Abstract
Small molecules of plant origin offer presumptively safe opportunities to prevent carcinogenesis, mutagenesis and other forms of toxicity in humans. However, the mechanisms of action of such plant-based agents remain largely unknown. In recent years the stress responsive transcription factor Nrf2 has been validated as a target for disease chemoprevention. Withania somnifera (WS) is a herb used in Ayurveda (an ancient form of medicine in South Asia). In the recent past, withanolides isolated from WS, such as Withaferin A (WA) have been demonstrated to be preventive and therapeutic against multiple diseases in experimental models. The goals of this study are to evaluate withanolides such as WA as well as Withania somnifera root extract as inducers of Nrf2 signaling, to probe the underlying signaling mechanism of WA and to determine whether prevention of acetaminophen (APAP)-induced hepatic toxicity in mice by WA occurs in an Nrf2-dependent manner. We observed that WA profoundly protects wild-type mice but not Nrf2-disrupted mice against APAP hepatotoxicity. WA is a potent inducer of Nrf2-dependent cytoprotective enzyme expression both in vivo and in vitro. Unexpectedly, WA induces Nrf2 signaling at least in part, in a Keap1-independent, Pten/Pi3k/Akt-dependent manner in comparison to prototypical Nrf2 inducers, sulforaphane and CDDO-Im. The identification of WA as an Nrf2 inducer that can signal through a non-canonical, Keap1-independent pathway provides an opportunity to evaluate the role of other regulatory partners of Nrf2 in the dietary and pharmacological induction of Nrf2-mediated cytoprotection.
Keywords: Nrf2, Withaferin A, stress response, hepatotoxicity, Pten
Graphical abstract
1. Introduction
Withania somnifera (WS) is a plant that has been used in traditional South Asian medicine for centuries against various ailments [1] [2]. Recent studies performed on WS plant extracts (roots and leaves) have revealed that medicinal properties of the plant arise from the presence of withanolides (a type of steroidal lactone). Out of the dozens of withanolides isolated from the WS plant, Withaferin A (WA) has been shown to be the most potent and the most abundant withanolide in WS roots. WA is effective as a cancer preventive [3] [4] [5] and therapeutic [6] [7] [8] agent in in vivo experimental models suggesting a potential to be developed as an anti-cancer compound in humans. While several molecular targets of WA have been identified, the mechanism underlying its cytoprotective responses remain unknown.
Nrf2 is a redox-responsive transcription factor that is a master regulator of cellular homeostasis. Enhanced Nrf2 signaling is associated with prevention of hepatotoxicity [9] [10] and protection against a wide array of diseases such as cancer [11], diabetes [12], sickle cell disease [13] and neurodegenerative disease [14] in animal models. Under basal conditions, Nrf2 is primarily regulated by the Keap1-Cul3 E3 ubiquitin ligase complex which mediates the proteasomal degradation of Nrf2. Under stress conditions, degradation of Nrf2 is disrupted allowing for enhanced nuclear translocation and accumulation, and subsequent transcriptional activation of antioxidant response element (ARE) genes. Small electrophilic molecules have been shown to specifically target reactive cysteine residues of Keap1 leading to a conformational change in the Nrf2-Keap1-Cul3 complex resulting in dampened marking of Nrf2 for proteolysis and thus enhanced Nrf2 signaling. This mechanism appears to account for the primary mode of action for many small molecules that induce this cytoprotective response.
Alternative mechanisms of Nrf2 regulation that are independent of Keap1 have also garnered interest recently. While Keap1 is thought to be the dominant repressor of Nrf2 under basal conditions, stress or disease conditions may allow for other proteins such as β-TrCP [15] [16] and Hrd1 [17] to participate significantly in the regulation of Nrf2. Furthermore, cross-talk between Nrf2 and other pathways such as Notch [18, 19] and more recently, Pten [20] [21] suggest the complexity of the Nrf2 signaling network. Although many reports to date have shown that small electrophilic molecules specifically target cysteine residues of Keap1 (canonical Nrf2 induction), some small molecules that target alternative elements of the signaling pathway (non-canonical Nrf2 induction) have been presented [22] [23] suggesting that both Keap1-dependent and independent mechanisms of Nrf2 regulation are targets to be considered when characterizing novel pharmacologic inducers of Nrf2 signaling.
We have examined the roles of both canonical and non-canonical Nrf2 induction pathways by WA. To assess the physiological significance of WA-mediated Nrf2 induction, we utilized an acetaminophen (APAP) hepatotoxicity model in genetically-engineered mice that exhibit different Nrf2 expression profiles. Our results demonstrate that WA is a potent inducer of Nrf2 signaling that protects against hepatotoxicity in an Nrf2-dependent manner and preferentially induces Nrf2 through a Keap1-independent, non-canonical mechanism that is modulated by the Pten/Pi3k/Akt axis.
2. Methods and materials
2.1. Animals and husbandry
Wild-type, 8–10 week old, male albino C57BL/6J mice (25–33 g) were purchased from Jackson Laboratories (Bar Harbor, ME). Systemic Nrf2-knockout mice (referred to as Nrf2-disrupted hereafter) [24] and Nrf2flox/flox mice [25] were maintained in the C57BL/6J background. Nrf2-disrupted mice also possess a knocked in LacZ reporter in the Nrf2 locus that allows for partial expression of Nrf2 and β-galactosidase fusion protein (Nrf2-β gal) [24] [26] [27]. Thus, Nrf2-disrupted mice will be referred to as Nrf2-reporter mice in the Nrf2-β gal immunostaining assay. Hepatocyte specific Nrf2-disrupted mice (referred to as Nrf2flox/flox::AlbCre hereafter) mice were genotyped as described previously [19]. Animals were fed a standard chow diet (Prolab Isopro RMH 3000, LabDiet, St Louis, MO) with access to ad libitum water except as noted under transient conditions of starvation. Experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at University of Pittsburgh.
2.2. In vivo experiments
Wild-type, Nrf2flox/flox, Nrf2flox/flox::AlbCre and Nrf2-disrupted mice were orally gavaged with 100 μl of either DMSO or 7 mg/kg WA. Animals were euthanized and tissues were harvested 20 hours later. RNA was isolated from livers and other organs. APAP (Sigma Aldrich, St. Louis, MO) was dissolved in warm saline prior to administration. For the APAP dosing regimen, mice were pre-treated with 7 mg/kg WA followed by a 22 hour starvation period. Starvation has been shown to result in lowered glutathione levels in the liver [28] and therefore can provide more uniformity in hepatotoxicity data. APAP (250 mg/kg, i.p., in 500 μl saline) was administered and animals were sacrificed 6 hours later. For serum alanine aminotransferase (ALT) measurements, blood was drawn by cardiac puncture and for histological analyses, livers were fixed in formalin.
2.3. Cell lines and reagents
Mouse embryonic fibroblasts (MEFs) from wild-type, Nrf2-disrupted, Keap1-disrupted and Keap1 & Nrf2 double-disrupted mice were generated as described previously [29]. MCF7 cells stably transfected with pTA-Nrf2-Luciferase reporter gene containing 4 tandem repeats of Nrf2 binding sites was obtained from Signosis (SL-0010-NP, Santa Clara, CA). Luciferase activity in MCF7 cells was calculated by normalizing the Firefly luciferase activity readout to the number of cells in each sample. WA (Enzo Lifesciences, Farmingdale NY) was dissolved in DMSO at 20 mg/ml. Withania somnifera root extract (WRE) was provided by Dr. Adam Marcus (Emory University, Atlanta, GA). Yang et al. characterized the withanolides composition of this extract preparation to be 5.88 mg/ml WA (79.03%), 0.14 mg/ml WLA (1.83%), 0.013 mg/ml WN (0.18%), 1.06 mg/ml 12-Deoxywithastramonolide (14.25%) and 0.36 mg/ml Withanoside V (4.78%) [30]. Withanolide A (WLA) and Withanone (WN) were obtained from ChromaDex (Irvine, CA). CDDO-Im was a generous gift from Dr. Michael Sporn (Dartmouth Medical School, Hanover, NH). Sulforaphane was obtained from LKT laboratories (St Paul, MN). LY294002 (Cell Signaling, Danvers, MA) and SF1670 (Sigma Aldrich, St. Louis, MO) were also dissolved in DMSO as per manufacturer recommendations. Control and shPten stably transfected MEFs were provided by Dr. Carola Neumann (University of Pittsburgh, Pittsburgh PA) [31]. PTEN plasmid constructs were kindly provided by Dr. Miho Iijima (Johns Hopkins University, Baltimore MD) [32].
2.4. In vitro experimental design
MEFs were cultured in Iscove’s Modified Dulbecco Medium with 10% fetal bovine serum (FBS). MCF7 reporter cells were cultured in Dulbecco’s Modified Eagle Medium with 10% FBS. All cells were seeded at 5 × 105 cells/well in a 6-well dish on the day prior to the experiment unless otherwise noted. For WA dose-response experiments, cells were treated with either 0.02% DMSO control or final concentrations of 0.1, 0.3, 0.7 or 1 μM WA in DMSO. RNA was isolated 20 hours later. CDDO-Im and sulforaphane (at noted concentrations) were utilized as positive control Nrf2 inducing agents. In pharmacologic inhibition experiments, cells were pre-treated with DMSO control/inhibitor (25 μM LY294002) for 1 hour followed by co-treatment with DMSO control/inhibitor and DMSO control/1 μM WA for a further 8 hours. RNA isolation or luciferase assays were performed 20 hours later.
2.5. Transfection studies and luciferase assays
Keap1 & Nrf2 double-disrupted MEF were seeded at 5 × 105 cells/well in 6-well plates on day 0. Plasmid DNA (5 ng pCMV Nrf2, 10 ng pRLTK- ΔARE, 100 ng pCMV NQO1-ARE-Luc, 2.5 ng pCMV WT Keap1) was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) on day 1. pCMV mock vector was utilized to maintain total DNA transfected at 400 ng. Cells were treated with inducers (3 μM WA, 10 μM sulforaphane, 25 nM CDDO-Im) 24 hours after transfection (day 2). For Pten silencing or overexpression experiments, scrambled/siPTEN (SC-36326, Santa Cruz, Dallas, TX) or wild-type PTEN (500 ng) constructs were transfected 24 hours prior to transfection with Nrf2 expression plasmid. Dual Luciferase Assay (Promega, Madison, WI) was carried out 24 hours later (day 3). Luminescence was measured using a Glomax Multi Jr Detection system (Promega, Madison, WI).
2.6. RNA isolation and Real time PCR (RT-PCR)
RNA from mouse organ homogenates and cell cultures was extracted and purified using an RNA extraction kit (5-PRIME, Gaithersburg, MD). For organs that had high lipid content, TRIzol (Invitrogen, Carlsbad, CA) was used. RNA integrity was confirmed by gel electrophoresis. Quantification of RNA concentration was performed using UV spectrophotometry at 260 nm. Absorbance ratio of 260/280 was utilized to determine the purity of RNA. RNA (1 μg) was used to synthesize cDNA with the qScript system (Quanta Biociences, Beverly, MA). Primer sequences were obtained from Primer Bank or previous publications (supplementary data, Table S1). Primer annealing temperatures were determined by semi-quantitative PCR with gradient temperature. Real time PCR was performed on a Bio-Rad My-IQ with SYBR green (Bio-Rad, Hercules, CA). PCR efficiency was determined using a standard curve and the Pfaffl method [33] was used for quantifications of fold changes.
2.7. Immunoblotting
MEFs were seeded at 1.0×106 cells/plate in 100 mm dishes and serum-starved overnight for 16 hours with fresh medium containing 0.02% FBS. Cells were treated with DMSO, 1 μM WA or 25 nM CDDO-Im for either 1 or 3 hours in the low-serum medium. Cells were lysed using ice cold RIPA buffer. Nuclear and cytosolic fractions were separated using a Nuclear Extraction Kit (Abcam, Cambridge, MA). For Pi3k signaling experiments, cells were pre-treated with WA for varying time points. 500 nM SF1670 and 1 μM insulin (Sigma Aldrich, St Louis, MO) were used as positive controls. Protein concentrations were determined by Bradford Assay (Bio-Rad, Hercules CA). Protein lysate (20 μg) was loaded into each well. The antibodies used were: Nrf2 (Santa Cruz, SC-722), Lamin B (Santa Cruz, SC-6216), p-Akt (phosphorylated Akt at Ser473) (Cell Signaling, 9271S), Akt (Cell Signaling, 9272S), Pten (D4.3) (Cell Signaling, 9188S), Gapdh (Novus Biologicals, NB-300-221). Imaging and quantification of blots was performed by Chemi Doc XRS imaging system (BioRad, Hercules, CA).
2.8. Immunostaining
Nrf2-reporter mice were orally administered DMSO vehicle, WA (7 mg/kg) or CDDO-Im (30 μmol/kg in 10% DMSO, 10% Cremophor-EL and PBS). Three hours later livers were removed and fixed in 4% paraformaldehyde. For fluorescence staining, liver sections were stained with rabbit anti-β-galactosidase antibody (Millipore, Billerica MA; AB986) at a 1:150 dilution. Fluorescence-conjugated secondary antibody (Life Technologies, Carlsbad CA; Alexa Fluor 546; A11035) was used at a 1:200 dilution. Sections were counterstained with DAPI (Molecular Probes, Eugene OR; D1306) and imaged with an Olympus Fluoview 1000 confocal microscope.
2.9. Statistical analysis
Values are expressed as mean ± SEM. Unpaired Student’s t-test was utilized for statistical comparisons between 2 groups and one-way analysis of variance (ANOVA) with Tukey’s post-test was utilized for multiple comparisons. Significance was determined according to p<0.05 criteria.
3. Results
3.1. Withania somnifera root extract and select withanolides are potent inducers of Nrf2 signaling
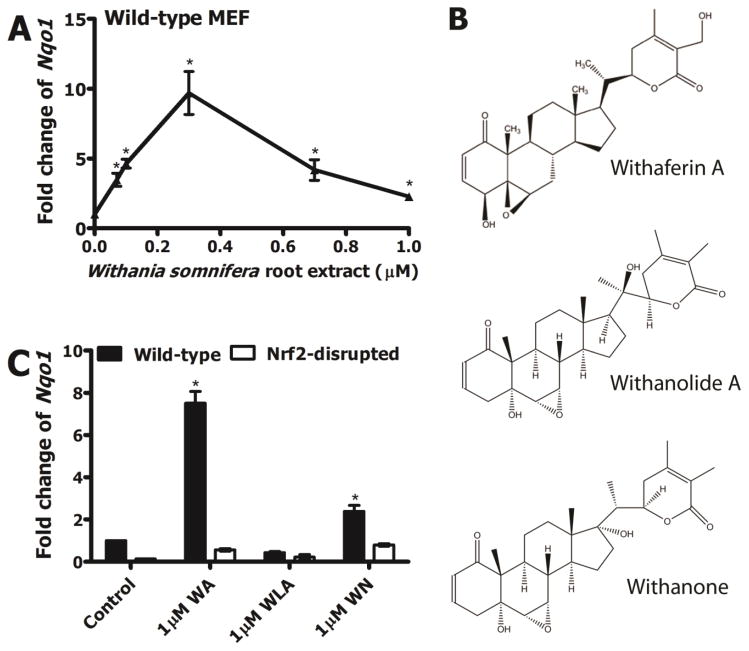
In order to determine whether the ethanolic root extract of this medicinal plant activated Nrf2 signaling, we evaluated induction of Nqo1 transcripts after treating wild-type MEF with graded doses of WA in WRE. A dose-dependent increase in Nqo1 transcripts was observed with WRE treatment which peaked (9.7-fold) at 0.3 μM WA concentration in WRE (Figure 1A). A gradual decline of Nqo1 transcripts was seen at concentrations >0.3 μM, possibly related to cytotoxicity elicited by the treatment. An in vitro cell viability assay revealed that WRE elicits cytotoxicity to WT MEF at high concentrations (supplementary data, Figure S1). The doubling concentration (CD value) of ~60 nM (based on WA content) suggested high potency of this WRE preparation (containing 79% WA of total withanolide concentration) in inducing Nqo1 transcripts. Given that WRE is comprised of multiple different bioactive components, including several types of withanolides we evaluated whether select withanolides induced Nqo1 expression in MEF in an Nrf2-dependent manner. WA, WLA and WN (structures shown in Figure 1B) were considered. WA treatment resulted in ~8-fold induction of Nqo1 transcripts in wild-type MEF which was almost completely absent in Nrf2-disrupted MEF (Figure 1C). WLA treatment did not show a significant induction in wild-type MEF but WN showed a ~2-fold induction of Nqo1 transcripts in wild-type MEF which appeared to be Nrf2-dependent. These data collectively suggested that withanolides found in WRE, particularly WA, are compounds that can induce cytoprotective enzyme expression in murine cells in an Nrf2-dependent manner.
Figure 1.
A) Nqo1 transcript induction in WT MEF by Withania somnifera root extract (WRE) standardized to WA. B) Structures and names of select withanolides (Withaferin A; WA, Withanolide A; WLA, Withanone; WN) present in Withania somnifera plant parts. C) Nqo1 transcript induction in WT and Nrf2-disrupted MEF by 1 μM WA, 1 μM WLA and 1 μM WN. Values are mean ± SEM (n=3). *p<0.05.
3.2. WA profoundly protects mice against APAP hepatotoxicity in an Nrf2-dependent manner
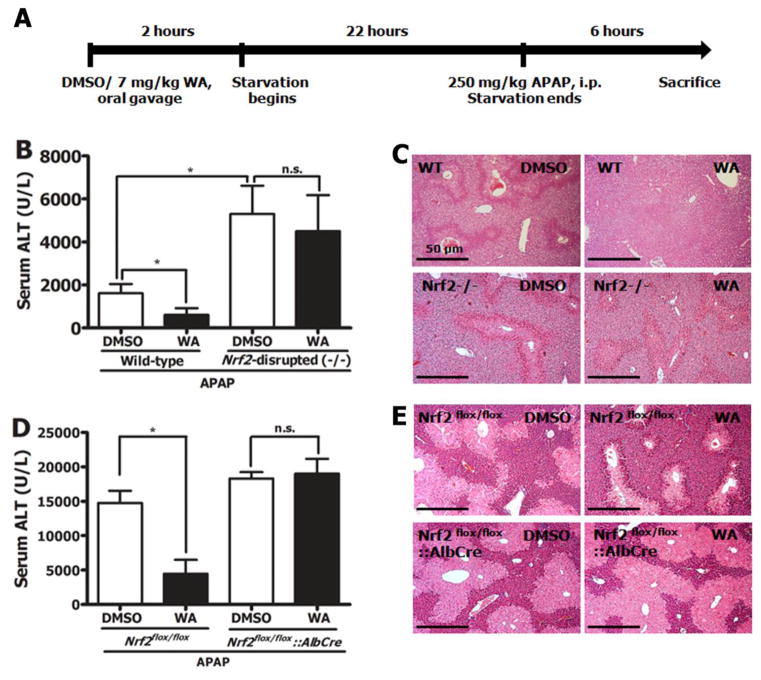
The in vivo study design is presented in Figure 2A. Pre-treating wild-type mice with 7 mg/kg WA (p.o.) substantially protected them against liver damage as observed with significantly lowered serum ALT levels (Figure 2B). WA treatment also protected against hepatocyte necrosis in the pericentral regions of the liver in these mice (Figure 2C). This protection was not observed in Nrf2-disrupted mice suggesting that the absence of Nrf2 abrogates a key mechanism by which WA provides hepatoprotection against APAP. To determine whether hepatocyte-specific Nrf2 disruption affects WA-mediated prevention of hepatotoxicity, Nrf2flox/flox and Nrf2flox/flox::AlbCre mice were subjected to the same dosing protocols. Nrf2flox/flox mice were protected, much like the wild-type mice but the Nrf2flox/flox::AlbCre mice were not (Figure 2D & 2E). WA treatment reduced the spike in serum ALT levels by 60% in wild-type mice and to a similar degree (70%) in Nrf2flox/flox mice. The seemingly higher sensitivity of DMSO treated Nrf2flox/flox mice (ALT level of ~ 15000 U/L) compared to DMSO treated wild-type mice (ALT level of ~2000 U/L) is likely related to inter-experimental variations in APAP solubility in warmed saline. Control experiments demonstrated that in wild-type, Nrf2-disrupted, Nrf2flox/flox and Nrf2flox/flox::AlbCre mice, WA alone did not affect serum ALT levels (data not shown). Overall, these data collectively suggested that WA provides profound protection that is directly mediated by Nrf2, largely in a hepatocyte-specific manner, against liver damage caused by APAP.
Figure 2.
A) Schematic of APAP dosing protocol. Mice were treated with either DMSO or 7 mg/kg WA (p.o.), followed by 22 hours of starvation before administration of 250 mg/kg APAP (i.p.) in saline. B) Serum ALT levels in WT (n=6) and Nrf2-disrupted (−/−) (n=15) mice sacrificed 6 hours after APAP treatment. Values are mean ± SEM. C) Representative H&E staining of livers of mice in B. D) Serum ALT levels in Nrf2flox/flox and Nrf2flox/flox::AlbCre mice. All values are mean ± SEM (n=5–10). E) Representative H&E staining of livers of mice in D. Statistical significance (*) of all ALT values was determined by p<0.05 (one-way Anova and Tukey’s multiple comparison test) * compared to DMSO control; n.s. not significant compared to DMSO control. Histology images are 10X magnification and are representative of all animals within each group. Scale bar = 50 μm.
3.3. Induction of cytoprotective enzymes by WA is Nrf2-dependent
To further investigate the involvement of Nrf2 in WA-mediated cytoprotection, induction of transcripts of enzymes important in APAP metabolism that are largely regulated by Nrf2 (Nqo1, Gclc, Sult1a1, Gstp1, Ugt1a1, Gsta1, Gsta4, Gsr1) as well as transcripts of Nrf2 in the mouse liver post-WA treatment were evaluated by RT-PCR (Table 1). Data revealed that under basal conditions, wild-type and Nrf2flox/flox mice showed comparable Nrf2 target gene expression with the exception of Gsta4 which showed a 2.3-fold induction in Nrf2flox/flox mice. In comparison, Nrf2flox/flox::AlbCre and Nrf2-disrupted mice showed low basal expression of the tested transcripts indicating that Nrf2 plays an indispensable role in regulating the basal expression of these genes. Wild-type and Nrf2flox/flox mice exhibited enhanced mRNA expression of Nqo1, Gclc, Gstp1, Ugt1a1, Gsta1, Gsr1 and Nrf2 with WA treatment. In comparison, the Nrf2-disrupted mice did not show significant induction of these cytoprotective enzymes after WA treatment indicating that WA-mediated upregulation of these genes is Nrf2-dependent. Interestingly, Nrf2flox/flox::AlbCre mice showed statistically significant induction of Nqo1, Gclc, Sult1a1, Gstp1, Ugt1a1, Gsta4 and Gsr1 after WA treatment, likely due to the induction of Nrf2 through liver cells other than hepatocytes such as Kupffer cells, sinusoidal endothelial cells and stellate cells. However, the sensitivity of Nrf2flox/flox::AlbCre to APAP treatment and the absence of hepatoprotection by WA against APAP in these mice suggests that upregulation of select target genes in non-parenchymal cells by WA is insufficient to protect these animals against hepatotoxicity. Moreover, it was observed that WA treatment resulted in significant induction of Nqo1 and Gclc in the mouse brain, small intestinal epithelium, lung and whole colon in addition to the liver in wild-type mice (supplementary data, Table S2) suggesting that WA can potentially be utilized to prevent and/or treat conditions that affect a large spectrum of organs.
Table 1.
Relative mRNA expression of representative Nrf2 target genes in WT, Nrf2flox/flox, Nrf2flox/flox::AlbCre and systemic Nrf2-disrupted mouse livers under basal (DMSO) and induced (7 mg/kg WA) conditions.
| Gene | Wild-type | Nrf2flox/flox | Nrf2flox/flox::AlbCre | Nrf2-disrupted | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| DMSO | WA | DMSO | WA | DMSO | WA | DMSO | WA | |
| Nqo1 | 1.0±0.2 | 2.9±0.2* | 1.6±0.6 | 3.1±0.3*,a | 0.7±0.1 | 1.4±0.1b | 0.1±0.02 | 0.2±0.02 |
| Gclc | 1.0±0.1 | 3.4±0.9* | 1.3±0.3 | 7.7±2.1*,a | 0.9±0.3 | 3.1±0.3*,b | 1.7±0.4 | 0.6±0.2c |
| Sult1a1 | 1.0±0.2 | 2.4±0.8 | 0.9±0.2 | 2.7±0.8 | 0.8±0.3 | 3.5±0.7*,b | 2.2±0.5 | 2.5±1.3 |
| Gstp1 | 1.0±0.1 | 2.8±0.5* | 0.9±0.2 | 4.9±1.2*,a | 0.4±0.1 | 3.1±0.2*,b | 1.7±0.5 | 1.3±0.6 |
| Ugt1a1 | 1.0±0.3 | 2.2±0.3* | 0.8±0.2 | 2.0±0.5a | 0.7±0.2 | 1.5±0.3b | 0.9±0.2 | 1.3±0.7 |
| Gsta1 | 1.0±0.2 | 3.2±1.2* | 0.8±0.2 | 7.3±2.1*,a | 0.2±0.1 | 0.8±0.2 | 0.8±0.2 | 0.6±0.2 |
| Gsta4 | 1.0±0.1 | 3.8±1.4* | 2.3±0.7* | 5.4±1.4* | 0.6±0.1 | 1.9±0.3b | 1.6±0.3 | 0.8±0.2 |
| Gsr1 | 1.0±0.1 | 3.4±0.9* | 0.8±0.1 | 4.9±1.6*,a | 0.9±0.3 | 4.7±0.7*,b | 1.1±0.5 | 0.9±0.5 |
| Nrf2 | 1.0±0.3 | 4.8±2.8* | 1.4±0.7 | 5.2±1.7*,a | 0.1±0.02 | 0.5±0.1 | ND | ND |
Values are mean ± SEM (n=3). p<0.05.
compared to WT DMSO,
compared to Nrf2flox/flox DMSO group,
compared to Nrf2flox/flox::AlbCre DMSO group,
compared to Nrf2-disrupted DMSO group.
ND: Not detected.
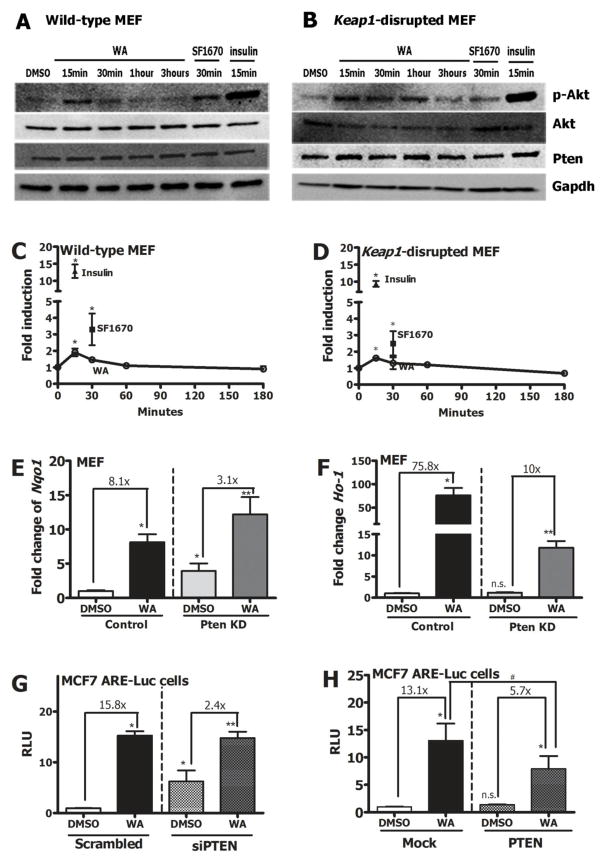
3.4. WA induces Nrf2 signaling in vivo and in vitro
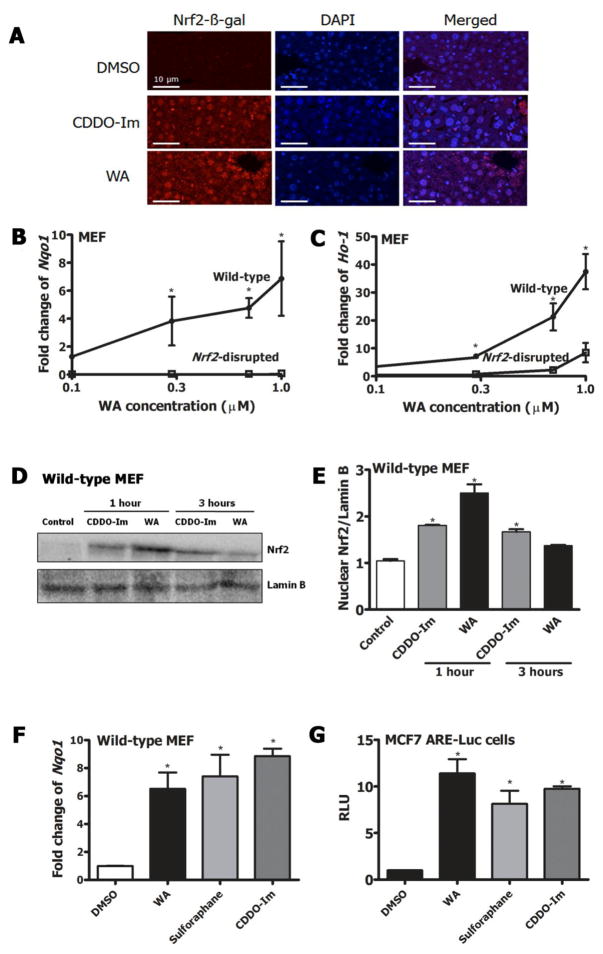
Nrf2 reporter mice gavaged with 7 mg/kg WA showed nuclear accumulation of Nrf2 in the liver after 3 hours as assessed by Nrf2-β-gal staining (Figure 3A). DMSO and CDDO-Im were used as negative and positive controls, respectively. Treatment of wild-type MEF with graded doses of WA for 20 hours showed a dose-dependent induction of Nqo1 and Ho-1 transcripts (Figure 3B & 3C). WA induced Nqo1 transcripts in wild-type MEF with a CD value of 200 nM indicative of relatively high potency. This induction was not observed in Nrf2-disrupted MEF suggesting that WA enhanced expression of cytoprotective enzymes in an Nrf2-dependent manner. Ho-1 induction by 1 μM WA in wild-type MEF was >40-fold compared to DMSO control, where 8-fold induction was observed in Nrf2-disrupted MEF as well. Nuclear translocation of Nrf2 was triggered by 1 μM WA compared to DMSO (Figure 3D & 3E) as measured by blotting for Nrf2 expression in the nuclear fraction of wild-type MEF. The positive control, CDDO-Im showed a more persistent nuclear accumulation, similar to what has been reported previously [34].
Figure 3.
A) Immunofluorescent analyses of liver sections of Nrf2-reporter mice orally treated with either DMSO, 30 μmol/kg CDDO-Im or 7 mg/kg WA for 3 hours by anti-β-galactosidase antibody. Nrf2-LacZ fusion protein expression and nuclei were stained by Alexa Fluor 546 and DAPI, respectively. Scale bars=10 μm. Transcript induction of B) Nqo1 C) Ho-1 in WT and Nrf2-disrupted MEF treated with graded doses of WA (0–1 μM) for 20 hours. D) Immunoblot of Nrf2 protein in the nuclear fraction of WT MEF treated with pharmacologic agents; DMSO, 25 nM CDDO-Im after 1 hour, 1 μM WA after 1 hour, 25 nM CDDO-Im after 3 hours, 1 μM WA after 3 hours. Lamin B was used as the loading control. E) Quantification of densitometry of 3 replicate western blots representing C. F) Nqo1 transcript induction by 1 μM WA, 25 nM CDDO-Im or 10 μM sulforaphane in WT MEF 20 hours post treatment. G) Relative luciferase activity in MCF7 ARE-luciferase reporter cells treated with 1 μM WA, 25 nM CDDO-Im or 10 μM sulforaphane for 20 hours. Gapdh was used as the normalization control for RT-PCR. All values are mean ± SEM (n=3). *p<0.05.
Complete characterization of an agent includes the comparison of its activity to other known agents that elicit a similar biological response. Thus, to further compare the Nrf2 induction capacity of WA to well-characterized Nrf2 inducers, wild-type MEF and MCF7 ARE-luciferase reporter cells were treated with 1 μM WA, 25 nM CDDO-Im and 7 μM sulforaphane for 20 hours. These optimal concentrations of the agents were determined based on previous publications [35]. Comparable induction levels of Nqo1 transcripts were observed with all 3 inducers; wild-type MEF; WA (6.5-fold), sulforaphane (7.4-fold), CDDO-Im (8.4-fold) (Figure 3F). A similar trend was observed in luciferase activity for all 3 inducers in MCF7 ARE Luciferase reporter cells; WA (11.4-fold), sulforaphane (8.1-fold), CDDO-Im (9.7-fold) (Figure 3G). Overall, these results indicate that WA is an inducer of the Nrf2-mediated stress response pathway in vitro and in vivo and that its efficacy is comparable to the highly characterized Nrf2 inducers, sulforaphane and CDDO-Im.
3.5. WA induces Nrf2 signaling in a Keap1-independent, Pi3k-dependent manner
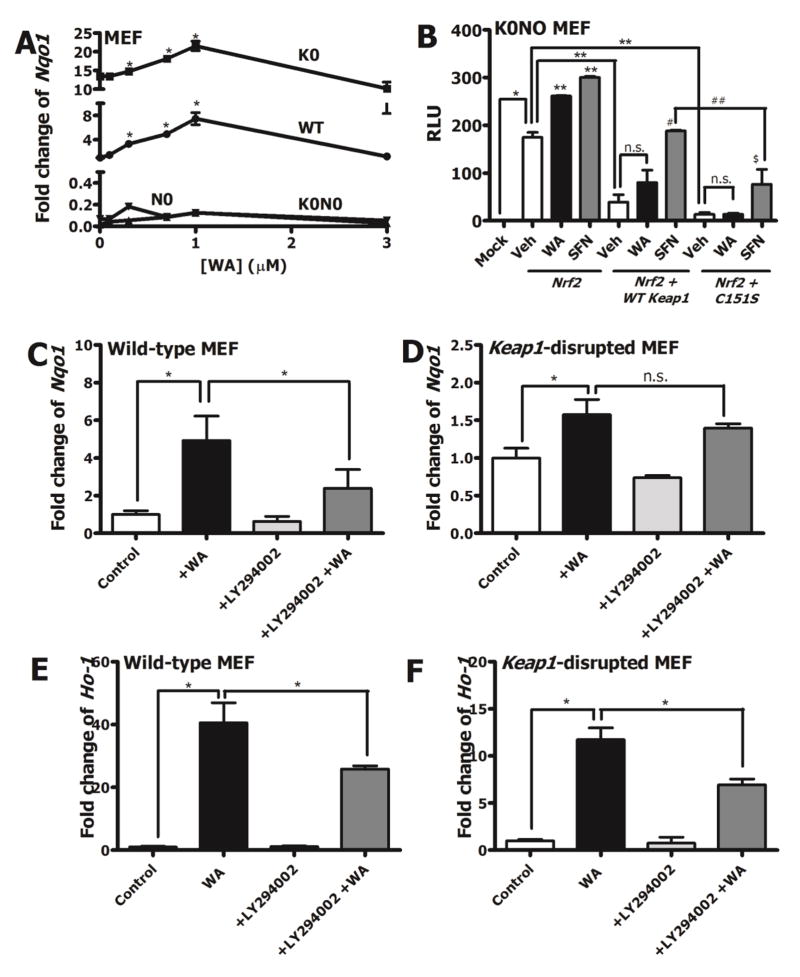
Given that multiple pharmacologic agents induce Nrf2 signaling via directly targeting reactive cysteines of Keap1, it was important to evaluate whether WA-mediated Nrf2 induction was Keap1-dependent or independent. Induction of Nqo1 transcripts by WA was compared in wild-type MEF, Nrf2-disrupted MEF, Keap1-disrupted MEF and Keap1 & Nrf2 double-disrupted MEF. Induction of Nqo1 transcripts was observed in both wild-type MEF and Keap1-disrupted MEF (Figure 4A). In Keap1-disrupted MEF, basal Nqo1 transcripts were higher than in wild-type, similar to what has been observed by others in Keap1flox/flox::AlbCre mice [36] and Keap1-disrupted MEF [37]. Consequently, WA (1 μM) induced Nqo1 mRNA expression by approximately 1.5-fold in Keap1-disrupted MEF (14-fold to 22-fold relative to wild-type) as compared to 6.5-fold in wild-type MEF (compared to DMSO). Corroborating the notions that Nqo1 induction by WA was Nrf2-dependent but Keap1-independent, induction was not observed in either Nrf2-disrupted MEF or in Keap1 & Nrf2 double-disrupted MEF. To determine how prototypical Nrf2 inducers such as sulforaphane compare to WA in a reconstituted system, transfection studies were conducted in Keap1 & Nrf2 double-disrupted MEF. Transfecting 5 ng of Nrf2 expression vector alone elevated NQO1-ARE-Luciferase activity in the absence of Keap1 (Figure 4B). Interestingly, both WA (1.5-fold) and sulforaphane (1.8-fold) resulted in induction of NQO1-luciferase activity in the absence of Keap1, suggesting that under Keap1-disrupted conditions, Keap1-independent pathways of Nrf2 regulation play important roles. Induction imparted by Nrf2 expression vector transfection was reduced by 80% when 2.5 ng Keap1 expression vector was introduced, suggesting the prominent role for Keap1 in inhibiting Nrf2 signaling in this reconstitution system. In the presence of Keap1, sulforaphane was able to reverse the suppressive effect of Keap1 as seen by elevated luciferase activity compared to the vehicle control. However, the induction by sulforaphane was largely lost when mutant Keap1 (C151S) expression vector was transfected, in accordance with the well-established role of C151 as the primary cysteine sensor for sulforaphane [38] [39] [40]. Conversely, WA did not abrogate the repression by Keap1. These data further suggested that WA, unlike sulforaphane, was working independent of Keap1 to induce ARE activity. Given the Keap1-independent induction mechanism of Nrf2 by WA, other pathways of Nrf2 regulation were considered in order to understand the mode of action of WA. The involvement of Pi3k signaling in Nrf2 induction by small molecules has been evaluated by others [41] [42]. Therefore, we tested WA against a pharmacologic inhibition model of Pi3k signaling with LY294002. In wild-type MEF, pre-treatment with 25 μM LY294002 for 1 hour prior to co-treatment of 25 μM LY294002 and 1 μM WA resulted in approximately 50% lower induction of Nqo1 transcripts compared to pre-treatment with DMSO control and co-treatment with DMSO control and WA (Figure 4C). In Keap1-disrupted MEF, a similar trend was observed, although not statistically significant (Figure 4D). This is likely due to the already high levels of Nqo1 present at basal state in these cells. Fold change of Ho-1 transcripts in both wild-type and Keap1-disrupted MEF were about 50% lower with pre-treatment of LY294002 (Figure 4E & F). No significant induction or inhibition of Nqo1 or Ho-1 resulted from the treatment of LY294002 alone in either cell type. Pharmacological inhibition of Pi3k with LY294002 in both wild-type and Keap1-disrupted MEF resulted in dampening of p-Akt expression indicating that LY294002 was in fact targeting the Pi3k-Akt pathway at the tested concentration (supplementary data, Figure S2). Further bolstering the notion that WA modulates the Pi3k/Akt/Gsk3 axis to enhance Nrf2 signaling, immunoblotting data revealed that phospho-GSK3β expression increased after 15 minutes of WA treatment in WT MEF (supplementary data, Figure S3). Collectively, these results implied that WA modulated Pi3k signaling in order to induce the Nrf2 response. However, the fact that LY294002 treatment did not completely abrogate Nrf2 target gene induction by WA argued for the possibility that Pi3k signaling was only partially responsible for regulating WA-mediated Nrf2 induction.
Figure 4.
Mechanisms of Nrf2 induction by WA. A) Nqo1 transcript induction in Keap1-disrupted (K0), WT, Nrf2-disrupted (N0) and Keap1 & Nrf2 double-disrupted (K0N0) MEF with graded doses of WA (0–3 μM), 20 hours post-treatment. B) Relative luciferase activity in Keap1 & Nrf2 double-disrupted MEF transfected with Nqo1-ARE luciferase reporter vector and either 5 ng pCMV Nrf2 alone or with 5 ng pCMV Nrf2 and 2.5 ng pCMV WT Keap1/pCMV C151S Keap1. The reconstituted cells were then treated with DMSO (Veh), 3 μM WA or 10 μM sulforaphane (SFN) for 20 hours. Transfection efficiencies were quantified by normalization to Renilla luciferase plasmid vector. Statistical comparisons: [*compared to mock control. **compared to + Nrf2 + Veh control. #compared to + Nrf2 + WT Keap1 + Veh control. ##compared to + Nrf2 + WT Keap1 + SFN. $compared to + Nrf2 + C151S Keap1 + Veh control]. Effects of LY294002 pre-treatment on WA-mediated Nqo1 induction in C) WT and D) Keap1-disrupted MEF. Effects of LY294002 pre-treatment on WA-mediated Ho-1 induction in E) WT and F) Keap1-disrupted MEF. Gapdh was used as the normalization control for RT-PCR. All values are mean ± SEM (n=3). *p<0.05.
3.6. Pten plays an important role in WA-mediated Nrf2 signaling induction
WA treatment enhanced p-Akt expression (by about 2-fold) 15 minutes post-treatment suggesting that activation of Akt, presumably via inactivation of Pten was an early event of the signaling cascade in both wild-type and Keap1-disrupted MEF (Figure 5A, B, C & D). SF1670 (pharmacologic inhibitor of Pten) and insulin were used as positive controls for p-Akt expression. Protein expression of Akt and Pten were observed to be unchanged by WA treatment. Interestingly, Pten transcripts were observed to be modestly higher (~1.5-fold) in Keap1-disrupted MEF compared to WT MEF, likely due to compensatory effects of alternative Nrf2 regulators (such as Pten) under conditions of Keap1 absence (supplementary data, Figure S4A). Given the important regulatory role of Pten in the Pi3k-Akt signaling cascade, the effects of Pten silencing and overexpression on WA-mediated Nrf2 induction were explored. In MEFs that were stably transfected with a shRNA construct against Pten (PTEN KD), the inducibility of Nqo1 and Ho-1 transcripts by WA was dampened compared to MEFs transfected with mock vector (Figure 5E & F). Nqo1 transcripts were induced by 8.1-fold by WA in control MEF while in Pten KD cells, Nqo1 transcripts were only induced by 3.1-fold by WA. Ho-1 transcripts were induced by 75.8-fold by WA in control MEF while in Pten KD cells, Ho-1 transcripts were only induced by 10-fold by WA. Interestingly, Pten suppression alone resulted in induction of Nqo1 transcripts but not Ho-1 transcripts. Approximately 45% lower expression of Pten was detected in shPten MEF compared to control MEFs (supplementary data, Figure S4B). A similar trend was observed in MCF7 ARE-Luc reporter cells transiently transfected with siPTEN, where the fold induction by WA under conditions of PTEN silencing (2.4-fold) was much lower compared to control (15.8-fold) (Figure 5G). PTEN transcripts were significantly lower in siPTEN transfected cells (supplementary data, Figure S4C). To determine the effect of PTEN overexpression on Nrf2 activation by WA, reporter cells were transfected with a wild-type PTEN expression plasmid prior to WA treatment. Treatment of PTEN overexpressing cells with WA resulted in the loss of induction of the ARE-luciferase reporter by WA (Figure 5H) suggesting that PTEN status of the cell is an important determinant of the Nrf2 inducer function of WA. Pten expression was enhanced by transfection of PTEN expression vector as expected (supplementary data, Figure S4D).
Figure 5.
Protein expression of p-Akt, Akt and Pten after 1 μM WA treatment of A) WT and B) Keap1-disrupted MEF. Quantification of p-Akt expression during WA time course in C) WT and D) Keap1-disrupted MEF. SF1670 (500 nM) and insulin (1 μM) were used as positive controls. Transcript induction of E) Nqo1 F) Ho-1 in MEFs stably transfected either control vector or shRNA against Pten (Pten KD) after a 20 hour 1 μM WA treatment. [*compared to control MEF treated with DMSO, **compared to Pten KD cells treated with DMSO, n.s.compared to control MEF treated with DMSO]. Fold change compared to DMSO control within each group is denoted as a multiple. G) Relative luciferase activity in MCF7 ARE-Luc cells treated with WA after transient transfection of either scrambled or siRNA against PTEN. H) Relative luciferase activity induction by WA in MCF7 reporter cells after cells were transfected with either mock vector or PTEN overexpression plasmid vector. [*compared to scrambled siRNA or mock vector transfected cells treated with DMSO, **compared to siPTEN or PTEN expression vector transfected cells treated with DMSO, #compared to mock vector transfected cells treated with WA, n.s.compared to mock vector transfected cells treated with DMSO]. Fold change compared to DMSO control within each group is denoted as a multiple. All values are mean ± SEM (n=3). *p<0.05.
4. Discussion
Given the important place Withania somnifera holds in traditional South Asian medicine as an herb that can be used safely and effectively for various ailments, studies conducted on its constituents, particularly withanolides such as WA can provide insight into its mode of action and potentially broaden its use in preventive medicine. The safety of WS plant extracts have been established in pre-clinical models and has been reviewed elsewhere [1] [43]. Cysteine reactivity of WA has been identified in the past via direct binding events with multiple key proteins [44] [45] [46]. Furthermore, the α,β-unsaturated carbonyl group of WA has been described to be thiol-reactive [47] suggestive of the possible structural moiety responsible for the interaction between WA and reactive cysteines of target proteins. A large array of biological pathways have been shown to be altered by WA including autophagy [48] [49], inflammation [50] [51] as well as the heat shock response [52] [47] [53]. While peak plasma concentration of WA after dietary exposure has not been assessed, Thaiparambil et al. showed that peak plasma concentration of WA following 4 mg/kg (i.p) was 2 μM [54]. In addition to the well-established therapeutic and preventive role of WA in cancer, it has also been shown to have therapeutic effects against hepatotoxicity [55] [56], diabetes [57] and cardiac fibrosis [58]. Results from our hepatotoxicity experiments are in line with the protective and therapeutic phenotypes exhibited by WA in other models. Importantly, it provides direct and detailed evidence, by use of knockout mouse models, for the role of Nrf2 signaling in the hepatoprotection elicited by WA.
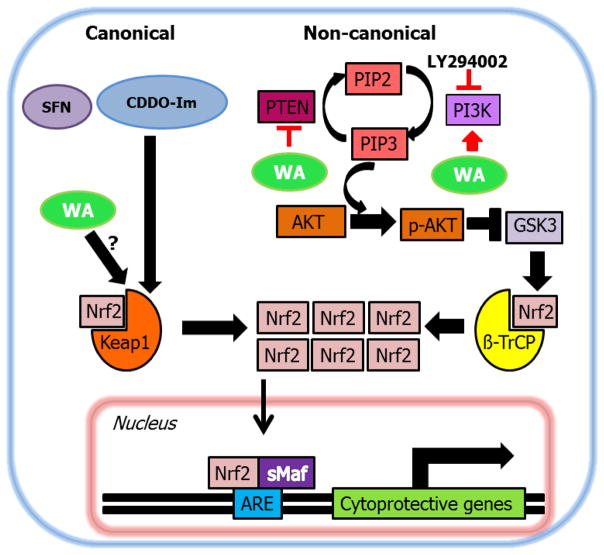
Non-canonical mechanisms of Nrf2 activation have been suggested for multiple electrophilic molecules including nordihydroguaiaretic acid [22] (Keap1-independent); nitro-fatty acids [59] and arsenic [23] (Keap1 C151-independent). In a recent report, Saito et al. categorized several pharmacologic agents according to their dependence on modification of C151, C273 and C288 amino acids of Keap1 [60]. Interestingly, some compounds were found to be independent of all 3 of the noted cysteines suggesting that either other cysteines of Keap1 may be involved or they work independent of Keap1, much like WA, to induce Nrf2 signaling. Alternative pathways have been shown to be modulated by several compounds that ultimately activate Nrf2 signaling [42] [61]. Pleiotropic actions of well-known Nrf2 inducers have also been reported. While Keap1 is thought to be the major target of sulforaphane, it has also been shown to modulate Pi3k/Akt and Mek/Erk [62], Egfr [63] as well as p38 Mapk [64]. An effective anticarcinogen and well-established inducer of Nrf2 signaling, 1,2-dithione-3-thione, has been shown to also target Erk1/2 [65]. CDDO-Im has been shown to target Pten at higher concentrations than required for the activation of the Nrf2 pathway [66]. Whether these signaling cascades are modulated independent of Keap1 or whether they ultimately feed into the Keap1-Nrf2 signaling pathway was not assessed in the studies. It is also important to decipher whether the dosage of the pharmacologic agent defines its actions wherein low concentrations of some agents typically target Keap1 while at higher concentrations the targets become more promiscuous. However, our data suggests that at relatively low concentrations, Keap1 independent mechanisms play an important role in WA-mediated Nrf2 signaling induction unlike other Nrf2 inducers such as sulforaphane.
Throughout our studies, we attempted to determine the specific role of Keap1-dependent and independent pathways of Nrf2 activation by WA. A recent study by Heyninck et al. suggested that WA interacts with Keap1 in endothelial cells [67]. While a direct biochemical interaction is possible, our results suggest that this WA-Keap1 interaction is not a determinant of Nrf2 induction in a functional context. According to our data, treating Nrf2 and Keap1 reconstituted Keap1 & Nrf2 double-disrupted MEFs with sulforaphane resulted in an enhanced or “rescued” ARE response which was not observed with WA treatment, suggesting that WA-mediated ARE induction was not dependent on Keap1 status of the cell. In addition, WA treatment resulted in significant induction of Nrf2 target genes suggesting that under Keap1-disrupted conditions, WA still retained its capacity to increase Nrf2 signaling. Furthermore, it is certainly possible that inducers of Nrf2 act through preferential pathways depending on the cell type. It is likely that endothelial cells, given their intrinsic properties of pro-angiogenic character and nitric oxide production, may respond differentially to pharmacologic inducers of Nrf2. Such questions warrant further investigations.
Pten is an essential molecule with phosphatase activity that blocks Pi3k signaling by inhibiting Pip3-dependent processes such as Akt activation and thereby inhibiting cell survival, growth and proliferation. From the standpoint of cancer and tumorigenesis, Pten silencing plays a detrimental role as seen with prenatal death of Pten-knockout mice and the observation that more than 2700 PTEN mutations have been seen in 28 types of human tumors [68]. However, under normal conditions, Pten seems to be playing an important role in regulating cellular homeostasis by acting as a redox sensing switch that either activates or deactivates Pi3k signaling. This finding directly supports the notion that short-term suppression of Pten could potentially be valuable in certain conditions, such as nerve injury [69]. Oxidants such as H2O2 inactivate Pten to in turn activate downstream signaling cascades driven by Pi3k [70]. The mechanism is likely related to H2O2 reversibly inactivating Pten via the formation of disulfide bonds between Cys71 and Cys124 [71]. Furthermore, overexpression of antioxidant enzymes such as SOD and catalase have been associated with production of H2O2 and activated Akt signaling mediated by oxidized Pten [72]. The redox-sensitive interplay between Pten and Pi3k signaling is also involved in the pathogenesis of conditions such as hypertension [73]. Age-dependent down regulation of Pi3k signaling is associated with increased susceptibility to oxidative stress [74]. Collectively, data by others supports the concept that Pten plays an important role as a stress sensory molecule allowing for the activation of pro-survival pathways under conditions of acute stress or injury. Given the indispensible function of Nrf2 as a pro-survival factor, it is valuable to understand the cross-talk between Pten and Nrf2 and whether electrophilic tuning of Pten can be utilized to modulate downstream Nrf2 signaling to enhance cellular survival potential under conditions of acute stress (Figure 6). Of course, longer term effects of Pten inhibition may render a survival disadvantage given the tumor suppressor activity of the molecule. Thus, studies need to be conducted to determine the effects of chronic administration of pharmacologic agents such as WA. Furthermore, these are studies that have to be conducted for all Nrf2 inducers that move forward into clinical trials, regardless of their mechanisms of action, especially given the recent reports of Nrf2’s dual role in cancer [75] [76] [77].
Figure 6.
Proposed scheme for Nrf2 induction by WA. While prototypical target genes are largely thought to be working via Keap1-dependent pathways, it is plausible that other mechanisms can be manipulated by select compounds such as WA to induce Nrf2-mediated cytoprotective responses.
The important role of Pten in regulation of Nrf2 signaling has recently received attention. An approach to determining the role played by negative regulators of Nrf2 (such as Pten and even Keap1) is to determine if the silencing or inhibition of these regulators lead to intrinsic activation of the Nrf2 signaling pathway. Pten inhibitor bpV(HOpic) at 3 μM was shown to significantly induce Nrf2 target genes in both wild-type and Keap1-disrupted MEFs [21]. In the same report, it was shown that overexpression of Pten reduced Nrf2 induction by tert-butylhydroquinone, suggesting that pharmacologic agents may rely on Pten to activate the Nrf2 pathway. Our data suggests that silencing of Pten selectively induces only some cytoprotective enzymes. In shPten MEF cells, Nqo1 transcripts were induced by 3.9-fold where as Ho-1 did not show any statistically significant induction (1.2-fold) compared to control MEF (Figure 5E & F). This is likely linked to biological relationships between these enzymes and Pten activity. Ho-1 expression has been shown to be negatively correlated with Pten activity in a certain subset of human prostate cancer cases [78]. Furthermore, Ho-1 has been shown to be regulated by multiple transcription factors including NFkB and AP-2 [79]. To determine the exact role of Nrf2 in this phenomenon, a larger array of target genes need to be analyzed under conditions of disrupted Pten signaling. Another mechanism by which PTEN modulates NRF2 and the ARE response was described by Sakamoto et al. [80]. Here, the authors showed that tert-butylhydroquinone induced FERRITIN H and NQO1 expression in PTEN silenced K562 cells compared to control cells, a response that was associated with histone modifications. Whether WA exerts a similar effect on the ARE is currently unknown.
As a molecule of plant origin that has strong roots in traditional South Asian medicine, WA – and perhaps WRE itself - has strong potential to move forward into human disease prevention clinical trials. Our in vivo data directly suggests that Nrf2 plays an indispensible role in WA-mediated cytoprotection and hepatoprotection. Such action has been implied by others [56] but to our knowledge, this is the first report to clearly document the involvement of Nrf2 in WA-mediated cytoprotection using knockout mouse models coupled with characterization of the underlying mechanism of action. The recognition that WA induces Nrf2 signaling via a Keap1-independent, Pten/Pi3k/Akt-dependent mechanism indicates that additional signal sensors may serve as useful clinical targets for Nrf2-directed disease prevention and therapy.
Supplementary Material
Highlights.
Withaferin A is an inducer of Nrf2 signaling both in vitro and in vivo
Withaferin A protects mice against acetaminophen hepatotoxicity in an Nrf2-dependent manner
The mechanism of Nrf2 induction by WA is at least in part Keap1-independent and is mediated by the Pten/Pi3k/Akt signaling axis
Acknowledgments
The authors would like to thank Dr. Shyam Biswal (Johns Hopkins University, Baltimore, MD) for providing the Nrf2flox/flox mice, Dr. Adam Marcus (Emory University, Atlanta, GA) for sharing the WS root extract, Dr. Carola Neumann (University of Pittsburgh, Pittsburgh, PA) for sharing shPten MEF cells and Dr. Miho Iijima (Johns Hopkins University, Baltimore, MD) for graciously providing us with PTEN expression constructs. This work was supported by NIH grant R35 CA197222 (TWK), R01CA142604 (SVS) and P30CA047904.
Abbreviations
- Akt (PKB)
protein kinase B
- APAP
acetaminophen
- ALT
alanine aminotransferase
- ARE
antioxidant response element
- β-Trcp
beta-transducin repeat containing protein
- CDDO-Im
1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
- Cul3
cullin-3
- DMSO
dimethyl sulfoxide
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- Gclc
glutamate-cysteine ligase catalytic subunit
- Gst
glutathione s transferase
- Gsr1
glutathione reductase 1
- Ho-1
heme oxygenase 1
- Hrd1
HMG-CoA Reductase Degradation 1
- H2O2
hydrogen peroxide
- Keap1
Kelch-like ECH-associated protein 1
- MEF
mouse embryonic fibroblast
- Nrf2
nuclear factor erythroid 2 [NF-E2]-related factor 2
- Nqo1
NAD(P)H quinone oxidoreductase 1
- Pi3k
phosphatidylinositol-4,5-bisphosphate 3-kinase
- Pten
phosphatase and tensin homolog
- SFN
sulforaphane
- Sult1a1
sulfotransferase family 1A member 1
- Ugt1a1
UDP glucuronosyltransferase family 1 member A1
- WA
Withaferin A
- WLA
Withanolide A
- WN
Withanone
- WRE
Withania somnifera root extract
- WS
Withania somnifera
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palliyaguru DL, Singh SV, Kensler TW. Withania somnifera: from prevention to treatment of cancer. Mol Nutr Food Res. 2016;60(6):1342–53. doi: 10.1002/mnfr.201500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirjalili MH, Moyano E, Bonfill M, Cusido RM, Palazon J. Steroidal lactones from Withania somnifera, an ancient plant for novel medicine. Molecules. 2009;14:2373–93. doi: 10.3390/molecules14072373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahm ER, Lee J, Kim SH, Sehrawat A, Arlotti JA, Shiva SS, et al. Metabolic alterations in mammary cancer prevention by withaferin A in a clinically relevant mouse model. J Natl Cancer Inst. 2013;105:1111–22. doi: 10.1093/jnci/djt153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manoharan S, Panjamurthy K, Menon VP, Balakrishnan S, Alias LM. Protective effect of Withaferin-A on tumour formation in 7,12-dimethylbenz[a]anthracene induced oral carcinogenesis in hamsters. Indian J Exp Biol. 2009;47:16–23. [PubMed] [Google Scholar]
- 5.Li W, Zhang C, Du H, Huang V, Sun B, Harris JP, et al. Withaferin A suppresses the up-regulation of acetyl-coA carboxylase 1 and skin tumor formation in a skin carcinogenesis mouse model. Mol Carcinog. 2015 doi: 10.1002/mc.22423. [DOI] [PubMed] [Google Scholar]
- 6.Stan SD, Hahm ER, Warin R, Singh SV. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 2008;68:7661–9. doi: 10.1158/0008-5472.CAN-08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munagala R, Kausar H, Munjal C, Gupta RC. Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis. 2011;32:1697–705. doi: 10.1093/carcin/bgr192. [DOI] [PubMed] [Google Scholar]
- 8.Nagalingam A, Kuppusamy P, Singh SV, Sharma D, Saxena NK. Mechanistic elucidation of the antitumor properties of withaferin a in breast cancer. Cancer Res. 2014;74:2617–29. doi: 10.1158/0008-5472.CAN-13-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reisman SA, Buckley DB, Tanaka Y, Klaassen CD. CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol Appl Pharmacol. 2009;236:109–14. doi: 10.1016/j.taap.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 11.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, et al. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99:7610–5. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uruno A, Furusawa Y, Yagishita Y, Fukutomi T, Muramatsu H, Negishi T, et al. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol. 2013;33:2996–3010. doi: 10.1128/MCB.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keleku-Lukwete N, Suzuki M, Otsuki A, Tsuchida K, Katayama S, Hayashi M, et al. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proc Natl Acad Sci U S A. 2015;112:12169–74. doi: 10.1073/pnas.1509158112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106:2933–8. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–81. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–33. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–22. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, et al. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, Noda K, et al. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol. 2014;34:653–63. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taguchi K, Hirano I, Itoh T, Tanaka M, Miyajima A, Suzuki A, et al. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol. 2014;34:900–13. doi: 10.1128/MCB.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojo AI, Rada P, Mendiola M, Ortega-Molina A, Wojdyla K, Rogowska-Wrzesinska A, et al. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal. 2014;21:2498–514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojo AI, Medina-Campos ON, Rada P, Zuniga-Toala A, Lopez-Gazcon A, Espada S, et al. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: Role of glycogen synthase kinase-3. Free Radic Biol Med. 2012;52:473–87. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, et al. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol. 2013;33:2436–46. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 25.Kong X, Thimmulappa R, Craciun F, Harvey C, Singh A, Kombairaju P, et al. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am J Respir Crit Care Med. 2011;184:928–38. doi: 10.1164/rccm.201102-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–91. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 27.Yagishita Y, Fukutomi T, Sugawara A, Kawamura H, Takahashi T, Pi J, et al. Nrf2 protects pancreatic beta-cells from oxidative and nitrosative stress in diabetic model mice. Diabetes. 2014;63:605–18. doi: 10.2337/db13-0909. [DOI] [PubMed] [Google Scholar]
- 28.McClain CJ, Price S, Barve S, Devalarja R, Shedlofsky S. Acetaminophen hepatotoxicity: An update. Curr Gastroenterol Rep. 1999;1:42–9. doi: 10.1007/s11894-999-0086-3. [DOI] [PubMed] [Google Scholar]
- 29.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–5. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Garcia A, Xu S, Powell DR, Vertino PM, Singh S, et al. Withania somnifera root extract inhibits mammary cancer metastasis and epithelial to mesenchymal transition. PLoS One. 2013;8:e75069. doi: 10.1371/journal.pone.0075069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao J, Schulte J, Knight A, Leslie NR, Zagozdzon A, Bronson R, et al. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28:1505–17. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen HN, Yang JM, Afkari Y, Park BH, Sesaki H, Devreotes PN, et al. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc Natl Acad Sci U S A. 2014;111:E2684–E2693. doi: 10.1073/pnas.1409433111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thimmulappa RK, Fuchs RJ, Malhotra D, Scollick C, Traore K, Bream JH, et al. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid Redox Signal. 2007;9:1963–70. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–9. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, et al. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–31. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–34. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaya K, Suzuki T, Motohashi H, Onodera K, Satomi S, Kensler TW, et al. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med. 2012;53:817–27. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayes JD, Chowdhry S, Dinkova-Kostova AT, Sutherland C. Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of beta-TrCP and GSK-3. Biochem Soc Trans. 2015;43:611–20. doi: 10.1042/BST20150011. [DOI] [PubMed] [Google Scholar]
- 41.Li MH, Cha YN, Surh YJ. Peroxynitrite induces HO-1 expression via PI3K/Akt-dependent activation of NF-E2-related factor 2 in PC12 cells. Free Radic Biol Med. 2006;41:1079–91. doi: 10.1016/j.freeradbiomed.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, et al. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem. 2004;279:8919–29. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 43.Prabu PC, Panchapakesan S. Prenatal developmental toxicity evaluation of Withania somnifera root extract in Wistar rats. Drug Chem Toxicol. 2015;38:50–6. doi: 10.3109/01480545.2014.900073. [DOI] [PubMed] [Google Scholar]
- 44.Antony ML, Lee J, Hahm ER, Kim SH, Marcus AI, Kumari V, et al. Growth arrest by the antitumor steroidal lactone withaferin A in human breast cancer cells is associated with down-regulation and covalent binding at cysteine 303 of beta-tubulin. J Biol Chem. 2014;289:1852–65. doi: 10.1074/jbc.M113.496844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bargagna-Mohan P, Hamza A, Kim YE, Khuan Abby HY, Mor-Vaknin N, Wendschlag N, et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem Biol. 2007;14:623–34. doi: 10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heyninck K, Lahtela-Kakkonen M, Van d, Haegeman VG, Vanden Berghe W. Withaferin A inhibits NF-kappaB activation by targeting cysteine 179 in IKKbeta. Biochem Pharmacol. 2014;91:501–9. doi: 10.1016/j.bcp.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7:340–9. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahm ER, Singh SV. Autophagy fails to alter withaferin A-mediated lethality in human breast cancer cells. Curr Cancer Drug Targets. 2013;13:640–50. doi: 10.2174/15680096113139990039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rah B, Ur RR, Nayak D, Yousuf SK, Mukherjee D, Kumar LD, et al. PAWR-mediated suppression of BCL2 promotes switching of 3-azido withaferin A (3-AWA)-induced autophagy to apoptosis in prostate cancer cells. Autophagy. 2015;11:314–31. doi: 10.1080/15548627.2015.1017182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee W, Kim TH, Ku SK, Min KJ, Lee HS, Kwon TK, et al. Barrier protective effects of withaferin A in HMGB1-induced inflammatory responses in both cellular and animal models. Toxicol Appl Pharmacol. 2012;262:91–8. doi: 10.1016/j.taap.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 51.Kaileh M, Vanden Berghe W, Heyerick A, Horion J, Piette J, Libert C, et al. Withaferin a strongly elicits IkappaB kinase beta hyperphosphorylation concomitant with potent inhibition of its kinase activity. J Biol Chem. 2007;282:4253–64. doi: 10.1074/jbc.M606728200. [DOI] [PubMed] [Google Scholar]
- 52.Yu Y, Hamza A, Zhang T, Gu M, Zou P, Newman B, et al. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem Pharmacol. 2010;79:542–51. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wijeratne EM, Xu YM, Scherz-Shouval R, Marron MT, Rocha DD, Liu MX, et al. Structure-activity relationships for withanolides as inducers of the cellular heat-shock response. J Med Chem. 2014;57:2851–63. doi: 10.1021/jm401279n. [DOI] [PubMed] [Google Scholar]
- 54.Thaiparambil JT, Bender L, Ganesh T, Kline E, Patel P, Liu Y, et al. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int J Cancer. 2011;129:2744–55. doi: 10.1002/ijc.25938. [DOI] [PubMed] [Google Scholar]
- 55.Sudhir S, Budhiraja RD. Comparison of the protective effect of Withaferin-‘A’ and hydrocortisone against CCL4 induced hepatotoxicity in rats. Indian J Physiol Pharmacol. 1992;36:127–9. [PubMed] [Google Scholar]
- 56.Jadeja RN, Urrunaga NH, Dash S, Khurana S, Saxena NK. Withaferin-A Reduces Acetaminophen-Induced Liver Injury in Mice. Biochem Pharmacol. 2015;97:122–32. doi: 10.1016/j.bcp.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J, Liu J, Feng X, Salazar Hernandez MA, Mucka P, Ibi D, et al. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat Med. 2016;22:1023–32. doi: 10.1038/nm.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Challa AA, Vukmirovic M, Blackmon J, Stefanovic B. Withaferin-A reduces type I collagen expression in vitro and inhibits development of myocardial fibrosis in vivo. PLoS One. 2012;7:e42989. doi: 10.1371/journal.pone.0042989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kansanen E, Bonacci G, Schopfer FJ, Kuosmanen SM, Tong KI, Leinonen H, et al. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J Biol Chem. 2011;286:14019–27. doi: 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, Suda H, et al. Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol Cell Biol. 2015;36(2): 271–84. doi: 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Na HK, Kim EH, Jung JH, Lee HH, Hyun JW, Surh YJ. (−)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–7. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Jakubikova J, Sedlak J, Mithen R, Bao Y. Role of PI3K/Akt and MEK/ERK signaling pathways in sulforaphane- and erucin-induced phase II enzymes and MRP2 transcription, G2/M arrest and cell death in Caco-2 cells. Biochem Pharmacol. 2005;69:1543–52. doi: 10.1016/j.bcp.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 63.Abbaoui B, Riedl KM, Ralston RA, Thomas-Ahner JM, Schwartz SJ, Clinton SK, et al. Inhibition of bladder cancer by broccoli isothiocyanates sulforaphane and erucin: characterization, metabolism, and interconversion. Mol Nutr Food Res. 2012;56:1675–87. doi: 10.1002/mnfr.201200276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, et al. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–13. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 65.Manandhar S, Cho JM, Kim JA, Kensler TW, Kwak MK. Induction of Nrf2-regulated genes by 3H-1, 2-dithiole-3-thione through the ERK signaling pathway in murine keratinocytes. Eur J Pharmacol. 2007;577:17–27. doi: 10.1016/j.ejphar.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 66.Pitha-Rowe I, Liby K, Royce D, Sporn M. Synthetic triterpenoids attenuate cytotoxic retinal injury: cross-talk between Nrf2 and PI3K/AKT signaling through inhibition of the lipid phosphatase PTEN. Invest Ophthalmol Vis Sci. 2009;50:5339–47. doi: 10.1167/iovs.09-3648. [DOI] [PubMed] [Google Scholar]
- 67.Heyninck K, Sabbe L, Chirumamilla CS, Szarc Vel SK, Vander VP, Lemmens KJ, et al. Withaferin A induces heme oxygenase (HO-1) expression in endothelial cells via activation of the Keap1/Nrf2 pathway. Biochem Pharmacol. 2016;109:48–61. doi: 10.1016/j.bcp.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 68.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: the long and the short of it. Trends Biochem Sci. 2014;39:183–90. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spinelli L, Lindsay YE, Leslie NR. PTEN inhibitors: an evaluation of current compounds. Adv Biol Regul. 2015;57:102–11. doi: 10.1016/j.jbior.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 70.Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–10. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–42. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 72.Connor KM, Subbaram S, Regan KJ, Nelson KK, Mazurkiewicz JE, Bartholomew PJ, et al. Mitochondrial H2O2 regulates the angiogenic phenotype via PTEN oxidation. J Biol Chem. 2005;280:16916–24. doi: 10.1074/jbc.M410690200. [DOI] [PubMed] [Google Scholar]
- 73.Wu KL, Wu CA, Wu CW, Chan SH, Chang AY, Chan JY. Redox-sensitive oxidation and phosphorylation of PTEN contribute to enhanced activation of PI3K/Akt signaling in rostral ventrolateral medulla and neurogenic hypertension in spontaneously hypertensive rats. Antioxid Redox Signal. 2013;18:36–50. doi: 10.1089/ars.2011.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin Q, Jhun BS, Lee SH, Lee J, Pi Y, Cho YH, et al. Differential regulation of phosphatidylinositol 3-kinase/Akt, mitogen-activated protein kinase, and AMP-activated protein kinase pathways during menadione-induced oxidative stress in the kidney of young and old rats. Biochem Biophys Res Commun. 2004;315:555–61. doi: 10.1016/j.bbrc.2004.01.093. [DOI] [PubMed] [Google Scholar]
- 75.Satoh H, Moriguchi T, Saigusa D, Baird L, Yu L, Rokutan H, et al. NRF2 intensifies host defense systems to prevent lung carcinogenesis, but after tumor initiation accelerates malignant cell growth. Cancer Res. 2016;76(10):3088–96. doi: 10.1158/0008-5472.CAN-15-1584. [DOI] [PubMed] [Google Scholar]
- 76.Rolfs F, Huber M, Kuehne A, Kramer S, Haertel E, Muzumdar S, et al. Nrf2 Activation Promotes Keratinocyte Survival during Early Skin Carcinogenesis via Metabolic Alterations. Cancer Res. 2015;75:4817–29. doi: 10.1158/0008-5472.CAN-15-0614. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8:334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 78.Li Y, Su J, DingZhang X, Zhang J, Yoshimoto M, Liu S, et al. PTEN deletion and heme oxygenase-1 overexpression cooperate in prostate cancer progression and are associated with adverse clinical outcome. J Pathol. 2011;224:90–100. doi: 10.1002/path.2855. [DOI] [PubMed] [Google Scholar]
- 79.Lavrovsky Y, Schwartzman ML, Abraham NG. Novel regulatory sites of the human heme oxygenase-1 promoter region. Biochem Biophys Res Commun. 1993;196:336–41. doi: 10.1006/bbrc.1993.2253. [DOI] [PubMed] [Google Scholar]
- 80.Sakamoto K, Iwasaki K, Sugiyama H, Tsuji Y. Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol Biol Cell. 2009;20:1606–17. doi: 10.1091/mbc.E08-07-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.