of the mPGMA-ConA and mPGMA-HDMA-ConA beads from human serum were investigated in a batch system and compared to each other by using enzyme-linked immuno sorbent assay (ELISA). Albumin and immunoglobulin G free diluted serum samples of patients with prostate cancer were incubated with 10 mg of each type of MBs for 2 h at 25 °C at a stirring rate of 100 rpm. The binding capacities of each type of MBs were calculated from the difference between initial and final tPSA and fPSA concentrations by using the Equation 4 [105].
| Equation 4 |
where Q is the amount of PSA bound onto unit mass of the mPGMA-ConA and the mPGMA-HMDA-Con A beads (ng/g), Ci and Cf are the initial and final PSA concentrations (ng/mL), respectively, V is the volume of aqueous phase (mL) and m is the amount of adsorbent used (g) [105].
The binding of tPSA and fPSA increased significantly by the attachment of spacer-arm on the mPGMA beads. It was determined that tPSA and fPSA binding of the Con A beads with the spacer were higher than that one without it. Maximum tPSA binding capacity was obtained by using the mPGMA-HDMA-Con A beads and calculated to be 91.2 ng/g. The mPGMA-HDMA-Con A beads could be reused without a remarkable decrease in the binding capacities after 5 binding-desorption cycles. The mPGMA-HDMA-Con A beads could be useful for the detection of PSA and suggested as a model system for other glycoprotein biomarkers [105].
Platinating agents are commonly prescribed anticancer drug damaging DNA. A ligands fishing trap was made of damaged plasmids by one of three different anticancer platinum drugs (cisplatin, oxaliplatin or the satraplatin metabolite JM118) attached to magnetic beads and exposed to HeLa (cervical cancer cell line) and MDA-MB231 (breast cancer cell line) cell nuclear extracts. Beads without DNA and beads grafted with undamaged plasmids were used as controls to discriminate between interesting candidates and non-specific proteins identified in the proteomic experiments. Retained proteins were identified by nanoLC-MS/MS. This approach identified 38 proteins interacting with DNA adducts that were validated by immunoassays and SPRi (Surface Plasmon Resonance imaging). Identified proteins may improve the understanding of molecular and cellular responses to this particular type of anticancer drugs [106].
Another interesting approach using latex beads instead of magnetic beads was developed using inhibitor-based affinity chromatography where the molecule of interest is tethered to the solid support (pull-down method) [107]. TAS-103 is an anticancer drug that probably exerts its effect on tumor cell viability by inhibiting topoisomerase activity [108]. Since a direct target of TAS-103 remain unclear, latex beads coated with a TAS-103 derivative, TAS-1–3383, which has an additional amino group for the coupling reaction with the carboxyl groups of the beads were prepared to search for other TAS-103 binding protein(s) in HeLA cell extracts The method allowed to “fish out” a 54kDa protein which specifically bounded to TAS-1–3383 on beads and latter was identified using Western blot analysis as being SRP54, a SPR subunit which mediates the proper delivery of secretory proteins in cells [109].
The broad spectrum of application for fishing assays ensure their potential towards the design of new methods targeting anti-cancer related proteins. On the other hand, when combined with other analytical tools, such as LC-MS, NMR, etc., these methods becomes a powerful bioanalytical tool for the screening of binders in complex matrices accelerating the discovery of new leads.
3. CONCLUSION
Affinity-based chromatography methods involving frontal and zonal elution, as well the ligands fishing approach, grants a crucial tool in the discovery of new active anti-cancer agents, and are an essential bioanalytical platform for medicinal and biochemistry analysts. Considering the traditional bioassays using the isolated biological target, the discussed procedures presents several advantages, including the reuse of the same amount of protein in several assays, possibility of automation and the identification of bioactive compounds directly in complex matrices, as natural products extracts and combinatory libraries.
Acknowledgments
We would like to thank the National Council for Scientific and Technological Development (CNPq), the São Paulo State Research Foundation (FAPESP – Grant 201301710-1) and the Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ) for the financial support. This work was supported in part by the Intramural Research Progam National Instiute on Aging, NIH (RM).
Biography

Marcela C. de Moraes
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
DISCLAIMER: The above article has been published in Epub (ahead of print) on the basis of the materials provided by the author. The Editorial Department reserves the right to make minor modifications for further improvement of the manuscript.
REFERENCES
- 1.Stepanenko AA, Dmitrenko VV. Pitfalls of the MTT assay: Direct and off-target effects of inhibitors can result in over/underestimation of cell viability. Gene. 2015;574:193–203. doi: 10.1016/j.gene.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Singh NS, Habicht KL, Dossou KSS, Shimmo R, Wainer IW, Moaddel R. Multiple protein stationary phases: A review. J Chromatography B-Analytical Technol Biomed Life Sci. 2014;968:64–68. doi: 10.1016/j.jchromb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NIH - National Cancer Institute - Targeted Cancer Therapies. 2016 [Google Scholar]
- 4.de Moraes MC, Vanzolini KL, Cardoso CL, Cass QB. New trends in LC protein ligand screening. J Pharm Biomed Anal. 2014;87:155–166. doi: 10.1016/j.jpba.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso CL, de Moraes MC, Cass QB. Immobilization of the Enzymes on Chromatographic Supports: A Tool to Research of Inhibitor Compounds. Quimica Nova. 2009;32:175–187. [Google Scholar]
- 6.de Moraes MC, Santos JB, dos Anjos DM, et al. Prion protein-coated magnetic beads: Synthesis, characterization and development of a new ligands screening method. J Chromatography A. 2015;1379:1–8. doi: 10.1016/j.chroma.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calleri E, Fracchiolla G, Montanari R, et al. Frontal affinity chromatography with MS detection of the ligand binding domain of PPAR receptor: Ligand affinity screening and stereoselective ligand-macromolecule interaction. J Chromatography A. 2012;1232:84–92. doi: 10.1016/j.chroma.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 8.de Moraes MC, Ducati RG, Donato AJ, et al. Capillary bioreactors based on human purine nucleoside phosphorylase: A new approach for ligands identification and characterization. J Chromatography A. 2012;1232:110–115. doi: 10.1016/j.chroma.2011.10.056. [DOI] [PubMed] [Google Scholar]
- 9.Hage DS, Anguizola JA, Bi C, et al. Pharmaceutical and biomedical applications of affinity chromatography: Recent trends and developments. J Pharm Biomed Anal. 2012;69:93–105. doi: 10.1016/j.jpba.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moaddel R, Lu LL, Baynham M, Wainer IW. Immobilized receptor- and transporter-based liquid chromatographic phases for online pharmacological and biochemical studies: a mini-review. J Chromatography B-Analytical Technol Biomed Life Sci. 2002;768:41–53. doi: 10.1016/s0378-4347(01)00484-4. [DOI] [PubMed] [Google Scholar]
- 11.Meiby E, Knapp S, Elkins JM, Ohlson S. Fragment screening of cyclin G-associated kinase by weak affinity chromatography. Analytical Bioanalytical Chem. 2012;404:2417–2425. doi: 10.1007/s00216-012-6335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong-Thi MD, Bergstrom M, Fex T, Isaksson R, Ohlson S. High-Throughput Fragment Screening by Affinity LC-MS. J Biomolecular Screening. 2013;18:160–171. doi: 10.1177/1087057112459271. [DOI] [PubMed] [Google Scholar]
- 13.Meiby E, Simmonite H, le Strat L, et al. Fragment Screening by Weak Affinity Chromatography: Comparison with Established Techniques for Screening against HSP90. Analytical Chem. 2013;85:6756–6766. doi: 10.1021/ac400715t. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg EM, Sicard C, Brennan JD. Solid-Phase Biological Assays for Drug Discovery. Ann Rev Analytical Chem. 2014;7:337–359. doi: 10.1146/annurev-anchem-071213-020241. [DOI] [PubMed] [Google Scholar]
- 15.de Moraes MC, Cardoso CL, Cass QB. Immobilized purine nucleoside phosphorylase from Schistosoma mansoni for specific inhibition studies. Analytical Bioanalytical Chem. 2013;405:4871–4878. doi: 10.1007/s00216-013-6872-7. [DOI] [PubMed] [Google Scholar]
- 16.Vanzolini KL, Vieira LCC, Correa AG, Cardoso CL, Cass QB. Acetylcholinesterase Immobilized Capillary Reactors-Tandem Mass Spectrometry: An On-Flow Tool for Ligand Screening. J Med Chem. 2013;56:2038–2044. doi: 10.1021/jm301732a. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues MVN, Correa RS, Vanzolini KL, Santos DS, Batista AA, Cass QB. Characterization and screening of tight binding inhibitors of xanthine oxidase: an on-flow assay. Rsc Advances. 2015;5:37533–3758. [Google Scholar]
- 18.Vanzolini KL, Jiang ZJ, Zhang XQ, et al. Acetylcholinesterase immobilized capillary reactors coupled to protein coated magnetic beads: A new tool for plant extract ligand screening. Talanta. 2013;116:647–652. doi: 10.1016/j.talanta.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jozwiak K, Ravichandran S, Collins JR, Wainer IW. Interaction of noncompetitive inhibitors with an immobilized alpha 3 beta 4 nicotinic acetylcholine receptor investigated by affinity chromatography, quantitative-structure activity relationship analysis, and molecular docking. J Med Chem. 2004;47:4008–4021. doi: 10.1021/jm0400707. [DOI] [PubMed] [Google Scholar]
- 20.de Moraes MC, Temporini C, Calleri E, et al. Evaluation of capillary chromatographic supports for immobilized human purine nucleoside phosphorylase in frontal affinity chromatography studies. J Chromatography A. 2014;1338:77–84. doi: 10.1016/j.chroma.2014.02.057. [DOI] [PubMed] [Google Scholar]
- 21.Sanghvi M, Moaddel R, Frazier C, Wainer IW. Synthesis and characterization of liquid chromatographic columns containing the immobilized ligand binding domain of the estrogen related receptor alpha and estrogen related receptor gamma. J Pharm Biomed Anal. 2010;53:777–780. doi: 10.1016/j.jpba.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wainer IW, Zhang YX, Xiao YX, Keller KJ. Liquid chromatographic studies with immobilized neuronal nicotinic acetylcho-line receptor stationary phases: effects of receptor subtypes, pH and ionic strength on drug-receptor interactions. J Chromatography B. 1999;724:65–72. doi: 10.1016/s0378-4347(98)00579-9. [DOI] [PubMed] [Google Scholar]
- 23.Kasai K-I, Oda Y, Nishikata M, Ishii S-I. Frontal affinity chromatography: Theory for its application to studies on specific interactions of biomolecules. J Chromatography B: Biomed Sci Appl. 1986;376:33–47. doi: 10.1016/s0378-4347(00)80822-1. [DOI] [PubMed] [Google Scholar]
- 24.Schriemer DC. Peer Reviewed: Biosensor Alternative: Frontal Affinity Chromatography. Analytical Chem. 2004;76:440 A–448 A. doi: 10.1021/ac041684m. [DOI] [PubMed] [Google Scholar]
- 25.Calleri E, Temporini C, Caccialanza G, Massolini G. Target-Based Drug Discovery: the Emerging Success of Frontal Affinity Chromatography Coupled to Mass Spectrometry. ChemMedChem. 2009;4:905–916. doi: 10.1002/cmdc.200800436. [DOI] [PubMed] [Google Scholar]
- 26.Singh NS, Jiang Z, Moaddel R. Analyzing Biomolecular Interactions by Mass Spectrometry. Wiley-VCH Verlag GmbH & Co. KGaA; 2015. Frontal and Zonal Affinity Chromatography Coupled to Mass Spectrometry; pp. 241–270. [Google Scholar]
- 27.Zhu L, Chen L, Luo H, Xu X. Frontal Affinity Chromatography Combined On-Line with Mass Spectrometry: A Tool for the Binding Study of Different Epidermal Growth Factor Receptor Inhibitors. Analytical Chem. 2003;75:6388–6393. doi: 10.1021/ac0341867. [DOI] [PubMed] [Google Scholar]
- 28.Fort Sb, Kim H-S, Hindsgaul O. Screening for Galectin-3 Inhibitors from Synthetic Lacto-N-biose Libraries Using Microscale Affinity Chromatography Coupled to Mass Spectrometry. J Organic Chem. 2006;71:7146–7154. doi: 10.1021/jo060485v. [DOI] [PubMed] [Google Scholar]
- 29.Sanghvi M, Moaddel R, Wainer IW. The development and characterization of protein-based stationary phases for studying drug-protein and protein-protein interactions. J Chromatogr A. 2011;1218:8791–8798. doi: 10.1016/j.chroma.2011.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan NWC, Lewis DF, Rosner PJ, Kelly MA, Schriemer DC. Frontal affinity chromatography-mass spectrometry assay technology for multiple stages of drug discovery: applications of a chromatographic biosensor. Analytical Biochem. 2003;319:1–12. doi: 10.1016/s0003-2697(03)00193-3. [DOI] [PubMed] [Google Scholar]
- 31.Slon-Usakiewicz JJ, Dai J-R, Ng W, et al. Global Kinase Screening. Applications of Frontal Affinity Chromatography Coupled to Mass Spectrometry in Drug Discovery. Analytical Chem. 2005;77:1268–1274. doi: 10.1021/ac048716q. [DOI] [PubMed] [Google Scholar]
- 32.Temporini C, Massolini G, Marucci G, et al. Development of new chromatographic tools based on A2A adenosine receptor subtype for ligand characterization and screening by FAC-MS. Analytical Bioanalytical Chem. 2013;405:837. doi: 10.1007/s00216-012-6353-4. [DOI] [PubMed] [Google Scholar]
- 33.Ohta A, Gorelik E, Prasad SJ, et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci. 2006;103:13132–13137. doi: 10.1073/pnas.0605251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slon-Usakiewicz JJ, Ng W, Foster JE, Dai J-R, Deretey E, Toledo-Sherman L, Redden PR, Pasternak A, Reid N. Frontal Affinity Chromatography with MS Detection of EphB2 Tyrosine Kinase Receptor. 1. Comparison with Conventional ELISA. J Med Chem. 2004;47:5094–5100. doi: 10.1021/jm049733a. [DOI] [PubMed] [Google Scholar]
- 35.Bhatia P, Bernier M, Sanghvi M, et al. Breast cancer resistance protein (BCRP/ABCG2) localises to the nucleus in glioblastoma multiforme cells. Xenobiotica. 2012;42:748–755. doi: 10.3109/00498254.2012.662726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habicht KL, Frazier C, Singh N, Shimmo R, Wainer IW, Moaddel R. The synthesis and characterization of a nuclear membrane affinity chromatography column for the study of human breast cancer resistant protein (BCRP) using nuclear membranes obtained from the LN-229 cells. J Pharm Biomed Analysis. 2013;72:159–162. doi: 10.1016/j.jpba.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian L, Li Q, Ji X. Binding of angiogenesis inhibitor kringle 5 to its specific ligands by frontal affinity chromatography. J Chromatography A. 2015;1401:42–51. doi: 10.1016/j.chroma.2015.04.058. [DOI] [PubMed] [Google Scholar]
- 38.Tetala KKR, Chen B, Visser GM, van Beek TA. Single step synthesis of carbohydrate monolithic capillary columns for affinity chromatography of lectins. J Separation Sci. 2007;30:2828–2835. doi: 10.1002/jssc.200700356. [DOI] [PubMed] [Google Scholar]
- 39.Tetala KKR, Chen B, Visser GM, et al. Preparation of a monolithic capillary column with immobilized alpha-mannose for affinity chromatography of lectins. J Biochem Biophys Methods. 2007;70:63–69. doi: 10.1016/j.jbbm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura K, Higashida K, Hata Y, et al. Comparative analysis of oligosaccharide specificities of fucose-specific lectins from Aspergillus oryzae and Aleuria aurantia using frontal affinity chromatography. Analytical Biochem. 2009;386:217–221. doi: 10.1016/j.ab.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 41.Hodgson RJ, Chen Y, Zhang Z, et al. Protein-Doped Monolithic Silica Columns for Capillary Liquid Chromatography Prepared by the Sol-Gel Method: Applications to Frontal Affinity Chromatography. Analytical Chem. 2004;76:2780–2790. doi: 10.1021/ac0352124. [DOI] [PubMed] [Google Scholar]
- 42.Ravichandran S, Singh N, Donnelly D, et al. Pharmacophore model of the quercetin binding site of the SIRT6 protein. J Mol Graphics Model. 2014;49:38–46. doi: 10.1016/j.jmgm.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh N, Ravichandran S, Norton DD, Fugmann SD, Moaddel R. Synthesis and characterization of a SIRT6 open tubular column: Predicting deacetylation activity using frontal chromatography. Analytical Biochem. 2013;436:78–83. doi: 10.1016/j.ab.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi T, Tamura M, Nishiyama K, et al. Mammalian galectins bind galactoseb1-4fucose disaccharide, a unique structural component of protostomial N-type glycoproteins. Biochemical and biophysical research communications. 2013;436:509–513. doi: 10.1016/j.bbrc.2013.05.135. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Leonessa F, Clarke R, Wainer IW. Development of an immobilized P-glucoprotein stationary phase for on-line liquid chromatographic determination of drug-binding affinities. J Chromatography B. 2000;739:33–37. doi: 10.1016/s0378-4347(99)00384-9. [DOI] [PubMed] [Google Scholar]
- 46.Temporini C, Ceruti S, Calleri E, Ferrerio S, Moaddel R, Abbrac-chio MP, Massolini G. Development of an immobilized GPR17 receptor stationary phase for binding determination using frontal affinity chromatography coupled to mass spectrometry. Analytical Biochem. 2009;384:123–129. doi: 10.1016/j.ab.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Marszall MP, Moaddel R, Jozwiak K, Bernier M, Wainer IW. Initial synthesis and characterization of an immobilized heat shock protein 90 column for online determination of binding affinities. Anal Biochem. 2008;373:313–321. doi: 10.1016/j.ab.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toledo-Sherman L, Deretey E, Slon-Usakiewicz JJ, et al. Frontal Affinity Chromatography with MS Detection of EphB2 Tyrosine Kinase Receptor. 2. Identification of Small-Molecule Inhibitors via Coupling with Virtual Screening. J Med Chem. 2005;48:3221–3230. doi: 10.1021/jm0492204. [DOI] [PubMed] [Google Scholar]
- 49.Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD. Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov. 2008;7:221–230. doi: 10.1038/nrd2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaiken IM. Analytical affinity Chromatography. Boca Raton, FL: CRC Press; 1987. [Google Scholar]
- 51.Chaiken IM. Analytical affinity chromatography in studies of molecular recognition in biology: A review. J Chromatography B: Biomed Sci Appl. 1986;376:11–32. doi: 10.1016/s0378-4347(00)80821-x. [DOI] [PubMed] [Google Scholar]
- 52.Hage DS, Tweed SA. Recent advances in chromatographic and electrophoretic methods for the study of drug-protein interactions. J Chromatography B: Biomed Sci Appl. 1997;699:499–525. doi: 10.1016/s0378-4347(97)00178-3. [DOI] [PubMed] [Google Scholar]
- 53.Schiel J, Joseph K, Hage D. Biointeraction Affinity Chromatography: General Principles and Recent Developments. In: Grushka E, Grinberg N, editors. Advances in Chromatography. Boca Raton, FL: 2010. pp. 145–193. [PubMed] [Google Scholar]
- 54.Hage DS, Chen J. Quantitative Affinity Chromatography: Practical Aspects. In: Hage D, editor. Handbook of Affinity Chromatography. Boca Raton: Taylor & Francis Group; 2006. pp. 596–628. [Google Scholar]
- 55.Jozwiak K, Haginaka J, Moaddel R, Wainer IW. Displacement and Nonlinear Chromatographic Techniques in the Investigation of Interaction of Noncompetitive Inhibitors with an Immobilized a3b4 Nicotinic Acetylcholine Receptor Liquid Chromatographic Stationary Phase. Analytical Chem. 2002;74:4618–4624. doi: 10.1021/ac0202029. [DOI] [PubMed] [Google Scholar]
- 56.Moaddel R, Wainer IW. Development of immobilized membrane-based affinity columns for use in the online characterization of membrane bound proteins and for targeted affinity isolations. Analytica Chimica Acta. 2006;564:97–105. doi: 10.1016/j.aca.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 57.Jia D, Chen X, Cao Y, et al. On-line comprehensive two-dimensional HepG2 cell membrane chromatographic analysis system for charactering anti-hepatoma components from rat serum after oral administration of Radix scutellariae: A strategy for rapid screening active compounds in vivo. J Pharm Biomed Analysis. 2016;118:27–33. doi: 10.1016/j.jpba.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 58.Landry Y, Gies J-P. Drugs and their molecular targets: an updated overview. Fundamental Clin Pharmacol. 2008;22:1–18. doi: 10.1111/j.1472-8206.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- 59.Pastore S, Mascia F, Mariani V, Girolomoni G. The Epidermal Growth Factor Receptor System in Skin Repair and Inflammation. J Invest Dermatol. 2008;128:1365–1374. doi: 10.1038/sj.jid.5701184. [DOI] [PubMed] [Google Scholar]
- 60.Lecca D, Abbraccio M. Deorphanisation of G protein-coupled receptors: A tool to provide new insights in nervous system pathophysiology and new targets for psycho-active drugs. Neurochem Int. 2008;52:339–351. doi: 10.1016/j.neuint.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 61.Tsai C-J, Nussinov R. The molecular basis of targeting protein kinases in cancer therapeutics. Seminars Cancer Biol. 2013;23:235–242. doi: 10.1016/j.semcancer.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moaddel R, Wainer IW. The preparation and development of cellular membrane affinity chromatography columns. Nat. Protocols. 2009;4:197–205. doi: 10.1038/nprot.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fruh V, Ijzerman AP, Siegal G. How to Catch a Membrane Protein in Action: A Review of Functional Membrane Protein Immobilization Strategies and Their Applications. Chem Rev. 2011;111:640–656. doi: 10.1021/cr900088s. [DOI] [PubMed] [Google Scholar]
- 65.Moaddel R, Rosenberg A, Spelman K, et al. Development and characterization of immobilized cannabinoid receptor (CB1/CB2) open tubular column for on-line screening. Analytical Biochem. 2011;412:85–91. doi: 10.1016/j.ab.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ciesla L, Okine M, Rosenberg A, et al. Development and characterization of the a3b4a5 nicotinic receptor cellular membrane affinity chromatography column and its application for on line screening of plant extracts. J Chromatography A. 2016;1431:138–144. doi: 10.1016/j.chroma.2015.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Cao Y, Lv D, Zhu Z, Zhang J, Chai Y. Comprehensive two-dimensional HepG2/cell membrane chromatography/monolithic column/time-of-flight mass spectrometry system for screening anti-tumor components from herbal medicines. J Chromatography A. 2012;1242:67–74. doi: 10.1016/j.chroma.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 68.Wang S, Sun M, Zhang Y, Du H, He L. A new A431/cell membrane chromatography and online high performance liquid chromatography/mass spectrometry method for screening epidermal growth factor receptor antagonists from Radix sophorae flavescentis. J Chromatography A. 2010;1217:5246–5252. doi: 10.1016/j.chroma.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 69.Wang S, Sun M, Zhang Y, Zhang J, He L. EGFR/cell membrane chromatography-online-high performance liquid chromatography/mass spectrometry method for screening EGFR antagonists from Radix Angelicae Pubescentis. Science China-Chemistry. 2010;53:2357–2362. [Google Scholar]
- 70.Yuan B-x, Hou J, He L-c, Yang G-d. Evaluation of drug-muscarinic receptor affinities using cell membrane chromatography and radioligand binding assay in guinea pig jejunum membrane. Acta Pharmacol Sin. 2005;26:113–116. doi: 10.1111/j.1745-7254.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- 71.Bing-Xiang Y, Jin H, Guang-De Y, Li-mei Z, Lang-Chong H. Comparison of Determination of Drug–Muscarinic Receptor Affinity by Cell-Membrane Chromatography and by Radioligand-Binding Assay with the Cerebrum Membrane of the Rat. Chromatographia. 2005;61:381. [Google Scholar]
- 72.Hou J. Evaluation of drug-muscarinic receptor affinities using cell membrane chromatography and radioligand binding assay in guinea pig jejunum membrane. Acta Pharmacol. Sin. 2005;26:13–116. doi: 10.1111/j.1745-7254.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- 73.Medina-Franco JL, Giulianotti MA, Welmaker GS, Houghten RA. Shifthing from the single to the multitarget paradigm in drug discovery. Drug Discovery Today. 2013;18:495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Firempong CK, Cao X, Tong S, Yu J, Xu X. Prospects for multi-target lipid-raft-coated silica beads: a remarkable online biomaterial for discovering multitarget antitumor lead compounds. Rsc Advances. 2015;5:49330–49342. [Google Scholar]
- 75.Tong S, Fu M, Cao X, et al. Lipid Raft Stationary Phase Chromatography for Screening Anti-tumor Components from Galla chinensis. Chromatographia. 2014;77:419–429. [Google Scholar]
- 76.Sun M, Guo Y, Dai B, Wang C, He L. High-expression EGFR/cell membrane chromatography-online-high-performance liquid chromatography/mass spectrometry: rapid screening of EGFR antagonists from Semen Strychni. Rapid Communications in Mass Spectrometry. 2012;26:2027–2032. doi: 10.1002/rcm.6318. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Wang S, Zhang Y, He L. An online coupled cell membrane chromatography with LC/MS method for screening compounds from Aconitum carmichaeli Debx. acting on VEGFR-2. J Pharmaceutical and Biomedical Analysis. 2010;53:1063–1069. doi: 10.1016/j.jpba.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 78.Ciardiello F, Tortora G. Drug therapy: EGFR antagonists in cancer treatment. New England J Medicine. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 79.Jiang BH, Jiang GQ, Zheng JZ, Lu ZM, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor. Cell Growth & Differentiation. 2001;12:363–369. [PubMed] [Google Scholar]
- 80.Mendelsohn J. Targeting the Epidermal Growth Factor Receptor for Cancer Therapy. J Clin Oncol. 2002;20:1s–13s. [PubMed] [Google Scholar]
- 81.Scaltriti M, Baselga J. The Epidermal Growth Factor Receptor Pathway: A Model for Targeted Therapy. Clin Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 82.Ciardiello F, Tortora G. Epidermal growth factor receptor (EGFR) as a target in cancer therapy: understanding the role of receptor expression and other molecular determinants that could influence the response to anti-EGFR drugs. Eur J Cancer. 2003;39:1348–1354. doi: 10.1016/s0959-8049(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 83.Wang S, Wang C, Zhao X, Mao S, Wu Y, Fan G. Comprehensive two-dimensional high performance liquid chromatography system with immobilized liposome chromatography column and monolithic column for separation of the traditional Chinese medicine Schisandra chinensis. Analytica Chimica Acta. 2012;713:121–129. doi: 10.1016/j.aca.2011.10.062. [DOI] [PubMed] [Google Scholar]
- 84.Han S, Huang J, Hou J, Wang S. Screening epidermal growth factor receptor antagonists from Radix et Rhizoma Asari by two-dimensional liquid chromatography. J Separation Sci. 2014;37:1525–1532. doi: 10.1002/jssc.201400236. [DOI] [PubMed] [Google Scholar]
- 85.Sun M, Zhang YM, Zhang J, Wang S-C, He LC. A high expression EGFR/cell membrane chromatography and online high performance liquid chromatography/mass spectrometry method for screening EGFR antagonists from Rhizoma Polygoni Cuspidati Acta Pharm Sin B. 2011;1:115–120. [Google Scholar]
- 86.Sun M, Ren J, Du H, et al. A combined A431 cell membrane chromatography and online high performance liquid chromatography/mass spectrometry method for screening compounds from total alkaloid of Radix Caulophylli acting on the human EGFR. J Chromatography B-Analytical Technol Biomed Life Sci. 2010;878:2712–2718. doi: 10.1016/j.jchromb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 87.Wang SC, Sun M, Zhang YM, Du H, He LC. A new A(431)/cell membrane chromatography and online high performance liquid chromatography/mass spectrometry method for screening epidermal growth factor receptor antagonists from Radix sophorae flavescentis. J Chromatography A. 2010;1217:5246–5252. doi: 10.1016/j.chroma.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 88.Tong S, Sun C, Cao X, et al. Development and thermodynamic evaluation of novel lipid raft stationary phase chromatography for screening potential antitumor agents. Biomed Chromatography. 2014;28:1615–1623. doi: 10.1002/bmc.3189. [DOI] [PubMed] [Google Scholar]
- 89.Zheng X, Zhao X, Yang R, Wang S, Wei Y, Zheng J. beta(2)-Adrenoceptor affinity chromatography and its application in the screening of the active compounds from Semen Armeniacae Amarum. Chinese Sci Bull. 2008;53:842–847. [Google Scholar]
- 90.Wang L, Ren J, Sun M, Wang S. A combined cell membrane chromatography and online HPLC/MS method for screening compounds from Radix Caulophylli acting on the human alpha(1A)-adrenoceptor. J Pharm Biomed Analysis. 2010;51:1032–1036. doi: 10.1016/j.jpba.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 91.Ohlson S. Designing transient binding drugs: A new concept for drug discovery. Drug Discov Today. 2008;13:433–439. doi: 10.1016/j.drudis.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 92.Hubbard RE, Murray JB. EXPERIENCES IN FRAGMENT-BASED LEAD DISCOVERY. In: Kuo LC, editor. Fragment-Based Drug Design: Tools, Practical Approaches, and Examples. San Diego: Elsevier Academic Press Inc; 2011. pp. 509–531. [Google Scholar]
- 93.Giamas G, Man YL, Hirner H, et al. Kinases as targets in the treatment of solid tumors. CellSignal. 2010;22:984–1002. doi: 10.1016/j.cellsig.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsai CJ, Nussinov R. The molecular basis of targeting protein kinases in cancer therapeutics. Seminars Cancer Biol. 2013;23:235–242. doi: 10.1016/j.semcancer.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 96.Catapano LA, HK M. Kinases as drug targets in the treatment of bipolar disorder. Drug Discov Today. 2008;13:295–302. doi: 10.1016/j.drudis.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 97.Wubshet SG, Brighente IMC, Moaddel R, Staerk D. Magnetic Ligand Fishing as a Targeting Tool for HPLC-HRMS-SPE-NMR: a-Glucosidase Inhibitory Ligands and Alkylresorcinol Glycosides from Eugenia catharinae. J Natural Products. 2015;78:2657–2665. doi: 10.1021/acs.jnatprod.5b00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xue X, Li L, Chen X, Hu S, Bai X. Hollow fiber cell fishing with high performance liquid chromatography for screening bioactive compounds from traditional Chinese medicines. J Chromatogr A. 2013;1280:75–83. doi: 10.1016/j.chroma.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 99.Liu X, Hu S, Chen X, Bai XH. Hollow fiber cell fishing with high-performance liquid chromatography for rapid screening and analysis of an antitumor-active protoberberine alkaloid group from Coptis chinensis. J Pharmaceutical and Biomedical Analysis. 2014;98:463–475. doi: 10.1016/j.jpba.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 100.Zhang LS, Chen X, Hu S, Sheng X, Bai XH. Rapid Screening of Different Types of Antitumor Compound Groups from Traditional Chinese Medicine by Hollow Fiber Cell Fishing with High Performance Liquid Chromatography. Combinatorial Chem High Throughput Screening. 2014;17:827–836. doi: 10.2174/1386207317666141031101709. [DOI] [PubMed] [Google Scholar]
- 101.Marszall MP, Moaddel R, Kole S, Gandhari M, Bernier M, Wainer IW. Ligand and protein fishing with heat shock protein 90 coated magnetic beads. Anal Chem. 2008;80:7571–7575. doi: 10.1021/ac801153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carinelli S, Marti M, Alegret S, Pividori MI. Biomarker detection of global infectious diseases based on magnetic particles. N Biotechnol. 2015;32:521–532. doi: 10.1016/j.nbt.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 103.Singh N, Ravichandran S, Spelman K, Fugmann SD, Moaddel R. The identification of a novel SIRT6 modulator from Trigonella foenum-graecum using ligand fishing with protein coated magnetic beads. J Chromatography B-Analytical Technologies in the Biomed Life Sci. 2014;968:105–111. doi: 10.1016/j.jchromb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vanzolini KL, Vieira LCC, Correa AG, Moaddel R, Cass QB. Acetylcholinesterase immobilized on modified magnetic beads as a tool for screening a compound library. Microchim Acta. 2015;182:2209–2213. [Google Scholar]
- 105.Idil N, Percin I, Karakoc V, Yavuz H, Aksoz N, Denizli A. Concanavalin A immobilized magnetic poly(glycidyl methacrylate) beads for prostate specific antigen binding. Colloids Surfaces B-Biointerfaces. 2015;134:461–468. doi: 10.1016/j.colsurfb.2015.06.050. [DOI] [PubMed] [Google Scholar]
- 106.Bounaix Morand du Puch C, Barbier E, Kraut A, et al. TOX4 and its binding partners recognize DNA adducts generated by platinum anticancer drugs. Arch Biochem Biophys. 2011;507:296–303. doi: 10.1016/j.abb.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 107.Szardenings K, Li B, Ma L, Wu M. Fishing for targets: novel approaches using small molecule baits. Drug Discov Today Technol. 2004;1:9–15. doi: 10.1016/j.ddtec.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 108.Azuma R, Urakawa A. Simultaneous determination of a novel anticancer drug, TAS-103, and its N-demethylated metabolite in monkey plasma by high-performance liquid chromatography using solid-phase extraction. J Chromatogr B Biomed Sci Appl. 1997;691:179–185. doi: 10.1016/s0378-4347(96)00449-5. [DOI] [PubMed] [Google Scholar]
- 109.Yoshida M, Kabe Y, Wada T, Asai A, Handa H. A new mechanism of 6-((2-(dimethylamino)ethyl)amino)-3-hydroxy-7H-indeno(2,1-c)quinolin-7-one dihydrochloride (TAS-103) action discovered by target screening with drug-immobilized affinity beads. Mol Pharmacol. 2008;73:987–994. doi: 10.1124/mol.107.043307. [DOI] [PubMed] [Google Scholar]