Summary
The prospect that the structures of protein assemblies, small and large, can be determined using cryo-electron microscopy (cryo-EM) is beginning to transform the landscape of structural biology and cell biology. Great progress is being made in determining 3D structures of biological assemblies ranging from icosahedral viruses and helical arrays to small membrane proteins and protein complexes. Here, we review recent advances in this field, focusing especially on the emerging use of cryo-EM in mapping the binding of drugs and inhibitors to protein targets, an application that requires structure determination at the highest possible resolutions. We discuss methods used to evaluate the information contained in cryo-EM density maps and consider strengths and weaknesses of approaches currently used to measure map resolution.
Introduction
Recent developments in single particle cryo-electron microscopy (cryo-EM) are taking the world of structural biology by storm [1,2]. This transformation, largely driven by the development of direct electron detectors [3,4], has enabled determination of structures of protein complexes spanning a broad range of sizes, many of which have been intractable to crystallographic analysis. The implications of these advances for drug discovery could be significant, as knowledge of the detailed interaction of protein targets with various small molecule modulators can yield critical information for the design and development of novel and improved therapeutics. This information generally requires high resolution (typically 3 Å or better) [1,5], enabling determination of the detailed placement of individual amino acids and how they interact with small molecules that alter the function of the protein.
Until very recently, only nuclear magnetic resonance spectroscopy (NMR) and X-ray crystallography have been capable of producing structural information at this type of resolution. Early efforts using cryo-EM took advantage of 2-dimensional (2D) crystals to achieve high resolution, exemplified by the determination of the structure of the membrane bacteriorhodopsin [6,7] or the determination of the binding site of taxol on tubulin [8]. This analysis, unlike single particle cryo-EM, relies on the packing of the proteins in an ordered assembly. Since 2013, however, changes in microscope and detector technology, especially the development of dose fractionation during image acquisition, have now made it possible for cryo-EM analyses of non-crystalline, single particle specimens to be used to directly derive atomic models. Increasing numbers of structural studies using cryo-EM are now reaching resolutions in the 3 Å range. Not only have these studies produced structures at near-atomic resolution, but in numerous instances, have also allowed analysis of multiple, functionally-relevant conformations present in mixtures. In this review, we focus specifically on an emerging theme in the potential use of cryo-EM for drug discovery applications. In this context, we also address some of the practical issues involved in analyzing density maps obtained using cryo-EM and consider some of the issues associated with assessing the resolution and information content present in these maps.
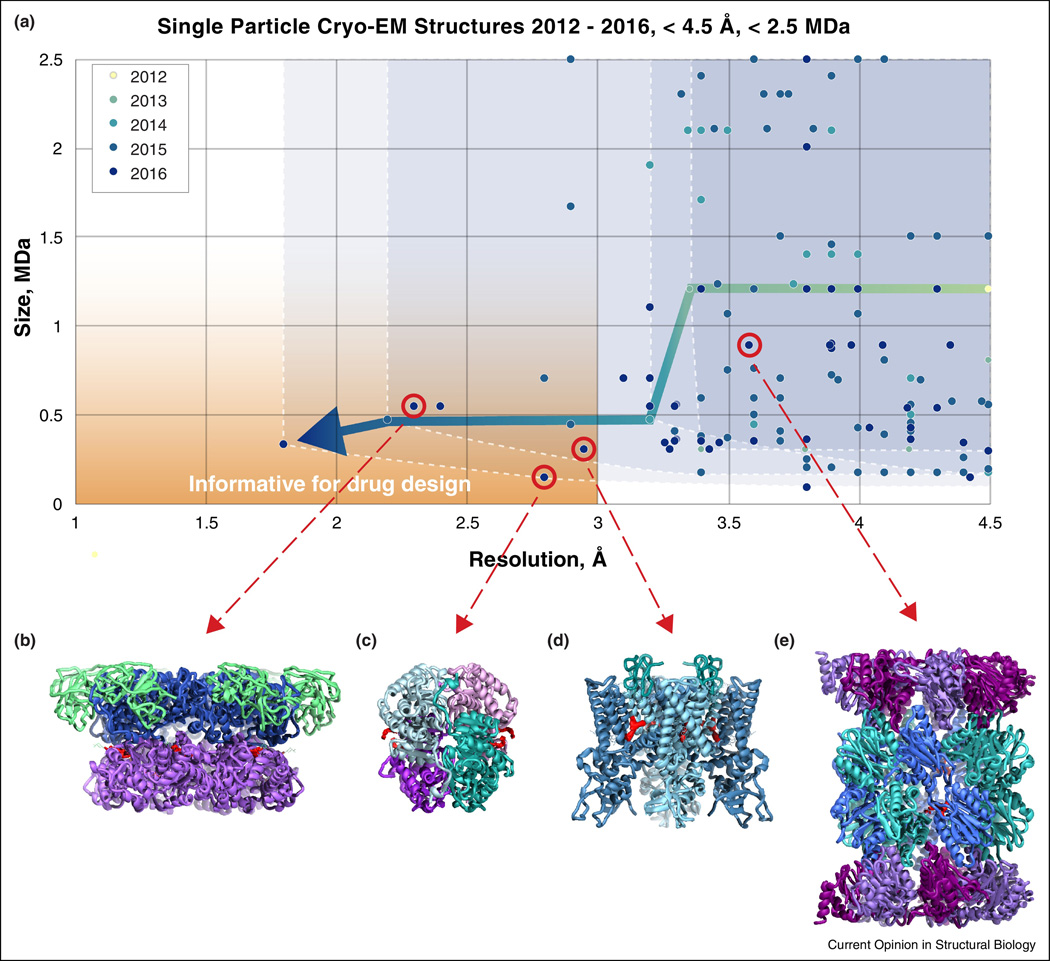
Single particle cryo-EM at high resolution
Some of the first forays into single particle cryo-EM at near-atomic resolution were with icosahedral viruses [9,10], a logical initial choice given that the large size of the particles and their high symmetry greatly facilitate accurate alignment of projection images to generate 3D reconstructions of the structure. Early examples included structures of the cytoplasmic polyhedrosis virus [9] and the human adenovirus [11], and have been followed by work on many other viruses such as the Sputnik viriophage [10], the Zika virus [12] and also on other protein assemblies with high symmetry [13]. The use of single particle cryo-EM to obtain near-atomic resolution information from smaller, lower symmetry complexes is a relatively recent phenomenon. Starting in 2013, the introduction of direct electron detectors such as the Falcon camera (from FEI), the K2 Summit (from Gatan), and the DE camera (from Direct Electron Inc.) has now enabled determination of numerous near-atomic resolution structures of small protein complexes. In Figure 1A, we show a plot of entries in the EM Data Bank reporting the single particle cryo-EM structures of proteins with size < 2.5 MDa at resolutions better than 4.5 Å, the resolution at which β-sheet secondary structural elements can be visualized unambiguously. As seen in the plot, the initial explosion of entries in this resolution range was dominated by ribosome structures in 2014, but has rapidly expanded to include much smaller complexes in the last couple of years [3,14–16]. Many of these are of small complexes with low- or medium-symmetry, highly relevant for drug design, and at resolutions that allow direct determination of specific ligand-protein interactions. As of now, there are only a handful of structures that are smaller than 1 MDa and at resolutions of 3 Å or better, although there is little doubt this number will increase rapidly in the coming years.
Figure 1.
Cryo-EM progress towards drug discovery. (A) Chart showing entries deposited in the Electron Microscopy Data Bank each year since 2012 with reported resolutions better than 4.5 Å for protein complexes smaller than 2.5 MDa. Four select high resolution structures (red circles) determined with bound small molecule drugs (density highlighted in red) are shown in (B–E) with ribbon diagrams of: (B) p97 (EMD-3295, PDB-5ftj) [49]; (C) lactate dehydrogenase (EMD-8191, PDB-5k0z) [48]; (D) TRPV1 (EMD-8117, PDB-5irx) [18]; and (E) Plasmodium falciparum 20S proteasome (EMD-3231, PDB-5fmg) [40].
Membrane proteins, and particularly ion channels, have been one class of proteins where cryo-EM structure determination has been especially useful both for understanding molecular mechanisms underlying protein function and the structural basis of small molecule action. The first of these proteins solved to near-atomic resolution was the TRPV1 channel in 2013 [14,17], followed by a higher resolution report this year of the structure of the channel in lipid nanodiscs [18]. Another early structure of a membrane protein at medium resolution was that of γ-secretase [19]; recent structures of the same protein have recently been improved to approach the 3 Å range [20,21] with sufficient resolution to resolve the location of a bound inhibitor. Rapid progress is also being made with challenging membrane proteins such as glutamate receptors and the ryanodine receptor RyR1, which have proven difficult to crystallize as native, full length proteins. Even at medium resolution, cryo-EM structures of the AMPA glutamate receptors showed how stabilizing mutations used to crystallize the proteins resulted in perturbations of the native structure [22]. Several additional studies of glutamate receptors at similar resolutions have been published in the past year, although with one exception [23], most have only reached resolutions in the 6 Å range, reflecting in part the high inherent flexibility of these dynamic protein assemblies [24–26]. The case of the RyR1 complex is also similar; the presence of mobile domains appears to have lowered the overall resolution, but still allows detailed analysis of the stable core region of the protein [27,28]. Yet another instance exemplifying the utility of cryo-EM to probe function in an ion channel is from recent work on the CorA magnesium ion channel. Here, the cryo-EM structure of stable, 5-fold symmetric closed conformation was obtained at near-atomic resolution, while the novel structures of the highly flexible, asymmetric open conformations were limited to medium resolution, but nevertheless offered an unexpected mechanism for magnesium gating of the channel [29]. Near-atomic resolution structures of the glycine receptor, a pentameric ligand-gated choride channel, bound to the agonist glycine, the antagonist strychnine, or with a combination glycine and the channel modulator ivermectin have yielded significant insights into conformational changes occurring during channel gating [30]. Other successes of analyzing soluble protein complexes include work on ribosomes [15,31–33], and smaller, more challenging complexes such as the spliceosome [34–37], the proteasome [38–40] and viral fusion proteins [41–43].
Major advances have also been made in the area of protein and nucleoprotein polymers, where cryo-EM and helical reconstruction have enabled structural analysis of these complexes. While icosahedral viruses and asymmetric complexes such as the ribosome can be crystallized, many proteins in cells and viruses are found in the form of helical polymers. Helical polymers can never be crystallized in their polymeric form due to the symmetry mismatch with any crystal space group. Recent examples of helical complexes determined by cryo-EM at better than 4 Å resolution include actomyosin [44], microtubules [45], bacterial pili [46], and an archaeal rod-like virus [47].
While all of the examples discussed above have approached resolutions in the 3 Å to 4 Å range, or revealed mechanistic insights into protein function even at lower resolutions, the use of cryo-EM to directly visualize small molecules interacting with the binding pocket is still relatively rare. Four examples of this (Figures 1B–1E), all published in 2016, are discussed in more detail below.
Visualizing small molecule ligands with cryo-EM
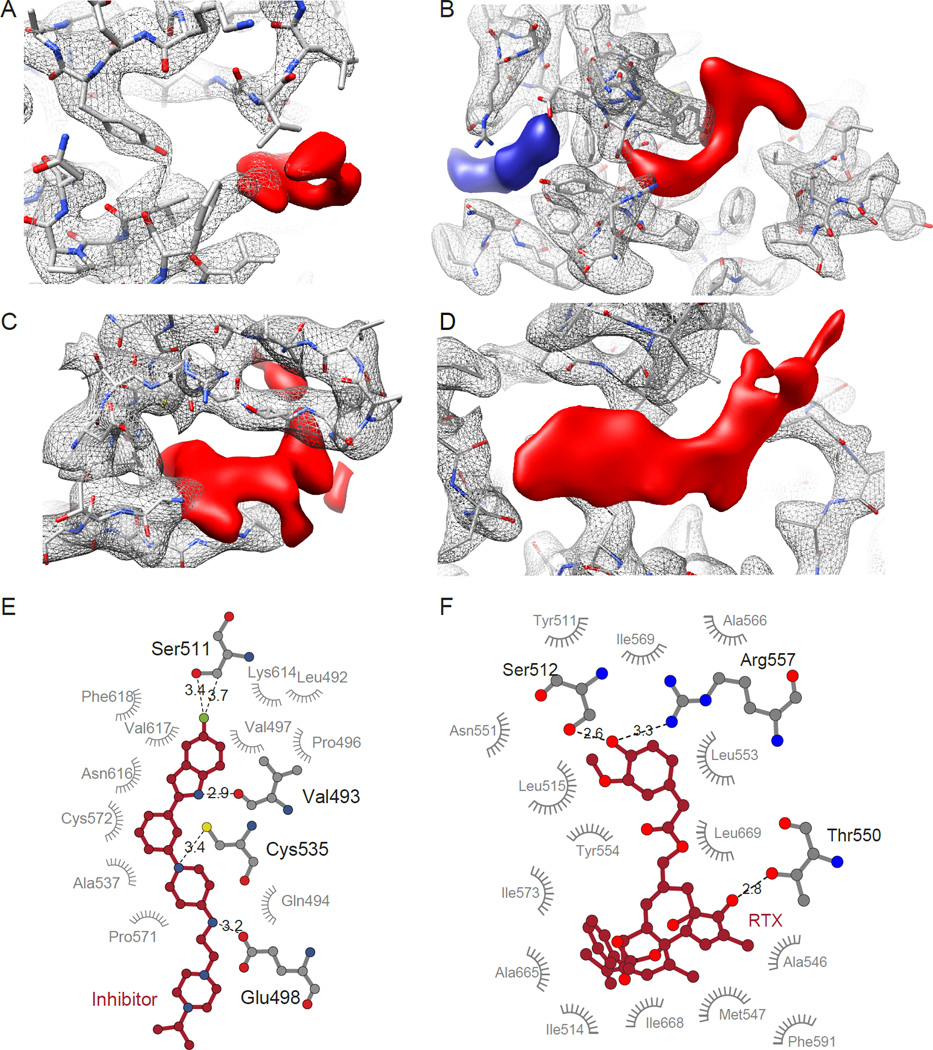
One of the smallest proteins whose structure has been determined at high resolution by cryo-EM is lactate dehydrogenase, a 145 kDa tetrameric protein whose function can be regulated by a variety of small molecule inhibitors (Figure 1C). While a number of crystal structures of this complex are available, the 2.8 Å resolution cryo-EM structure of this protein [48] reveals specific density for a novel inhibitor (Figure 2A), which binds to the outside edge of the structure. Cryo-EM analysis at 3.8 Å resolution of an even smaller protein, the 93 kDa isocitrate dehydrogenase complexed to ML309, a drug under consideration for cancer treatment, provides further encouragement for the use of cryo-EM both for understanding drug localization and effects of binding on protein conformation [48].
Figure 2.
Visualizing drug density in cryo-EM maps. (A–D) Close-up views of protein (grey mesh) and drug (red solid) densities, fit with the atomic model of the protein. (A) Lactate dehydrogenase bound to the inhibitor GSK2837808A (EMD-8191, PDB-5k0z) [49]. (B) TRPV1 ion channel bound to double-knot toxin (DkTx) and resiniferatoxin (RTX) (EMD-8117, PDB-5irx) [18]. (C) p97 bound to the inhibitor UPCDC30245 (EMD-3295, PDB-5ftj) [49]. (D) Plasmodium falciparum 20S proteasome bound to WLW vinyl sulfone (EMD-3231, PDB-5fmg) [40]. (E, F) LIGPLOT diagrams showing binding pocket interactions of inhibitor bound p97, as shown in (A) and RTX bound to TRPV1, as shown in (B).
The structure of the TRPV1 channel was already reported at near-atomic resolution in 2013 [14], but a new follow-up study of its structure [18] is of particular interest for several reasons. First, the use of lipid nanodiscs rather than amphipols (which can artificially stabilize transmembrane regions) allowed for the visualization of lipid directly interacting with the transmembrane helices. Second, two structures reported in this study at 3.4 Å and 3.0 Å resolution include density for the drugs capsazepine and resiniferatoxin (RTX), respectively. The density for RTX within its binding site in TRPV1, which is shown in Figure 2B, was sufficient to visualize individual interactions with amino acids and to delineate the drug binding site (Figure 2F).
Another recent example of the use of cryo-EM to visualize inhibitor binding to protein complexes is that of p97, an AAA ATPase that is of critical interest as a cancer drug target. Because of its flexible nature, it has not been possible to obtain crystal structures of the full-length p97 complex in its activated state, while crystal structures of individual domains are not useful for inhibitors that bind at interfaces between individual domains. Banerjee et al. [49] not only reported the binding site and mechanism of drug action, but also revealed specific interactions between different domains and the drug molecule. Further, this study showed that as protein domains in p97 undergo large movements during its ATPase reaction cycle, separating three distinct co-existing conformations at resolutions of ~ 3 Å by 3D classification. The inhibitor is localized to the interface between the D1 and D2 ATP binding domains, with highly specific interactions between the protein binding pocket at one end of the drug (Figures 2C, 2E). In instances such as this where 3D crystallization is not an option, cryo-EM analyses provide a significant advantage for accelerating the identification of more effective small molecule inhibitors by determination of structures of candidate compounds with native proteins.
A recent study of the Plasmodium falciparum proteasome offers another example of using structural information from cryo-EM analysis in the context of drug design. Li et al. [40] report visualization of the P. falciparum proteasome 20S core in complex with WLW vinyl sulfone (WLW-vs), an inhibitor modified to interact specifically with the P. falciparum, but not the human proteasome. While at 3.6 Å resolution, it is not possible to identify specific interactions with the binding site, the inhibitor can be placed in the binding pocket of the proteasome unambiguously (Figure 2D), and comparisons suggest that this inhibitor cannot bind the analogous binding site on the structure of the human proteasome.
The ability to place small molecules with precision requires maps at sufficiently high resolution. In structures determined by X-ray crystallography, a minimum resolution of ~ 2.5 Å to 3 Å is typically required for this purpose, and a similar effective resolution is probably required for cryo-EM structures as well. However, the definition of resolution in cryo-EM is a much more nebulous entity than in X-ray crystallography, and the most commonly used method of reporting resolution using a plot of the Fourier Shell Correlation (FSC) is fraught with problems. Another critical issue is that because of the almost indefinite number of ways in which raw data can be processed to obtain final reconstructions, it is important that raw data can be made available in publicly accessible archives for independent validation of the results. Both of these issues are discussed in the next section.
Cryo-EM map resolution and validation
Cryo-EM density maps or movies showing the map quality are now often provided routinely during manuscript review and a growing number of raw data sets are now available through EMPIAR (https://www.ebi.ac.uk/pdbe/emdb/empiar) enabling independent validation of published results, and have also led to informative data challenge competitions (http://challenges.emdatabank.org/?q=2015_map_challenge). Despite these efforts, the question of defining resolution by a single number remains problematic. The main criterion that is currently used to assess the resolution of a cryo-EM map is the Fourier Shell Correlation (FSC) coefficient [50]. This metric, which seeks to compare the correlation between two independent reconstructions as a function of resolution in Fourier space is not a true measure of resolution, but rather is a measure of self-consistency in the imaging and reconstruction process. In the early years of the development in the cryo-EM field, computing the FSC involved comparing two maps, each generated from separate halves of the data (“half-maps”), split just before the final reconstruction was generated. But these half-maps are not really independent when both sets of images are aligned to the same reference. This then led to the idea of a “Gold Standard” FSC, which is computed from two independently created half-map reconstructions, overcoming the bias introduced by a common reference. Even so, such reconstructions are not truly independent, since they are both generated with many shared assumptions and bias introduced by the software.
The problems with the FSC become most obvious when one considers objects with internal symmetry, whether it be helical, icosahedral or point-group symmetry, but these same problems exist for objects with no internal symmetry. One can impose the wrong symmetry in the reconstruction process, and as long as the same wrong symmetry is imposed in two independent reconstructions, there is excellent correlation between the two half-maps that has nothing to do with the real resolution [51]. One example that highlights this is the case of the microtubule, which consists of two homologous proteins, α- and β-tubulin. At low resolution these proteins are indistinguishable, but at high resolution one can observe the sequence differences and the slightly different folds. When α-tubulin subunits in the microtubule are correctly averaged with other α-tubulin subunits the FSC does not improve (in fact, becomes slightly worse by traditional measures) over when α-tubulin subunits are incorrectly averaged with β-tubulin subunits [52]. However, the map becomes more interpretable when the averaging is done correctly, showing that the FSC is not measuring resolution or interpretability but self-consistency.
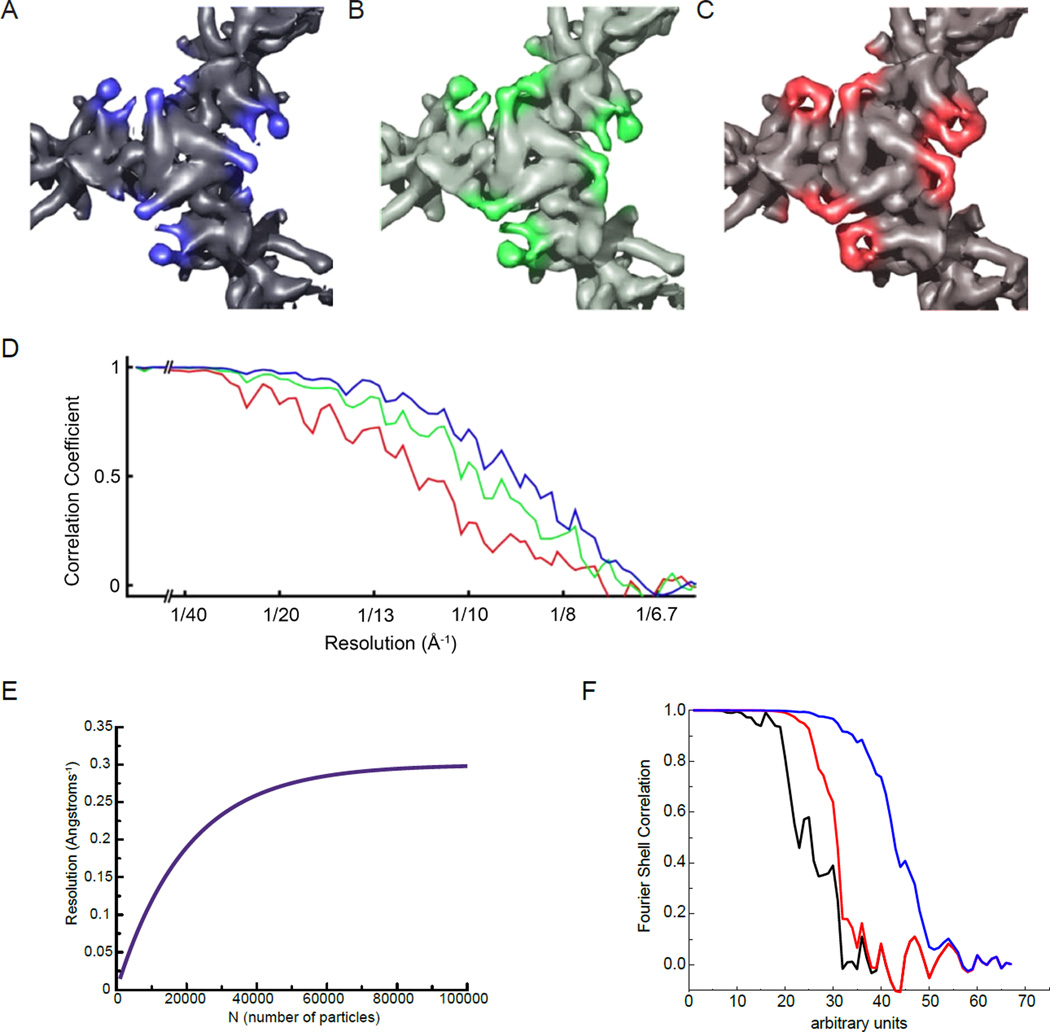
The idea that map quality and FSC can be anti-correlated was first pointed out by Borgnia et al. [53] in cryo-EM structural studies of the core domain of pyruvate dehydrogenase, where they demonstrated that while map quality could be improved by using progressively fewer numbers of better molecular images in a given data set (Figures 3A– 3C), the resolution estimated by the FSC criterion for these maps was inversely related to map quality because of the use of smaller numbers of images (Figure 3D). This problem stems primarily from the fact that the FSC approach of comparing half-maps is a biased measurement, dependent upon the number (N) of particles. This is illustrated in Figure 3E, where we show a theoretical curve for resolution as a function of sample size N, based upon the observations that this curve tends to follow a log-linear relationship [54] and becomes asymptotic at some large value of N, where an increase in N will lead to no further improvement in resolution. If, when N=100,000, the resolution is asymptotic and is actually 3.3 Å for the full map, then the comparison of the two half-maps will yield a resolution of 3.6 Å (the resolution for 50,000 particles or helical segments). For a map that has 40,000 particles or segments, a resolution of 3.8 Å for the full map will show a resolution of 5.3 Å when comparing two half-maps, significantly worse than the resolution of the full map that we are trying to estimate.
Figure 3.
Fourier Shell Correlation (FSC) can be an inaccurate measurement of resolution in cryo-EM maps. (A–D) From [53], top views of the pyruvate dehydrogenase E2 catalytic domains, contoured to the same sigma levels, reconstructed using the best 963 (A), 395 (B) or 139 (C) particles. Although features of the map become better resolved as fewer, better particles are used, the reported FSC resolution (D) decreases. Colors of the FSC plots in (D) correspond to map colors in (A–C). (E) An idealized curve for resolution as a function of sample size N, which tends to follow a log-linear relationship [54]. (F) An actual FSC curve between two independent half-map reconstructions of a helical polymer is shown in black (the scale of the abscissa is arbitrary). In red the same FSC is computed, but after adding an artifact to the reconstruction, a Gaussian cylinder of density along the axis with a standard deviation in the radius of the cylinder of 2 pixels. The FSC is improved (it is shifted to the right) and at high resolution the black and red curves are completely superimposed. In the blue curve the Gaussian cylinder is even narrower, with a standard deviation in radius of 1.5 pixels, which greatly improves the overall FSC and still matches the “true” FSC (the black curve) at the highest resolution.
A further problem with the FSC is that it is quite susceptible to artifacts and manipulation. We show in Figure 3F how FSC improves when one introduces spurious density into a map. Of course, this artifact would be apparent to any observer who actually looked carefully at the two maps being compared, and not just at the outer surface where this artifact would be invisible. But it illustrates how the FSC is not immune to such artifacts, and in the present example the greater the artifact, the better the resolution.
Given these problems with the FSC, how can one assess the actual resolution of a cryo-EM reconstruction? For maps at high enough resolution, a reliable starting point is to take advantage of our extensive prior knowledge of protein and nucleic acid stereochemistry and compute a FSC curve between the observed map and the atomic model built into the density. This approach of computing the FSC between the map and the model overcomes many, although not all of the challenges inherent to FSC measurements. For maps with relatively uniform B-factors across the entire polypeptide, the FSC plot of model vs. map is an excellent measure of map resolution and quality. However, this measure has other problems that are worth keeping in mind. An increasing number of cryo-EM structures are now being reported from dynamic protein assemblies, where there is variability in the amount of flexibility in different regions of the polypeptide. As a result, the cryo-EM density map in such cases has a lower effective resolution in the more flexible regions, and a higher effective resolution in the more ordered regions. While there are tools such as RESMAP [55] and blocres [56] that can provide a way to describe relative changes in resolution, they are not necessarily reliable as absolute measures of resolution. An additional challenge is that from the same data, the use of different image processing packages for 3D reconstruction, use of different B-factors, and the use of different subsets of exposure (recorded using the movie mode of direct detectors) can lead to small variations in map quality, and further compound the challenges involved in FSC measurements.
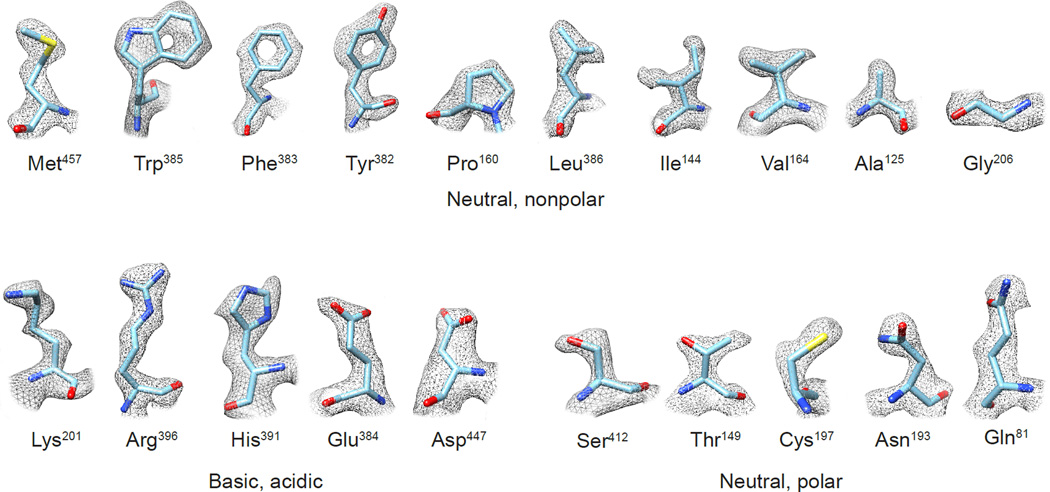
Ultimately, there can be no substitute for map validation by careful visual inspection. The highest resolution single particle cryo-EM reconstruction reported so far is that of the 334 kDa glutamate dehydrogenase [48], a protein complex with an ordered core and flexible outer domains. In the ordered core, the quality of this 1.8 Å resolution map is good enough to visualize every type of amino acid at a level of detail (Figure 4) that is comparable to that observed in maps obtained from X-ray crystallography [57]. The map quality is also adequate to visualize densities for hundreds of water molecules. In an earlier report of a structure at 2.2 Å resolution of β-galactosidase [58], the map quality was also adequate to visualize density for a single water molecule between the bound inhibitor and protein. The ability to obtain density maps at this kind of resolution will be critical for truly enabling the application of cryo-EM to drug discovery applications. Relying on single numbers for resolution will not be adequate as already well-appreciated in the field [55,56], and visual inspection of map quality in key regions will be needed to establish the validity of the conclusions about local structure and conformation. We believe that once achieving cryo-EM map resolutions in the 2 Å to 3 Å range becomes routine, the single number assigned to map resolution will not be very important, but the ability to unambiguously discern geometry of individual residues and the conformation of bound ligands will emerge as the new “platinum standard” for map quality.
Figure 4.
Experimental densities for selected examples of the 20 amino acid types from the 1.8 Å resolution cryo-EM structure of glutamate dehydrogenase (EMDB-8194; PDB-5k12) [48]. Features such as holes in aromatic rings, as well as the “zigzag” structure of extended Arg and Lys sidechains, are visible in the density maps.
Highlights.
There is rapid growth in number of structures determined using cryo-EM methods
Single particle cryo-EM can now achieve resolutions better than 2 Å
Drug discovery applications are emerging and look promising
Improved measures are needed for assessing map resolution for dynamic complexes
Acknowledgments
This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (NIH) and by NIH GM035269 (to E.H.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egelman EH. The Current Revolution in Cryo-EM. Biophys J. 2016;110:1008–1012. doi: 10.1016/j.bpj.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. This paper presents the use of direct detectors and motion correction to achieve near-atomic resolutions of the Thermoplasma acidophilum 20S proteosome, outlining methods that helped drive the subsequent revolution advances in cryo-EM.
- 4.Faruqi AR, Cattermole DM, Henderson R, Mikulec B, Raeburn C. Evaluation of a hybrid pixel detector for electron microscopy. Ultramicroscopy. 2003;94:263–276. doi: 10.1016/s0304-3991(02)00336-4. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 7.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 8.Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. The binding conformation of Taxol in beta-tubulin: a model based on electron crystallographic density. Proc Natl Acad Sci U S A. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryoelectron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Sun S, Xiang Y, Wong J, Klose T, Raoult D, Rossmann MG. Structure of Sputnik, a virophage, at 3.5-A resolution. Proc Natl Acad Sci U S A. 2012;109:18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. The structure of the Zika virus, obtained by cryo-EM, will be a powerful foundation for developing drugs and vaccines against Zika, just as structural studies of HIV have played an important role in developing drugs against that virus.
- 13.Allegretti M, Mills DJ, McMullan G, Kuhlbrandt W, Vonck J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. Elife. 2014;3:e01963. doi: 10.7554/eLife.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 17.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. This paper shows how lipid nanodiscs can be used to solublize integral membrane proteins for cryo-EM, at the same time avoiding the potential artifacts associated with detergents and amphipols.
- 19.Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, et al. Three-dimensional structure of human gamma-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SH, Shi Y. An atomic structure of human gamma-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyerson JR, Chittori S, Merk A, Rao P, Han TH, Serpe M, Mayer ML, Subramaniam S. Structural basis of kainate subtype glutamate receptor desensitization. Nature. 2016 doi: 10.1038/nature19352. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herguedas B, Garcia-Nafria J, Cais O, Fernandez-Leiro R, Krieger J, Ho H, Greger IH. Structure and organization of heteromeric AMPA-type glutamate receptors. Science. 2016;352:aad3873. doi: 10.1126/science.aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S, Stein RA, Yoshioka C, Lee CH, Goehring A, McHaourab HS, Gouaux E. Mechanism of NMDA Receptor Inhibition and Activation. Cell. 2016;165:704–714. doi: 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Z, Bai XC, Yan C, Wu J, Li Z, Xie T, Peng W, Yin CC, Li X, Scheres SH, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, Marks AR. Structure of a mammalian ryanodine receptor. Nature. 2015;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthies D, Dalmas O, Borgnia MJ, Dominik PK, Merk A, Rao P, Reddy BG, Islam S, Bartesaghi A, Perozo E, et al. Cryo-EM Structures of the Magnesium Channel CorA Reveal Symmetry Break upon Gating. Cell. 2016;164:747–756. doi: 10.1016/j.cell.2015.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife. 2014;3:e03080. doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 37.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 38.Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife. 2015;4:e06380. doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.da Fonseca PC, Morris EP. Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat Commun. 2015;6:7573. doi: 10.1038/ncomms8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H, O'Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PC, Bogyo M. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530:233–236. doi: 10.1038/nature16936. Being able to selectively target Plasmodium proteasomes and not human ones will be an important step in developing anti-malarial drugs.
- 41.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls AC, Tortorici MA, Bosch BJ, Frenz B, Rottier PJ, DiMaio F, Rey FA, Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ecken JV, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hospenthal MK, Redzej A, Dodson K, Ukleja M, Frenz B, Rodrigues C, Hultgren SJ, DiMaio F, Egelman EH, Waksman G. Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling. Cell. 2016;164:269–278. doi: 10.1016/j.cell.2015.11.049. Pili from uropathogenic E. coli allow bacteria to maintain their attachment to the urinary tract epithelial lining despite large shear forces, and this paper provides the structural explanation for this property in atomic detail.
- 47. DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348:914–917. doi: 10.1126/science.aaa4181. The helical virus described in this paper may use a similar mechanism as bacterial spores to protect DNA against the most aggressive environments.
- 48. Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JL, et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. This paper reports crossing two important perceived barriers in the cryo-EM field by reporting a structure at better than 2 Å resolution and another of a protein smaller than 100 kDa.
- 49. Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. This study of the cancer target p97 shows that cryoEM analysis can define, at near atomic resolutions, how small molecule regulators and drug candidate molecules shift proteins between several conformational states, information that can inform drug design and reveal potential mechanisms of action.
- 50.Harauz G, Vanheel M. Exact Filters for General Geometry 3-Dimensional Reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 51.Egelman EH. Ambiguities in helical reconstruction. Elife. 2014;3:e04969. doi: 10.7554/eLife.04969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang R, Nogales E. A new protocol to accurately determine microtubule lattice seam location. J Struct Biol. 2015;192:245–254. doi: 10.1016/j.jsb.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borgnia MJ, Shi D, Zhang P, Milne JL. Visualization of alpha-helical features in a density map constructed using 9 molecular images of the 1.8 MDa icosahedral core of pyruvate dehydrogenase. J Struct Biol. 2004;147:136–145. doi: 10.1016/j.jsb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 54.Stagg SM, Noble AJ, Spilman M, Chapman MS. ResLog plots as an empirical metric of the quality of cryo-EM reconstructions. J Struct Biol. 2014;185:418–426. doi: 10.1016/j.jsb.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesk AM. Introduction to protein science: architecture, function, and genomics. 3rd. Oxford: Oxford University Press; 2016. [Google Scholar]
- 58. Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, Subramaniam S. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. The resolution levels attained in this paper enabled visualization of a water molecule associated with an inhibitor compound present in the enzyme active site, showing that structural information obtained by single particle cryo-EM can be used to direct drug discovery without crystallization.