RAB43 is a vesicular transport protein unique to CD8α+ DCs that is localized to the Golgi. Kretzer et al. show that RAB43 is necessary for optimal cross-presentation of cell-associated antigens by CD8α+ DCs in vitro and in vivo but that it is dispensable for cross-presentation by in vitro monocyte-derived DCs.
Abstract
In this study, to examine cross-presentation by classical dendritic cells (DCs; cDCs), we evaluated the role of RAB43, a protein found to be selectively expressed by Batf3-dependent CD8α+ and CD103+ compared with other DC subsets and immune lineages. Using a specific monoclonal antibody, we localized RAB43 expression to the Golgi apparatus and LAMP1− cytoplasmic vesicles. Mice with germline or conditional deletion of Rab43 are viable and fertile and have normal development of cDCs but show a defect for in vivo and in vitro cross-presentation of cell-associated antigen. This defect is specific to cDCs, as Rab43-deficient monocyte-derived DCs showed no defect in cross-presentation of cell-associated antigen. These results suggest that RAB43 provides a specialized activity used in cross-presentation selectively by CD8α+ DCs but not other antigen-presenting cells.
Introduction
In cross-presentation, exogenous antigens taken up by APCs are processed for presentation by MHCI (Bevan, 1976; Ackerman and Cresswell, 2004; Rock and Shen, 2005). In vivo, cross-presentation of cell-associated antigen has been attributed primarily to a subset of splenic DCs identified by expression of CD8α (den Haan et al., 2000). The dominant role of CD8α+ DCs in cross-presentation appears not to result from more efficient antigen capture but, rather, specialized pathways of processing that are active in CD8α+ DCs but not in CD8α− DCs (Schnorrer et al., 2006). CD8α+ DCs express cell-surface receptors, such as CLEC9A (Poulin et al., 2012), that can deliver antigens to cross-presentation pathways (Schulz and Reis e Sousa, 2002). Additionally, DC lysosomes have decreased protease activity, compared with macrophages, which improves antigen presentation and persistence in lymphoid organs (Delamarre et al., 2005). RAB27A (Jancic et al., 2007) and RAC2 (Savina et al., 2009) have been linked to this process through recruitment of NOX2 to phagosomes, which delays their acidification by producing reactive oxygen species that consume protons generated by V-ATPase in phagosomal membranes (Savina et al., 2006).
Peptides derived from exogenous proteins can be loaded onto MHCI either in the ER or in endocytic compartments where components of the peptide-loading complex have been recruited (Cebrian et al., 2011). Cross-presentation may involve movement of antigens from endosomes through transporters such as SEC61 into the cytosol where they are degraded (Zehner et al., 2015). Alternatively, cross-presentation may involve a strictly vacuolar pathway in which antigens are digested by cathepsins within vesicles (Shen et al., 2004) and directly loaded onto MHCI molecules that are present in the same compartment (Guermonprez et al., 2003). This pathway may be facilitated by the fusion of phagosomes with the ER–Golgi intermediate complex, a process enabled by SEC22B (Cebrian et al., 2011).
Much of the work done to identify these cross-presentation pathways has relied on in vitro generated monocyte-derived DCs (moDCs) and, therefore, not described pathways that are unique to CD8α+ DCs in vivo (Cebrian et al., 2011; Nair-Gupta et al., 2014; Zehner et al., 2015). We identified RAB43 as a small GTPase that is specifically expressed in CD8α+ DCs and is necessary for optimal cross-presentation of cell-associated and soluble antigens by CD8α+ classical DCs (cDCs) but not moDCs. We show that RAB43 is localized to the Golgi and cytoplasmic vesicles distinct from lysosomes and that it is not essential for normal Golgi development, in contrast to previous suggestions (Haas et al., 2007).
Results and discussion
Rab43 is highly expressed in CD8α+ DCs
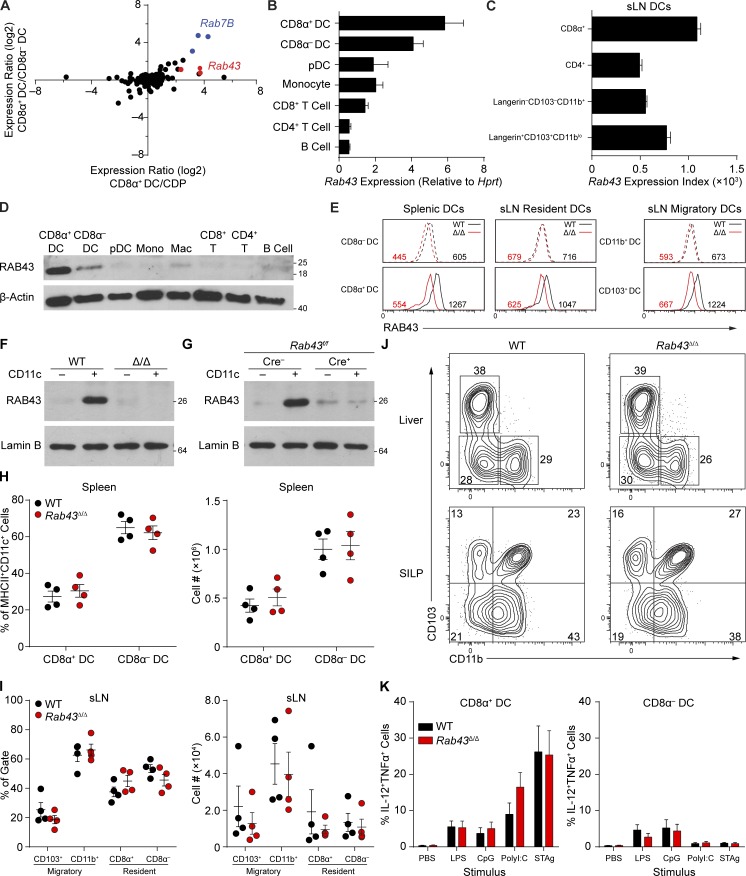
We evaluated expression of all RAB family members present in mouse genome 430 2.0 microarrays (Satpathy et al., 2012; Kc et al., 2014) for expression in CD8α+ DCs, CD8α− DCs, and common DC progenitors (CDPs; Fig. 1 A; Naik et al., 2007). Rab43 was among the most highly expressed RAB proteins in CD8α+ DCs compared with CDPs and CD8α− DCs. Mouse and human RAB43 are 95% identical (Fig. S1), suggesting evolutionary conservation. Using RT-PCR, we directly confirmed that Rab43 was most highly expressed in CD8α+ DCs compared with CD8α− DCs, plasmacytoid DCs (pDCs), monocytes, T cells, and B cells (Fig. 1 B). In the skin-draining LN (sLN), CD8α+ resident and CD103+ migratory DCs express the highest levels of Rab43 (Fig. 1 C; Heng et al., 2008).
Figure 1.
Rab43is highly and selectively expressed in CD8α+ cDCs and does not impact cDC development. (A) Sorted WT CD8α+ DCs, CD8α− DCs, and BM CDPs analyzed by gene expression microarray. Expression levels were determined for a list of Rab proteins in mouse 430 2.0 and plotted based on CD8α+ DC/CDP (x axis) versus CD8α+ DC/CD8α− DC (y axis) expression ratios. Each dot indicates an individual probe set. Data are representative of at least two independent experiments with three pooled mice. (B) Expression value (in arbitrary units) of Rab43 mRNA normalized to Hprt by quantitative RT-PCR for the indicated cell populations. Data from three independently sorted replicates of three WT mice displayed as mean ± SEM are shown. (C) Immgen data showing expression of Rab43 in the indicated populations from sLN. Data are displayed as mean ± SEM with three measurements per sample. (D) Western analysis of RAB43 and β-actin for the indicated spleen or BM populations from WT mice. Data are representative of at least three independent experiments. Mac, macrophage; Mono, monocyte. (E) Intracellular staining for RAB43 in the indicated cells from spleen and sLN from WT and Rab43Δ/Δ B6 mice. The numbers represent the mean fluorescence intensity of RAB43 staining for the indicated cells. Data are representative of two independent experiments. (F) Western analysis for RAB43 and lamin B from WT or Rab43Δ/Δ 129 (Δ/Δ) splenocytes. CD11c-negative (−) or CD11c-positive (+) splenocytes were isolated using CD11c microbeads. Data are representative of at least two experiments. (G) Western analysis for RAB43 and lamin B from CD11c-negative (−) or CD11c-positive (+) B6 splenocytes isolated as in A derived from Rab43f/f mice that were either CD11cCre− (Cre-) or CD11cCre+ (Cre+) as indicated. Data are representative of at least two experiments. (D, F, and G) Scales indicate molecular weight in kD. (H) Percentage (left) and absolute number (right) of DC subpopulations from spleen of WT and Rab43Δ/Δ B6 mice. (I) Percentage (left) and absolute number (right) of DC subpopulations from sLN of WT and Rab43Δ/Δ mice. Cells gated based on resident (B220−MHCIIintCD11chi) and migratory (B220−MHCIIhiCD11cint/lo) populations are shown. (H and I) Data from three independent experiments are shown. Each dot represents a single mouse. (J) Contour plots of tissue DCs from the small intestine lamina propria (SILP) or liver of WT or Rab43Δ/Δ 129 mice pregated on B220−CD45.2+MHCII+CD11c+. Data are representative of at least two experiments. (K) Percentage of IL-12– and TNF-positive cells after incubation of FLT3L-cultured BM cells from WT and Rab43Δ/Δ 129 mice with LPS, CpG, polyI:C, or STAg. Data from two independent experiments displayed as mean ± SEM are shown.
To analyze RAB43 at the protein level, we generated a monoclonal antibody, 2E6, directed to amino acids 179–203, a region of RAB43 that is highly divergent from other RAB family members. Using 2E6 for Western analysis, we confirmed that RAB43 protein was specifically expressed in CD8α+ cDCs at levels that were substantially higher than in CD8α− DCs, pDCs, monocytes, T cells, and B cells (Fig. 1 D). Intracellular staining with biotinylated 2E6 also showed that RAB43 is most abundant in CD8α+ resident and CD103+ migratory DCs in the sLN compared with other DC subsets, similar to what is observed in the spleen (Fig. 1 E).
Rab43-deficient mice are viable and have normal cDC development
RAB43 has only been examined by overexpression or shRNA-mediated knockdown in transformed cell lines in vitro (Haas et al., 2005, 2007; Fuchs et al., 2007). A potential dominant-negative RAB43 (T32N) mutation reportedly induced Golgi fragmentation in HeLa cells (Haas et al., 2007), suggesting that RAB43 may function in Golgi development or maintenance. However, these conclusions were based on in vitro morphological analysis of HeLa and hTERT-RPE1 cells, in which endogenous levels of RAB43 protein were not determined. No studies have yet analyzed the in vivo role of RAB43 or its function in CD8α+ DCs where it is most highly expressed.
To test the in vivo role of RAB43, we generated Rab43f/f mice that allow conditional deletion of the Rab43 exon 2, which encodes critical residues of the Rab domain (Fig. S2). Conditional deletion in cDCs was achieved by crossing to CD11c (Itgax)-Cre (Caton et al., 2007) to produce Rab43cKO mice, and constitutive germline Rab43 deletion (Rab43Δ/Δ) was achieved on 129SvEv and C57BL/6 backgrounds by crossing to germline-expressing Cre-deleter strains.
Rab43Δ/Δ and Rab43cKO mice were viable and produced fertile offspring at normal Mendelian frequencies. To confirm that RAB43 protein was absent from DCs in Rab43Δ/Δ and Rab43cKO mice, we performed Western analysis for RAB43 using 2E6 on CD11c− or CD11c+ splenocytes (Fig. 1 F). In WT mice, RAB43 protein was detected in CD11c+ splenocytes but not in CD11c− splenocytes (Fig. 1 F), as expected. In Rab43Δ/Δ mice, RAB43 was not detectable in either CD11c+ splenocytes or CD11c− splenocytes, indicating that germline deletion of Rab43 exon 2 is sufficient to eliminate RAB43 protein (Fig. 1 F). In Rab43f/f mice, RAB43 was detectable in CD11c+ splenocytes as expected, but RAB43 was nearly undetectable in Rab43cKO mice (Fig. 1 G). This result indicates that exon 2 deletion by CD11c-Cre can efficiently eliminate RAB43 protein in cDCs.
Loss of RAB43 in Rab43Δ/Δ mice caused no changes in the development of mature B cells, CD4+ T cells, CD8+ T cells, BM monocytes, or pDCs (not depicted). Furthermore, there was no impact on DC development in Rab43Δ/Δ mice, which showed normal populations of splenic cDCs (Fig. 1 H) and normal migratory and resident DCs in sLNs (Fig. 1 I). Populations of DCs were normal in nonlymphoid tissues such as the liver and small intestine lamina propria (Fig. 1 J). RAB43-deficient DCs also produced normal levels of cytokines upon stimulation by LPS, CpG, polyinosinic:polycytidylic acid (polyI:C), and soluble tachyzoite antigen (STAg; Fig. 1 K). Thus, RAB43 is selectively expressed in CD8α+ cDCs but is not required for their development or response to stimuli.
RAB43 is localized to the Golgi and LAMP1− vesicles in CD8α+ DCs
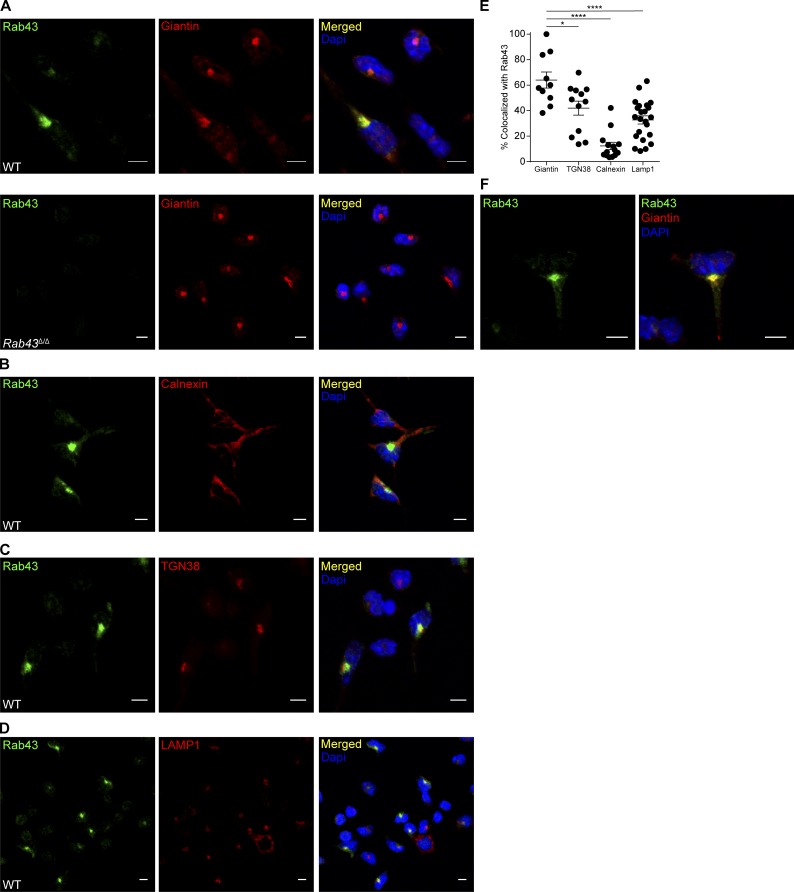
To determine the localization of RAB43 within DCs, we performed immunofluorescence microscopy on FLT3L-cultured BM cells using 2E6 antibody along with markers of various organelles (Fig. 2). RAB43 was expressed in brightly staining perinuclear organelles that colocalized with giantin, a cis-Golgi marker (Fig. 2, A and E). 2E6 staining was specific for RAB43 because no 2E6 signal was seen in Rab43Δ/Δ DCs (Fig. 2 A, bottom). RAB43 did not colocalize with the ER marker calnexin, (Fig. 2, B and E) and showed only limited colocalization with the trans-Golgi marker TGN38 (Fig. 2, C and E).
Figure 2.
RAB43 is abundant in Golgi and vesicles of Batf3-dependent DCs. (A) Day 10 FLT3L-cultured BM from WT (top) or Rab43Δ/Δ (bottom) 129 mice was treated with LPS for 4 h (to improve attachment), allowed to attach to coverslips, fixed, stained with 2E6 (anti-RAB43; green) and anti-giantin (red), and attached to slides using Prolong Gold antifade with DAPI (blue). (B–D) WT cells as described in A untreated with LPS were stained with 2E6 (green) and anti-calnexin (red; B), anti-TGN38 (red; C), or anti-LAMP1 (red; D). Coverslips were attached to slides using Prolong Gold antifade with DAPI (blue). (E) Percentage of organelle stain that is colocalized with RAB43 stain for the indicated organelles. Each dot represents a single cell with 10–23 cells analyzed per stain. Data were obtained using Imaris Coloc2. Statistics were analyzed using ANOVA. *, P < 0.05; ****, P < 0.0001. (F) WT cells prepared as described in A shown at increased zoom to highlight vesicular staining. All microscopy data are representative of at least two independent experiments. Bars, 5 µm.
Specific RAB43 staining was also evident in small cytoplasmic vesicles throughout the DCs’ long dendritic branches (Fig. 2 F). No vesicular staining was seen in Rab43Δ/Δ DCs (Fig. 2 A). 2E6 staining did not colocalize with LAMP1, suggesting that the RAB43-expressing vesicles are not lysosomes (Fig. 2, D and E). In summary, RAB43 appears to be expressed on cis-Golgi and in a nonlysosomal vesicular compartment.
RAB43-deficient CD8α+ DCs have an intact Golgi apparatus
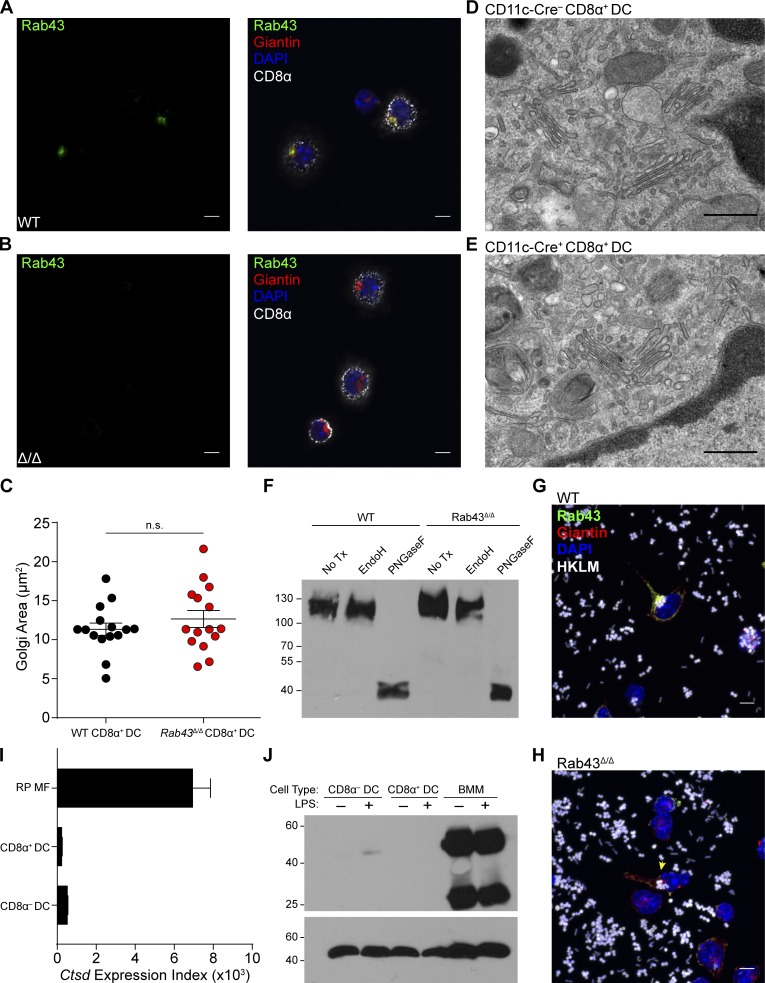
Because RAB43 was suggested to control Golgi integrity in HeLa cells (Haas et al., 2007), we asked whether the Golgi was intact in DCs isolated from WT and Rab43Δ/Δ mice (Fig. 3, A and B). RAB43 colocalized with giantin in CD8α+ cDCs freshly isolated from WT mice (Fig. 3 A), in agreement with analysis of in vitro generated cDCs (Fig. 2). Freshly isolated CD8α− cDCs were negative for 2E6 staining (Fig. 3 A), consistent with RAB43 expression assessed by Western analysis (Fig. 1 D) and intracellular staining (Fig. 1 E). The area of giantin staining is similar in both WT and Rab43Δ/Δ DCs, suggesting that the absence of RAB43 protein had no apparent impact on its distribution (Fig. 3 C).
Figure 3.
Rab43Δ/ΔCD8α+ DCs have normal Golgi development. (A and B) Sorted Ly6G−Ly6C−Thy1.2−B220−DX5−F4/80−CD11c+ splenocytes from WT (A) or Rab43Δ/Δ (Δ/Δ; B) 129 mice were allowed to attach to coverslips, fixed, and stained for CD8α (white), giantin (red), and RAB43 (green). Coverslips were attached to slides using ProLong Gold antifade with DAPI (blue). Data are representative of at least three independent experiments. Bars, 5 µm. (C) Area of Golgi staining from WT and Rab43Δ/Δ DCs. Each dot represents a single cell from staining in A and B, with 15 cells analyzed per genotype. Area was obtained using ImageJ. Statistics were analyzed by a two-tailed Mann-Whitney U test with P > 0.5. (D and E) Electron microscopy of sorted B220−CD11c+MHCII+CD24+CD172a− splenocytes from Rab43f/f-CD11cCre− (D) or Rab43f/f-CD11cCre+ (E) B6 mice showing normal Golgi development. Data are from analysis of cells from five pooled mice. Bars, 500 nm. (F) Western analysis for LAMP1 in splenocyte protein lysate given no treatment (No Tx), EndoH, or PNGaseF. Data are representative of three independent experiments. The scale indicates molecular weight in kD. (G and H) Sorted SiglecH−CD11b−Sirpa−Bst2− cells from FLT3 cultures of WT (G) or Rab43Δ/Δ (H) B6 BM were incubated with Alexa Fluor 647–labeled HKLM-OVA (white) and then prepared as in B without CD8α stain (Videos 1 and 2). The yellow arrowhead indicates a RAB43-deficient DC that has taken up HKLM-OVA. Images are representative of three experiments. Bars, 5 µm. (I) Immgen expression data for cathepsin D in red pulp macrophages (RP MF), CD8α+ DCs, and CD8α− DCs, displayed as mean ± SEM with three to four measurements per cell type. (J) Western analysis for cathepsin D (top) and β-actin (bottom) in BM macrophages (BMM), CD8α+ DCs, and CD8α− DCs from Flt3L or M-CSF cultures either treated or untreated with LPS for 24 h. The scale indicates molecular weight in kD.
Furthermore, we used electron microscopy to examine CD8α+ DCs purified from Rab43f/f and Rab43cKO mice. There were no notable differences in morphology or density of Golgi membranes (Fig. 3, D and E). Glycoprotein analysis of LAMP1 shows that Golgi-mediated glycosylation is normal in Rab43Δ/Δ DCs (Fig. 3 F). It has also been shown that overexpression of RN-tre, a possible GTPase-activating protein for RAB43 (Fuchs et al., 2007), does not alter ER to Golgi transport of vesicular stomatitis virus G in HeLa cells (Haas et al., 2007). Therefore, RAB43 appears to be dispensable for development and function of the Golgi apparatus in cDCs.
To analyze the localization of RAB43 after antigen uptake, we incubated in vitro Flt3L-derived DCs with heat-killed Listeria monocytogenes expressing OVA (HKLM-OVA). We found that RAB43 still localizes to the Golgi after uptake of antigen and also that HKLM-OVA localized to a perinuclear region near the Golgi apparatus in both WT and Rab43Δ/Δ DCs (Fig. 3, G and H; and Videos 1 and 2). Therefore, RAB43 localization is not dependent on the uptake of antigen.
A previous study has suggested that RAB43 is involved in the recruitment of cathepsin D to phagosomes (Seto et al., 2011). Because cathepsin D has been implicated in cross-presentation in human moDCs (Fonteneau et al., 2003), we examined whether the mechanism of RAB43 action could involve regulation of cathepsin D. We found that cathepsin D was not expressed in either cDC population at either the transcript or protein level and that this did not change upon activation by LPS (Fig. 3, I and J). This suggests that the mechanism of action of RAB43 in CD8α+ DCs does not involve the regulation of cathepsin D.
RAB43-deficient CD8α+ cDCs have reduced cross-presentation both in vitro and in vivo
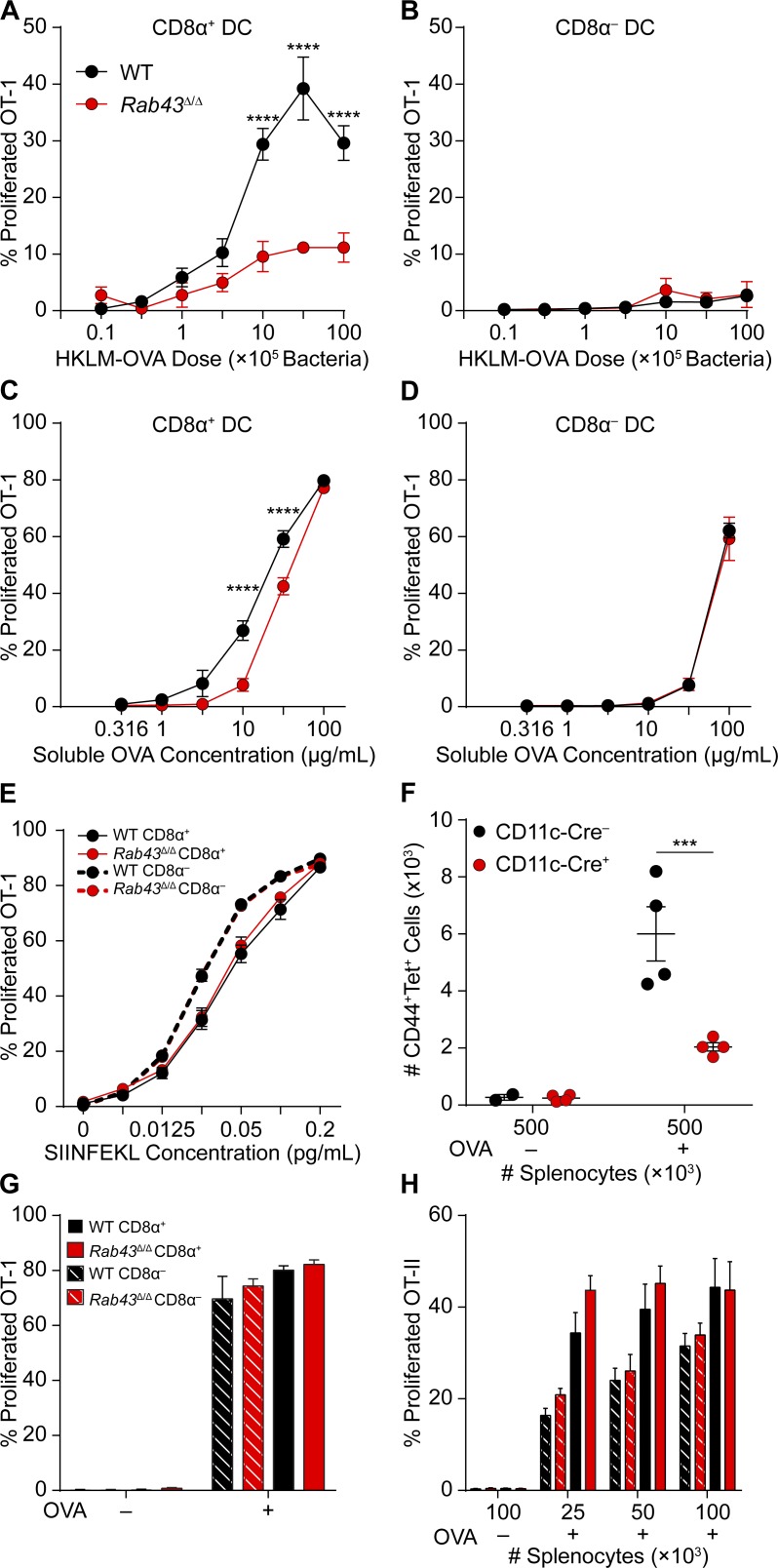
Because trafficking of intracellular vesicular compartments is necessary for antigen cross-presentation and CD8α+ DCs are efficient for this activity (den Haan et al., 2000; Iyoda et al., 2002; Schnorrer et al., 2006; Hildner et al., 2008), we asked whether RAB43 might have a role in antigen presentation by DCs. CD8α+ DCs and CD8α− DCs purified from WT and Rab43Δ/Δ mice were tested for their ability to cross-present HKLM-OVA to OT-1 T cells in vitro (Fig. 4, A and B). WT CD8α+ DCs cross-presented HKLM-OVA in a dose-dependent manner; however, CD8α+ DCs from Rab43Δ/Δ mice induced significantly less OT-1 proliferation at both low and high doses of bacteria (Fig. 4 A). Cross-presentation of HKLM-OVA by CD8α− DCs from WT or Rab43Δ/Δ mice was not observed at any dose tested (Fig. 4 B).
Figure 4.
CD8α+ DCs from Rab43Δ/Δ mice are defective in cross-presentation of cell-associated and soluble antigens. (A and B) Sorted CD8α+ (A) or CD8α− (B) DCs from WT (black) or Rab43Δ/Δ (red) B6 mice were cultured for 3 d with CFSE-labeled OT-I T cells and different doses of HKLM-OVA and assayed for OT-I proliferation and activation (CFSE−CD44+). (C and D) Sorted CD8α+ (C) or CD8α− (D) DCs from WT (black) or Rab43Δ/Δ (red) mice were cultured and analyzed as in A with various doses of soluble OVA as antigen. (E) Sorted CD8α+ (continuous lines) or CD8α− (dashed lines) DCs from WT (black) or Rab43Δ/Δ (red) mice were cultured and analyzed as in A with the indicated amounts of SIINFEKL peptide as antigen. (F) Rab43f/f-CD11cCre− (black) or Rab43f/f-CD11cCre+ (red) mice were injected with the indicated numbers of PBS (OVA−)- or OVA (OVA+)-loaded irradiated MHCI TKO splenocytes and analyzed 8 d later for the number of Kb SIINFEKL tetramer+ (tet+) T cells that were CD44+CD62L−. Each dot represents cells obtained from one mouse. (G) Sorted CD8α+ (solid) or CD8α− (dashed) DCs from WT (black) or Rab43Δ/Δ (red) mice were osmotically loaded with OVA, cultured, and analyzed as in A. (H) Sorted CD8α+ (solid) or CD8α− (dashed) DCs from WT (black) or Rab43Δ/Δ (red) mice were cultured for 3 d with CFSE-labeled OT-II T cells and irradiated splenocytes osmotically loaded with OVA. T cells were assayed for proliferation and activation (CFSE−CD44+). All data are displayed as mean ± SEM from at least two independent experiments. Statistics were analyzed using two-way ANOVA. ***, P < 0.001; ****, P < 0.0001.
OT-1 T cells also proliferated in response to soluble antigen presented by WT CD8α+ DCs in a dose-dependent manner (Fig. 4 C). Using CD8α+ DCs from Rab43Δ/Δ mice, OT-1 proliferation was significantly reduced at intermediate doses of soluble OVA but not at the maximal dose (Fig. 4 C). CD8α− DCs were able to cross-present soluble OVA, but this required a higher concentration than that needed for OT-1 stimulation by CD8α+ DCs, and there was no difference between WT and Rab43Δ/Δ DCs (Fig. 4 D). These results suggest that CD8α+ DCs have a unique cross-presentation program that is Rab43 dependent. This pathway is involved in the presentation of cell-associated but less so soluble antigens and may not be necessary for presentation of high soluble antigen loads. As a control, presentation of SIINFEKL peptide was equal between CD8α+ DCs and CD8α− cDCs from either WT or Rab43Δ/Δ mice, suggesting MHCI levels are normal on Rab43Δ/Δ DCs (Fig. 4 E).
We next tested the ability of Rab43cKO mice to prime T cells in vivo using cell-associated antigen (Carbone and Bevan, 1990). The number of active (CD44+CD62L−) Kb SIINFEKL tetramer-positive T cells in the spleen was analyzed 8 d after injection of irradiated MHCI TKO (Kb−/−Db−/−β2m−/−) splenocytes loaded with OVA or PBS. We found that Rab43cKO mice had a reduced ability to prime CD8 T cells against cell-associated antigen in vivo (Fig. 4 F).
The defect in T cell priming was specific to cross-presentation because direct presentation of intracellular OVA and MHCII presentation were not affected in Rab43Δ/Δ mice (Fig. 4, G and H). In summary, RAB43 functions in a cross-presentation–specific pathway that is involved in the presentation of cell-associated and low-dose soluble antigen by CD8α+ DCs.
moDCs from RAB43-deficient mice show no defect in cross-presentation
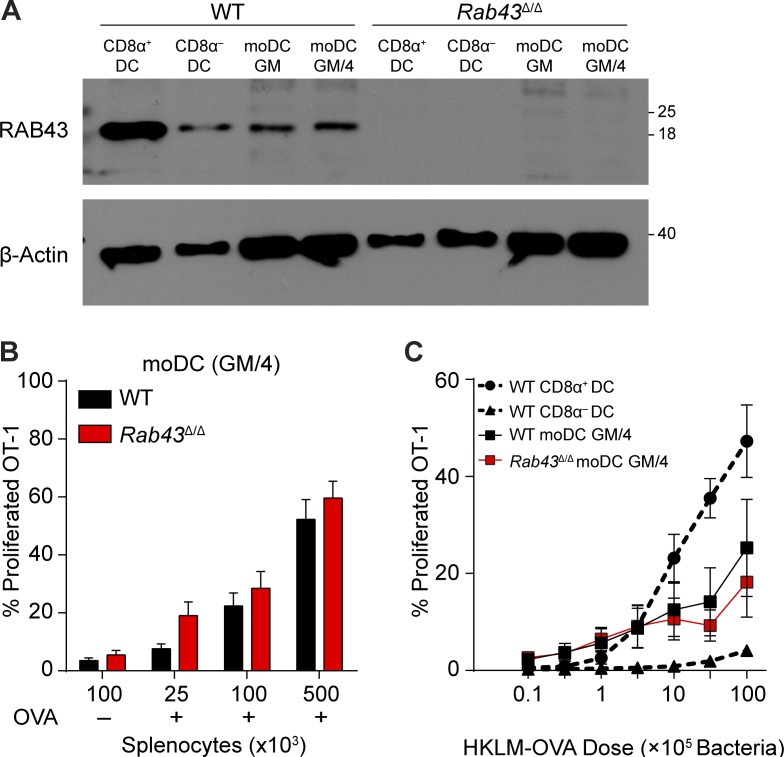
moDCs generated with GM-CSF and IL-4 can cross-present cell-associated antigen to a similar efficiency as CD8α+ DCs but use a distinct transcriptional program to acquire this capacity (Briseño et al., 2016). We asked whether cross-presentation by moDCs is Rab43 dependent (Fig. 5). Western analysis using 2E6 showed that RAB43 is expressed at a low level in moDCs generated with GM-CSF only (moDC GM) or with GM-CSF and IL-4 (moDC GM/4; Fig. 5 A). OT-1 priming by Rab43Δ/Δ moDC GM/4 was equal to that of WT moDC GM/4 at all antigen doses for both presentation of OVA-loaded irradiated splenocytes and HKLM-OVA (Fig. 5, B and C). Thus, cross-presentation by moDCs uses a Rab43-independent pathway distinct from the Rab43-dependent pathway in CD8α+ DCs. Similarly, cross-presentation of soluble OVA by moDCs and cDCs can occur through distinct pathways (Segura et al., 2009).
Figure 5.
moDCs from Rab43Δ/Δ mice show no defect in cross-presentation. (A) Western analysis of RAB43 and β-actin in CD8α+ and CD8α− cDCs compared with moDCs cultured for 4 d with either GM-CSF alone (GM) or GM-CSF + IL-4 (GM/4). Data are representative of at least two experiments. The scale indicates molecular weight in kD. (B) GM-CSF + IL-4–cultured moDCs from WT (black) or Rab43Δ/Δ (red) 129 monocytes were cultured with the indicated numbers of PBS (OVA−)- or OVA (OVA+)-loaded irradiated MHCI TKO splenocytes and OT-I T cells. After 3 d, cultures were assayed for OT-I proliferation and activation (CFSE−CD44+). (C) Sorted CD8α+ and CD8α− DCs and in vitro generated moDC GM-CSF + IL-4 from WT (black) or Rab43Δ/Δ (red) B6 mice were cultured for 3 d with CFSE-labeled OT-I T cells and different doses of HKLM-OVA and assayed for OT-I proliferation and activation (CFSE−CD44+). Cross-presentation data are displayed as mean ± SEM from at least three independent experiments.
Several in vitro studies have relied on moDCs cells to analyze cross-presentation (Cheong et al., 2010; Cebrian et al., 2011; Nair-Gupta et al., 2014; Zehner et al., 2015), even though in vivo cross-presentation appears specific to CD8α+ DCs (Iyoda et al., 2002; Schulz and Reis e Sousa, 2002; Sancho et al., 2009). Based on our data, moDCs may not be appropriate in vitro models for cross-presentation, and molecules analyzed using moDCs should be confirmed using primary CD8α+ DCs obtained from animal models.
In conclusion, RAB43 functions in a CD8α+ DC–specific pathway necessary for efficient cross-presentation of cell-associated antigen. Our data suggest that there are two separate pathways for soluble antigen presentation, and low-dose presentation was seen only in CD8α+ DCs and was dependent on RAB43. These results suggest that the form and amount of internalized antigen may impact how it is eventually processed and presented in CD8α+ DCs. This could be caused by differential endocytosis (i.e., receptor mediated versus micropinocytosis) or alternative methods of processing soluble proteins versus intact cells. The Golgi and vesicular localization of RAB43 suggests it may function in intracellular transport of antigen or the peptide-loading complex. Our data show that moDCs cross-present through mechanisms distinct from those used by CD8α+ DCs in vivo, suggesting that previous mechanisms proposed for cross-presentation that relied on moDCs may need to be reevaluated in the correct cell type. These studies will provide useful insight for designing vaccines that more selectively target efficient cross-presentation pathways.
Materials and methods
Mice
WT 129SvEv mice were purchased from Taconic. WT C57BL/6, B6.SJL-PtprcaPepcb/BoyJ (B6.SJL), and C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-1) mice were purchased from The Jackson Laboratory. Mice designated as WT were from in-house breeding of C57BL/6 or 129SvEv strains. Experiments done with Rab43f/f mice used littermate controls. MHCI KO mice (Kb−/−Db−/−β2m−/−; TKO) were a gift from H.W. Virgin and T. Hansen (Washington University, St. Louis, MO; Lybarger et al., 2003). Experiments were performed with age- and sex-matched mice between 6 and 12 wk of age. All mice were bred and maintained in specific pathogen–free facilities according to institutional guideline protocols approved by the Animal Studies Committee of Washington University in St. Louis.
Generation of the Rab43cKO mouse
The Rab43f/f-targeting construct was generated using Gateway recombination (Invitrogen) as follows. The entry vector, pENTR-lox-FRT-rNeo, was created by replacing the puromycin resistance gene in pENTR-lox-Puro (Iiizumi et al., 2006) with the neomycin resistance gene from vector pLNTK, and frt sites flanking each loxP site were added. The Rab43 5′ homology arm was amplified by PCR from 129SvEv-EDJ22 embryonic stem cell (ES cell) genomic DNA using oligonucleotides containing attB4 and attB1 sites, 5′-GGGGACAACTTTGTATAGAAAAGTTGGCAGACCTCCTACTCCAAAG-3′ and 5′-GGGGACTGCTTTTTTGTACAAACTTGCAAGGCTGCACTGAGGCTA-3′. The attB4-attB1 PCR fragment was inserted into the pDONR (P4-P1R) plasmid (Invitrogen) by the bp recombination reaction generating pENTR-RAB43-5HA. The Rab43 3′ homology arm was amplified by PCR from genomic DNA using the following oligonucleotides containing attB2 and attB3 sites: 5′-GGGGACAGCTTTCTTGTACAAAGTGGGAGCCAATTCCTTACCTCCA-3′ and 5′-GGGGACAACTTTGTATAATAAAGTTGTAGAAGGGACGGATGCAGC-3′. The attB2-attB3 PCR fragment was inserted into pDONR(P2R-P3) plasmid (Invitrogen) by the bp recombination reaction generating pENTR-RAB43-3HA. pENTR-RAB43-3HA was digested with Acc1 to produce an overhang where an overlapping oligonucleotide containing an orphan loxP site could anneal. The annealed oligonucleotides for insertion of the orphan loxP site were as follows: 5′-CTATAACTTCGTATAGCATACATTATACGAAGTTATGGATCC-3′ and 5′-AGGGATCCATAACTTCGTATAATGTATGCTATACGAAGTTAT-3′. After ligation, sequencing confirmed the insertion of the orphan loxP, and the resulting plasmid was called pENTRE-RAB43-3HA-Acc1. LR recombination reaction was performed using pENTR-RAB43-5HA, pENTR-RAB43-3HA-Acc1, pENTR-lox-FRT-rNeo, and pDEST-DTA-MLS to generate the final targeting construct.
The linearized vector was electroporated into EDJ22 ES cells (129SvEv background), and targeted clones were identified by Southern analysis with the 5′ probe and confirmed with the 3′ probe. Probes were amplified from genomic DNA using the following primers: RAB43_5P_F, 5′-GCCGATGTCCTCAGATCAAT-3′; RAB43_5P_R, 5′-GTAGAGCCCTCGCTCCTTCT-3′; RAB43_3P_F, 5′-GAAACAGGTTGGAGCCCATA-3′; and RAB43_3P_R, 5′-TGACTTGGAAAAGCCCATTC-3′.
Blastocyst injections were performed to generate male chimeras. Germline Rab43+/fl mice were crossed to 129S6-Tg(Prnp-GFP/Cre)1Blwd mice for constitutive germline deletion (RAB43Δ/Δ 129). Germline Rab43+/f were also crossed to B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/JRainJ (FLPeR) to delete the neomycin cassette, generating the Rab43fl allele. Rab43+/fl mice were backcrossed 10 generations to the C57BL/6J background (stock no. 029844). Rab43+/fl were either bred to B6.C-Tg(CMV-cre)1Cgn/J to generate Rab43Δ/Δ B6 mice (stock no. 029845) or bred to B6.Cg-Tg(Itgax-cre)1-1Reiz/J (CD11c-cre) mice from The Jackson Laboratory to generate conditional deletion of Rab43.
Genotyping PCR was conducted to confirm germline transmission and to confirm neo deletion from the FLPeR mouse. The primers used were the following: 5′-CACTGCCCAGTCTAGCTTCC-3′, 5′-GAGTGGCTCTCCCCTTAACC-3′, and 5′-GGGTGGGGTGGGATTAGATA-3′. To screen for the presence of the orphan loxP or constitutive deletion, we used the following primers: 5′-AGGCAGAAGCAAGCAGGTTT-3′, 5′-GTGATCTGGGCCAAAACGTA-3′, and 5′-CAAAGCTATCCGACCAGGAC-3′.
Antibodies and flow cytometry
Flow cytometry and cell-sorting experiments were completed on a FACS CantoII, FACSAriaII, or FACSAria Fusion instrument (BD) and analyzed using FlowJo analysis software (Tree Star). Staining was performed at 4°C in the presence of Fc block (clone 2.4G2; BD) in magnetic-activated cell-sorting (MACS) buffer (Dulbecco’s PBS [DPBS] + 0.5% BSA + 2 mm EDTA).The following antibodies were purchased from BD: anti-CD3 (145-2C11), anti-CD8 (53-6.7), anti-CD11c (HL3), anti-CD19 (1D3), anti-B220 (RA3-6B2), anti-CD172a (P84), anti-Ly6C (AL-21), anti-Ly6G (1A8), and anti–H-2Kb (AF6-88.5). The following antibodies were purchased from BioLegend: anti-CD4 (RM4-5), anti-CD11c (N418), anti-CD24 (M1/69), anti-CD115 (AFS98), anti-CD135 (A2F10), anti-MHCII (I-A/I-E; M5/114.15.2), and anti–TCR-Vα2 (B20.1). The following antibodies were purchased from eBioscience: anti-CD44 (IM7), anti-CD45.2 (104), anti-CD90.2 (53-2.1), anti-CD103 (2E7), anti-CD117 (2B8), anti-CD317 (eBio927), anti-F4/80 (BM8), anti-TER119 (TER-119), anti-CD62L (MEL-14), and anti-SiglecH (eBio440c). The following antibodies were purchased from Tonbo Biosciences: anti-CD11b (M1/70) and anti-CD45.1 (A20).
For immunofluorescence and confocal microscopy, the following antibodies were purchased from Abcam: rabbit polyclonal anti-giantin (ab24586) and rabbit polyclonal anti-calnexin (ab22595). The following antibody was purchased from Santa Cruz Biotechnology, Inc.: rabbit polyclonal anti-TGN38 (sc-33784). The following antibodies were purchased from Invitrogen: Alexa Fluor 488 goat anti–mouse IgG (H + L), Alexa Fluor 488 goat anti–rabbit IgG (H + L), Alexa Fluor 647 goat anti–rabbit IgG (H + L), and streptavidin, Alexa Fluor 555 conjugate.
For Western analysis, the following antibodies were purchased from Santa Cruz Biotechnology, Inc.: anti–β-actin (C4) and anti–lamin B (M-20). Anti-Lamp1 (ab24170) was from Abcam. Anti–cathepsin D (AF1029) was from R & D Systems. H2-Kb SIINFEKL tetramer was purchased from MBL International.
DC preparation
To harvest DCs from lymphoid tissue, organs were minced and digested with 250 µg/ml collagenase B (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 30–60 min at 37°C with stirring in 5 ml complete IMDM + 10% FCS (cIMDM). Erythrocytes (RBCs) were removed using ACK lysis buffer (150 mM ammonium chloride, 10 mM potassium bicarbonate, and 0.1 mM EDTA). Cells were passed through a 70-µm nylon mesh and counted with a Vi-CELL analyzer (Beckman Coulter). For FACS analysis, 5–10 × 106 cells were stained in MACS buffer. Before sorting, DCs were first enriched using CD11c microbeads (Miltenyi Biotec).
For peripheral tissue DCs, organs were digested in collagenase D (Roche) and DNase I (Sigma-Aldrich) for 1 h at 37°C with stirring in 5 ml cIMDM. Liver cells were separated using a Percoll gradient (Sigma-Aldrich). Small intestine suspensions were prepared as described previously (Satpathy et al., 2013).
BM culture with FLT3L
BM was flushed from mouse tibias and femurs, and RBCs were removed using ACK lysis buffer. BM cells were cultured at 2 × 106 cells/ml in cIMDM containing 100 ng/ml FLT3L for 8–10 d. Loosely adherent cells were harvested for analysis.
moDC preparation
To obtain BM monocytes, femurs, tibias, and hip bones were crushed using a mortar and pestle in MACS buffer. After passage through a 70-µm strainer, BM cells were isolated by gradient centrifugation using Histopaque 1119 (Sigma-Aldrich). Before sorting, B cells were depleted using B220 microbeads (Miltenyi Biotec). Up to one million sorted monocytes were cultured in 3 ml cIMDM with 20 ng/ml GM-CSF with or without 20 ng/ml IL-4 (PeproTech) for 4 d. Loosely adherent cells were obtained for analysis.
Cell sorting
Spleen DCs were sorted as B220−MHCII+CD11c+CD24+CD172− (CD8α+) and B220−MHCII+CD11c+ CD24−CD172+ (CD8α−) DCs. Populations from BM were sorted as Lin− CD117intCD135+CD115+CD11c−MHCII− (CDP), B220+CD317+SiglecH+ (pDC), or MHCII−CD11c−SiglecH−B220−c-kit−Ly6G−M-CSFR+CD11b+Ly6chi (BM monocytes). Other populations were sorted from spleens as auto+ F4/80+ (macrophages), CD90.2+CD8+ (CD8 T cells), CD90.2+CD4+ (CD4 T cells), or B220+ (B cells). OT-1 cells were sorted as B220−CD4−CD11c−CD45.1+CD8+Vα2+. OT-II cells were sorted as B220−CD4+CD11c−CD45.1+CD8−Vα2+.
In vitro TLR stimulation
2 × 105 FLT3L-cultured BM cells were plated into 96-well plates and stimulated with the following TLR agonists: 1 µg/ml LPS from Escherichia coli (055:B5; Sigma-Aldrich), 50 µg/ml polyI:C (Sigma-Aldrich), 63 nM ODN1826, class B CpG oligonugleotide (CpG; InvivoGen), and 2.5 µg/ml STAg (prepared as previously described; Subauste, 2012). Cells were allowed to incubate at 37°C and 8% CO2 for 1 h, and then, 1 µg/ml brefeldin A was added for an additional 4 h. Cells were then washed and prepared for surface and intracellular cytokine staining.
Antigen presentation assays
The in vivo cross-presentation assay has been previously described (den Haan et al., 2000). In brief, for both in vivo and in vitro cross-presentation assays, MHCI TKO splenocytes were osmotically loaded with 10 mg/ml OVA (Worthington Biochemical Corporation), irradiated at 1,350 rad, and injected i.v. at the indicated doses. After 8 d, the spleens from the mice were harvested and analyzed for tetramer+ T cells.
For in vitro cell-associated antigen cross-presentation assays by moDCs, 25,000 CFSE-labeled OT-1 T cells were plated with 25,000 moDCs from WT or Rab43Δ/Δ 129 mice and various doses of MHCI TKO PBS- or OVA-loaded and irradiated splenocytes in cIMDM in a 96-well plate. After incubating for 3 d in a 37°C and 8% CO2 incubator, CD45.1 OT-1 T cells were analyzed by FACS analysis for CFSE dilution and up-regulation of CD44.
HKLM-OVA (a gift from H. Shen, University of Pennsylvania, Philadelphia, PA) was grown in Brain-Heart Infusion broth at 37°C for 6 h and then frozen overnight after dilution plating for titer enumeration. Then, bacteria was thawed and washed three times with DPBS before heat killing at 80°C for 1 h and frozen at −80°C. 10,000 DCs from WT or Rab43Δ/Δ B6 mice were incubated with various doses of HKLM-OVA and 25,000 CFSE-labeled OT-1 T cells for 3 d in a 37°C 5% CO2 incubator and assayed for CFSE dilution and CD44 expression of OT-1 cells. Soluble OVA and peptide presentation assays used 10,000 DCs and 25,000 CFSE-labeled OT-1 T cells for 3 d. In peptide presentation assays, DCs were incubated with SIINFEKL for 45 min and then washed with cIMDM before incubation with 25,000 OT-1 T cells. Class II presentation assays were performed with 10,000 DCs incubated with OVA-loaded irradiated splenocytes and sorted OT-II T cells for 3 d and then analyzed for OT-II proliferation. Direct presentation assays were performed by sorting DCs and performing hypertonic loading with soluble OVA protein (Worthington Biochemical Corporation). After loading, 10,000 DCs were plated with 25,000 OT-1 cells for 3 d and then analyzed for OT-1 proliferation.
Generation of RAB43 monoclonal antibody
For immunization, we used Rab43−/− mice that were generated from Rab43tm1(KOMP)Wtsi ES cells (Knockout Mouse Project). Genotyping primers for Rab43−/− mice were: 5′-GCCATCACGAGATTTCGATT-3′, 5′-TCCCTTCCTACACAGCATCC-3′, and 5′-CCTGGCTGAGCACTATGACA-3′. Rab43−/− mice were immunized with KLH-conjugated RAB43 peptide, KLH-CIMRHGGPMFSEKNTDHIQLDSKDIA (Genscript). Sera and hybridoma supernatants were screened by ELISA using recombinant His-tagged RAB43 produced as follows: Rab43 was amplified from cDNA using the primers RAB43 Nde1 F (5′-ATTAAGATCTATGGCGGGCCCTGGCC-3′) and RAB43 Xho1 R (5′-ATTACTCGAGTCAGCACCCACAGCCCCA-3′) and cloned into the pET-28a+ vector (EMD Millipore). Recombinant protein was purified on Ni– nitrilotriacetic acid agarose (QIAGEN). Anti–mouse RAB43 antibody clone 2E6, an IgG1, was purified from hybridoma culture supernatant (Bio X Cell).
Intracellular staining
After staining for surface molecules, splenocytes were fixed in 4% paraformaldehyde in DPBS for 15 min at room temperature, washed, and suspended in permeabilization buffer (DPBS + 0.5% saponin + 2% FCS). Cells were incubated with anti–mouse RAB43 antibody biotinylated using NHS-dPEG4-Biotin (G-Biosciences) for 30–60 min at 4°C, washed with DPBS buffer containing + 0.05% saponin + 2% FCS, and incubated with streptavidin-FITC (BD).
Confocal microscopy
DCs from FLT3L-cultured BM were added to 24-well plates containing Alcian blue (Electron Microscopy Sciences)–coated coverslips at 500,000 cells/well in cIMDM either alone or with 1 µg/ml LPS (Sigma-Aldrich) and allowed to adhere for 3–4 h at 37°C and 8% CO2. Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in DPBS for 15 min at room temperature, quenched with 0.4 M glycine in DPBS, washed, and incubated with BlockAid (Invitrogen) in 0.2% saponin. Cells were stained with primary antibody in 2% FCS and 0.2% saponin in DPBS overnight at 4°C. After washing 3 × 5 min in DPBS with 0.2% saponin, cells were stained with secondary antibody in 2% FCS and 0.2% saponin in DPBS for 1–2 h at 4°C. Cells were washed and mounted in ProLong Gold Antifade with DAPI (Invitrogen) and allowed to cure overnight at room temperature before imaging. FLT3L-cultured DCs were imaged using a confocal microscope (LSM-510; ZEISS) with a 63× oil immersion objective and analyzed using ImageJ (National Institutes of Health).
Labeled HKLM-OVA was prepared by labeling 3 × 109 bacteria with 100 µl Alexa Fluor 647 succinimidyl ester (Thermo Fisher Scientific) for 1 h while vortexing. Then, bacteria were washed three times in PBS with reducing concentrations of BSA. HKLM-OVA was added to cells during attachment, and staining was performed as described in the previous paragraph.
Sorted Ly6G−CD90.2−B220−DX5−Ly6c−F4/80−CD11c+ splenocytes were plated at 100,000–300,000 cells/well in cIMDM onto anti-MHCII–coated coverslips in 24-well plates. After centrifuging at 1,000 rpm for 3 min, cells were allowed to adhere for 30–60 min at 37°C and 8% CO2, surface stained, and then fixed and stained similar to the FLT3L DCs.
Images of splenocytes and images after HKLM-OVA internalization were acquired on a confocal microscope (A1Rsi; Nikon) with a 60× oil immersion objective and analyzed using ImageJ. Colocalization analysis was performed using ImarisColoc. In brief, fluorescence was masked on a cell-by-cell basis for organelle stain, and percentage of Rab43 staining within that area was recorded. Golgi area was determined using ImageJ region of interest area calculations.
Western analysis
Isolated cells were washed in DPBS and resuspended in radioimmunoprecipitation assay lysis buffer with protease inhibitors at 107 cells/ml. Whole-cell extracts were obtained and denatured in Laemmli sample buffer at 95°C for 5 min. For glycosylation analysis, 2–3 × 106 cell equivalents of protein lysate were incubated with either EndoH Hf (New England Biolabs, Inc.) or PNGaseF (New England Biolabs, Inc.) for 1 h at 37°C as per the manufacturer’s protocol before Western analysis. 1 × 105–2.5 × 105 cell equivalents of whole-cell extract were run on a polyacrylamide gel and transferred onto a nitrocellulose membrane. Immunoblots were blocked in DPBS containing 3% BSA and 0.1% Tween 20 at room temperature for 1 h and then incubated with primary antibody overnight at 4°C. After extensive washing, membranes were incubated with goat anti–rabbit IgG (H + L) or goat anti–mouse IgG (H + L) conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature in 5% nonfat milk and 0.1% Tween 20 DPBS. Then, membranes were washed and developed with ECL Plus Western Blotting substrate (Thermo Fisher Scientific). Membranes were stripped using 37.5 mM Tris-HCl, pH 6.8, 1% SDS, and 0.1 M β-mercaptoethanol at 60°C for 30 min.
Quantitative RT-PCR
Cells were processed using an RNeasy Mini kit (QIAGEN) and Superscript III reverse transcription (Invitrogen) following the manufacturer’s protocol to obtain RNA and subsequent cDNA. A StepOnePlus Real-Time PCR system (Applied Biosystems) was used according to the manufacturer’s instructions with the Quantitation-Standard Curve method and HotStart-IT SYBR green quantitative PCR master mix (Affymetrix). PCR conditions were 95°C for 10 min, followed by 40 two-step cycles of 95°C for 15 s and 60°C for 1 min. The primers used for measurement of RAB43 were as follows: RAB43 quantitative PCR forward, 5′-ACTGGATCGAGGATGTGAGG-3′; and reverse, 5′-ACATTGCTGGAGTCCTTTGC-3′. Hprt primers were as follows: HPRT quantitative PCR forward, 5′-TCAGTCAACGGGGGACATAAA-3′; and reverse, 5′-GGGGCTGTACTGCTTAACCAG-3′.
Microarray analysis
Microarray data previously generated in our laboratory (GEO accession nos. GSE53312 and GSE37030; Satpathy et al., 2012; Kc et al., 2014) or by the Immgen Consortium (GEO accession no. GSE15907; Heng et al., 2008) were analyzed. Gene array data were processed using robust multiarray average quantile normalization, and data were modeled using ArrayStar software (DNASTAR).
Statistics
Statistical analyses were performed with Prism (GraphPad Software). Two-way ANOVA with multiple comparisons was performed for statistical significance.
Online supplemental material
Fig. S1 shows a sequence alignment between human and mouse RAB43. Fig. S2 is the targeting strategy used to generate Rab43Δ/Δ and Rab43cKO mice. Video 1 shows a rotating Z stack of a WT CD8α+ DC stained with RAB43, giantin, and DAPI after uptake of labeled HKLM-OVA. Video 2 shows a rotating Z stack of a Rab43Δ/Δ CD8α+ DC stained with RAB43, giantin, and DAPI after uptake of labeled HKLM-OVA.
Supplementary Material
Acknowledgments
We would like to thank Michael White for blastocyst injection and generation of mouse chimeras, Wandy Beatty for electron microscopy and confocal microscopy, Dennis Oakley at the Washington University Center for Cellular Imaging for confocal microscopy, and the Alvin J. Siteman Cancer Center at Washington University in St. Louis School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. We would like to thank David Fremont and Chris Nelson for advice on protein production and Bernd Zinselmeyer for help with microscopy analysis. The RAB43−/− ES cell used for this research project was generated by the trans–National Institutes of Health Knockout Mouse Project (KOMP) and obtained from the KOMP Repository.
This work was funded by the Howard Hughes Medical Institute. National Institutes of Health grants to Velocigene at Regeneron Inc. (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8,500 genes in the KOMP Program and archived and distributed by the KOMP Repository at University of California, Davis and Children’s Hospital Oakland Research Institute (U42RR024244). For more information or to obtain KOMP products go to www.komp.org.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- cDC
- classical DC
- CDP
- common DC progenitor
- DPBS
- Dulbecco’s PBS
- HKLM-OVA
- heat-killed Listeria monocytogenes expressing OVA
- MACS
- magnetic-activated cell sorting
- moDC
- monocyte-derived DC
- pDC
- plasmacytoid DC
- polyI:C
- polyinosinic:polycytidylic acid
- sLN
- skin-draining LN
- STAg
- soluble tachyzoite antigen
References
- Ackerman A.L., and Cresswell P.. 2004. Cellular mechanisms governing cross-presentation of exogenous antigens. Nat. Immunol. 5:678–684. 10.1038/ni1082 [DOI] [PubMed] [Google Scholar]
- Bevan M.J. 1976. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J. Exp. Med. 143:1283–1288. 10.1084/jem.143.5.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Haldar M., Kretzer N.M., Wu X., Theisen D.J., Kc W., Durai V., Grajales-Reyes G.E., Iwata A., Bagadia P., et al. 2016. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Reports. 15:2462–2474. 10.1016/j.celrep.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F.R., and Bevan M.J.. 1990. Class I-restricted processing and presentation of exogenous cell-associated antigen in vivo. J. Exp. Med. 171:377–387. 10.1084/jem.171.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch–RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L.F., Amigorena S., and Savina A.. 2011. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 147:1355–1368. 10.1016/j.cell.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G., et al. 2010. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell. 143:416–429. 10.1016/j.cell.2010.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delamarre L., Pack M., Chang H., Mellman I., and Trombetta E.S.. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 307:1630–1634. 10.1126/science.1108003 [DOI] [PubMed] [Google Scholar]
- den Haan J.M., Lehar S.M., and Bevan M.J.. 2000. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696. 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonteneau J.F., Kavanagh D.G., Lirvall M., Sanders C., Cover T.L., Bhardwaj N., and Larsson M.. 2003. Characterization of the MHC class I cross-presentation pathway for cell-associated antigens by human dendritic cells. Blood. 102:4448–4455. 10.1182/blood-2003-06-1801 [DOI] [PubMed] [Google Scholar]
- Fuchs E., Haas A.K., Spooner R.A., Yoshimura S., Lord J.M., and Barr F.A.. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143. 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., Van Endert P., and Amigorena S.. 2003. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature. 425:397–402. 10.1038/nature01911 [DOI] [PubMed] [Google Scholar]
- Haas A.K., Fuchs E., Kopajtich R., and Barr F.A.. 2005. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 7:887–893. 10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- Haas A.K., Yoshimura S., Stephens D.J., Preisinger C., Fuchs E., and Barr F.A.. 2007. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 120:2997–3010. 10.1242/jcs.014225 [DOI] [PubMed] [Google Scholar]
- Heng T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiizumi S., Nomura Y., So S., Uegaki K., Aoki K., Shibahara K., Adachi N., and Koyama H.. 2006. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 41:311–316. 10.2144/000112233 [DOI] [PubMed] [Google Scholar]
- Iyoda T., Shimoyama S., Liu K., Omatsu Y., Akiyama Y., Maeda Y., Takahara K., Steinman R.M., and Inaba K.. 2002. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J. Exp. Med. 195:1289–1302. 10.1084/jem.20020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic C., Savina A., Wasmeier C., Tolmachova T., El-Benna J., Dang P.M., Pascolo S., Gougerot-Pocidalo M.A., Raposo G., Seabra M.C., and Amigorena S.. 2007. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat. Cell Biol. 9:367–378. 10.1038/ncb1552 [DOI] [PubMed] [Google Scholar]
- Kc W., Satpathy A.T., Rapaport A.S., Briseño C.G., Wu X., Albring J.C., Russler-Germain E.V., Kretzer N.M., Durai V., Persaud S.P., et al. 2014. L-Myc expression by dendritic cells is required for optimal T-cell priming. Nature. 507:243–247. 10.1038/nature12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger L., Wang X., Harris M.R., Virgin H.W. IV, and Hansen T.H.. 2003. Virus subversion of the MHC class I peptide-loading complex. Immunity. 18:121–130. 10.1016/S1074-7613(02)00509-5 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Nair-Gupta P., Baccarini A., Tung N., Seyffer F., Florey O., Huang Y., Banerjee M., Overholtzer M., Roche P.A., Tampé R., et al. 2014. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell. 158:506–521. 10.1016/j.cell.2014.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin L.F., Reyal Y., Uronen-Hansson H., Schraml B.U., Sancho D., Murphy K.M., Håkansson U.K., Moita L.F., Agace W.W., Bonnet D., and Reis e Sousa C.. 2012. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 119:6052–6062. 10.1182/blood-2012-01-406967 [DOI] [PubMed] [Google Scholar]
- Rock K.L., and Shen L.. 2005. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol. Rev. 207:166–183. 10.1111/j.0105-2896.2005.00301.x [DOI] [PubMed] [Google Scholar]
- Sancho D., Joffre O.P., Keller A.M., Rogers N.C., Martínez D., Hernanz-Falcón P., Rosewell I., and Reis e Sousa C.. 2009. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 458:899–903. 10.1038/nature07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W., Wu X., Thomas S.R., Lee W.L., Turkoz M., et al. 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14:937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Duménil A.M., Seabra M.C., Raposo G., and Amigorena S.. 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 126:205–218. 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Savina A., Peres A., Cebrian I., Carmo N., Moita C., Hacohen N., Moita L.F., and Amigorena S.. 2009. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8+ dendritic cells. Immunity. 30:544–555. 10.1016/j.immuni.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Schnorrer P., Behrens G.M., Wilson N.S., Pooley J.L., Smith C.M., El-Sukkari D., Davey G., Kupresanin F., Li M., Maraskovsky E., et al. 2006. The dominant role of CD8+ dendritic cells in cross-presentation is not dictated by antigen capture. Proc. Natl. Acad. Sci. USA. 103:10729–10734. 10.1073/pnas.0601956103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., and Reis e Sousa C.. 2002. Cross-presentation of cell-associated antigens by CD8α+ dendritic cells is attributable to their ability to internalize dead cells. Immunology. 107:183–189. 10.1046/j.1365-2567.2002.01513.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Albiston A.L., Wicks I.P., Chai S.Y., and Villadangos J.A.. 2009. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc. Natl. Acad. Sci. USA. 106:20377–20381. 10.1073/pnas.0910295106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S., Tsujimura K., and Koide Y.. 2011. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 12:407–420. 10.1111/j.1600-0854.2011.01165.x [DOI] [PubMed] [Google Scholar]
- Shen L., Sigal L.J., Boes M., and Rock K.L.. 2004. Important role of cathepsin S in generating peptides for TAP-independent MHC class I crosspresentation in vivo. Immunity. 21:155–165. 10.1016/j.immuni.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Subauste C. 2012. Animal models for Toxoplasma gondii infection. Curr. Protoc. Immunol. Chapter 19:3.1–3.23. [DOI] [PubMed] [Google Scholar]
- Zehner M., Marschall A.L., Bos E., Schloetel J.G., Kreer C., Fehrenschild D., Limmer A., Ossendorp F., Lang T., Koster A.J., et al. 2015. The translocon protein Sec61 mediates antigen transport from endosomes in the cytosol for cross-presentation to CD8+ T cells. Immunity. 42:850–863. 10.1016/j.immuni.2015.04.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.