Abstract
Individuals with Down syndrome (DS, trisomy 21) exhibit a pro-oxidative cellular environment as well as mitochondrial dysfunction. Increased oxidative stress may damage the mitochondrial DNA (mtDNA). The coexistence of mtDNA variants in a cell or tissue (i.e., heteroplasmy) may contribute to mitochondrial dysfunction. Given the evidence on mitochondrial dysfunction and the relatively high incidence of multi-organic disorders associated with DS, we hypothesized that cardiac tissue from subjects with DS may exhibit higher frequencies of mtDNA variants in comparison to cardiac tissue from donors without DS. This study documents the analysis of mtDNA variants in heart tissue samples from donors with (n = 12) and without DS (n = 33) using massively parallel sequencing. Contrary to the original hypothesis, the study’s findings suggest that the cardiac mitochondrial genomes from individuals with and without DS exhibit many similarities in terms of a) total number of mtDNA variants per sample, b) the frequency of mtDNA variants, c) the type of mtDNA variants, and d) the patterns of distribution of mtDNA variants. In both groups of samples, the mtDNA control region showed significantly more heteroplasmic variants in comparison to the number of variants in protein- and RNA-coding genes (p < 1.00 × 10−4, ANOVA).
Keywords: Down syndrome, Trisomy 21, Heteroplasmy, Cardiac, Mitochondrial Variants, Massively Parallel Sequencing
INTRODUCTION
Mitochondria are organelles critical for various cellular functions including the synthesis of ATP by oxidative phosphorylation, maintenance of cellular calcium homeostasis, and detoxification of reactive oxygen species generated from cellular respiration (Nicholls, 2002). Each mitochondrion contains multiple copies of mitochondrial DNA (mtDNA), and there are hundreds to thousands of mtDNA copies per cell (Ye, et al., 2014). Human mtDNA is a double stranded circle of 16569 base pairs. The mitochondrial genome contains a non-coding control region (CR), 13 genes encoding for proteins of the oxidative phosphorylation system, 22 tRNA genes, and 2 rRNAs genes. Damage to mtDNA can result in mitochondrial dysfunction. Mitochondrial dysfunction contributes to a number of metabolic and degenerative diseases, and is also involved in cancer and ageing (Wallace, 2005). Metabolically active tissues such as heart and brain are especially susceptible to the bioenergetic deficits resulting from mitochondrial dysfunction (Wallace and Chalkia, 2013).
Individuals with Down syndrome (DS, trisomy 21) exhibit mitochondrial dysfunction (Arbuzova, et al., 2002; Coskun and Busciglio, 2012; Infantino, et al., 2011; Ogawa, et al., 2002; Roat, et al., 2007). Mitochondrial dysfunction has been associated with the etiology of early-onset dementia in patients with DS. Higher frequency of mutations in mtDNA, specifically in the CR, and decreased mtDNA copy numbers have been observed in brains of patients with DS and dementia in comparison to age-matched controls (Coskun, et al., 2010). The exact molecular events linking variation in mtDNA sequence to disease are not yet fully characterized (Dai, et al., 2012). Some individuals with DS display signs and symptoms of premature ageing in specific body systems including the endocrine, musculoskeletal, immunological, and neurological systems. The cardiovascular system seems to be less affected by ageing in the DS setting; for example, the incidence of hypertension and ischemic cardiac disease is relatively low in adults with DS (Zigman, 2013). However, patients with acute leukemias and DS may be more susceptible to the cardiotoxic effects of anticancer anthracycline drugs (Hefti and Blanco, 2016). A recent study suggests that in myocardial tissue from individuals with DS, the expression of certain anthracycline metabolizing enzymes (i.e., AKR7A2) and mtDNA content are important variables in determining the cardiac synthesis of toxic drug metabolites (Hoefer, et al., 2016). Increases in the frequency of specific mtDNA variants (i.e., heteroplasmy) in cardiac tissue may contribute to the development of cardiac dysfunction that does occur in some cases with DS (Balli, et al., 2016; Vetrano, et al., 2016). Recently, we documented a non-significant 30% decrease in the levels of cardiac mtDNA and a non-significant 30% increase in the frequency of the “common” mtDNA4977 deletion in heart tissue samples from donors with DS (n = 11) in comparison to samples from donors without DS (n = 31). We noted that the trend towards decreased mtDNA content in DS hearts seemed to be independent from the donor’s age (Hefti, et al., 2016). Thus, and given both the evidence on mitochondrial dysfunction and the relatively high incidence of multi-organic disorders associated with DS, we hypothesized that cardiac tissue from subjects with DS may exhibit higher numbers and frequencies of mtDNA variants in comparison to cardiac tissue from donors without DS (Glasson, et al., 2014; Head, et al., 2015; Hwang and Jea, 2013). With the exception of data on mtDNA heteroplasmy derived from brain tissue from subjects with DS and a report on tissular heteroplasmy from Neau et al, there is a noticeable paucity of reports describing the extent of mtDNA heteroplasmy in most tissues (Coskun and Busciglio, 2012; Naue, et al., 2015). This is especially true with regards to cardiac tissue from subjects with DS.
Massively parallel sequencing is a robust, high throughput technique that allows the accurate identification of heteroplasmic mtDNA variants. This method was pioneered by Tang and colleagues for identifying mitochondrial heteroplasmy in 2010. The analysis of mtDNA variants by massively parallel sequencing will contribute to expand our understanding of the molecular bases underlying mitochondrial dysfunction in various clinical settings, including DS. In this study, massively parallel sequencing was applied to the analysis and annotation of heteroplasmic mtDNA variants in a collection of heart tissue samples from donors with and without DS. These data provide the first insights into the extent of heteroplasmic mtDNA variation in the cardiac tissue of individuals with DS.
MATERIALS AND METHODS
Heart samples
The Institutional Review Board at the University at Buffalo approved this research. Heart samples from donors with (n = 12) and without DS (n = 33) were procured from The National Disease Research Interchange, The Cooperative Human Tissue Network, and the National Institute of Child Health and Human Development Brain and Tissue Bank. The postmortem to tissue recovery interval was ≤10 h. Samples (0.50 − 20 g, myocardium, left ventricle only) were frozen immediately after recovery and stored in liquid nitrogen until further processing. The main demographics from donors with and without DS are summarized in Table S1. Down syndrome status (yes/no) and relevant diagnoses (when available) were obtained from anonymous medical histories. Heart samples were processed following standardized procedures to isolate total cardiac DNA as described (Hefti, et al., 2016). Age matched groups (n = 10 for each group) were utilized for some comparisons to minimize the potential confounding effect of age on mtDNA variation (mean ageNDS = 46.50 years, SD = 22.40 versus mean ageDS = 46.6, SD = 21.83, p = 0.99, Student’s t-test) (Naue, et al., 2015; Sevini, et al., 2014).
Cardiac mtDNA amplification
The entire mitochondrial genome was amplified in two fragments using high-fidelity long-distance PCR (LD-PCR) with Takara LA (Clontech, California USA) as described by Tang et al. (Tang and Huang, 2010). Cardiac mtDNA fragment 1 (9289 bp in length) was amplified with the following primers: forward 5’-AACCAAACCCCAAAGA CACC-3’, reverse 5’-GCCAATAATGACGTGAAAGTCC-3’. Cardiac mtDNA fragment 2 (7626 bp in length) was amplified with the following primers: forward 5’-TCCCACTCCT AAACACATCC-3’, reverse 5’-TTTATGGGGTGATGTGAGCC-3’. The LD-PCR amplification conditions were as follows: 25 cycles at 94°C for 25 seconds and 68°C for 16 minutes. PCR amplification products were analyzed by electrophoresis on agarose gels and purified with suitable kits (Bio Basic, Ontario CA). DNA concentrations were determined with the PicoGreen Assay (Invitrogen, Massachusetts USA).
Massively parallel sequencing
cDNA libraries were prepared with Nextera sample preparation kits (Illumina, California USA). The cDNA libraries were quantified using a PicoGreen assay (Invitrogen) and Kapa qPCR library quantification kit (Kapa Biosystems, Massachusetts USA). Size and quality of cDNA libraries were confirmed using an Agilent Bioanalyzer 2100 (California, USA) DNA high sensitivity chip. The cDNA libraries were then pooled based on qPCR values. cDNA products were sequenced in rapid mode on an Illumina HiSeq 2500 DNA sequencing system at the University at Buffalo Genomics and Bioinformatics Core Facility (Buffalo, NY). The sequencing data were uploaded to GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The accession number for the data is SRP074312.
Sequencing output analysis
Paired sequence reads were demultiplexed using Illumina’s bcl2fastq version 1.8.4. Reads were then mapped to the mitochondrial genome NC_012920.1 (version GI: 251831106) utilizing the CLC Genomics Workbench version 8.0.1 alignment tool (Venlo Netherlands). After mapping, reads underwent variant calling using CLC Genomics Workbench (v8.0.1) requiring a minimum coverage of 1000X and a minimum frequency of 1.00 × 10−6. An average of 7.50 million reads were mapped for each sample. Imported reference data from dbSNP was used to identify mitochondrial variants, which were compiled into Microsoft Excel spreadsheets for further analysis. Data on mtDNA variants were submitted to the MITOMAP database (www.mitomap.org). Diagnostic single nucleotide polymorphisms (SNPs) available on the MITOMAP database were utilized to identify mtDNA haplogroups (Table S1). Distributions and frequencies of heteroplasmic mtDNA variants were mapped on a circular plot using the Circlize package in R.
Data analysis
The mean number of heteroplasmic mtDNA variants present in a locus was obtained from the average number of individual heteroplasmic variants present in the specified location from all samples within the groups being considered. The frequency represents the percentage of mitochondrial genomes that contain a specific variant. Mean frequency values present in a locus were obtained from the average frequency of all heteroplasmic mtDNA variants present in the specified location from all samples within the group. Frequency values were used to classify mtDNA variants after the application of a conservative 2% error rate (He, et al., 2010). mtDNA variants exhibiting frequency values between the 2% − 98% range were considered heteroplasmic. The D'Agostino and Pearson omnibus tests were used to ascertain the normality of datasets. Two-tailed Student’s t-test was used to compare the means of normally distributed datasets, and the Mann-Whitney U test was used to compare the means of non-normally distributed datasets. Correlation analyses were performed with Pearson’s product-moment test. Analysis of variance (ANOVA) with post-hoc Turkey’s test was used to compare mean number of variants and mean frequency. In all cases, differences between means were considered to be significant at p < 0.050.
RESULTS
Cardiac mtDNA variants in samples from donors with and without DS
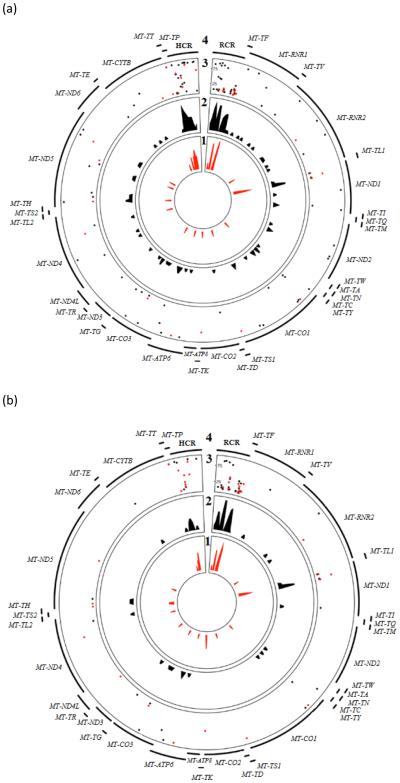
Fig. 1a shows the distribution and frequency of heteroplasmic mtDNA variants in heart samples from donors with (red, n=12) and without (black, n=33) DS. The CR and the MT-ND1 gene showed relative increases in the density of mtDNA variants in comparisons to other loci in the mitochondrial genome (Fig. 1a). The frequency values associated with the heteroplasmic variants in the CR and the MT-ND1 gene were highly variable and ranged from 2.03% to 95.96% for MT-ND1 and 2.01% to 97.80% for the CR.
Fig. 1.
(A) Heteroplasmic mtDNA variants in cardiac tissue from donors with and without DS. Rings 1 and 2 show the density of mtDNA variants in tissue samples from donors with (red, n = 12) and without DS (black, n = 33). Ring 3 shows the frequency of the corresponding variants. The closer the circle is to the outer region of ring 3, the higher the frequency of the variant. Ring 4 indicates the genes or regions of the mitochondrial genome. (B) Map of heteroplasmic mtDNA variant density and frequency in heart samples from age-matched groups of donors with and without DS (n = 10 for both groups).
There were 415 heteroplasmic mtDNA variants in the group of heart samples from subjects without DS (12.57 heteroplasmic variants per sample). According to the NCBI dbSNP database, 89 of the variants (21.44%) have been previously identified; however, these variants have not been yet associated to a particular disease state. Of the remaining mtDNA variants, 323 (77.83%) have not yet been previously identified, and 3 (0.72%) were identified as pathogenic or possibly pathogenic by NCBI dbSNP and MITOMAP databases. Specifically, the single nucleotide variants (SNV) rs200145866:T>C (Ile165Thr), rs1599988:T>C (Tyr304His), and rs28357980:A>G (Asn150Asp) have been associated with the pathogenesis of Leber’s optic neuropathy (Howell, et al., 1995; Johns and Berman, 1991; Yu-Wai-Man and Chinnery, 1993). The frequency values of these potentially pathogenic mtDNA variants in cardiac tissue were 97.25%, 95.97% and 96.71%. There were 73 non-synonymous SNVs in the group of samples from donors without DS (mean = 2.21, SD = 1.41 non-synonymous variants per sample).
Fig. 1a compiles the distribution and frequency values of mtDNA variants in 12 cardiac mitochondrial genomes from donors with DS. The CR of mitochondrial genomes from individuals with DS showed a relative increase in the density of variants in comparison to the other mtDNA regions (Fig. 1a). There was an area of increased mtDNA variant density in MT-ND1, similar to the NDS group. In general, heteroplasmic mtDNA variants were more prevalent in the CR and protein coding regions.
There were 127 mtDNA variants in the group of heart tissue samples from donors with DS (10.6 variants per sample). Twenty-nine variants (22.83%) were previously identified and are present in the NCBI dbSNP database. These variants have not yet been clinically associated with any disease state. Ninety-eight mtDNA variants (77.16%) are currently unidentified and are not present in the NCBI dbSNP database. There were no pathogenic or potentially pathogenic mtDNA variants in the group of cardiac mitochondrial genomes from individuals with DS. There were 25 non-synonymous heteroplasmic mtDNA variants, with an average of 2.08 (SD = 2.28) non-synonymous variants per sample in donors with DS.
Cardiac mtDNA variants in samples from age-matched donors with and without DS
Aging affects mtDNA heteroplasmy in tissues (Naue, et al., 2015; Sevini, et al., 2014). Thus, we compared cardiac heteroplasmic mtDNA variants in a sub-set of age-matched samples to control for this potential confounder. Fig. 1b shows the distribution and frequency of heteroplasmic mtDNA variants in age-matched heart samples from donors with (red) and without (black) DS. The distribution of variants and corresponding frequency values across the mitochondrial genomes from donors with and without DS were similar (Fig 1a-b).
Overall, the number of mtDNA variants per gene were similar between age-matched samples from donors with and without DS (Table S2, Table 1). In age-matched samples from donors with DS, there was a trend towards a decrease in the mean frequency (freq%) of mtDNA variants in the hypervariable control region (HCR) versus the regulatory control region (RCR) (mean freq%DS-HCR = 11.54, SD = 18.62 versus mean freq%DS-RCR = 30.45, SD = 7.32, p = 0.070, Mann Whitney U test). This trend was not apparent in the group of age-matched samples from donors without DS (mean freq%Non-DS-HCR = 35.97, SD = 42.94 versus mean freq%Non-DS-RCR = 30.33, SD = 7.28, p = 0.54, Mann Whitney U test). Samples from donors with and without DS also showed similar distributions of mtDNA variants along the binding motifs for essential mitochondrial transcription factors (Table S3).
Table 1.
Comparison of number and frequency of heteroplasmic mtDNA variants in control region, protein coding region, and RNA coding region in heart samples from age-matched donors with and without DS (n = 10 samples for both groups).
| Down Syndrome (Mean ± SD) |
Non Down Syndrome (Mean ± SD) |
p-valuea | ||
|---|---|---|---|---|
| Control Region Variants | Mean variant number | 7.20 ± 3.85 | 8.60 ± 2.46 | 0.40 |
|
| ||||
| Mean variant frequency (freq%) | 31.06 ± 7.23 | 33.62 ± 10.38 | 0.54 | |
|
| ||||
| Protein Coding Variants | Mean variant number | 2.30 ± 1.55 | 3.20 ± 1.40 | 0.11 |
|
| ||||
| Mean variant frequency (freq%) | 25.68 ± 30.62 | 18.61 ± 17.61 | 0.68 | |
|
| ||||
| RNA Coding Variants | Mean variant number | 0.80 ± 0.92 | 0.80 ± 0.63 | 0.77 |
|
| ||||
| Mean variant frequency (freq%) | 16.17 ± 30.01 | 16.95 ± 30.91 | 0.85 | |
Student’s t-test used for comparisons of normally distributed data, Mann Whitney U test used for non-normally distributed data.
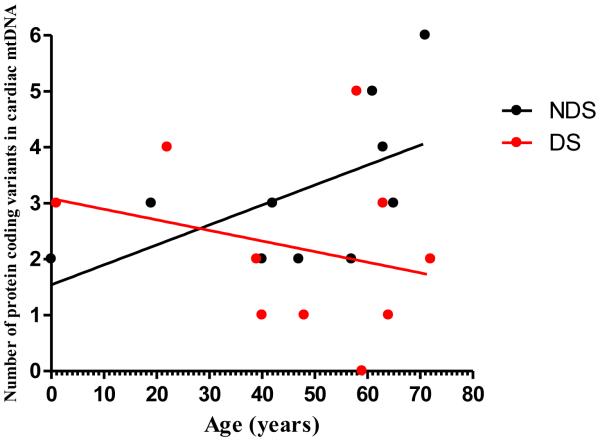
There was a trend towards a positive correlation between the total number of protein coding mtDNA variants and age in the group of age-matched samples from donors without DS (r = 0.57, p = 0.080, Pearson’s Product-Moment correlation). The trend was not evident in the group of age-matched samples from donors with DS (r = −0.27, p = 0.46, Pearson’s Product-Moment correlation. Fig. 2). There were no significant correlations between either the number or frequency of heteroplasmic mtDNA variants and age in both age-matched groups (Table 2).
Fig. 2.
Linear regression analysis of the number of protein coding variants in mtDNA versus age in heart samples from age-matched donors with and without DS (n = 10 samples for both groups. Non-DS = 0.57, p = 0.080 versus rDS = −0.27, p = 0.46, Pearson’s Product-Moment correlation). Each symbol represents the total number of protein coding variants per sample.
Table 2.
Correlation analysis of heteroplasmic mtDNA variant number and frequency with age in age-matched donors with and without DS (n = 10 samples for both groups)a.
| Down syndrome | Non Down syndrome | ||||
|---|---|---|---|---|---|
|
| |||||
| r | p-value | r | p-value | ||
| Control Region Variants vs. age | Mean variant number | 0.44 | 0.20 | 0.25 | 0.48 |
|
| |||||
| Mean variant frequency (freq%) | −0.15 | 0.68 | −0.016 | 0.96 | |
|
| |||||
| Protein Coding Variants vs. age | Mean variant number | −0.27 | 0.46 | 0.57 | 0.080 |
|
| |||||
| Mean variant frequency (freq%) | 0.10 | 0.79 | 0.16 | 0.66 | |
|
| |||||
| RNA Coding Variants vs. age | Mean variant number | 0.32 | 0.37 | 0.042 | 0.48 |
|
| |||||
| Mean variant frequency (freq%) | −0.77 | 0.072 | 0.20 | 0.70 | |
Pearson’s Product-Moment correlation analysis
There were no significant differences in RNA or protein coding non-synonymous, synonymous, or single nucleotide variant number or frequency between the age-matched groups (Table 3). There were no statistically significant differences in the number of RNA or protein coding insertions and deletions (INDELs) between the age-matched groups (Table 3). The coding INDELs observed in both groups ranged from 1 to 9 bp in length.
Table 3.
Comparison of SNVs, coding INDELS, and non-synonymous heteroplasmic mtDNA variants in heart samples from age-matched donors with and without DS (n = 10 samples for both groups).
| Down Syndrome (Mean ± SD) |
Non Down Syndrome (Mean ± SD) |
p-valuea | ||
|---|---|---|---|---|
|
Single Nucleotide
Variants (SNVs)* |
Mean variant number | 6.00 ± 2.94 | 7.60 ± 3.50 | 0.25 |
|
| ||||
| Mean variant frequency (freq%) | 24.50 ± 17.90 | 32.54 ± 21.33 | 0.32 | |
|
| ||||
|
Coding Insertions and
Deletions (INDELs) |
Mean variant number | 0.70 ± 1.34 | 1.20 ± 1.03 | 0.17 |
|
| ||||
| Mean variant frequency (freq%) | 6.19 ± 15.11 | 15.74 ± 21.56 | 0.12 | |
|
| ||||
| Non-synonymous SNVs | Mean variant number | 1.30 ± 1.25 | 2.10 ± 1.37 | 0.18 |
|
| ||||
| Mean variant frequency (freq%) | 8.37 ± 11.74 | 12.92 ± 15.44 | 0.40 | |
Total SNV in coding and non-coding regions.
Student’s t test used for comparisons of normally distributed data, Mann-Whitney U test used for non-normally distributed data.
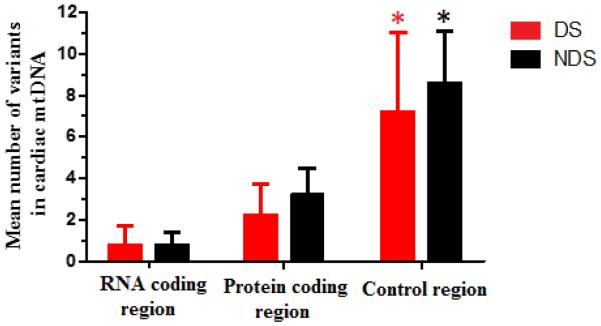
The mean number of mtDNA variants in the CR, protein and RNA coding regions were significantly different in both groups of age-matched samples (ANOVANon-DS and ANOVADS p < 1.00 × 10−4. Fig. 3). The number of heteroplasmic mtDNA variants located in RNA coding regions was lower than the number of variants in protein coding and CR, respectively (Fig. 3). In terms of protein coding mtDNA variants, both groups of samples showed a preponderance of non-synonymous over synonymous variants (non-DS: 84.38% and DS: 78.26%). The number of non-synonymous variants per sample was similar between both groups (Table 3).
Fig. 3.
Mean number of heteroplasmic mtDNA variants (protein coding, RNA coding, and control regions) in heart samples from age-matched donors with and without DS (n = 10 samples for both groups). Each bar represents the mean ± SD. * p < 0.050 (one-way ANOVA with post hoc Turkey test).
DISCUSSION
Massively parallel sequencing allows the examination of tissular mtDNA variants with a very high depth of coverage (>1000X). With the exception of data on mtDNA variants derived from brain tissue from subjects with DS, there is a noticeable paucity of reports describing the extent of mtDNA heteroplasmy in most tissues, including cardiac tissue, from subjects with DS. This is noteworthy because several lines of evidence suggest that widespread mitochondrial dysfunction contributes to many of the pathophysiological alterations associated with DS (Coskun and Busciglio, 2012; Pagano and Castello, 2012). Despite these pathophysiological alterations, the current life expectancy for individuals with DS has reached the sixth decade of life with some individuals (~10%) living up to 70 years of age, this increase is attributed to improved medical care for those with DS (Brown, et al., 2001; Head, et al., 2015). In this study, massively parallel sequencing was applied to the analysis of mtDNA heteroplasmy in a collection of heart tissue samples representative of the current life span for persons with DS. For comparison, mtDNA heteroplasmy was analyzed in a group of samples from subjects without DS, which included an age-matched sub-group.
Our initial hypothesis postulated that the cardiac mitochondrial genomes from subjects with and without DS would show differences in the number and/or distribution of mtDNA variants. However, the study’s main findings suggest that the cardiac mitochondrial genomes from individuals with and without DS exhibit many similarities in terms of a) total number of mtDNA variants per sample, b) the frequency of mtDNA variants, c) the type of mtDNA variants, and d) the patterns of distribution of mtDNA variants (Fig. 1 and Table S2). At first glance, these findings are surprising given that DS status has been associated with increased intracellular generation of damaging reactive oxygen species (ROS) combined with less effective mtDNA repair mechanisms (Druzhyna, et al., 1998; Roat, et al., 2007). However, studies that considered non-cardiac cells (e.g., DS astrocytes and lymphocytes) showed that trisomic samples displayed an “adaptive response” to the chronic increase in oxidative stress. This “adaptive response” results in an overall decrease in mitochondrial function in order to minimize the production of ROS and reduce further damage to the mtDNA (Helguera, et al., 2013; Perluigi and Butterfield, 2012). It is possible that this “adaptive response” is present in the myocardial tissue of individuals with DS through the entire lifespan. The result would be the preservation of the cardiac mitochondrial genome, as manifested by levels of mtDNA variation comparable to the non-DS setting, at the expense of relative decreases in mitochondrial function. Further comparative studies in cardiac tissue samples and/or cellular models (e.g., induced pluripotent stem cell derived cardiomyocytes) would be needed to determine whether the control of cardiac mtDNA heteroplasmy in the trisomic setting is associated with compensatory changes in mitochondrial function.
This study documented relatively high numbers of mtDNA variants in cardiac tissue (Tables 1-2). Cardiac tissue is mostly composed of post-mitotic cells, and the majority of the cardiomyocytes in an individual will not be replaced during a normal lifespan (Bergmann, et al., 2009). Thus, the low turnover of cardiac tissue may result in the accumulation and propagation of certain heteroplasmic mtDNA variants (Taylor and Turnbull, 2005; Tuppen, et al., 2010). Heart tissue samples from donors with and without DS showed a wide range of heteroplasmy (i.e., >2% – <98%), and mtDNA variants with frequency values near the upper limit may be in fact homoplasmic mtDNA variants. In general, there was good agreement between the donor’s ethnicity as listed in the available clinical reports (i.e., “white” or European American and “black” or African American) and corresponding mtDNA haplogroups from Africa and Europe (Table S1). However, in the absence of paired samples of maternal DNA it was not possible to: a) estimate the fraction of haplotype associated variants, and b) estimate the fractions of germline and “de novo” mtDNA variants (He, et al., 2010).
In both groups of samples, the variants with the highest degree of heteroplasmy tended to concentrate in the CR and the MT-ND1 gene. The CR is critical for transcription and regulation of the mitochondrial genome. The CR is the most variable region in the mitochondrial genome. Naue et al. examined mtDNA variants in the CR in nine different tissues from 100 individuals, and found similar numbers of mitochondrial point heteroplasmies in heart and brain, while skeletal muscle and liver exhibited the highest number of heteroplasmies (Naue, et al., 2015).Variation in the CR may affect mtDNA copy number (i.e., mtDNA content) (Coskun, et al., 2010). In a previous study involving this set of heart samples, we documented a non-significant 30% (p = 0.65) decrease in mtDNA content in samples with DS in comparison to samples without DS (Hefti, et al., 2016). Coskun et al. examined the frequency of mtDNA CR mutations in samples from cerebral frontal cortices from age-matched subjects with and without DS, and found that mean mutation frequencies were similar between both groups (Coskun, et al., 2010). In agreement, heart tissue samples from subjects with and without DS showed similar levels of heteroplasmy when considering the entire CR as well as distinct regulatory elements within the CR that are involved in the replication of mtDNA (Fig. 1a-b and Table S3).
Age-matched groups of cardiac samples from donors with and without DS displayed similar distribution and mean frequency values of heteroplasmic mtDNA variants in RNA and protein coding genes (Fig. 1b). Both groups displayed reduced numbers of RNA variants, as well as reduced numbers of protein coding variants in comparison to the number of variants in the CR (Fig. 3). The majority of the protein coding variants in both groups were non-synonymous; therefore, potentially damaging. In general, increased numbers and frequencies of mtDNA variants in protein coding and tRNA coding regions are more likely to be pathogenic compared to other mtDNA regions (Ye, et al., 2014). This pattern of regional variation in cardiac mtDNA is consistent with the recent conclusions made by Ye and colleagues after analyzing massively parallel sequencing data from the 1000 Genomes Project. The authors proposed that a purifying selection process may reduce the prevalence and accumulation of potentially pathogenic mtDNA variants in specific mitochondrial loci such as tRNAs and protein coding genes to avoid the propagation of mitochondrial dysfunction (Ye, et al., 2014).
There are limitations in the present study. The sample size was limited due to the scarcity of good quality cardiac tissue samples from donors with DS; this common problem impacts research on many of the issues concerning the pathobiology of DS (Oster-Granite, et al., 2011). Thus, it was not possible to detect relatively subtle differences in mtDNA frequencies (Table S4). The lack of matched maternal tissue samples precluded determining whether the cardiac mtDNA variants were either inherited or acquired (i.e., somatic) mtDNA variants. Thus, the observed frequency values may not fully reflect the impact of DS status on somatic mtDNA variants. Furthermore, the reliance on cadaveric tissue samples from donors with various clinical histories makes it almost impossible to control for potential clinical confounders. Furthermore, the resulting cross sectional nature of the comparisons should be considered when evaluating the potential impact of age and/or other putative co-variables on heteroplasmic mtDNA frequencies (Sevini, et al., 2014). Although massively parallel sequencing is a powerful method for the analysis of mtDNA variants in human tissues, it often cannot detect large deletions in mtDNA. For example, the “common” mtDNA 4977 bp deletion cannot be reliably detected by massively parallel sequencing because of its relatively low frequency in cardiac tissue (DS% frequency mtDNA4977deletion: 0.0086 ± 0.0166, non-DS% frequency mtDNA4977deletion: 0.0066 ± 0.0124; p= 0.514) (Hefti, et al., 2016; Tang and Huang, 2010).
The results from this study suggest similar distribution of heteroplasmic mtDNA variants in hearts from individuals with and without DS. The data presented in this study offer a glimpse into the patterns of heteroplasmic mtDNA variants in cardiac tissue from donors with DS. These data may advance our understanding of tissular mtDNA variation and its potential relation to the complex pathobiology of DS.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks to the University at Buffalo Genomics and Bioinformatics Core, New York State Center of Excellence in Bioinformatics and Life Sciences.
Grant Support:
-National Institute of General Medical Sciences (award GM073646)
-Eunice Kennedy Shriver National Institute of Child Health and Human Development (award HD076055)
-Mae Stone Goode Trust (award 767035)
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- Arbuzova S, Hutchin T, Cuckle H. Mitochondrial dysfunction and Down's syndrome. Bioessays. 2002;24(8):681–4. doi: 10.1002/bies.10138. [DOI] [PubMed] [Google Scholar]
- Balli S, Yucel IK, Kibar AE, Ece I, Dalkiran ES, Candan S. Assessment of cardiac function in absence of congenital and acquired heart disease in patients with Down syndrome. World J Pediatr. 2016 doi: 10.1007/s12519-016-0012-3. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R, Taylor J, Matthews B. Quality of life--ageing and Down syndrome. Downs Syndr Res Pract. 2001;6(3):111–6. doi: 10.3104/case-studies.101. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Busciglio J. Oxidative stress and mitochondrial dysfunction in Down's syndrome: relevance to aging and dementia. Curr Gerontol Geriatr Res. 20122012:383170. doi: 10.1155/2012/383170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. J Alzheimers Dis. 2010;20(Suppl 2):S293–310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110(8):1109–24. doi: 10.1161/CIRCRESAHA.111.246140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhyna N, Nair RG, LeDoux SP, Wilson GL. Defective repair of oxidative damage in mitochondrial DNA in Down's syndrome. Mutat Res. 1998;409(2):81–9. doi: 10.1016/s0921-8777(98)00042-1. [DOI] [PubMed] [Google Scholar]
- Glasson EJ, Dye DE, Bittles AH. The triple challenges associated with age-related comorbidities in Down syndrome. Journal of Intellectual Disability Research. 2014;58(4):393–398. doi: 10.1111/jir.12026. [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr., Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–4. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head E, Lott IT, Wilcock DM, Lemere CA. Aging in Down syndrome and the development of Alzheimer's disease neuropathology. Current Alzheimer Research. 2015;13(1):18–29. doi: 10.2174/1567205012666151020114607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti E, Blanco JG. Anthracycline-related cardiotoxicity in patients with acute myeloid leukemia and Down syndrome: a literature review. Cardiovasc Toxicol. 2016;16(1):5–13. doi: 10.1007/s12012-015-9307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti E, Quiñones-Lombraña A, Redzematovic A, Hui J, Blanco JG. Analysis of mtDNA, miR-155 and BACH1 expression in hearts from donors with and without Down syndrome. Mitochondrial DNA Part A. 2016;27(2):896–903. doi: 10.3109/19401736.2014.926477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera P, Seiglie J, Rodriguez J, Hanna M, Helguera G, Busciglio J. Adaptive downregulation of mitochondrial function in down syndrome. Cell Metab. 2013;17(1):132–40. doi: 10.1016/j.cmet.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefer CC, Blair RH, Blanco JG. Development of a CART model to predict the synthesis of cardiotoxic daunorubicinol in heart tissue samples from donors with and without Down syndrome. Journal of pharmaceutical sciences. 2016;105(6):2005–2008. doi: 10.1016/j.xphs.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Kubacka I, Halvorson S, Howell B, McCullough DA, Mackey D. Phylogenetic analysis of the mitochondrial genomes from Leber hereditary optic neuropathy pedigrees. Genetics. 1995;140(1):285–302. doi: 10.1093/genetics/140.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SW, Jea A. A Review of the Neurological and neurosurgical implications of Down syndrome in children. Clinical Pediatrics. 2013;52(9):845–856. doi: 10.1177/0009922813491311. [DOI] [PubMed] [Google Scholar]
- Infantino V, Castegna A, Iacobazzi F, Spera I, Scala I, Andria G, Iacobazzi V. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down's syndrome. Mol Genet Metab. 2011;102(3):378–82. doi: 10.1016/j.ymgme.2010.11.166. [DOI] [PubMed] [Google Scholar]
- Johns DR, Berman J. Alternative, simultaneous complex I mitochondrial DNA mutations in Leber's hereditary optic neuropathy. Biochemical and Biophysical Research Communications. 1991;174(3):1324–1330. doi: 10.1016/0006-291x(91)91567-v. [DOI] [PubMed] [Google Scholar]
- Naue J, Hörer S, Sänger T, Strobl C, Hatzer-Grubwieser P, Parson W, Lutz-Bonengel S. Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion. 2015;20:82–94. doi: 10.1016/j.mito.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Mitochondrial function and dysfunction in the cell: its relevance to aging and aging-related disease. The International Journal of Biochemistry & Cell Biology. 2002;34(11):1372–1381. doi: 10.1016/s1357-2725(02)00077-8. [DOI] [PubMed] [Google Scholar]
- Ogawa O, Perry G, Smith MA. The "Down's" side of mitochondria. Dev Cell. 2002;2(3):255–6. doi: 10.1016/s1534-5807(02)00139-9. [DOI] [PubMed] [Google Scholar]
- Oster-Granite ML, Parisi MA, Abbeduto L, Berlin DS, Bodine C, Bynum D, Capone G, Collier E, Hall D, Kaeser L. Down syndrome: national conference on patient registries, research databases, and biobanks. Mol Genet Metab. 2011;104(1-2):13–22. doi: 10.1016/j.ymgme.2011.07.005. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G, Castello G. Oxidative stress and mitochondrial dysfunction in Down syndrome. Adv Exp Med Biol. 2012;724:291–9. doi: 10.1007/978-1-4614-0653-2_22. [DOI] [PubMed] [Google Scholar]
- Perluigi M, Butterfield DA. Oxidative stress and Down syndrome: a route toward Alzheimer-like dementia. Curr Gerontol Geriatr Res. 2012;2012:724904. doi: 10.1155/2012/724904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roat E, Prada N, Ferraresi R, Giovenzana C, Nasi M, Troiano L, Pinti M, Nemes E, Lugli E, Biagioni O. Mitochondrial alterations and tendency to apoptosis in peripheral blood cells from children with Down syndrome. FEBS Lett. 2007;581(3):521–5. doi: 10.1016/j.febslet.2006.12.058. others. [DOI] [PubMed] [Google Scholar]
- Sevini F, Giuliani C, Vianello D, Giampieri E, Santoro A, Biondi F, Garagnani P, Passarino G, Luiselli D, Capri M. mtDNA mutations in human aging and longevity: Controversies and new perspectives opened by high-throughput technologies. Experimental Gerontology. 2014;56:234–244. doi: 10.1016/j.exger.2014.03.022. others. [DOI] [PubMed] [Google Scholar]
- Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 2010;48(4):287–96. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen HAL, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(2):113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Vetrano DL, Carfi A, Brandi V, L'Angiocola PD, Di Tella S, Cipriani MC, Antocicco M, Zuccala G, Palmieri V, Silveri MC. Left ventricle diastolic function and cognitive performance in adults with Down syndrome. Int J Cardiol. 2016;203:816–8. doi: 10.1016/j.ijcard.2015.11.041. others. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci U S A. 2014;111(29):10654–9. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Chinnery PF. In: Leber hereditary optic neuropathy. Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, editors. GeneReviews(R); Seattle (WA): 1993. others. [Google Scholar]
- Zigman WB. Atypical aging in down syndrome. Developmental Disabilities Research Reviews. 2013;18(1):51–67. doi: 10.1002/ddrr.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.