Abstract
Next-generation sequencing techniques, such as RNA-sequencing, have provided a wealth of genomic information for non-model species. Transcriptomic information can be used to quantify patterns of gene expression, which can identify how environmental differences invoke organismal stress responses and provide a gauge in predicting species adaptability. In our study, we used RNA-sequencing to characterize the first transcriptome from a naupliar tadpole shrimp (Triops newberryi) to identify the genes expressed during the early life history stages and which could be important for future genomic studies. RNA was extracted from naupliar T. newberryi that were reared in a laboratory controlled setting and in two different water types, a native and a non-native condition. A total of six replicates, three per condition, were sequenced with the Illumina Hi-Seq 2000 achieving 365 M 50nt reads. High quality reads were produced and de novo assembly was used to construct a T. newberryi transcriptome that was approximately 24.8M base pairs. More than 10,000 peptides were predicted from the assembly and genes were sorted into gene ontology categories. The use of different water conditions allowed for a preliminary differential gene expression analysis in order to compare changes in gene expression between conditions. There were 299 differentially expressed genes between water conditions that might serve as a focal point for future genomic studies of Triops acclimation to different environments. The Triops transcriptome could serve as vital genomic information for additional studies on Branchiopod crustaceans.
Keywords: RNA-sequencing, branchiopod, crustacean, gene regulation
Introduction
Genomic resources available for crustaceans, and in particular Branchiopods, are severely limited (Colbourne et al. 2011). The tadpole shrimp, Triops spp. (Branchiopoda: Notostraca), also known as a living fossil (Suno-Uchi et al. 1997), is a crustacean that inhabits ephemeral ponds in arid regions world-wide. Their unconventional life history and habitat as well as their prolonged evolutionary history make them a suitable candidate species to study genetic adaptation to environmental changes. Genes that confer ecoresponsiveness, allowing species to respond to changing environmental conditions, have been observed in another Branchiopod, Daphnia pulex (Colbourne et al. 2011).
Decreasing costs for genomic studies on non-model species has allowed a deeper understanding of gene expression differences during environmental changes. One way to quantify gene expression in different environments is transcriptome analysis via high-throughput sequencing of cDNA (RNA-Seq). This method has provided a more comprehensive measure of gene expression compared to other methods such as microarrays (Mortazavi et al. 2008). RNA-Seq can be performed on non-model organisms, including those without sequenced genomes, and can accurately represent differences in expression levels across various cell types (Mortazavi et al. 2008; Wang et al. 2009). Transcriptome analysis of aquatic organisms has been able to quantify how gene expression is modulated in response to changes in environmental parameters of aquatic systems such as salinity, pH, and thermal stress (Latta et al. 2012; Evans et al. 2013; Chu et al. 2014; Du et al. 2014). How the genome responds to changes in the environment, such as genomic stress responses, can indicate if organisms are locally adapted to native conditions (Kondrashov 2012; Chu et al. 2014; Mandic et al. 2014).
In this study, we use RNA-seq to sequence the transcriptome of naupliar Triops newberryi from southern New Mexico. By using the early developmental period, we aim to describe the genes expressed during this critical time period of morphological change leading to an adult body form (Møller et al. 2003). The naupliar developmental period is also important for the colonization success of the passively dispersed Triops cysts, therefore, we anticipate that the transcriptome will provide an important resource to identify genes related to acclimation to changing environments in Triops populations. To allow for downstream development of genomic markers (i.e. SNPs) to study ecoresponsiveness in Branchiopod crustaceans, the Triops nauplii samples used for RNA-sequencing were reared in a controlled laboratory setting in two different water conditions, termed native and non-native. Therefore, this study also acts as a pilot project to determine the feasibility of utilizing laboratory reared specimens under different environmental conditions, coupled with the transcriptome information to identify genes important for environmental acclimation and adaptation. Differential gene expression analysis of T. newberryi nauplii reared in the two different water conditions was performed to identify potential candidate genes to target in future genomic studies.
Materials and Methods
Sample preparation and RNA isolation
In order to collect sufficient naupliar T. newberryi for RNA-sequencing, the Triops used were reared in the laboratory. Triops newberryi cysts were isolated from soil collected from a dried playa lake in which T. newberryi is the only Triops species that occurs (PL-36; Horn et al. 2014). The collected cysts were incubated and reared in a controlled microcosm setting (deep petri dish) in two water conditions, a native condition and a non-native condition. The native condition consisted of playa lake water reconstituted in the lab using distilled water and dried soil from the same playa lake where the T. newberryi cysts were collected. The non-native condition is reconstituted playa lake water from a different playa in which it is known that T. newberryi does not occur (PL-09; Macdonald et al. 2011; Horn et al. 2014). Water chemistry was measured for the reconstituted PL-36 and PL-09 pond water to assess environmental differences between ponds and was replicated four times. Dissolved oxygen, pH and salinity were measured with a Hach model HQ 40d18 portable combination meter (Hach Company, Loveland, CO) and ammonia, nitrate-N (NO3-N), nitrite-N (NO2-N), phosphate, sulfate and sulfide were analyzed colorimetrically with a LaMotte Smart2 colorimeter (LaMotte Company, Chestertown, MD).
Fifty isolated T. newberryi cysts were placed into each microcosm with 75 mL of reconstituted pond water filtered through a screen of 80 µm mesh size. The experiment was replicated twice with 10 total microcosms; five containing T. newberryi cysts and native water and five containing T. newberryi cysts and non-native water. The microcosms were incubated in a water bath at a temperature of 22°C (±1°C) on a 12-hour light:dark photoperiod. Microcosms were checked three times a day for newly hatched individuals. Once cysts hatched, the naupliar stage was identified and samples were preserved in RNA-later (Qiagen, Valencia, CA).
All of the Triops naupliar life stages (stage I to post-naupliar) were collected from the microcosms (Møller et al. 2003). Stage III individuals were chosen for RNA-sequencing as this represented the middle of the nauplius developmental stages and provided sufficient mRNA for RNA-seq with minimal sample pooling. The second replicate of the experiment produced more of the stage III napulii, therefore, all samples used for RNA extraction came from the second experiment. Eight stage III individuals reared in the native or non-native conditions were pooled for subsequent RNA extraction. For each water condition, three biological replicates were prepared for a total of six samples for RNA-sequencing (three native, three non-native water). Total RNA was extracted using the RNeasy mini kit following manufacturer’s directions (Qiagen, Valencia, CA). RNA was quantified via a nanodrop to ensure sufficient RNA was present (>1100 ng/µl).
Library preparation and RNA-sequencing
Libraries for a total of six samples were made from approximately 2 µg total RNA, quantified by Qubit, using Illumina’s TruSeq RNA Sample Preparation Kit from Illumina (San Diego, CA). The constructed libraries were quality checked using Bioanlyzer (Agilent Technologies, Santa Clara, CA). The average insert size of each library was approximately 160 base pairs long. Libraries were sequenced on the Illumina HiSeq 2000 to obtain 2 × 50 base pair (bp) paired-end reads.
De novo assembly
Paired-end sequence reads from all six libraries were pooled together to generate a de novo transcriptome assembly. The raw sequence reads from the Illumina HiSeq 2000 sequencer were processed to remove Illumina adapters and primers. These post-processed sequence reads were further processed using SGA (Simpson and Durbin 2011) preprocess program for quality trimming (swinging average) at Q15. Sequence reads less than 25 bp after trimmings were discarded. Preprocessed sequence reads were assembled into contigs with ABySS v.1.3.3 (Simpson et al. 2009), using 20 unique k-mers between k=26 and k=50. ABySS was run requiring a minimum k-mer coverage of five, graph bubble popping at >0.9 branch identity, with the scaffolding flag disabled to avoid over reduction of divergent regions. Unitigs from all k-mer assemblies were combined and redundancies were removed using CD-HIT-EST (Li and Godzik 2006) with a clustering threshold of 0.98 identity. The OLC (Overlap-Layout-Consensus) assembler CAP3 (Huang and Madan 1999) was then used to identify minimum 100 bp overlaps between the resultant contigs and assemble larger sequences. The resulting contigs were paired-end scaffolded using ABySS (Simpson et al. 2009). Sequence read pairing information was used in GapCloser v. 1.10 (Li et al. 2008; part of SOAP de novo package) to walk in on gaps created during assembly scaffolding. Redundant sequences were again removed using CD-HIT-EST (Li and Godzik 2006) at a clustering threshold of 0.98 identity. In an attempt to remove incomplete sequences, the consensus contigs were filtered at a minimum length of 150 bp to produce the final set of contigs. The final assembly was used as a reference for subsequent annotation and gene expression analysis.
An assembly assessment for validation was performed for the de novo transcriptome assembly by mapping preprocessed Illumina sequence reads back to the assembly using Burrows Wheeler Aligner (BWA) (Li and Durbin 2009). A high percentage of reads mapped back to the assemblies (99% of the reads mapped back, 96% mapping uniquely and 3% mapping to more than one position in the transcriptome), validating the de novo transcriptome assembly process. CEGMA (Core Eukaryotic Genes Mapping Approach) software was applied to identify the presence of a highly conserved core gene set found in a wide range of eukaryotes (Parra et al. 2007). Transcriptome completeness can be assessed by the identification of these 248 genes expected to be represented in the transcriptome.
Transcriptome annotation
The final transcriptome scaffolds were utilized to predict coding sequences using ESTScan (Iseli et al. 1999; Lottaz et al. 2003) with the Daphnia pulex genome scoring matrix, as this is the most closely related species to Triops with a sequenced genome (Colbourne et al. 2011). Sequence reads were aligned back to the nucleotide motifs of the predicted coding sequences using BWA (Li and Durbin 2009). BLASTp (Altschul et al. 1990) was used to generate annotations of the coding sequences against the UniProtKB/Swiss-Prot database. Protein sequences were also functionally characterized using HMMER3 (Zhang and Wood 2003) against the Pfam-A (Finn et al. 2010), TIGRFAM (Haft et al. 2001), and SUPERFAMILY (Gough et al. 2001) databases.
The Swiss-Prot terms generated with BLASTp were converted to gene ontology (GO) terms in order to characterize the GO classes represented by the T. newberryi transcriptome. The program CateGOrizer (Hu et al. 2008) was used to group and categorize the GO terms into the three broad biological terms, ‘biological processes’, ‘molecular functions’ or ‘cellular components’, against the GO-Slim database.
Differential gene expression analysis
The preprocessed sequence reads used in the de novo transcriptome assembly were aligned to the final transcriptome assembly using BWA (Li and Durbin 2009). Gene expression was quantified as the total number of reads for each sample that uniquely aligned to the reference (i.e. the de novo transcriptome assembly) binned by transcript. The read counts for each biological replicate were used as input for the program edgeR (the empirical analysis of differentially expressed genes in R; Robinson et al. 2010), which is part of the Bioconductor project (Gentleman et al. 2004). EdgeR can detect differentially expressed genes even for those that are lowly expressed, or, if there is high variability between biological replicates (Zhang et al. 2014). The edgeR package used the read count data to determine if there were significant differences in expression between the native and non-native conditions. Differential gene expression in the non-native condition was compared to the native condition at a significance level of 0.05 and a false discovery rate (FDR) correction was applied (Benjamini and Hochberg 1995). The data were normalized using the trimmed means of M values (TMM; Robinson and Oshlack 2010), which excluded genes with high read counts or increased expression differences between conditions and then used a weighted average of the remaining genes. Genes that were lowly expressed (a count of < 6 or 7) were filtered out of the dataset as recommended by the edgeR manual (Robinson et al. 2010). Due to the inflation of reads from one replicate in the non-native condition (see results below), the differential gene expression analysis was performed without this replicate to ensure no bias was present. To have a balanced design, replicate one from the native condition was also dropped and edgeR was run with the same parameters as mentioned previously with the four remaining replicates.
The amount of dispersion between genes was calculated and a MA plot, which compares the log-counts-per-million (logCPM) to the log fold change between conditions, was generated. The annotations generated for the transcriptome were used to identify the differentially expressed genes or were annotated manually using the BLASTn algorithm for the nucleotide (nr/nt) database with an e-value threshold of 1e−5. A statistical overrepresentation test was performed of differentially expressed genes in PANTHER v. 10 (Mi et al. 2013, 2016).
Results
RNA-seq de novo assembly
The total singleton reads for all six samples sequenced was 365,168,688. Most of the replicates produced between 45 and 53 million (M) singleton sequence reads with the exception of one replicate within the non-native condition, which produced over 124M singleton reads (Table 1). Reads less than 25 base pairs (bp) after trimming were removed from assembly, which constituted only 0.34 – 0.43% of the total number of reads from the six replicates. Over 99.5% of the reads were then retained for de novo transcriptome assembly (Table 1). Additional de novo transcriptome assembly metrics are captured in Table 2 and are as follows. The assembly of RNA-seq reads from the 6 replicates produced a total transcriptome length of almost 24.8M bp. There were 15,273 total scaffolds with an average scaffold size of 1,623 bp. The maximum scaffold size was 20,812 bases and the minimum size was 150 bp. The number of contigs produced was 15,841 with an average contig size of 1,565 bp and an N50 value of 3,175. CEGMA analysis identified 247 (99.6%) out of 248 core genes as complete. Complete is defined as an alignment greater than 70% to a core gene suggesting a complete transcriptome assembly. The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GEHY00000000. The version described in this paper is the first version, GEHY01000000.
Table 1.
Details on the raw and quality filtered 2×50nt RNA-seq data for Triops newberryi. Reads that were < 25 base pairs after quality trimming were discarded from use in the transcriptome assembly.
| Sample | Paired-end Reads |
Total Singleton Reads |
Discarded | Percent Discarded |
Retained | Percent Retained |
|---|---|---|---|---|---|---|
| Native_a | 26,589,259 | 53,178,518 | 230,644 | 0.43 | 52,947,874 | 99.57 |
| Native_b | 23,095,438 | 46,190,876 | 180,708 | 0.39 | 46,010,168 | 99.61 |
| Native_c | 23,984,488 | 47,968,976 | 178,553 | 0.37 | 47,790,423 | 99.63 |
| NonNative_a | 62,244,591 | 124,489,182 | 454,719 | 0.37 | 124,034,463 | 99.63 |
| NonNative_b | 23,930,253 | 47,860,506 | 179,496 | 0.38 | 47,681,010 | 99.62 |
| NonNative_c | 22,740,315 | 45,480,630 | 156,828 | 0.34 | 45,323,802 | 99.66 |
Table 2.
Triops newberryi transcriptome assembly details (bp = base pair). Contigs refer to continuous lengths of sequence; scaffolds include both contigs and gaps.
| Assembly Metric | Base Pairs |
|---|---|
| Total bp length including gaps | 24,833,947 |
| Total bp length without gaps | 24,794,310 |
| Total # of contigs | 15,841 |
| Average contig size | 1,565.2 |
| Contig N50 | 3,175 |
| Total # of scaffolds | 15,273 |
| Average scaffold size including gaps | 1,626.0 |
| Average scaffold size without gaps | 1,623.4 |
| Maximum scaffold size | 20,812 |
| Minimum scaffold size | 150 |
| Scaffold N50 | 3,331 |
Functional annotation of the transcriptome
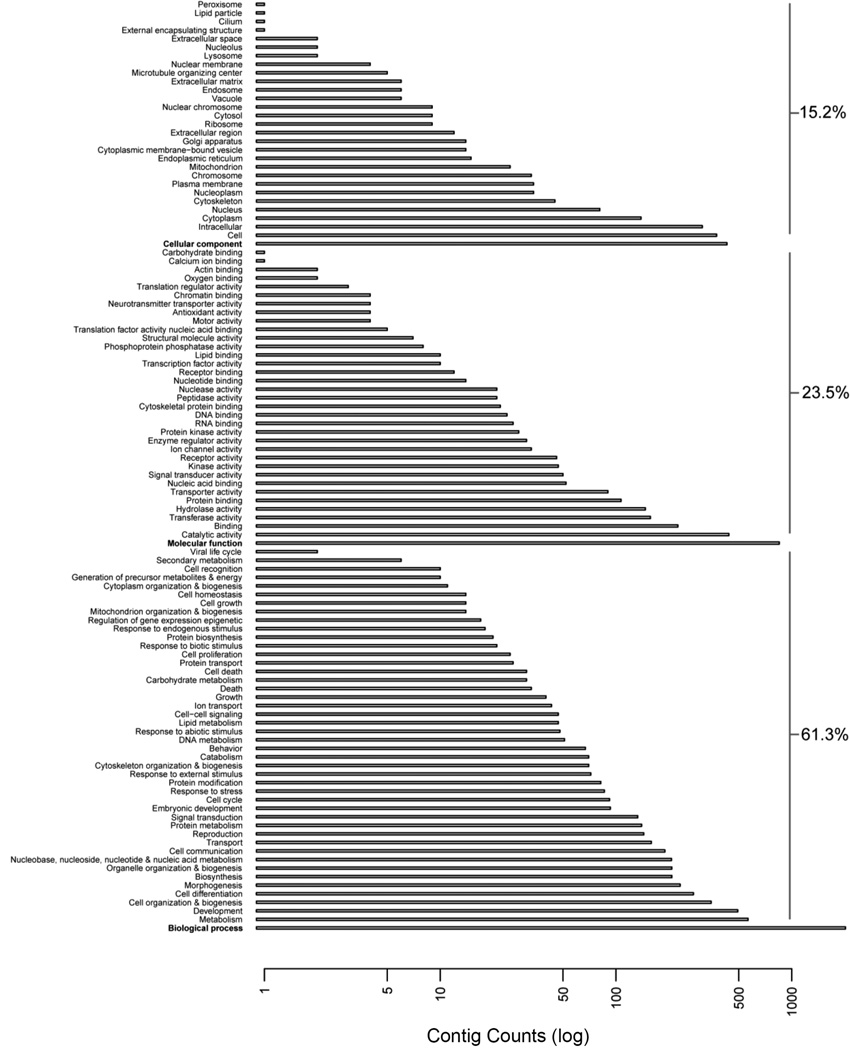
A total of 10,148 peptides greater than 30 amino acids in length were predicted from the scaffolds with ESTScan and annotated using the UniProtKB/Swiss-Prot database. Files for all functional characterizations from the different databases (Swiss-Prot, Pfam-A, TIGRFAM, SUPERFAMILY) are deposited in DataDryad [doi:10.5061/dryad.t3j7m]. Conversion of the Swiss-Prot accession numbers to GO terms returned 1,297 contigs that were associated with a GO term. The GO categorizations are shown in detail in Figure 1 and are summarized as follows. Most of the cellular components were present, comprising 15.2%. Molecular functions comprised 23.5% with ‘catalytic activity’, ‘binding’, ‘transferase activity’ and ‘hydrolase activity’ most represented. Biological processes were predominant at 61.3%, with the major categories encompassing ‘metabolism’, ‘development’, ‘cell organization and biogenesis’, ‘cell differentiation’ and ‘morphogenesis’.
Figure 1.
The gene ontology (GO) categories (y-axis) present in the Triops newberryi transcriptome. The number of read counts (log scale) used to infer expression level is on the x-axis. The percentage of read counts that fall into biological processes, molecular function and cellular components are on the right.
Hatching success and differential gene expression between native and non-native water conditions
In the first replicate of the experiment, the final hatching percentage (FHP) for T. newberryi cysts hatched in the native water was 45% compared to 36% hatching in the non-native water. In the second experiment, the FHP for T. newberryi was 44.7% in native water and 64.7% in the non-native water (Table S1, supporting information). Results of an ANOVA indicated no significant difference in FHP between the two experiments (P > 0.2), therefore, data from experiment one and two were pooled. There was no significant difference in the FHP of T. newberryi between the native and non-native waters. The water chemistry test did indicate significant differences (P < 0.05, t-test) between the native and non-native water in measures of phosphate, sulfate, pH and dissolved oxygen (Table 3).
Table 3.
Water chemistry measurements for PL-36 (native conditions) and PL-09 (non-native condition. There were four total replicates from each playa lake water and the average of those measurements is given.
| Ammonia | DO | Nitrate | Nitrite | pH | Phosphate | Salinity | Sulfate | Sulfide | |
|---|---|---|---|---|---|---|---|---|---|
| PL36-1 | 0.82 | 5.79 | 0.35 | 0.11 | 8.30 | 0.13 | 0.18 | 56 | 0.01 |
| PL36-2 | 0.89 | 6.04 | 12.4 | 2.59 | 7.83 | 0.09 | 0.18 | 56 | 0.01 |
| PL36-3 | 0.52 | 6.29 | 37.5 | 3.97 | 7.92 | 0.16 | 0.23 | 52 | 0.02 |
| PL36-4 | 0.81 | 6.58 | 17 | 3.38 | 8.03 | 0.10 | 0.19 | 39 | 0.00 |
| Average | 0.76 | 6.18* | 16.81 | 2.51 | 8.02* | 0.12* | 0.20 | 50.75* | 0.01 |
| PL09-1 | 0.84 | 1.31 | 0.02 | 0.00 | 7.61 | 0.67 | 0.28 | 2 | 0.01 |
| PL09-2 | 0.36 | 1.05 | 0.05 | 0.00 | 7.65 | 0.51 | 0.23 | 3 | 0.00 |
| PL09-3 | 0.38 | 3.60 | 0.10 | 0.00 | 7.81 | 0.93 | 0.17 | 2 | 0.00 |
| PL09-4 | 0.31 | 1.13 | 0.18 | 0.00 | 7.64 | 0.44 | 0.23 | 5 | 0.00 |
| Average | 0.47 | 1.77* | 0.09 | 0.00 | 7.68* | 0.64* | 0.23 | 1.41* | 0.00 |
A significant t-test between water chemistry measurements is denoted by an * in the ‘Average’ field.
There were 299 contigs that were differentially expressed in the non-native condition when compared to the native condition at an adjusted P-value, comprising 2.9% of the 10,148 coding sequences predicted from total transcriptome (see Fig. S1 and Table S2, supporting information). After removing two of the replicates to compensate for one replicate having a high read count, the edgeR program identified 219 differentially expressed contigs. In total, there were 174 identical contigs that were differentially expressed in both of the edgeR runs.
Of the 299 contigs differentially expressed, 75 were upwardly expressed and 224 were downwardly expressed in the non-native condition compared to the native condition. There were matches with the Swiss-Prot database for 131 of the 299 peptides and included mostly matches to ribosomal genes (Table S2). The remaining 168 coding sequences were screened against the nr/nt database in BLASTn. There were 34 of the 168 peptides that produced a BLASTn match with an e-value below the cut-off of 1e−5 (Table S2). About half of the sequences with BLASTn matches were identified as cuticle or other structural proteins, 10 coded for ribosomal proteins and other matches to the database are noted in Table S1. BLASTn identified 114 as uncharacterized or hypothetical proteins. The remaining 20 coding sequences produced BLAST matches that were above the e-value cut-off.
The statistical overrepresentation test identified nine GO terms that are enriched in the differential gene expression analysis (Table 4). Under ‘biological processes’, the GO terms of ‘translation’, ‘protein metabolic process’, and ‘metabolic process’ were significantly over-represented (P < 0.001). The GO terms ‘peroxidase activity’, ‘antioxidant activity’, ‘structural constituent of ribosome’, ‘structural molecule activity’, and ‘nucleic acid binding’ were over-represented in the ‘molecular functions’ category (P < 0.0003). ‘Ribosome’ was the only significantly over-represented term (P < 0.004) in the ‘cellular components’ GO category. These nine terms were also significantly over-represented in the differentially expressed genes identified when two of the six replicates were removed in addition to the GO terms ‘binding’ and ‘primary metabolic process’ (Table 4).
Table 4.
GO terms statistically over-represented in the differential gene expression analysis.
| GO Term | GO ID | Fold Enrichment | P-value |
|---|---|---|---|
| Translation | GO:0006412 | > 5 | 2.22E-09 |
| Protein metabolic process | GO:0019538 | > 5 | 4.75E-05 |
| Metabolic process | GO:0008152 | 2.44 | 1.25E-03 |
| Primary metabolic process | GO:0044238 | 2.86 | 1.90E-03 |
| Peroxidase activity | GO:0004601 | > 5 | 5.92E-04 |
| Antioxidant activity | GO:0016209 | > 5 | 9.55E-04 |
| Structural constituent of ribosome | GO:0003735 | > 5 | 1.58E-11 |
| Structural molecule activity | GO:0005198 | > 5 | 5.47E-09 |
| Nucleic acid binding | GO:0003676 | > 5 | 3.32E-04 |
| Binding | GO:0005488 | 3.69 | 7.07E-03 |
| Ribosome | GO:0005840 | > 5 | 3.99E-02 |
Discussion
This study characterizes the first transcriptome of a tadpole shrimp, Triops newberryi, during the naupliar stage of development. The small naupliar stage III of Triops was sufficient to provide high-quality RNA-seq data with over 99.5% of the reads retained for assembly and annotation. Despite not having a reference genome for transcriptome assembly, greater than 99% of the reads were properly mapped back to the transcriptome, providing validation for the de novo assembly. It was observed that many of the genes essential in development and growth are operational during this time. In addition, the preliminary differential gene expression analysis of T. newberryi reared in two environmental conditions indicated changes in gene expression despite no significant difference in the cyst hatching percentage between water types.
Triops newberryi transcriptome details
The RNA-seq provided high quality reads of the T. newberryi transcriptome allowing for successful de novo assembly. There was one replicate from the non-native condition that produced more than double the amount of singleton reads. Initial quality checks of the RNA concentration before library preps indicated no significant increase in RNA concentration for this one replicate. The final concentration of RNA from all replicates ranged from 47 – 92.5 ng/µL and the replicate in question had a concentration of 70.9 ng/µL. The six replicates were pooled in one lane on the Illumina HiSeq 2000, which has been noted as causing unbalance among the replicates (Zhang et al. 2014).
Despite successful de novo assembly of the RNA-seq reads, only about half of the contigs present in the T. newberryi transcriptome could be annotated. This is comparable to some other non-model species of crustacean, in which the number of annotated genes ranged from 29–45% (Barreto et al. 2011; Li et al. 2012; Schoville et al. 2012; Harms et al. 2013; Lenz et al. 2014). In addition, lineage specific genes are often difficult to annotate because their function is specific to the species and the environmental stress experienced (Asselman et al. 2015). Despite having low success with classifying predicted peptides into GO categories, other studies on non-model species of crustaceans have reported similar annotation rates (Li et al. 2012; Lenz et al. 2014). There were also consistencies in ontology analysis between T. newberryi and other crustacean transcriptome studies. For example, the majority of expressed transcripts in Triops newberryi fell into the GO category of biological processes. In other crustacean species, such as the Pacific white shrimp (Litopenaeus vannamei; Li et al. 2012), the copepod Calanus finmarchicus (Lenz et al. 2014), and the boreal spider crab Hyas araneus (Harms et al. 2013), biological processes also represent the largest category with transcripts mapping to this GO term. The category of molecular functions is the second largest in all studies including the present one and cellular components have the smallest percentage of transcripts expressed.
The transcriptome ontology in nauplii of T. newberryi had some overlap with the transcriptome ontology for insects in the developmental stage, including the whitefly (Benisia tabaci) and the milkweed bug (Oncopeltus fasciatus) (Wang et al. 2010; Ewen-Campen et al. 2011). Under the GO term ‘biological processes’, the studies on developing insects identified the sub-categories ‘cellular process’, ‘biological regulation’, ‘localization’, ‘multicellular organismal process’, ‘response to stimulus’, ‘development’ and ‘metabolism’ with the most mapped transcripts (Wang et al. 2010; Ewen-Campen et al. 2011). Within ‘biological processes’, the two categories with the most transcripts for T. newberryi were ‘development’ and ‘metabolism’ which overlap with observed categories in the developing insects, however, the other categories found in the whitefly and milkweed bug were not present in the T. newberryi GO analysis. In the broad category of ‘molecular processes’, there was more congruence as the terms ‘catalytic activity’ and ‘binding’ were the most prevalent for T. newberryi, the whitefly, milkweed bug, as well as several species of crustaceans (Wang et al. 2010; Ewen-Campen et al. 2011; Li et al. 2012; Lenz et al. 2014; Harms et al. 2013). Differences observed between T. newberryi and other species could be due to the low amount of predicted peptides with GO terms within the T. newberryi transcriptome data. Despite high quality reads and assembly, it was impossible to classify all of the genes differentially expressed during Triops naupliar development because annotations could not be made.
The gene ontology analysis indicated that a substantial proportion of the genes expressed in a stage III nauplius Triops were those related to development. Triops have five naupliar stages that commence immediately after hatching from the cyst and end when the adult form is reached at the post-naupliar stage (Møller et al. 2003). These developmental stages represent the majority of morphological changes that Triops undergo during its life cycle. Active genes during stage III included those that encode for the neurogenic locus notch protein, which operates during development to establish cellular communication in the central nervous system (Smoller et al. 1990), and a gene similar to the Drosophila developmental protein sprouty involved in cellular signaling (King et al. 2005). Genes involved in cell differentiation, such as the COP9 signalsome and caspases (Wei and Deng 2003; Lamkanfi et al. 2007), and cell proliferation (cad proteins; Grande-García et al. 2014) were also expressed. The gene encoding the protein tamozhennic was present in the Triops transcriptome, which in Drosophila has been shown to be active during development as an importer to the nucleus (Minakhina et al. 2003). Expressed transcripts also included those involved in the processes of gene regulation. Various genes encoding transcription proteins were active and included exonuclease, the general transcription factor IIF and the c-terminal binding protein (Schaeper et al. 1998). Also active were those genes involved in DNA processing (flap exonuclease; Liu et al. 2004) and translation (threonylcarbamoyladenosine tRNA methylthiotransferase, Arragain et al. 2010).
Potential implications and downstream applications
There was generally high agreement in the differential gene expression analysis between analyses with the full dataset and when removing the replicate with a high read count. The amount of differentially expressed genes decreased by 80 when the replicates were removed, however, removal of replicates can cause a decrease in the amount of differentially expressed genes even when the read counts among replicates are equal (Zhang et al. 2014). The same GO terms were statistically over-represented when all replicates were analyzed for differential gene expression and when two replicates were removed. This likely indicates that the replicate with the inflated read count did not bias the test for differences in gene expression; therefore, we discuss results based on the full dataset.
There was not a significant difference in the amount of cysts that hatched between the water types, but we did see differences in gene expression. Changes in gene expression can be the result of organisms coping with environmental pressures or indicative of populations that are locally adapted to native conditions (Schoville et al. 2012; De Wit and Palumbi 2013). One cellular reaction to a stressor is the expression of heat-shock proteins or other molecular chaperones (Feder and Hofmann 1999; Chu et al. 2014; Gleason and Burton 2015). There was only one instance of a heat-shock protein up-regulated within the non-native condition suggesting hatching of T. newberryi in a non-native water condition may not be causing protein unfolding, which would be expected to result in a need for heat shock proteins or molecular chaperons. The lack of molecular chaperone expression could be due to the preferential sampling of stage III nauplii; these stress proteins may be more abundant during early developmental stages before the individual as time to acclimate to the water conditions. Cellular stress can also be accompanied by the global repression of translation in order to avoid errors in gene regulation (Mayer and Grummt 2005; Shenton et al. 2006), and organisms often regulate transcription and translation genes in unison under stress (Lackner et al. 2012). For example, genes involved in transcription and translation, including ribosomal genes, were differentially expressed when exposing D. magna to cadmium, an insecticide and an herbicide, all of which affect Daphnia growth and development (Connon et al. 2008; Pereira et al. 2010). Changes in pH have been shown to effect cellular signaling, ion transportation and transcription (Evans et al. 2013); a significant difference in pH was measured between the native and non-native water conditions. The majority of annotated genes that were differentially expressed and over-represented between water conditions in T. newberryi were those involved in gene regulation, including the processes of transcription, translation and post-translational activities. Triops nauplii may have a general cellular stress response when exposed to non-native water conditions, however, more research is needed to confirm the downstream biological and developmental effects of the differential expression of these genes.
The other majority of differential expressed genes were related to cuticle protein and include the endocuticle structural glycoprotein and the enzyme chitotriosidase, or chitinase, which are responsible for breaking down chitin or cuticle protein during the intermolt portion of the molt cycle (Svitil et al. 1997; Merzendorfer and Zimoch 2003; Seear et al. 2010). The process of molting, in which the entire exoskeleton is shed, is a critical life point for crustaceans and is highly regulated through hormonal changes (Chang and Mykles 2011). The cuticle protein is a major constituent of the exoskeleton and is essential in the growth and development of crustaceans (Roer and Dillaman 1984). In the water flea Daphnia magna, both zinc (Poynton et al. 2007) and ibuprofen (Heckmann et al. 2008) have been shown to affect the regulation of chitinase genes, which had direct effects on growth and reproduction. Cadmium also triggered the regulation of molt related genes in Daphnia pulex, including those encoding cuticle proteins, causing significant differences in the overall body size and reproductive output (Shaw et al. 2007). Exposure to other chemicals, such as herbicides and insecticides affected molting in D. magna by either accelerating or delaying the molting process (Pereira et al. 2010). During the naupliar stages of development, Triops undergo massive changes in morphology and increases in size (Møller et al. 2003) that are facilitated by the process of molting (Fryer 1988). The molt related genes identified by transcriptome sequencing might be an important indication of stress response in Triops. Despite being able to hatch in non-native water, water chemical conditions might exist that cause differential gene expression of important molting related genes inhibiting proper development of nauplii. To fully understand the impact of differential expression of molting genes, Triops nauplii should be reared past stage III in different water conditions to assess if the adult body form is reached and if morphological differences or abnormalities are present among the Triops.
Further laboratory studies confirming the importance of the genes identified by differential expression analysis of the transcriptome will need to be conducted as well as expanding the various water types in which Triops are hatched and extending the work to include developmental stages beside stage III. Many of the differentially expressed genes were hypothetical or uncharacterized proteins, but might be important to the overall genomic responses of the organism to the environment as was documented in Daphnia (Asselman et al. 2015). Further testing of the expression of these uncharacterized genes in different conditions and the organismal responses will aid in identification of genes conferring ecoresponsiveness. Lastly, the utility of laboratory experiments with Triops will need to be confirmed in field situations.
Conclusions
The process of RNA-seq and de novo assembly has produced the first transcriptome data on a species of Triops. Annotation of gene transcripts present, however, proved difficult despite the high quality data obtained from the RNA-seq. It is anticipated that together with increased research on the characterization of genes with unknown functions and obtaining a complete Triops genome will facilitate understanding changes at the molecular level among Triops populations and identify genes critical to acclimation to various environmental conditions. This study also identified some genes that are differentially expressed between two water conditions and could serve as a starting point at which to begin more detailed genomic analysis of Triops. This study also serves as a foundation to further develop genomic resources, such as SNPs, and as a tool for marker development in other closely related Branchiopods. Future studies observing the growth of Triops in different water conditions, along with a more detailed analysis of the biotic and abiotic constituents of pond water will help clarify the changes in gene expression observed.
Supplementary Material
Acknowledgments
We thank K. Macdonald and R. Sallenave for help with monitoring the microcosms used to rear samples and A. Unk for use of his nanodrop machine. Funding for the RNA-seq and analysis portion of the study was provided through New Mexico-IDeA Network of Biomedical Research Excellence (NM-INBRE) Sequencing and Bioinformatics Pilot Project Award mechanism made possible through the National Institute of General Medical Sciences (5P20GM103451). The New Mexico Agricultural Experiment Station provided additional support.
Footnotes
Data Accessibility
RNA-sequencing data including assembled transcriptome: Genebank short read archiving facility under the accession number PRJNA314525. The accession numbers for each of the six replicates are SRX1617753, SRX1619604, SRX1619606, SRX1619610, SRX1619611, SRX1619612). The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GEHY00000000. The version described in this paper is the first version, GEHY01000000.
Annotation results for the transcriptome, direct output from CateGOrizer, the read counts, and the R script used to run edgeR program: DRYAD entry doi:10.5061/dryad.t3j7m.
Author Contributions
RLH reared and collected samples, extracted and quantified the RNA, performed the differential expression and gene ontology analysis, and prepared the manuscript; ND performed the transcriptome annotation and assisted in manuscript preparation; TR assembled the transcriptome and assisted in manuscript preparation; FDS designed the RNA-seq and analysis experiment, managed its execution and data generation at NCGR and assisted in manuscript review; DEC conceived the study, reared and collected samples and provided comments on the manuscript. All authors approved the final manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arragain S, Handelman SK, Forouhar F, et al. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-Methylthio-N6-threonylcarbamoyladenosine in tRNA. The Journal of Biological Chemistry. 2010;285:2825–28433. doi: 10.1074/jbc.M110.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselman J, Pfrender ME, Lopez JA, et al. Conserved transcriptional responses to cyanobacterial stressors are mediated by alternate regulation of paralogous genes in Daphnia. Molecular Ecology. 2015;24:1844–1855. doi: 10.1111/mec.13148. [DOI] [PubMed] [Google Scholar]
- Barreto FS, Moy GW, Burton RS. Interpopulation patterns of divergence and selection across the transcriptome of the copepod Tigriopus californicus. Molecular Ecology. 2011;20:560–572. doi: 10.1111/j.1365-294X.2010.04963.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Chang ES, Mykles DL. Regulation of crustacean molting: a review and our perspectives. General and Comparative Endocrinology. 2011;172:323–330. doi: 10.1016/j.ygcen.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Chu ND, Miller LP, Kaluziak ST, Trussell GC, Vollmer SV. Thermal stress and predation risk trigger distinct transcriptomic responses in the intertidal snail Nucella lapillus. Molecular Ecology. 2014;23:6104–6113. doi: 10.1111/mec.12994. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon R, Hooper HL, Sibly RM, et al. Linking molecular and population stress responses in Daphnia magna exposed to cadmium. Environmental Science and Technology. 2008;42:2181–2188. doi: 10.1021/es702469b. [DOI] [PubMed] [Google Scholar]
- De Wit P, Palumbi SR. Transcriptome-wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Molecular Ecology. 2013;22:2884–2897. doi: 10.1111/mec.12081. [DOI] [PubMed] [Google Scholar]
- Du X, Li L, Zhang S, Meng F, Zhang G. SNP identification by transcriptome sequencing and candidate gene-based association analysis for heat tolerance in the bay scallop Argopecten irradians. PLoS ONE. 2014;9:e104960. doi: 10.1371/journal.pone.0104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TG, Chan F, Menge BA, Hofmann GE. Transcriptome responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Molecular Ecology. 2013;22:1609–1625. doi: 10.1111/mec.12188. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Shaner N, Panfilio KA, Suzuki Y, Roth S, Extavour CG. The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC Genomics. 2011;12:61. doi: 10.1186/1471-2164-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annual Review of Physiology. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer G. Studies on the functional morphology and biology of the Notostraca (Crustacea: Branchiopoda) Philosophical Transactions of the Royal Society of London B. 1988;321:27–124. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason LU, Burton RS. RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Molecular Ecology. 2015;24:610–627. doi: 10.1111/mec.13047. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. Journal of Molecular Biology. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Grande-García A, Lallous N, Díaz-Tejada C, Ramón-Maiques S. Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD. Structure. 2014;22:185–198. doi: 10.1016/j.str.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, et al. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Research. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms L, Frickenhaus S, Schiffer M, et al. Characterization and analysis of a transcriptome from the boreal spider crab Hyas araneus. Comparative Biochemistry and Physiology, Part D. 2013;8:344–351. doi: 10.1016/j.cbd.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Heckmann L-H, Sibly RM, Connon R, et al. Systems biology meets stress ecology: linking molecular and organismal stress responses in Daphnia magna. Genome Biology. 2008;9:R40. doi: 10.1186/gb-2008-9-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn RL, Kuehn R, Drechsel V, Cowley DE. Discriminating between the effects of founding events and reproductive mode on the genetic structure of Triops populations (Branchiopoda: Notostraca) PLoS ONE. 2014;9:e97473. doi: 10.1371/journal.pone.0097473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z-L, Bao J, Reecy JM. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online Journal of Bioinformatics. 2008;9:108–112. [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings of the International Conference on Intelligent Systems for Molecular Biology. 1999:138–148. [PubMed] [Google Scholar]
- King JAJ, Straffon AFL, D’Abaco GM, et al. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochemical Journal. 2005;388:445–454. doi: 10.1042/BJ20041284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proceedings of the Royal Society B. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Schmidt MW, Wu S, Wolf DA, Bähler J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biology. 2012;13:R25. doi: 10.1186/gb-2012-13-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Gerghe TV, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death and Differentiation. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Latta LC, Weider LJ, Colbourne JK, Pfrender ME. The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecology Letters. 2012;15:794–802. doi: 10.1111/j.1461-0248.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- Lenz PH, Roncalli V, Hassett RP, et al. De novo assembly of a transcriptome for Calanus finmarchicus (Crustacea, Copepoda) - the dominant zooplankter of the North Atlantic Ocean. PLoS ONE. 2014;9:e88589. doi: 10.1371/journal.pone.0088589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Weng S, Chen Y, et al. Analysis of Litopenaeus vannamei transcriptome using the next-generation DNA sequencing technique. PLoS ONE. 2012;7:e47442. doi: 10.1371/journal.pone.0047442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;25:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kao H-I, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annual Review of Biochemistry. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Lottaz C, Iseli C, Jongeneel CV, Bucher P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics. 2003;19:ii103–ii112. doi: 10.1093/bioinformatics/btg1067. [DOI] [PubMed] [Google Scholar]
- Macdonald KS, III, Sallenave R, Cowley DE. Morphologic and genetic variation in Triops (Branchiopoda: Notostraca) from ephemeral waters of the northern Chihuahuan desert of North America. Journal of Crustacean Biology. 2011;31:468–484. [Google Scholar]
- Mandic M, Ramon ML, Gracey AY, Richards JG. Divergent transcriptional patterns are related to differences in hypoxia tolerance between the intertidal and the subtidal sculpins. Molecular Ecology. 2014;23:6091–6103. doi: 10.1111/mec.12991. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. The Journal of Experimental Biology. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature Protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Research. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhina S, Yang J, Steward R. Tamo selectively modulates nuclear import in Drosophila. Genes to Cells. 2003;8:299–310. doi: 10.1046/j.1365-2443.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- Møller OS, Olesen J, Høeg JT. SEM studies on the early larval development of Triops cancriformis (Bosc) (Crustacea: Branchiopoda, Notostraca) Acta Zoologica. 2003;84:267–284. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pereira JL, Hill CJ, Sibly RM, et al. Gene transcription in Daphnia magna: effects of acute exposure to a carbamate insecticide and an acetanilide herbicide. Aquatic Toxicology. 2010;97:268–276. doi: 10.1016/j.aquatox.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Poynton HC, Varshavsky JR, Chang B, et al. Daphnia magna ecotoxicogenomics provides mechanistic insights into metal toxicity. Environmental Science and Technology. 2007;41:1044–1050. doi: 10.1021/es0615573. [DOI] [PubMed] [Google Scholar]
- Roer R, Dillaman R. The structure and calcification of the crustacean cuticle. American Zoologist. 1984;24:893–909. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U, Subramanian T, Lim L, Boy JM, Chinnadurai G. Cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. The Journal of Biological Chemistry. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- Schoville SD, Barreto FS, Moy GW, Wolff A, Burton RS. Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evolutionary Biology. 2012;12:170. doi: 10.1186/1471-2148-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seear PJ, Tarling GA, Burns G, et al. Differential gene expression during the moult cycle of Antarctic krill (Euphausia superba) BMC Genomics. 2010;11:582. doi: 10.1186/1471-2164-11-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JR, Colbourne JK, Davey JC, et al. Gene response profiles for Daphnia pulex exposed to the environmental stressor cadmium reveals novel crustacean metallothioneins. BMC Genomics. 2007;8:477. doi: 10.1186/1471-2164-8-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, et al. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. The Journal of Biological Chemistry. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Research. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson T, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Research. 2011;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller D, Friedel C, Schmid A, Bettler D, Lam L, Yedvobnick B. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes & Development. 1990;4:1688–1700. doi: 10.1101/gad.4.10.1688. [DOI] [PubMed] [Google Scholar]
- Suno-Uchi N, Sasaki F, Chiba S, Kawata M. Morphological stasis and phylogenetic relationships in Tadpole shrimps, Triops (Cructacea: Notostraca) Biological Journal of the Linnean Society. 1997;61:439–457. [Google Scholar]
- Svitil AL, Ní Chadhain SM, Moore JA, Kirchman DL. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Applied and Environmental Microbiology. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Review Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-W, Luan J-B, Bao J-M, Zhang C-X, Liu S-S. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics. 2010;11:400. doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. The COP9 signalosome. Annual Review of Cell and Developmental Biology. 2003;19:261–268. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wood WI. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics. 2003;19:307–308. doi: 10.1093/bioinformatics/19.2.307. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Jhaveri DJ, Marshall VM, et al. A comparative study of techniques for differential expression analysis on RNA-seq data. PLoS ONE. 2014;9:e103207. doi: 10.1371/journal.pone.0103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.