Abstract
By sequencing the exomes of distantly related individuals in multiplex families, rare mutational and structural changes to coding DNA can be characterized and their relationship to disease risk can be assessed. Recently, several rare single nucleotide variants (SNVs) were associated with an increased risk of non-syndromic oral cleft, highlighting the importance of rare sequence variants in oral clefts and illustrating the strength of family-based study designs. However, the extent to which rare deletions in coding regions of the genome occur and contribute to risk of non-syndromic clefts is not well understood. To identify putative structural variants underlying risk, we developed a pipeline for rare hemizygous deletions in families from whole exome sequencing and statistical inference based on rare variant sharing. Among 56 multiplex families with 115 individuals, we identified 53 regions with one or more rare hemizygous deletions. We found 45 of the 53 regions contained rare deletions occurring in only one family member. Members of the same family shared a rare deletion in only 8 regions. We also devised a scalable global test for enrichment of shared rare deletions.
Keywords: copy number, oral clefts, multiplex families, rare variants, structural variants
Introduction
Appreciable genetic heterogeneity must be expected in complex diseases such as nonsyndromic oral clefts. One component of heterogeneity at the DNA level is single nucleotide variants (SNVs). SNVs that are private to affected individuals in a single multiplex family or appear in only a few multiplex families may be responsible for association signals detected with common variant analyses and have the potential to implicate new regions not previously linked to disease (Cirulli et al., 2010). In the context of non-syndromic oral clefts, we recently identified rare variants in the gene ADAMTS9, a gene encoding a member of the ADAMTS protein family and located in a region known to be lost in hereditary renal tumors, and CDH1, a known tumor suppressor whose down-regulation decreases cellular adhesion (Bureau, Parker, et al., 2014; Bureau, Younkin, et al., 2014). Structural changes to the DNA copy number that include deletions and amplifications of small sections of the genome can also influence risk to oral clefts, but these have not been systematically evaluated using whole exome sequencing (WES) data.
Copy number methodologies relevant to the study of rare germline deletions include CoNIFER, XHMM, and CLAMMS (Fromer et al., 2012; Krumm et al., 2012; Packer et al., 2016), but in general these methods are not tailored to rare deletions shared among family members. CoNIFER normalizes exon-level reads per kilobase per million (RPKM) by singular value decomposition. After removing the components from the standardized RPKM scores, these adjusted scores provide a relative measure of expression for copy number. XHMM follows an approach similar to CoNIFER where exon-level read coverage are normalized by principal components analysis. A hidden Markov model with states for copy number gain and loss is used to identify CNVs (Fromer et al., 2012). Unlike CoNIFER and XHMM, CLAMMS proproses a Lattice Aligned Mixture model for both rare and common CNVs and is scalable to thousands of samples (Packer et al., 2016).
Methodologies to evaluate the association between rare variants and disease are largely based on SNVs. In non-family based designs, rare variants are often grouped and statistical models for association are based on a linear combination of protective and risk alleles, possibly using a weighted score (Asimit & Zeggini, 2010; Coombes, Basu, Guha, & Schork, 2015; Kim, Lee, & Sun, 2015; Madsen & Browning, 2009; Wu et al., 2011). The idea of grouping rare variants has been extended to family-based studies (S. Feng et al., 2015), while others have proposed statistical tests for sib-pairs (Epstein et al., 2015; Lin & Zöllner, 2015). We recently proposed an exact test for the statistical significance of a single rare sequence variant shared by distant relatives in multiplex families (Bureau, Parker, et al., 2014). The probability from this exact test is referred to as a sharing probability. A critical assumption of our approach is that the variant is sufficiently rare so copies in the sequenced relatives are almost certainly identical by descent (IBD).
This paper delineates hemizygous deletions identified from WES in multiplex families of individuals with non-syndromic oral cleft. A combination of bioinformatic and model-based filters identify rare deletions, including several shared within families. We then extend analyses of shared rare SNVs to assess the statistical significance of shared rare deletions. In particular, we compute the probability that distant relatives share the same rare deletion under the a priori null hypothesis of no linkage or association (Bureau, Younkin, et al., 2014). We introduce potential sharing probabilities in the context of shared deletions as a means to control the false discovery rate. Last, we also devise a scalable global test for enrichment of rare deletion sharing.
Methods
Library preparation, exome sequence capture, and read alignment
Exome sequencing and genotyping was performed at the Center for Inherited Disease Research (CIDR). The Agilent SureSelect Human All Exon Target Enrichment system kit S0297201 was used for exon capture, yielding ≈ 51 Mb of targeted sequence capture per sample. For DNA sequencing, the Illumina HiSeq 2500 instrument was run using standard protocols for 100-bp paired-end reads. Six samples were run per flowcell, where 92% of exons received at least 8x coverage and the mean exon coverage was 84×. Illumina HiSeq reads were processed through Illumina’s Real-Time Analysis software and resulting reads were aligned to the human hg19 reference genome using the Burrows Wheeler Aligner (BWA; Li & Durbin, 2009). Additional details regarding library preparation, exome sequencing, and alignment have been previously described (Bureau, Parker, et al., 2014).
Processing of aligned reads
Normalized bin counts
Adjacent or partially overlapping exons for the known genes in hg19 were merged to generate 242,600 non-overlapping genomic intervals spanning 85Mb. The number of single end reads aligning to each bin was counted using the countBam function in the R package Rsamtools. We added 1 read to each bin to avoid numerical issues, and log2 transformed the resulting counts.
As the alignment is highly dependent on the complexity of the sequence and may confound read-depth based counts of copy number, we employed a number of filters to remove exomic regions with low DNA complexity. Surrogates of DNA complexity included mappability (Derrien et al., 2012; Koehler, Issac, Cloonan, & Grimmond, 2011), a score on the interval [0,1] indicating how unique a 100mer sequence is in the genome (0 is highly repetitive and 1 is unique), and the percent GC content of the bin (Fromer et al., 2012; Teo, Pawitan, Ku, Chia, & Salim, 2012; van Heesch et al., 2013). We removed bins with average mappability less than 0.75, as well as bins with %GC less than 0.1 or greater than 0.85 (Cabanski et al., 2013). In addition, we removed autosomal bins for which 5 or more subjects (4 percent or greater) in the study had a log-transformed count less than 3 median absolute deviations (MADs) from the median (Supplementary Figure 1).
After mappability and GC content exclusions, the remaining 176,912 autosomal bins spanning 65Mb were adjusted for GC composition and bin size. In particular, a local regression smoother (loess) with a span of 0.75 was fit independently to each sample to model the non-linear relationship between log ratios and GC content. The residuals from this GC-loess were then adjusted for size (using log10 transformed bin sizes) with a loess smoother of the same span (Supplementary Figures 2 and 3). Finally, we center each bin by its median across all samples. This final step reduces unmodeled bin-to-bin variation in copy number while preserving rare changes. We denote the normalized log ratios by M (Supplementary Figure 4).
Quality control statistics for the M values included the autosomal lag 10 autocorrelation (ACF10) and MAD. High autocorrelations indicate a spatial dependence along the genome often due to technical sources of variation (Marioni et al., 2007). Similarly, high MADs indicate low quality data, commonly giving rise to false positive CNV calls in subsequent seqmentation analyses. Upper limits for the acceptable range of ACF10 and MAD in this study were chosen as 0.2 and 0.3, respectively (Supplementary Figure 5).
Identification of hemizygous deletions
Candidate boundaries of copy number alterations were identified by circular binary segmentation (CBS) using the R package DNAcopy (Olshen, Venkatraman, Lucito, & Wigler, 2004). Segments with mean M values less than −0.5 and greater than −2 represented candidate hemizygous deletions. Segments with mean M less than −2 were presumed to be homozygous deletions. Regions with one or more homozygous deletions (i.e. not rare) were excluded from further analyses. To remove regions among the candidates that were (i) not rare or (ii) likely false positives, we fit Bayesian normal mixture models implemented in the R package CNPBayes. Specifically, we fit 4 mixture models with fixed means θ for the M values: (i) θ = (0) representing all samples being copy-neutral, (ii) θ = (θ1, θ2) = (−1, 0) representing a population of samples with 1 and 2 DNA copies, (iii) representing a population of samples with 2 and 3 DNA copies, and (iv) representing a population of samples with 1, 2 and 3 DNA copies. We assume the mixture components have equal variance. For each of the 4 models, we compute the marginal likelihood as described by Chib (1995) using the correction factor suggested by Neal (1999). The ratio of the maximum marginal likelihood for models (ii) and (iv) to the maximum marginal likelihoods for models (i) and (iii) becomes the Bayes factor for a hemizygous deletion model. Regions were excluded from further study if the logarithm of the Bayes factor was less than 2 or if deletions were identified in 6 or more of the multiplex families. For regions in which the log Bayes factor exceeded 2, hemizygous deletions were identified as those samples with a posterior probability for the hemizygous state exceeding 0.9.
Implementation of alternative methods for whole exome deletion analysis
XHMM, CoNIFER, and CLAMMS were implemented using default parameter settings where possible using the same set of genomic intervals described above. Briefly, we followed the on-line tutorial for XHMM version 1.0 (Fromer & Purcell, 2014). The XHMM hidden Markov model was fit to principal component normalized coverage estimates using a parameter file available from the tutorial (http://atgu.mgh.harvard.edu/xhmm/tutorial.shtml). CLAMMS version 1.0 was implemented as per instructions on the GitHub website using default parameters (https://github.com/rgcgithub/clamms). CoNIFER version 0.2.2 was fit using default parameters described in their tutorial http://conifer.sourceforge.net/tutorial.html.
PCR-based validation of putative hemizygous deletions
Selected hemizygous deletions for a region involving gene DUSP22 on chr6 were experimentally verified by qPCR. We used the TaqMan™ Copy Number Assays kit Hs01284455_cn (ThermoFisher, PN 4400291) that aligns to exon 6 of this gene with TaqMan Copy Number Reference Assay RNAse (PN 4403326). DNA was extracted and prepared in accordance with TaqMan™ protocol (PN 4397425D). Following qPCR, copy number estimates were obtained using Applied Biosystems CopyCaller™ Software v2.0.
Statistical significance of shared deletions
Exclusion of deletions
As common deletions are less likely to be shared IBD (required in our statistical approach, see below), we excluded deletions if 80% or more of the width of the deleted allele was identified in ≥ 2% of the 1000 Genomes study (1000G) participants (Auton et al., 2015). In addition, we excluded regions if ≥ 80% or more of the implicated deletion was identified as a homozygous deletion in any 1000G study participant.
Sharing probabilities
We previously developed a method to compute the exact probabilities that multiple affected relatives share an observed rare allele (nucleotide variant) given the pedigree structure (Bureau, Younkin, et al., 2014). In this procedure, we compute the exact probability a rare allele is shared by all sequenced relatives in a family, given it occurred in any one of them, under the null hypothesis of complete absence of linkage and association. Our approach requires sharing of a specific rare allele. For pairs of relatives, these sharing probabilities can easily be expressed using kinship coefficients or degree of relatedness (T. Feng, Elston, & Zhu, 2011). For two family members with genetic distance D, the rare allele sharing probability is (Bureau, Younkin, et al., 2014). By contrast, the IBD sharing probability used in linkage analysis is . Rare allele sharing probabilities are always smaller than IBD sharing probabilities, approaching a factor of 4 in the limit. Our approach extends rare allele sharing probabilities to families with multiple affected relatives, and does not require estimates of allele frequencies in the population to calculate the sharing probabilities. The key assumption is that the alleles tested are sufficiently rare such that identity-by-state implies identity-by-descent. However, we do use estimates of allele frequencies from published reference data sets such as the 1000G study to filter alleles of appreciable frequencies in non-affected subjects. In this application, we applied this method to sharing of hemizygous deletions. When two deletions overlap, we define their intersection as a shared deletion allele and calculate its sharing probability using the RVsharing package.
P-values
For deletions seen in only one family, the sharing probability can be interpreted directly as a p-value from a Bernoulli trial. For deletions seen in M families and shared by affected relatives in some of them, the appropriate p-value can be obtained as the sum of the probability of events as or more extreme than the observed sharing event. Mathematically, as described in Bureau, Younkin, et al. (2014), let pm denote the sharing probability between the subjects in family m, and let So be the set of families that share this deletion. The p-value for the observed sharing across families is
where V is the subset of family sets Sv such that
Potential p-values
The lowest possible (“potential”) p-value for any rare deletion, achieved if all family members share the deletion, depends on the number of families in which the deletion was observed and the pedigree structures. If found in only one or very few families, the sharing probabilities and thus the potential p-value for a rare deletion may be high. For example, the potential p-value is for a grandparent-grandchild pair. We test the null hypothesis only for rare deletions having a sufficiently low potential p-value after multiple comparison correction. These potential p-values are independent of the actual sharing pattern among affected relatives, and therefore of the subsequent testing of deletion sharing (i.e. the type I error is protected). We obtain this subset of rare deletions by ordering potential p-values of all rare deletions in decreasing order, and stopping at the last potential p-value lower than the type I error level 0.05 divided by the rank t of that p-value. The critical threshold then becomes 0.05/t, assuring a family-wise error rate of at most 0.05 (Bureau, Younkin, et al., 2014).
Global enrichment test
We also conducted an overall test for enrichment of sharing, addressing whether we observe more sharing of hemizygous deletions than expected under the global null hypothesis of complete absence of linkage and association. A critical assumption of this test is that rare deletions are independent. We denote the collection of hemizygous deletions that could potentially be shared in a family as D1, …, DK. Note, the same region observed in multiple families would enter multiple times. The global enrichment test statistic is the probability
where pk denotes the sharing probability of deletion Dk. Similar to Fisher’s exact test based on the hypergeometric distribution, we calculate the significance of this test statistic using the enumeration of all 2K possible sharing patterns across D1, …, DK, denoted Π1, …, Π2K, ranging from complete sharing of all K deletions (Π1) to no sharing (Π2K) (Supplementary Figure 6). For each of these patterns we calculate
and the p-value is the sum of the probabilities of all patterns that are not more likely than the one observed, i.e.
The calculation of this p-value can be computationally expensive with large K. We have implemented a binary tree representation of this algorithm that allows for significant pruning to expedite computation (see Supplementary Materials).
Results
Families were recruited by separate research groups under protocols reviewed and approved by their respective institutional review boards as previously described (Bureau, Parker, et al., 2014). Two or three affected second, third, and higher degree relatives from 56 families (n=115 individuals) were whole exome sequenced to an average depth of 60× coverage. Ethnic groups represented in this study are 19 families of German ancestry (n=38), 12 Indian families (n=26), 11 Filipino families (n=22), 10 Syrian families (n=22), 2 European-American families (n=3), one Chinese family (n=2), and one Taiwanese family (n=2).
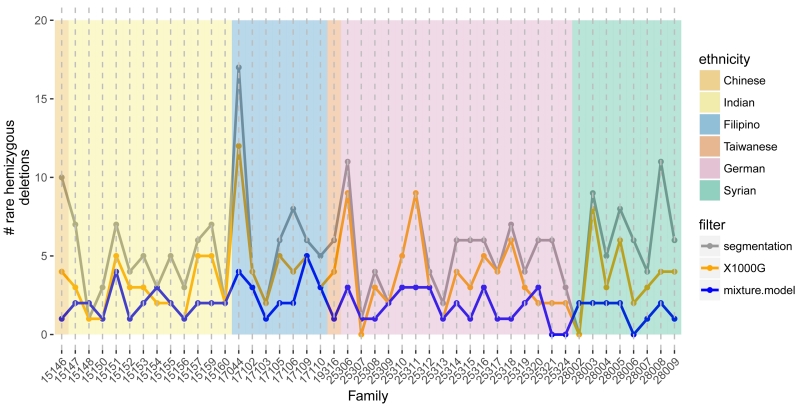
Following alignment to the hg19 reference genome by BWA, we defined 242,600 non-overlapping bins of the exome by merging the full set of exons. A total of 59,279 bins with low GC content, poor mappability, or low coverage were subsequently removed. The autosomal M-values were approximately Gaussian with a median ACF10 of 0.03 and median MAD of 0.17 (Supplementary Figure 5). Four samples with ACF10 greater than 0.2 and 3 additional samples with MADs greater than 0.3 were excluded from further analyses. While a family must have at least two members to assess sharing, at this stage we included all genomes with high quality WES data. Segmentation of the M values by CBS identified an initial set of 252 segments among 95 participants with an average M consistent with a hemizygous deletion. We excluded regions where hemizygous deletions were identified in 6 or more families (≈ 10 percent) and regions where a homozygous deletion was identified in any oral cleft participant or was previously reported in any 1000G participant. The remaining 169 candidate hemizygous deletions comprised 100 distinct, non-overlapping genomic regions. Using Bayes Factors to compare normal fixed mean mixture models, we identified 88 deletions from 53 regions, spanning 12Mb of the exome (Supplementary Figure 7). The median number of rare hemizygous deletions identified per multiplex family was 2 with an interquartile range of 1.0 – 2.8 (Figure 1).
Figure 1.
The number of autosomal hemizygous deletions (y-axis) identified among 95 participants across 46 mulitiplex families (x-axis). Candidate deletions were first identified by segmentation of M values (gray). Excluding deletions overlapping with homozygous deletions and copy number polymorphisms in the 1000G project, we obtained an initial estimate of the frequency of rare, autosomal hemizygous deletions per family (orange). At each region with a potentially rare deletion, we fit Bayesian mixture models with and without a mixture component for the hemizygous copy number state to the average M values. For regions where the log Bayes factor comparing the model with deletion to the model without deletion was at least 2, a sample was considered hemizygous if the posterior probability for the hemizygous component was at least 0.9. Excluding regions with more than 5 families identified as hemizygous under this mixture model, a total of 88 rare deletions were identified in the 46 families with a median frequency per family of 2 (blue).
The assumption that the 100 deletions are rare depends on estimates of deletion frequencies in the 1000G study. While there exists heterogeneity of CNV frequencies among subpopulations in the 1000G study, the deletions identified in this study are rare either because very few individuals with CNVs have been identified in any of the 1000G subjects or because their size is substantially larger than previously identified CNVs in these regions (Supplementary Figure 8).
To gauge performance of our approach (hereafter, RV) relative to existing pipelines for whole exome copy number analysis, we applied the algorithms XHMM, CoNIFER, and CLAMMS to the oral cleft study. Overall, 68 of the 88 (77%) rare deletions detected by RV were identified by at least one other method. Specifically, XHMM and CoNIFER identified 61 (69%) and 53 (60%) of the rare deletions, while CLAMMS identified 32 (36%). None of the alternative methods identified the rare deletion shared by distant relatives on chromosome 6, a region subsequently validated by qPCR (Supplementary Figure 9). In addition, adapting XHMM and CoNIFER to the identification of rare deletions is not possible since these methods do not distinguish between hemizygous and homozygous deletions. For nearly all homozygous deletions identified by RV and called as deletions by XHMM or CoNIFER irrespective of rarity status, the signal to noise ratio of the normalized coverage estimates is more than 2-fold higher in RV (Supplementary Figure 10). Normalized copy number estimates were comparable in CLAMMS and RV, differing mainly in scale (Supplementary Figure 11).
Among the 46 multiplex families used in the sharing analysis, three families each had three members, and the remaining 43 families had two members. Family 15157 had a deletion shared in 2 of 3 affected members, but no three-member family had a deletion shared by all members. For the two-member families, we identified 8 shared deletions (median size is 46 kb). The most frequent deletion meeting our rarity criteria occurred in DUSP22 (chr6: 292,101-393,098bp) in two individuals from one family and three individuals from three other families. Deletions involving DUSP22 have been reported as causal for Duane retraction syndrome which can occur with oral clefts (Bedoyan et al., 2011; Pilon, 2009), although deletions involving this segment of chr.6p25.3 may be critical (Chen et al., 2013). Chromosomal aberrations on the short arm of chr6 have been previously observed in children with oral clefts, suggesting the presence of an orofacial clefting locus (denoted OFC1) near 6p24 (Davies et al., 1995). Five of the six samples with called hemizygous deletions for gene DUSP22 were confirmed by qPCR, including the two first cousins 17110_01 and 17110_19 who share this deletion.
In addition to DUSP22, the top-ranked region (chr13:53,078,416 - 53,158,768bp) also contains a shared deletion (Figure 2). While nominally statistically significant (p=0.004), this shared deletion spanning pseudogene TPTE2P3 was not statistically significant after multiple testing adjustment nor has this region been previously implicated in clefting. To further investigate the sequence complexity of this region, we extracted the sequence of 15 regions (targets) captured by the Agilent SureSelect kit spanning this deletion. We aligned the target sequences to the human reference genome using BLAT (Kent, 2002). These BLAT alignments revealed other, off-target regions of the genome for which these sequences match with near perfect fidelity (Supplementary Figure 12).
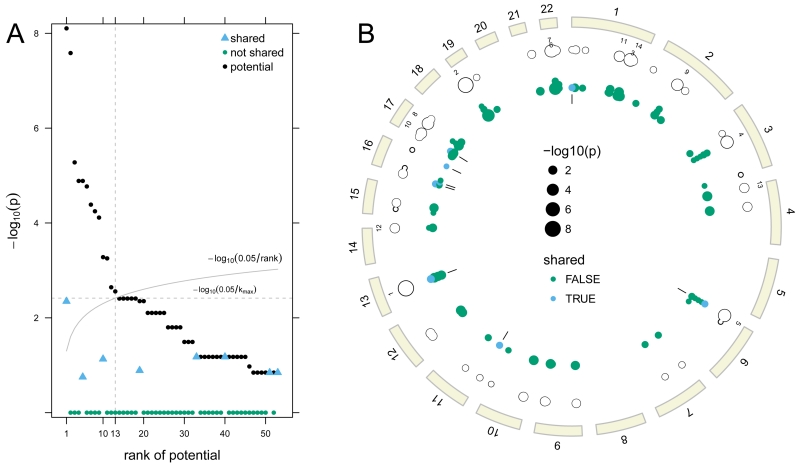
Figure 2.
Ranks of the potential p-values are plotted against the −log10 potential p-value (A). Of the 53 regions with one or more rare deletion alleles, the first 13 ranked regions have potential for a statistically significant association with oral cleft. Observed sharing probabilities for the first 13 regions were less than their potential p-values and are not statistically significant. A circos plot displays these data for each deleted region by genomic position (B). The tracks starting from the outermost ring are the ideograms (beige), the top 13 ranks of the potential sharing probabilities, the potential sharing probabilities (unfilled circles), and the contribution of each family to the potential sharing probabilities (solid circles). Families with a shared deletion are indicated in blue with tick marks on the innermost track highlighting the 8 regions with shared deletions.
Ordering the 53 regions by their potential p-values yielded 13 regions included in the list of formal hypothesis tests. Four of the 13 regions were shared in some but not all families, but none of these regions was statistically significant (Figure 2). We recorded a total of 88 hemizygous deletions that could potentially be shared in a family. The mean separation between rare deletions in this study was 21.5Mb (minimum across all autosomes: ≈ 29kb). Stratified by subject, there were only two individuals in which a rare deletion occurred on the same chromosome. For these two regions on chromosome 17 and 22, the distance between the rare deletions was 34.8Mb and 13.2Mb, respectively. As many individuals had only one rare deletion and all but two individuals had rare deletions on different chromosomes, the assumption that all rare deletions are independent for the global enrichment test is highly plausible. Of the 88 regions, 8 were shared within families and 72 were not (Figure 2). Our global test for the total number of shared deletions was not statistically significant (p = 0.84).
Discussion
We present an exome-wide map of 88 rare hemizygous deletions at 53 regions from 56 multiplex oral cleft families. These families were recruited by separate groups originally for linkage analysis. Probands were examined to establish they had an apparent non-syndromic oral cleft, and affected relatives were recruited and examined when possible. While firmly establishing multiplex families as truly non-syndromic is difficult and some families were known to be inbred, the WES data used here came from distant affected relatives (second or higher degree) and we deliberately screened common deletions. The majority of rare deleted regions (45/53) were not shared by members of the same family. Of the 8 shared deletions, four occurred in a single family. For each of these regions, the potential p-value exceeded the cutoff needed to control the family-wise error rate at 5 percent. Interestingly, one of the top ranked regions in this study occurs on chr6p, a region previously implicated in clefting and containing at least one reported orofacial locus (OFC1). The deleted region identified here spans DUSP22, approximately 6Mb from OFC1. Deletions involving DUSP22 have been associated with a disorder of eye movement (Duane retraction syndrome), although oral clefts can occur in individuals with this complex and heterogenous disorder. Sharing of the DUSP22 hemizygous deletion occurred in only one of the four families where it was identified in this sample of multiplex families, underscoring the genetic complexity of oral clefts.
Our study builds on the work of others for identifying rare deletions. The idea of combining segmentation and mixture models to identify copy number alterations was originally described for arrays (Barnes et al., 2008; Cardin, Holmes,, Donnelly, & Marchini, 2011; Marioni et al., 2007; Picard et al., 2011; Scharpf et al., 2012). Here, mixture models account for heterogeneity of the precision between exomic bins used to estimate coverage. The Bayesian mixture model increases the specificity of our approach by removing false positives at high variance regions. At other regions, the Bayesian mixture model identifies additional hemizygous samples not originally detected by the segmentation, increasing sensitivity. Finally, the methodology for modeling the statistical significance of rare deletions shared by members within extended families is a natural extension of the rare SNV association models proposed by Bureau, Younkin, et al. (2014).
Our analysis removes all homozygous deletions as these are very likely to occur for common deletions. Discriminating between hemizygous and homozygous deletions is critical for the analysis of rare deletion sharing, but this is not currently available in many whole exome copy number analysis tools such as XHMM and CoNIFER.
For a highly inbred family, a rare deletion can occur in homozygous form due to inbreeding alone. In both the SNV and CNV approaches, all founders are assumed to be unrelated, and violating this assumption would lead to inflated statistical significance. For families with low levels of kinship between founders (cryptic relatedness), Bureau et al. 2014b propose a correction of the sharing probabilities based on empirical estimates of kinship among founders obtained from genome-wide marker data. Integration of genome-wide markers with the deletion analysis described here to estimate cryptic relatedness and its corresponding sharing probabilities for homozygous deletions in the Syrian families is a future direction of investigation.
Considerable genetic heterogeneity must be expected with complex diseases. Rare variants may only explain part of the “missing heritability”. In a family where cases cluster, one possible explanation is that affected members carry the same rare but highly penetrant variant (Cirulli et al., 2010; Wijsman, 2012), although other explanations such as high genetic burden also apply (Gonzaga-Jauregui et al., 2015; Loohuis et al., 2015). Our variant sharing approach specifically targets the former scenario, and thus can only be successful for families where such a single rare but highly penetrant variant segregates. Our method does not assume complete penetrance of the variant, but requires that every sequenced affected member is a carrier of the variant (i.e. no phenocopies). Further, our deletion sharing probabilities are calculated under the assumption that a single deletion allele exists among the founders such that IBS cannot occur without IBD. The true sharing probabilities depend on the unknown deletion frequency in the population, with higher deletion frequencies resulting in larger sharing probabilities. The assumption of IBD is crucial, and sensitivity analyses with respect to the population deletion frequency are recommended to assess when deletion sharing in a pedigree cannot be explained by random chance (see Bureau, Parker, et al. (2014); Bureau, Younkin, et al. (2014)).
For the study of rare disorders such as oral clefts, affected probands from multiple study sites are needed to attain large sample sizes. In such studies, genetic differences across populations and racial groups further complicate the identification of rare, highly penetrant risk variants. Here, family-based designs offer an important advantage over case-control studies of unrelated individuals. In extended families with several affected members, there is a high probability that affected relatives carry the same rare, high-penetrance risk variant if such a variant is found in one affected individual. We expect the methodology for identifying rare deletions and evaluating the probability that rare deletions are shared will be useful for other family-based studies of complex traits, opening new avenues of epidemiologic investigation.
Supplementary Material
Acknowledgements
We thank the members of the families who participated in this study and the field and laboratory staff who mad this analysis possible. This work was supported by the National Institutes of Health (NIH) (R01-DE-014581 and U01-DE-018993 to T.H.B., R01-DE-016148 to M.L.M, P50-DE-016215 to J.C.M., and R03-DE-02579 to I.R. and R.B.S), with additional support from X01 HG006177 to T.H.B., M.L.M., and J.C.M. for whole exome sequencing at the Center for Inherited Disease Research, which is funded through a federal contract from the NIH to Johns Hopkins University (contract no. HHSN268200782096C). J.E.B-W. is supported by the Intramural Program of the National Human Genome Research Institute, NIH. The qPCR was conducted at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine, Baltimore, MD. We thank Laura Kasch-Semenza and Roxann Ashworth for their assistance.
Footnotes
The authors have no competing interests to declare.
References
- Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annu Rev Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. Retrieved from http://dx.doi.org/10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Abecasis GR. A global reference for human genetic variation. Nature. 2015 Oct;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C, Plagnol V, Fitzgerald T, Redon R, Marchini J, Clayton D, Hurles ME. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008 Oct;40(10):1245–1252. doi: 10.1038/ng.206. Retrieved from http://dx.doi.org/10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoyan JK, Lesperance MM, Ackley T, Iyer RK, Innis JW, Misra VK. A complex 6p25 rearrangement in a child with multiple epiphyseal dysplasia. Am J Med Genet A. 2011 Jan;155A(1):154–163. doi: 10.1002/ajmg.a.33751. Retrieved from http://dx.doi.org/10.1002/ajmg.a.33751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau A, Parker MM, Ruczinski I, Taub MA, Marazita ML, Murray JC, Beaty TH. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics. 2014 Jul;197(3):1039–1044. doi: 10.1534/genetics.114.165225. Retrieved from http://dx.doi.org/10.1534/genetics.114.165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau A, Younkin SG, Parker MM, Bailey-Wilson JE, Marazita ML, Murray JC, Ruczinski I. Inferring rare disease risk variants based on exact probabilities of sharing by multiple affected relatives. Bioinformatics. 2014 Aug;30(15):2189–2196. doi: 10.1093/bioinformatics/btu198. Retrieved from http://dx.doi.org/10.1093/bioinformatics/btu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanski CR, Wilkerson MD, Soloway M, Parker JS, Liu J, Prins JF, Hayes DN. BlackOPs: increasing confidence in variant detection through mappability filtering. Nucleic Acids Res. 2013 Oct;41(19):e178. doi: 10.1093/nar/gkt692. Retrieved from http://dx.doi.org/10.1093/nar/gkt692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin N, Holmes C, W. T. C. C. C. Donnelly P, Marchini J. Bayesian hierarchical mixture modeling to assign copy number from a targeted CNV array. Genet Epidemiol. 2011 Sep;35(6):536–548. doi: 10.1002/gepi.20604. Retrieved from http://dx.doi.org/10.1002/gepi.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-P, Lin S-P, Chern S-R, Wu P-S, Su J-W, Wang W. A boy with cleft palate, hearing impairment, microcephaly, micrognathia and psychomotor retardation and a microdeletion in 6p25.3 involving the DUSP22 gene. Genet Couns. 2013;24(2):243–246. [PubMed] [Google Scholar]
- Chib S. Marginal likelihood from the Gibbs output. Journal of the American Statistical Association. 1995;90(432):1313–1321. [Google Scholar]
- Cirulli ET, Kasperaviciūte D, Attix DK, Need AC, Ge D, Gibson G, Goldstein DB. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010 Jul;18(7):815–820. doi: 10.1038/ejhg.2010.2. Retrieved from http://dx.doi.org/10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes B, Basu S, Guha S, Schork N. Weighted score tests implementing model-averaging schemes in detection of rare variants in case-control studies. PLoS One. 2015;10(10):e0139355. doi: 10.1371/journal.pone.0139355. Retrieved from http://dx.doi.org/10.1371/journal.pone.0139355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AF, Stephens RJ, Olavesen MG, Heather L, Dixon MJ, Magee A, Ragoussis J. Evidence of a locus for orofacial clefting on human chromosome 6p24 and STS content map of the region. Hum Mol Genet. 1995 Jan;4(1):121–128. doi: 10.1093/hmg/4.1.121. [DOI] [PubMed] [Google Scholar]
- Derrien T, Estellé J, Marco Sola S, Knowles DG, Raineri E, Guigó R, Ribeca P. Fast computation and applications of genome mappability. PLoS One. 2012;7(1):e30377. doi: 10.1371/journal.pone.0030377. Retrieved from http://dx.doi.org/10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duncan R, Ware EB, Jhun MA, Bielak LF, Zhao W, Satten GA. A statistical approach for rare-variant association testing in affected sibships. Am J Hum Genet. 2015 Apr;96(4):543–554. doi: 10.1016/j.ajhg.2015.01.020. Retrieved from http://dx.doi.org/10.1016/j.ajhg.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Pistis G, Zhang H, Zawistowski M, Mulas A, Zoledziewska M, Abecasis GR. Methods for association analysis and meta-analysis of rare variants in families. Genet Epidemiol. 2015 May;39(4):227–238. doi: 10.1002/gepi.21892. Retrieved from http://dx.doi.org/10.1002/gepi.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Elston RC, Zhu X. Detecting rare and common variants for complex traits: sibpair and odds ratio weighted sum statistics (SPWSS, ORWSS) Genet Epidemiol. 2011 Jul;35(5):398–409. doi: 10.1002/gepi.20588. Retrieved from http://dx.doi.org/10.1002/gepi.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Purcell SM. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012 Oct;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. Retrieved from http://dx.doi.org/10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7.23.1–7.2321. doi: 10.1002/0471142905.hg0723s81. Retrieved from http://dx.doi.org/10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, Lupski JR. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015 Aug;12(7):1169–1183. doi: 10.1016/j.celrep.2015.07.023. Retrieved from http://dx.doi.org/10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002 Apr;12(4):656–664. doi: 10.1101/gr.229202. Retrieved from http://dx.doi.org/10.1101/gr.229202.ArticlepublishedonlinebeforeMarch2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee K, Sun H. Statistical selection strategy for risk and protective rare variants associated with complex traits. J Comput Biol. 2015 Oct; doi: 10.1089/cmb.2015.0091. Retrieved from http://dx.doi.org/10.1089/cmb.2015.0091. [DOI] [PubMed] [Google Scholar]
- Koehler R, Issac H, Cloonan N, Grimmond SM. The uniqueome: a mappability resource for short-tag sequencing. Bioinformatics. 2011 Jan;27(2):272–274. doi: 10.1093/bioinformatics/btq640. Retrieved from http://dx.doi.org/10.1093/bioinformatics/btq640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012 Aug;22(8):1525–1532. doi: 10.1101/gr.138115.112. Retrieved from http://dx.doi.org/10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. Retrieved from http://dx.doi.org/10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K-H, Zöllner S. Robust and powerful affected sibpair test for rare variant association. Genet Epidemiol. 2015 Jul;39(5):325–333. doi: 10.1002/gepi.21903. Retrieved from http://dx.doi.org/10.1002/gepi.21903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loohuis LMO, Vorstman JAS, Ori AP, Staats KA, Wang T, Richards AL, Ophoff RA. Genome-wide burden of deleterious coding variants increased in schizophrenia. Nat Commun. 2015;6:7501. doi: 10.1038/ncomms8501. Retrieved from http://dx.doi.org/10.1038/ncomms8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009 Feb;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. Retrieved from http://dx.doi.org/10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Thorne NP, Valsesia A, Fitzgerald T, Redon R, Fiegler H, Hurles ME. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8(10):R228. doi: 10.1186/gb-2007-8-10-r228. Retrieved from http://dx.doi.org/10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal RM. Erroneous results in marginal likelihood from the Gibbs output. minmeo, University of Toronto; 1999. [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004 Oct;5(4):557–72. doi: 10.1093/biostatistics/kxh008. Retrieved from http://dx.doi.org/10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Packer JS, Maxwell EK, O’Dushlaine C, Lopez AE, Dewey FE, Chernomorsky R, Reid JG. Clamms: a scalable algorithm for calling common and rare copy number variants from exome sequencing data. Bioinformatics. 2016 Jan;32(1):133–135. doi: 10.1093/bioinformatics/btv547. Retrieved from http://dx.doi.org/10.1093/bioinformatics/btv547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Lebarbier E, Hoebeke M, Rigaill G, Thiam B, Robin S. Joint segmentation, calling, and normalization of multiple CGH profiles. Biostatistics. 2011 Jul;12(3):413–428. doi: 10.1093/biostatistics/kxq076. Retrieved from http://dx.doi.org/10.1093/biostatistics/kxq076. [DOI] [PubMed] [Google Scholar]
- Pilon AF. Midline orofacial cleft defects in association with type 1 Duane’s retraction syndrome. Clin Exp Optom. 2009 Mar;92(2):133–136. doi: 10.1111/j.1444-0938.2008.00311.x. Retrieved from http://dx.doi.org/10.1111/j.1444-0938.2008.00311.x. [DOI] [PubMed] [Google Scholar]
- Scharpf RB, Beaty TH, Schwender H, Younkin SG, Scott AF, Ruczinski I. Fast detection of de novo copy number variants from SNP arrays for case-parent trios. BMC Bioinformatics. 2012 Dec;13(1):330. doi: 10.1186/1471-2105-13-330. Retrieved from http://dx.doi.org/10.1186/1471-2105-13-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics. 2012 Nov;28(21):2711–2718. doi: 10.1093/bioinformatics/bts535. Retrieved from http://dx.doi.org/10.1093/bioinformatics/bts535. [DOI] [PubMed] [Google Scholar]
- van Heesch S, Mokry M, Boskova V, Junker W, Mehon R, Toonen P, Guryev V. Systematic biases in DNA copy number originate from isolation procedures. Genome Biol. 2013 Apr;14(4):R33. doi: 10.1186/gb-2013-14-4-r33. Retrieved from http://dx.doi.org/10.1186/gb-2013-14-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet. 2012 Oct;131(10):1555–1563. doi: 10.1007/s00439-012-1190-2. Retrieved from http://dx.doi.org/10.1007/s00439-012-1190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011 Jul;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. Retrieved from http://dx.doi.org/10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.