Baron et al. show that mycolactone, an immunosuppressive macrolide produced by the pathogen Mycobacterium ulcerans, operates by targeting the Sec61 translocon. This identifies the most potent Sec61 inhibitor reported to date and the potential of inhibiting Sec61 for immune modulation.
Abstract
Mycolactone, an immunosuppressive macrolide released by the human pathogen Mycobacterium ulcerans, was previously shown to impair Sec61-dependent protein translocation, but the underlying molecular mechanism was not identified. In this study, we show that mycolactone directly targets the α subunit of the Sec61 translocon to block the production of secreted and integral membrane proteins with high potency. We identify a single–amino acid mutation conferring resistance to mycolactone, which localizes its interaction site near the lumenal plug of Sec61α. Quantitative proteomics reveals that during T cell activation, mycolactone-mediated Sec61 blockade affects a selective subset of secretory proteins including key signal-transmitting receptors and adhesion molecules. Expression of mutant Sec61α in mycolactone-treated T cells rescued their homing potential and effector functions. Furthermore, when expressed in macrophages, the mycolactone-resistant mutant restored IFN-γ receptor–mediated antimicrobial responses. Thus, our data provide definitive genetic evidence that Sec61 is the host receptor mediating the diverse immunomodulatory effects of mycolactone and identify Sec61 as a novel regulator of immune cell functions.
Introduction
Mycobacterium ulcerans, the causative agent of Buruli ulcers (BUs), infects and destroys human skin without alerting the host immune system (Demangel et al., 2009). The lack of inflammatory infiltrates in ulcerative lesions is a striking histopathological feature of BU (Guarner et al., 2003). Moreover, BU patients display systemic defects in cellular immune responses, such as a reduced capacity of peripheral blood T cells to produce cytokines upon ex vivo stimulation (Phillips et al., 2009). These defects are independent of the activation stimulus and resolve upon treatment of the disease, showing their association with M. ulcerans. Bacterial virulence relies on the production of mycolactone, a polyketide-derived macrolide with ulcerative properties in the skin (George et al., 1999). Although bacteria remain primarily at the site of infection, mycolactone diffuses into mononuclear blood cells, LNs, and spleen (Hong et al., 2008; Sarfo et al., 2011), allowing it to exert immunosuppressive effects at the systemic level. Intraperitoneal delivery of mycolactone protects mice against chemically induced skin inflammation (Guenin-Macé et al., 2015). It prevents peripheral blood lymphocyte homing to draining LNs and expansion upon antigenic stimulation (Guenin-Macé et al., 2011). Finally, M. ulcerans strains deficient for mycolactone production do not induce functional defects in peripheral blood T cells of infected mice (Hong et al., 2008). Therefore, mycolactone has the intrinsic capacity to block the development of innate and adaptive immune responses in vivo.
In vitro mycolactone blunts the capacity of immune cells to produce selected cytokines, chemokines, and homing receptors without inducing cellular stress or cytotoxicity (Hall and Simmonds, 2014). Mycolactone operates posttranscriptionally and independently of mammalian target of rapamycin (mTOR) and, as such, represents a novel type of natural immunosuppressor. Hall et al. (2014) showed that mycolactone blocks the translocation of inflammatory mediators (TNF and Cox2) as well as model secretory proteins into the ER, with subsequent degradation of these proteins by the ubiquitin–proteasome system. Using cell-free systems, McKenna et al. (2016) later identified the translocation stage that mycolactone inhibits and highlighted differences in mycolactone-mediated inhibition of co-translationally versus posttranslationally inserted Sec61 secretory substrates. In eukaryotes, co-translational protein translocation is initiated by recognition of signal peptides or nascent polypeptide anchor domains by the signal recognition particle (SRP). The SRP then targets the ribosome-nascent polypeptide complex to the Sec61 translocon for insertion into the ER lumen (Park and Rapoport, 2012). McKenna et al. (2016) provided biochemical evidence that mycolactone induces a conformational change in the pore-forming subunit of the translocon, Sec61α. Although Sec61, SRP-receptor, and SRP are sufficient for minimal translocation to occur, accessory components such as Sec62/63, translocating chain-associated membrane protein (TRAM), translocon-associated protein (TRAP) complex, and binding immunoglobulin protein (BiP) facilitate the process. What the precise molecular target of mycolactone is and how mycolactone’s ability to prevent protein translocation connects with reduced cellular immune responses remained critical open questions.
Results and discussion
Mycolactone targets the Sec61 translocon
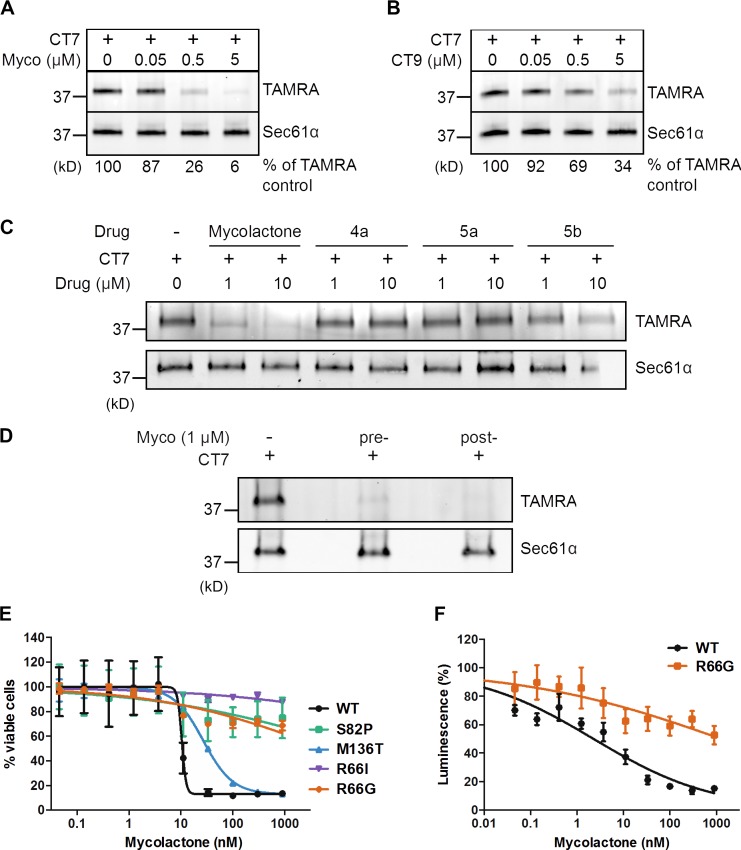
Among known inhibitors of protein translocation, three have been formally shown to act by directly targeting Sec61α: the cyclic heptadepsipeptide HUN-7293/cotransin/CT8, decadepsipeptide decatransin, and cyanobacterial product apratoxin A (Garrison et al., 2005; Maifeld et al., 2011; MacKinnon et al., 2014; Junne et al., 2015; Paatero et al., 2016). All of these drugs target a partially overlapping site in the pore-forming Sec61α subunit. However, unlike decatransin and apratoxin A, CT8 inhibits Sec61 in a substrate-selective manner. To test the hypothesis that mycolactone and CT8 use similar mechanisms of action, we performed competitive Sec61α-binding assays with a structural variant of CT8 that covalently cross-links to Sec61α upon photoactivation (Fig. S1 A; MacKinnon et al., 2007, 2014). ER microsomes were incubated with CT7 in the presence or absence of increasing amounts of mycolactone and then photolyzed and denatured. The presence of CT7 cross-linked to Sec61α was then quantitatively assessed by click chemistry and in-gel fluorescent scanning. Mycolactone competed dose dependently with CT7 for binding to Sec61α (Fig. 1 A), similarly as the potent cotransin analogue CT9 (Fig. 1 B and Fig. S1 A). Importantly, mycolactone displaced CT7 slightly more efficiently than CT9, indicating that it binds Sec61α with comparable or higher affinity and may share a coinciding binding site on Sec61α.
Figure 1.
Mycolactone targets the Sec61 translocon. (A and B) CRMs were preincubated with increasing concentrations of mycolactone (Myco; A) or CT9 (B) at the indicated concentrations, followed by 100 nM CT7. Covalent CT7/Sec61α adduct was detected using click chemistry between the alkyne group in CT7 and rhodamine-azide (TAMRA). (C) As in A but comparing the competitive activity of mycolactone to that of synthetic subunits of the intact molecule. (D) CRMs were incubated with a saturating concentration of mycolactone (10 µM) either before (pre-) or after (post-) extensive washing. CT7 photo–cross-linking was performed after final CRM pelleting. (E) HEK293-FRT TRex cells stably expressing WT or mutant Sec61α were treated with increasing concentrations of mycolactone for 72 h, and cell viability was analyzed by the Alamar blue assay (Mean ± SEM; n = 4). (F) HEK293-FRT TRex cells stably expressing WT Sec61α or R66G-Sec61α were transfected for inducible expression of a secreted Gaussia luciferase and then treated with increasing concentrations of mycolactone for 24 h. Data are luminescence values (mean ± SEM; n = 2) measured from culture supernatants. (A–F) Data shown are from one of two independent experiments, which gave similar results.
Mycolactone consists of a lactone ring and two polyketide chains branched in the north and south positions (Fig. S1 A). We reported previously that variant 5b lacking the northern side chain partially retains the immunosuppressive activity of mycolactone, whereas subunits lacking the southern or both side chains (4a and 5a, respectively) are biologically inert (Guenin-Macé et al., 2015). Consistently, 5b competed with CT7 with an ∼10-fold reduced potency, whereas 4a and 5a showed no competitive activity (Fig. 1 C). No difference in ability of mycolactone to compete with CT7 for Sec61α binding was observed after extensive washing of microsomes (Fig. 1 D), indicating that mycolactone binds tightly to the translocon and has a slow dissociation rate.
A previous genetic screen identified several point mutations in Sec61α (R66I, R66G, S82P, and M136T) that reduce CT8 binding without major effects on channel function (MacKinnon et al., 2014). Given that mycolactone and CT8 likely have overlapping binding sites, we tested whether these mutations confer resistance to mycolactone. For this purpose, we treated HEK293-FRT cells overexpressing WT or mutant Sec61α constructs with increasing concentrations of mycolactone. The viability of cells expressing WT Sec61α was potently reduced by mycolactone (IC50 = 10 nM; Fig. 1 E). In contrast, cells expressing the R66I-, R66G-, and S82P-mutant alleles were highly desensitized (IC50 > 1,000 nM). Interestingly, these mutations cluster near the lumenal plug of Sec61α (Fig. S1 B), suggesting that this region forms the mycolactone interaction site. This finding was fully consistent with the observation by McKenna et al. (2016) that mycolactone alters protease sensitivity of Sec61α in vitro. Focusing on the R66G construct, we investigated whether this single–amino acid mutation confers resistance to mycolactone-mediated blockade of protein secretion. HEK293-FRT cells stably expressing WT or R66G-Sec61α were transfected with a secreted Gaussia luciferase construct and then subjected to a 24-h mycolactone treatment that did not alter cell viability. Although mycolactone efficiently blocked luciferase secretion in cells expressing WT Sec61α (IC50 = 3 nM), cells expressing the R66G-Sec61α mutant proved highly resistant (IC50 > 1,000 nM; Fig. 1 F). In addition to providing additional evidence that mycolactone binds to Sec61α, these data revealed the critical importance of the Sec61α R66 residue for mycolactone’s inhibitory activity on protein translocation.
Mycolactone is a broad-spectrum inhibitor of Sec61
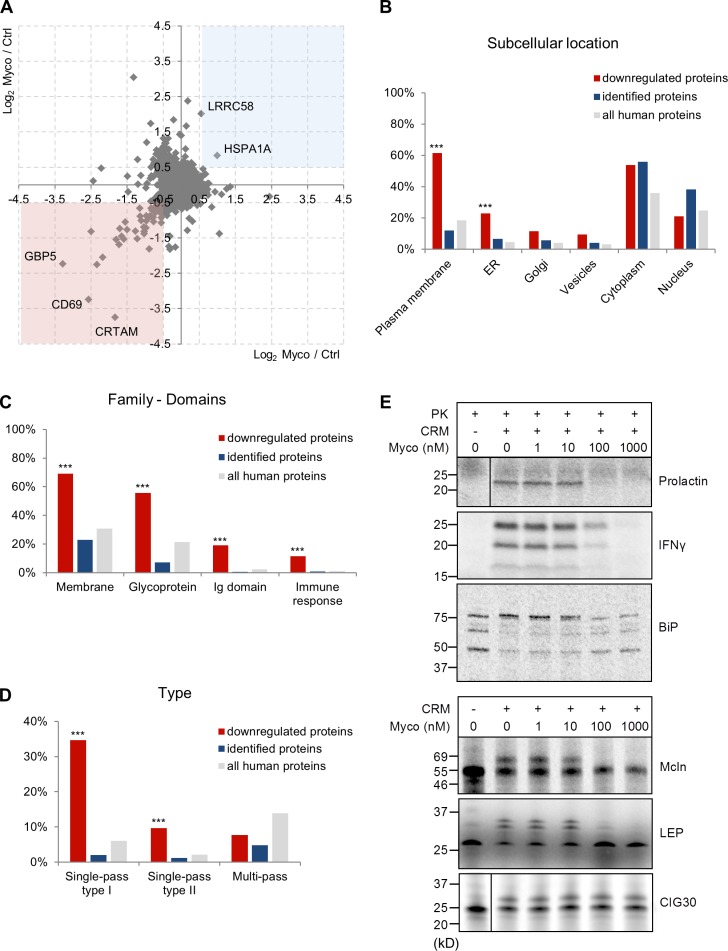
A distinguishing feature of CT8 is its ability to prevent the translocation of only a minor subset of Sec61 clients (Besemer et al., 2005; Garrison et al., 2005; Maifeld et al., 2011). To determine whether mycolactone shares this property, we compared the effects of mycolactone and CT8 on the production of known Sec61 clients by human immune cells (namely TNF production by monocyte-derived macrophages and IFN-γ, IL-2, and L-selectin production by peripheral blood-derived CD4+ T cells). Cotransin showed a highly variable inhibitory activity toward the different substrates (IC50 between 20 and 1,050 nM; not depicted). In contrast, mycolactone prevented the production of all tested proteins with IC50 between 4.5 and 12 nM, suggesting that it is a more potent and less selective Sec61 inhibitor. Next, we used global proteome analysis of SILAC (stable-isotope labeling with amino acids in cell culture) T cells to gain a broader view of mycolactone activity and identify the proteins impacted by Sec61 inhibition during T cell activation. Jurkat T cells were grown in light or heavy SILAC medium for five cell divisions and then treated with 40 nM mycolactone or vehicle for 1 h before activation with PMA and ionomycin (IO) for 6 h. These conditions induced full cell activation, bypassing a potential inhibitory effect of mycolactone on TCR expression (Boulkroun et al., 2010). Cells were then lysed, and equal amounts of light- and heavy-labeled protein extracts were mixed. Proteins were trypsin digested, and peptide mixtures were analyzed by liquid chromatography–tandem mass spectrometry (MS; LC-MS/MS). The SILAC analysis was repeated with reversed labeling conditions, allowing the reliable identification and quantification of 6,503 proteins (hereafter referred to as identified proteins). Among these, 4,636 proteins were quantified in both labeling conditions. SILAC analyses were performed on cell extracts, and consequently, most secreted proteins were not detected. Notably, 52 proteins were consistently down-regulated in mycolactone-treated cells (log2 mycolactone/control ratio <−0.5), whereas only two proteins (putative E3 ubiquitin–protein ligase LRRC58 and Hsp70 chaperone HSPA1A) were up-regulated (log2 mycolactone/control ratio >0.5; Fig. 2 A and Table S1). Fig. 2 B compares the distribution of mycolactone–down-regulated, identified, and all human proteins across the different subcellular compartments. In contrast to cytoplasmic and nuclear proteins, the incidence of plasma membrane– and ER-located proteins was increased in mycolactone–down-regulated proteins, compared with identified proteins (Fig. 2 B), indicating a selective down-regulation of these proteins by mycolactone. A key word analysis confirmed this observation and revealed an additional enrichment in glycoproteins, immunoglobulin domain–containing proteins, and proteins involved in the immune response among mycolactone–down-regulated proteins (Fig. 2 C).
Figure 2.
Mycolactone is a broad-spectrum inhibitor of Sec61. (A) Scatter plot showing the log2 SILAC ratios for individual proteins quantified in analysis 1 on the x axis (light condition: 40 nM mycolactone [Myco]; heavy condition: vehicle control [Ctrl]) and analysis 2 on the y axis (reversed conditions). Proteins with a log2 ratio <−0.5 (pink square) or >0.5 (blue square) in both analyses were considered modulated by mycolactone. CRTAM, cytotoxic and regulatory T cell molecule. (B and C) Gene ontology cellular component (B) and SwissProt Protein Information resource keywords (C) annotation analyses of the proteins that were reproducibly down-regulated in mycolactone-exposed T cells (red; n = 52), compared with identified proteins (blue; n = 6,503) and all human proteins in UniProt (gray; n = 20,204). (D) Distribution of downregulated (red), identified (blue), and all human proteins (gray) over different categories of membrane proteins. (B–D) Statistics were calculated by Fisher exact tests comparing downregulated versus identified proteins. ***, P < 0.001. (E) IVT assays of various Sec61 clients in the presence of increasing concentrations of mycolactone. ER translocation of nonglycosylated proteins (prolactin, IFN-γ, and BiP) was assessed by treatment with proteinase K (PK), with resistance to proteinase K indicating correct translocation into the ER lumen. Detergent-treated controls are shown in Fig. S1 C. It should be noted that BiP is largely protease resistant and, upon proteinase K treatment, forms shorter fragments. Translocation of glycosylated proteins (Mcln, LEP, and CIG30) was assessed by analyzing the change in migration in SDS-PAGE and autoradiography. Glycosidase-treated controls are shown in Fig. S1 C. The data shown are from one of two independent experiments, which gave similar results.
Consistent with Sec61 inhibition, 42 of the 52 mycolactone–down-regulated proteins contained a signal sequence or transmembrane domain directing newly synthesized proteins to the translocon (Table S1). The mycolactone–down-regulated subset was significantly enriched in single-pass type I/II membrane proteins (Fig. 2 D), indicating that such proteins are particularly susceptible to Sec61 inhibition by mycolactone. In contrast, the incidence of multipass membrane proteins was comparable between down-regulated proteins and identified proteins, suggesting that some multipass membrane proteins may bypass mycolactone-mediated blockade of Sec61. To test this hypothesis, mRNAs for various Sec61 substrates were translated in a reconstituted mammalian translation system in the presence of canine rough microsomes (CRMs), [35S]methionine, and increasing concentrations of mycolactone. In accordance with previously reported in vitro translation (IVT) assays of Sec61-dependent secretory and type II transmembrane protein TNF (Hall et al., 2014; McKenna et al., 2016), ER translocation of secreted prolactin and IFN-γ was efficiently and dose-dependently suppressed by mycolactone (Fig. 2 E and Fig. S1 C). Translocation of ER-resident BiP (also known as HSPA5) was also affected, confirming the SILAC data (Fig. 2 E and Table S1). BiP being a critical mediator of Sec61-dependent translocation, its depletion may contribute indirectly to the defective biogenesis of Sec61 clients in mycolactone-exposed cells. Multipass membrane proteins mucolipin 1 (Mcln) and a synthetic multipass membrane protein derived from Escherichia coli leader peptidase (LEP; Lundin et al., 2008) were also susceptible to mycolactone in IVT assays. In contrast, the multipass ER membrane protein CIG30 (Monné et al., 1999) was consistently resistant to mycolactone concentrations up to 1 µM (Fig. 2 E and Fig. S1 C). The SILAC and in vitro assays of protein translocation are thus fully consistent with mycolactone being a broad-acting inhibitor of Sec61 client production, with a more selective activity on multipass membrane proteins.
Sec61 blockade affects IFN-γ signaling in Jurkat T cells
Among the 52 proteins found to be down-regulated by mycolactone in PMA/IO-stimulated Jurkat T cells, 10 did not contain a signal sequence or transmembrane domain identifying them as a Sec61 client (Table S1). They were all encoded by IFN-stimulated genes (nine by IFN-γ and one by IFN-α), leading us to examine the effects of mycolactone on both the production of IFNs and the cell’s response to exogenous IFNs. Consistent with a previous study, exposing Jurkat T cells to mycolactone for 1 h before PMA/IO activation efficiently prevented IFN-γ production (Fig. 3 A), despite robust IFNG mRNA induction (Fig. 3 B; Phillips et al., 2009). Moreover, mycolactone-treated cells rapidly lost the ability to respond to IFN-γ: T cells exposed to mycolactone for >20 min before 20-min stimulation with IFN-γ showed reduced STAT1 phosphorylation (Fig. 3 C). The IFN-γ receptor (IFNGR) was not detected by our SILAC analysis, likely because protein level was below the detection limit. Yet, using flow cytometry, we found that a 6-h exposure to mycolactone led to a 60% reduction in T cell surface expression of IFNGR1 (Fig. 3 D). Because the level of IFNGR1 transcripts was not altered in mycolactone-treated cells (Fig. 3 E), the loss of IFNGR1 likely results from Sec61 blockade. We conclude that both the reduced IFN-γ production and the loss of IFNGR1 impair the IFN-γ autocrine loop in PMA/IO-activated T cells exposed to mycolactone. This was further indicated by the reduced accumulation of IFN-γ–inducible GBP2 at the mRNA level (Fig. 3 F) and a Western blot analysis validating our SILAC observation that mycolactone down-regulates GBP2 protein levels in activated Jurkat T cells (Fig. 3 G and Table S1). We also measured T cell responses to IFN-α. In Jurkat T cells exposed for 6 h to mycolactone, the surface level of the type I IFN receptor subunits (IFNAR1 and IFNAR2) was also reduced but to a lesser extent than IFNGR1 (Fig. 3 D). Consistently, phosphorylation of STAT1/3 was barely affected in T cells exposed to mycolactone for 6 h and then pulsed with IFN-α for 20 min. However, after a 24-h exposure, IFN-α signaling declined considerably (Fig. 3 H). Altogether, our SILAC data show that the magnitude and kinetics of mycolactone effects vary between Sec61 substrates, likely reflecting differences in protein turnover rates.
Figure 3.
Mycolactone targets primarily the IFN-γ signaling pathway in Jurkat T cells. (A) Production of IFN-γ by Jurkat T cells treated with 20 nM mycolactone (Myco) or vehicle (Ctrl) for 6 h (Resting) or for 1 h before 6 h of activation with PMA/IO. (B) Quantitation of IFNG mRNAs in Jurkat T cells treated with mycolactone or vehicle for 6 h or for 1 h before 3 or 6 h of activation with PMA/IO. (C) Western blot analysis of tyrosine phosphorylated (STAT1-P) and total STAT1 in Jurkat T cells treated with mycolactone or vehicle for the indicated times before activation with 1 ng/ml IFN-γ for 20 min or left unstimulated (Unst). (D) Flow cytometric analysis of surface expression of IFNGR1, IFNAR1, and IFNAR2 by Jurkat T cells incubated with or without mycolactone for 6 h. (E) Quantification of IFNGR1 mRNAs in Jurkat T cells treated as in B. (F and G) Quantitation of GBP2 mRNAs (F) and total GBP2 protein (G) in Jurkat T cells treated as in B. (H) Western blot analysis of phosphorylated (STAT1-P, STAT2-P, and STAT3-P) and total STAT1, STAT2, and STAT3 with β-actin as the loading control in Jurkat T cells treated with mycolactone or vehicle for 24 h before activation with 1 ng/ml IFN-γ or IFN-α for 20 min or left unstimulated (Unst). (A and D) Data are mean IFN-γ levels or mean fluorescence intensity, respectively, ± SEM of one experiment performed in triplicate, relative to vehicle controls. (B, E, and F) Data are mean fold-changes ± SEM of one experiment performed in duplicate, compared with resting controls. Similar results were obtained in independent experiments. (A, C, G, and H) Data are from one of two independent experiments, which gave similar results.
The R66G mutation in Sec61α confers broad resistance to mycolactone
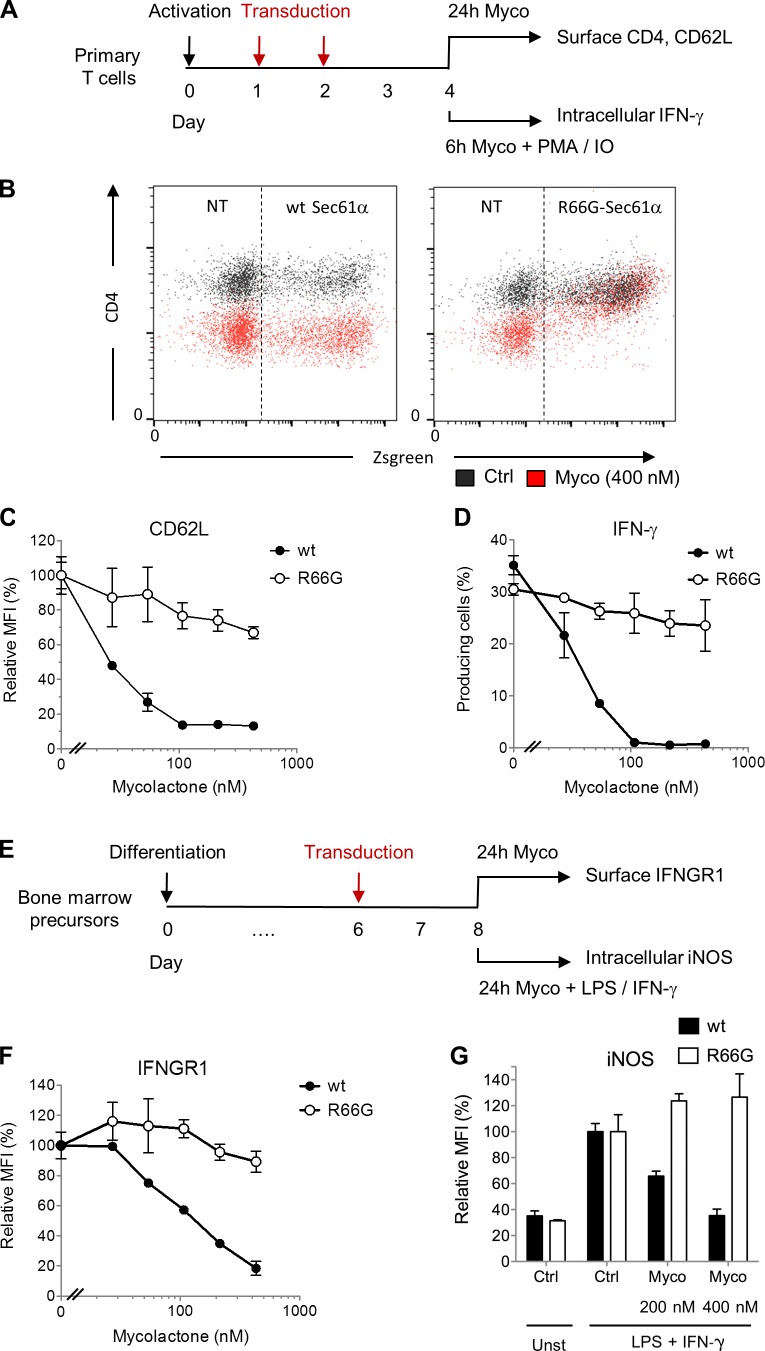
Production of IFN-γ by T cells and IFN-γ–driven expression of inducible nitric oxide synthase (iNOS) in infected macrophages are both essential for control of mycobacterial infection (Flynn and Chan, 2001). M. ulcerans is no exception, as shown by the reduced capacity of IFN-γ knockout mice to kill intracellular bacilli during the early intramacrophage growth phase of the bacteria (Bieri et al., 2016). To evaluate the contribution of Sec61 to mycolactone virulence, we examined whether mycolactone-resistant Sec61 mutants rescued the generation of antimycobacterial immune responses. Primary T cells isolated from mouse lymphoid organs were transduced with retroviral vectors for overexpression of WT Sec61α or R66G-Sec61α and fluorescent reporter protein Zsgreen (Fig. 4 A). Nontransduced and WT Sec61α–transduced cells were equally susceptible to mycolactone treatment, as demonstrated by the comparable inhibition of CD4 expression in mycolactone-exposed Zsgreen+ and Zsgreen− cells (Fig. 4 B). Strikingly, expression of R66G-Sec61α conferred resistance to mycolactone-induced defects in CD4 expression (Fig. 4 B). We reported previously that mycolactone efficiently down-regulates the expression of CD62L at the surface of naive T cells (Guenin-Macé et al., 2011). Similar to CD4, CD62L expression resisted mycolactone treatment in T cells expressing R66G-Sec61α but not WT Sec61α (Fig. 4 C). Further, in T cells transduced with R66G-Sec61α and stimulated with PMA/IO, the production of IFN-γ was unaffected by mycolactone treatment (Fig. 4 D). This demonstrated that defects in cytokine production are also fully corrected by expression of R66G-Sec61α. A similar approach was used to assess the functional impact of Sec61 inhibition in macrophages (Fig. 4 E). Transduction of R66G-Sec61α, but not WT Sec61α, in bone marrow–derived macrophages conferred resistance to mycolactone-mediated inhibition of IFNGR1 expression (Fig. 4 F). This reestablished the bactericidal capacity of macrophages, as LPS + IFN-γ–driven production of iNOS was restored in macrophages expressing R66G-Sec61α but not WT Sec61α (Fig. 4 G). Thus, by inhibiting Sec61 activity, mycolactone prevents both IFN-γ production by T cells and macrophage responsiveness to IFN-γ stimulation.
Figure 4.
The R66G mutation in Sec61α confers resistance to mycolactone. (A) Primary mouse T cells were activated with anti-CD3/CD28 antibodies and then transduced with WT Sec61α or R66G-Sec61α before exposure to mycolactone (Myco) in resting or PMA/IO-stimulated conditions. (B) Differential effect of mycolactone (24 h at 400 nM) on the CD4 surface expression of WT Sec61α– or R66G-Sec61α–transduced (Zsgreen+) cells. Data are mean fluorescence intensity (MFI) from one of two independent experiments, which gave similar results. Ctrl, vehicle control; NT, nontransduced. (C) Dose-dependent effect of mycolactone on the CD62L surface expression of WT Sec61α– or R66G-Sec61α–transduced (Zsgreen+-gated) cells. (D) Effect of a 1-h pretreatment with increasing doses of mycolactone on the PMA/IO-induced production of IFN-γ by primary T cells transduced with WT Sec61α or R66G-Sec61α (Zsgreen+-gated) cells. (E) Bone marrow–derived macrophages were transduced with WT Sec61α or R66G-Sec61α before exposure to mycolactone in resting or LPS + IFN-γ–stimulated conditions. (F) Dose-dependent effect of mycolactone on the IFNGR1 surface expression of WT Sec61α– or R66G-Sec61α–transduced (Zsgreen+-gated) cells. (G) Dose-dependent effect of mycolactone on the LPS + IFN-γ–induced production of iNOS by WT Sec61α– or R66G-Sec61α–transduced (Zsgreen+-gated) cells. (C, D, F, and G) Data are mean fluorescence intensity or mean cell percentages ± SEM of triplicates, relative to vehicle controls. They are from one of two independent experiments, which gave similar results.
Mycolactone suppresses Sec61 activity in T cells in vivo
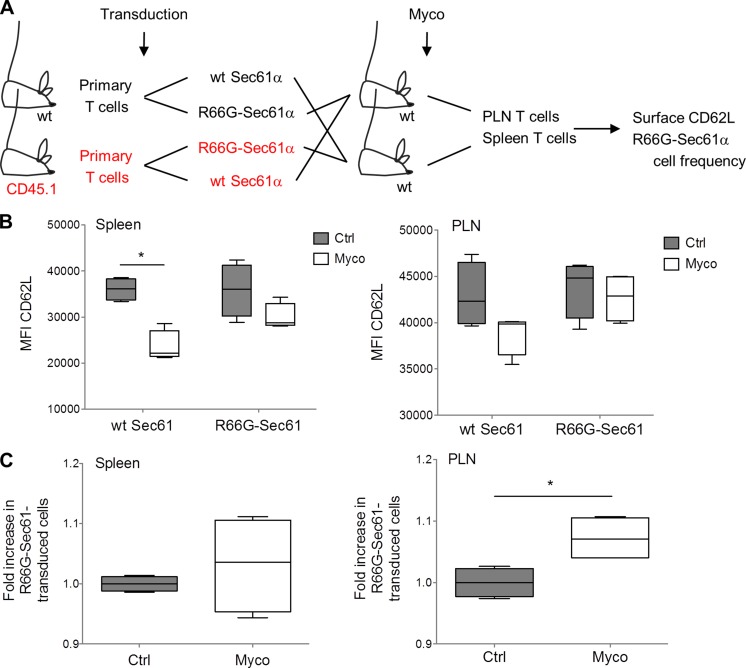
The data in Fig. 4 C show that CD62L expression by mouse primary T cells is highly susceptible to mycolactone-induced inhibition of Sec61. Using this membrane receptor as a readout, we next investigated whether systemically delivered mycolactone impacts Sec61 activity in adoptively transferred T cells. Because mycolactone-induced loss of CD62L impairs T cell capacity to reach peripheral LNs (PLNs; Guenin-Macé et al., 2011), we also examined whether Sec61 blockade results in impaired homing properties. Primary T cells isolated from WT (C57BL/6J and CD45.2+) and congenic CD45.1 mice were transduced with WT Sec61α or R66G-Sec61α (Fig. 5 A). CD45.2+ WT Sec61α–transduced cells were then mixed with CD45.1+ R66G-Sec61α–transduced cells in equal proportions and vice versa. Each mix of cells was then injected intravenously into WT recipient mice. Concomitantly, mice were given an intraperitoneal injection of 1 mg/kg mycolactone, a treatment previously shown to induce antiinflammatory effects in vivo (Guenin-Macé et al., 2015). After 24 h, the mean surface expression of CD62L and the relative proportions of WT Sec61α– and R66G-Sec61α–transduced cells in PLN and spleen were determined by FACS analysis. In mycolactone-injected mice, the expression of CD62L was reduced in WT Sec61α– but not R66G-Sec61α–transduced T cells from the spleen (Fig. 5 B, left). A similar trend was observed in T cells from the PLN (Fig. 5 B, right). This experiment demonstrated that mycolactone modulates T cell expression of CD62L in vivo in a Sec61-dependent manner. Notably, R66G-Sec61α–transduced T cells were recovered from PLNs at significantly higher frequencies than WT Sec61α–transduced T cells (Fig. 5 C, right). These frequencies were instead comparable in the spleen, consistent with CD62L not being critical for T cell homing to this organ (Fig. 5 C, left). Therefore, mycolactone down-regulates both Sec61-dependent expression of CD62L and CD62L-dependent lymphocyte homing in vivo.
Figure 5.
Mycolactone suppresses Sec61 activity in T cells in vivo. (A) Primary T cells isolated from WT (C57BL/6J and CD45.2+) and congenic CD45.1 mice were transduced with WT Sec61α or R66G-Sec61α and mixed, as depicted. Each cell mix was injected intravenously into four recipient mice, two of which received concomitantly an intraperitoneal injection of mycolactone (Myco) and, the other two, vehicle as control. (B) CD62L surface expression on WT Sec61α and R66-Sec61α T cells (CD45.1+ or CD45.1−; Zsgreen+ gated) recovered from the spleen and PLN. Ctrl, vehicle control; MFI, mean fluorescence intensity. (C) Relative proportion of R66G-Sec61α cells, compared with WT Sec61α cells, in the spleen and PLN. (B and C) Data are mean fluorescence intensity (B) and mean cell numbers (C) in each experimental group, presented as box and whiskers (*, P ≤ 0.05, Mann-Whitney test, each box corresponding to four experimental values). They are representative of two independent experiments giving similar results.
In conclusion, we have shown that mycolactone-induced Sec61 blockade is caused by a direct interaction with Sec61α, which determines mycolactone’s ability to prevent the generation of innate and adaptive immune responses. These data provide a molecular explanation for the immunological defects of BU patients. More generally, they highlight the critical importance of Sec61 activity for immune cell function, migration, and communication. Compared with CT8, mycolactone was more cytotoxic in human primary dermal fibroblasts and equally poorly cytotoxic in Jurkat T cells (Fig. S2 A). It was more effective than CT8 at inhibiting the production of cytokines and homing receptors by immune cells, and our on-going investigations suggest that mycolactone is also more potent than apratoxin A in these bioassays (not depicted). Among known inhibitors of Sec61, mycolactone is therefore the first produced by a human pathogen and likely the most potent.
Mycolactone was previously reported to bind and activate N–Wiskott–Aldrich syndrome protein (N-WASP) and type 2 angiotensin II receptor (AT2R) to mediate skin ulceration and analgesia, respectively (Guenin-Macé et al., 2013; Marion et al., 2014). Silencing of N-WASP/WASP or AT2R in relevant cell models did not modify the inhibitory effect of mycolactone on the production of secreted and membrane proteins (Fig. S2, B and C), showing that the immunomodulatory properties of mycolactone are independent of these proteins. It is nevertheless possible that Sec61 inhibition mediates or at least contributes to the ulcerative and analgesic properties of mycolactone.
Altogether, our data reveal a novel mechanism of immune evasion evolved by pathogenic mycobacteria that targets host cell protein translocation. Inhibition of Sec61 activity efficiently prevented the production of key mediators of innate and adaptive immune responses against intracellular pathogens, as we demonstrated for IFN-γ and IFN-γ receptor. The discovery that mycolactone inhibits Sec61 opens novel perspectives beyond the field of inflammation. Because CT8 was effective at limiting proteostasis of enveloped viruses (Heaton et al., 2016), mycolactone may similarly show broad antiviral activity. It may also prove useful in the treatment of pathologies associated with elevated secretory protein synthesis. Genetically modifying Sec61 demonstrated the specificity of mycolactone binding to the translocon. Because Sec61 clients are expressed in a cell type–specific manner, mycolactone-mediated inhibition of protein translocation into the ER could underpin the variety of its effects in different cell types and the distinctive features of BU.
Materials and methods
Reagents and expression vectors
All experiments using mycolactone were done with natural mycolactone A/B purified from M. ulcerans bacteria (strain 1615; 35840; ATCC) and then quantified by spectrophotometry (λmax = 362 nm; log ε = 4.29; Spangenberg and Kishi, 2010). Synthetic modules of mycolactone (4a, 5a, and 5b) were generated as previously described (Chany et al., 2011). Stock solutions were prepared in either ethanol or DMSO and then diluted 1,000× in culture medium for cellular assays or 10× in PBS before injection in mice. CT7, CT8, and CT9 were prepared as previously described (MacKinnon et al., 2007; Maifeld et al., 2011). Sec61 WT or mutant sequences were cloned upstream of an internal ribosome entry site (IRES) of the pRetroX–IRES-Zsgreen retroviral vector (Takara Bio Inc.) for simultaneous translation of Sec61α and Zsgreen in mouse primary T cells and macrophages.
SDS-PAGE, autoradiography, and Western blotting
Cell lysates were resolved on NuPAGE Bis-Tris gels and transferred to nitrocellulose membranes (Thermo Fisher Scientific). For autoradiography, dried Tris-tricine gels were exposed to a storage phosphorus screen (GE Healthcare) and imaged on a Typhoon Trio phosphorimager (GE Healthcare). Protein detections used the following antibodies: WASP F-8 (sc-365859; Santa Cruz Biotechnology, Inc.), N-WASP 30D10 (no. 4848; Cell Signaling Technology), pSTAT1 Y701 (no. 9171L; Cell Signaling Technology), pSTAT2 Y689 (no. 07-224; EMD Millipore), pSTAT3 Y705 (no. 9131L; Cell Signaling Technology), STAT1 (no. 06-501; EMD Millipore), STAT2 (06-502; EMD Millipore), STAT3 (sc-7179; Santa Cruz Biotechnology, Inc.), AT2R (sc-9040; Santa Cruz Biotechnology, Inc.), β-actin (no. 3700; Cell Signaling Technology), GAPDH (no. 2118; Cell Signaling Technology), and Sec61α (NB120-15575; Novus Biologicals). Here, complexes were revealed with ECL Prime detection reagent (GE Healthcare) and chemiluminescence reading on a luminescent image analyzer (LAS-4000; Fujifilm).
Photoaffinity labeling
Protocols for CRM preparation and CT7 photoaffinity labeling and click chemistry were described previously (Walter and Blobel, 1983; MacKinnon et al., 2007). In brief, CRMs equivalent to 100 nM Sec61 were treated with 1 or 10 µM mycolactone or DMSO for 30 min at 0°C, followed by incubation with 100 nM CT7 for 10 min at room temperature. Samples were then photolyzed for 10 min, and cross-linked proteins were detected by click chemistry, SDS-PAGE, and in-gel fluorescence scanning. In Fig. 1 D, 50 µl of photoaffinity-labeling reactions were treated with 10 µM mycolactone on ice before (pre) or after (post-) three rounds of membrane pelleting. A third sample was treated after pelleting with an equal volume DMSO. All the samples were further incubated for 30 min on ice prior to CT7 photoaffinity labeling.
IVT assays
Protein translocation assays were performed as described previously (Sharma et al., 2010): DNA templates encoding the indicated constructs were transcribed with T7 or SP6 polymerase (New England Biolabs, Inc.) for 1–2 h at 37°C and used in subsequent translation/translocation reactions. The reactions were assembled at 0°C in the presence of mycolactone or an equivalent volume of solvent. Reactions included [35S]methionine (2 µCi per 10 µl translation; PerkinElmer) and CRM. The amount of CRM was optimized to be 0.25 µl per 10 µl reaction volume. Translation was initiated by transferring the reactions to 32°C for 30 or 60 min and stopped by returning reactions onto ice. Translocation of nonglycosylated proteins was assessed by treating the samples with proteinase K for 1 h at 0°C. An aliquot was incubated in the presence of TX-100 to demonstrate protection by CRMs. Proteinase digestion was stopped with PMSF and boiling in the presence of SDS. After TCA precipitation, the remaining, protected proteins/protein fragments were analyzed with SDS-PAGE and autoradiography. The translocation of glycosylated proteins was analyzed by SDS-PAGE and autoradiography. Alternatively, the control samples were first denatured and treated with endoglycosidase H (EndoH) to demonstrate that differences in gel migration are based on glycosylation.
Cell cultures
Jurkat T cells (E6.1 clone; no. 88042803; European Collection of Authenticated Cell Cultures [ECACC]), HeLa cells (no. 93021013; ECACC), human primary dermal fibroblasts (C-013-5C; Thermo Fisher Scientific), and HEK293-FRT TRex cells stably expressing WT or mutant Sec61α were cultured in RPMI GlutaMAX (Jurkat) or DMEM GlutaMAX (other cells) from Thermo Fisher Scientific, supplemented with 10% heat-inactivated FCS (Invitrogen), 100 U/ml penicillin, and 100 µg/ml streptomycin. Human primary T cells were isolated from blood donors by Ficoll density gradient centrifugation and CD4+ T cell purification by negative depletion (Miltenyi Biotec). Human primary macrophages were obtained from peripheral blood-derived monocytes, isolated by adhesion to tissue culture plasticware, and cultured with 10 ng/ml human GM-CSF (PeproTech) for 7–12 d. Mouse CD3+ primary T cells were isolated from spleens and LNs by negative selection using the Pan T cell isolation kit (Miltenyi Biotec) and then placed in RPMI medium supplemented with 10% heat-inactivated FCS, 10 mM Hepes, 1 mM pyruvate, and 25 µM 2-mercaptoethanol. Bone marrow–derived macrophages were obtained by a 7-d differentiation of mouse progenitors in DMEM supplemented with 20% heat-inactivated horse serum (Gibco) and 30% L929-conditioned medium as a source of M-CSF.
SILAC labeling and LC-MS/MS analysis
For SILAC labeling, Jurkat T cells were cultured in DMEM medium without l-lysine, l-arginine, or l-glutamine (Silantes Gmbh) supplemented with 10% heat-inactivated FCS (Invitrogen), 2 mM GlutaMAX, and either natural l-arginine HCl and l-lysine HCl (light labeling; Sigma-Aldrich) or [13C6][15N2] l-lysine HCl and [13C6]l-arginine HCl (heavy labeling; Silantes Gmbh). l-Lysine HCl was added at its normal concentration in DMEM (146 mg/L), but the concentration of l-arginine HCl was reduced to 30 mg/L (36% of the normal concentration in DMEM) to prevent metabolic conversion of arginine to proline. Cells were kept for at least six population doublings to ensure complete incorporation of the labeled lysine and arginine. Light (L) and heavy (H) SILAC-labeled Jurkat T cells were treated with 40 nM mycolactone or vehicle as control for 1 h and then activated with PMA/IO for 6 h. Two experiments were performed in reverse labeling conditions, yielding four samples. From each condition, 5 × 106 cells were harvested and washed twice with PBS, and cell pellets were frozen at −80°C until further use. Each pellet was resuspended in 500 µl lysis buffer (9 M urea in 20 mM Hepes, pH 8.0), sonicated (three bursts of 15 s at an amplitude of 20%) and centrifuged for 15 min at 16,000 g at 4°C to remove insoluble material. The protein concentration in the supernatants was measured using a Bradford assay (Bio-Rad Laboratories), and equal protein amounts of mycolactone-treated and untreated cell lysates were mixed to obtain two replicate samples with reversed SILAC for further analysis, each containing 5.6 mg total protein (sample 1: vehicle [H] + mycolactone [L]; sample 2: vehicle [L] + mycolactone [H]). Proteins in each sample were reduced with 5 mM dithiothreitol and incubation for 30 min at 30°C and then alkylated by addition of 100 mM chloroacetamide for 15 min at room temperature in the dark. Both samples were further diluted with 20 mM Hepes, pH 8.0, to a final urea concentration of 2 M, and proteins were digested with 50 µg trypsin (1/113, wt/wt; Promega) overnight at 37°C. Peptides were then purified on a Sep-Pak C18 cartridge (Waters), and 500 µg of peptides of each sample was redissolved in 10 mM ammonium acetate, pH 5.5, in water/acetonitrile (98/2, vol/vol) and injected on a capillary reversed phase high-performance liquid chromatography column (Zorbax 300SB-C18; 2.1 mm internal diameter and 150 mm length; Agilent Technologies) using a high-performance liquid chromatography system (1200 Series; Agilent Technologies). Peptides were separated by a linear gradient of acetonitrile (from 2% to 70% in 100 min in 10 mM ammonium acetate, pH 5.5), and peptides that eluted between 20 and 92 min were collected in 72 fractions of 1 min each. Fractions with 12-min difference in retention time were pooled to obtain total of 12 fractions for LC-MS/MS per sample. Peptides in each fraction were dried and redissolved in 12 µl of solvent A (0.1% formic acid in water/acetonitrile; 98:2, vol/vol), of which 5 µl was injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano system (Thermo Fisher Scientific) in-line connected to a Q Exactive mass spectrometer with a Nanospray Flex Ion source (Thermo Fisher Scientific). Trapping was performed at 10 µl/min for 3 min in solvent A on a PepMap C18 column (0.3 mm inner diameter × 5 mm; Dionex), and after back flushing from the trapping column, the sample was loaded on a reverse-phase column (made in house; 75 µm inner diameter × 500 mm; 1.9 µm beads C18 Reprosil-Pur; Dr. Maisch GmbH). Peptides were eluted by an increase in solvent B (0.08% formic acid in water/acetonitrile; 2:8, vol/vol) in linear gradients from 5% to 20% in 47 min, then from 20% to 40% in 150 min, and finally from 40% to 55% in 30 min, all at a constant flow rate of 300 nl/min. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 15 most abundant ion peaks per MS spectrum. Full-scan MS spectra (300–2,000 m/z) were acquired at a resolution of 70,000 after accumulation to a target value of 1,000,000 with a maximum fill time of 100 ms. The 15 most intense ions above a threshold value of 100,000 were isolated (window of 2.5 Th) for fragmentation by collision-induced dissociation at a normalized collision energy of 27% after filling the trap at a target value of 100,000 for a maximum of 160 ms with an underfill ratio of 0.1%. The S-lens radio frequency level was set at 55, and we excluded precursor ions with single, unassigned, and charge states above six from fragmentation selection.
Data processing and gene ontology terms enrichment analysis
Data analysis was performed with MaxQuant software (version 1.4.1.2; Cox and Mann, 2008) using the Andromeda search engine (Cox et al., 2011) with default search settings including a false discovery rate set at 1% on both the peptide and protein levels. Spectra were searched against the human proteins in the UniProt/SwissProt database (database release version of January 2014 containing 20,272 human protein sequences) with a mass tolerance for precursor and fragment ions of 4.5 and 20 ppm, respectively, during the main search. To enable the identification of SILAC-labeled peptides, the multiplicity was set to two with Lys8 and Arg6 settings in the heavy channel, allowing for a maximum of three labeled amino acids per peptide. Enzyme specificity was set as C-terminal to arginine and lysine, also allowing cleavage at proline bonds and a maximum of two missed cleavages. Variable modifications were set to oxidation of methionine residues and acetylation of protein N termini. Carbamidomethyl formation of cysteine residues was set as a fixed modification. In total, 6,503 proteins were identified in both samples, of which 4,636 proteins were quantified. For each quantified protein, the log2 values of the normalized mycolactone/untreated ratio in both samples were plotted against each other to generate the scatter plot depicted in Fig. 2 and Table S1. Proteins with log2 (mycolactone/untreated ratios) <−0.5 in both samples were considered as specific mycolactone targets that are down-regulated upon treatment. Proteomic data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository under accession no. PXD002971. Gene Ontology and SwissProt Protein Information resource terms enrichment analyses were performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources (Huang et al., 2009). Information on the topology of membrane proteins were retrieved from the UniProt/SwissProt database.
Flow cytometry
Staining of mouse cells was performed using anti-CD4 (no. 553051; BD), anti-CD62L (l-selectin; no. 553162; BD), anti-CD3 (no. 553064; BD), anti-CD19 (no. 550992; BD), anti-CD45.1 (no. 5061788; BD), anti-IFNGR1 (130-104-988; Miltenyi Biotec), anti–IFN-γ (no. 554412; BD), and anti–IL-2 (no. 554429; BD). For intracellular staining of cytokines, cells were treated with mycolactone for 1 h and then activated with PMA/IO. GolgiStop (BD) was added 2 h later. After 6 h, cells were fixed with 4% (wt/vol) paraformaldehyde during 20 min at room temperature and then stained with PE-conjugated anti–IFN-γ antibodies (BD) in 100 µl PBS + 0.1% BSA + 0.5% saponin for 30 min at room temperature. For intracellular staining of iNOS, macrophages were fixed with Lyse/Fix solution (no. 558049; BD) for 10 min at 37°C and then permeabilized with Perm Buffer III (no. 558050; BD) for 20 min at 4°C. Staining was performed with goat anti-NOS2 (sc-650-G; Santa Cruz Biotechnology, Inc.) followed by an anti–goat secondary antibody (no. 96938; Abcam). Staining of Jurkat was performed using anti-IFNGR1 (no. 558937; BD), IFNAR1 (no. 550331; BD), and IFNAR2 (no. 1080-08; SouthernBiotech). In brief, human cells were stained with IFNAR1 or IFNAR2, washed twice with PBS, incubated with biotin-conjugated rat anti–mouse IgG (no. 415-065-166; Jackson ImmunoResearch Laboratories, Inc.), washed, and then incubated with R-PE–conjugated streptavidin (PNIM0557; Beckman Coulter). All FACS acquisition was performed on a FACS Accuri C6 flow cytometer (BD), and data were analyzed using FlowJo software (Tree Star).
Retroviral transduction
Platinum-E ecotropic packaging cells (plat E; Biolabs) transfected with pRetroX–IRES-Zsgreen plasmids containing Sec61α sequences were used to produce retroviral particles. Immediately after isolation from mouse organs, CD3+ T cells were activated with Dynabeads Mouse T-activator CD3/CD28 (Miltenyi Biotec) with 1 bead/cell in RPMI medium supplemented with 10% heat-inactivated FCS, 10 mM Hepes, 1 mM pyruvate, and 25 µM 2-mercaptoethanol (complete medium). 24 h later, cells were centrifuged at 1,200 rpm, and the supernatant (conditioned medium) was saved. Cells were resuspended at 4 × 106 cells/ml in viral supernatant freshly collected from plat E cells and supplemented with 10 µg/ml polybrene (EMD Millipore) and distributed at 1 ml/well in a 6-well plate and spin infected for 1 h at 2,800 rpm and 32°C. The cell supernatant was then removed and replaced with conditioned medium. After 48 h, spin infection was repeated, and T cells were resuspended in complete medium containing 50% conditioned medium. Bone marrow–derived macrophages were plated in 12- or 24-well plates (2–4 × 105 cells/well) for 20 h. Fresh viral supernatant collected from plat E cells and 10 µg/ml polybrene were added before spin infection for 1 h at 2,800 rpm and 32°C. The cell supernatant was then removed and replaced with fresh DMEM supplemented with 20% horse serum.
Bioassays
The cytopathic effect of mycolactone on HEK293-FRT cells was assessed after 72 h of exposure with the Alamar blue assay (Thermo Fisher Scientific). Its effect on secretory protein production was assessed with the Gaussia Glow-Juice Luciferase kit (PJK GmbH) as follows. HEK293-FRT cell lines expressing WT Sec61α or R66G-Sec61α were grown on a 6-well plate and then transfected with a plasmid encoding a signal sequence-containing Gaussia luciferase using Fugene 6 reagent (Promega). The expression of both the luciferase and Sec61α was induced 5 h later by addition of 1 µg/ml doxycycline. On the next day, 200,000 cells/well were plated in 96-well plates and treated 5 h later with increasing concentrations of mycolactone. Luciferase activity in culture media was measured 24 h later with an EnSpire Multimode plate reader (PerkinElmer). Assays of mycolactone inhibition on cytokine production and homing receptors by mouse and human immune cells have been described previously (Boulkroun et al., 2010; Guenin-Macé et al., 2011, 2015).
WASP/N-WASP and AT2R silencing
siRNAs were ON-Target plus SMARTpools (GE Healthcare) targeting human WASP (L-028294-00-0005), N-WASP (L-006444-00-0005), or AT2R (L-005429-00-005) or were nontargeting SMARTpool (D0018101005) as controls. 107 Jurkat T cells were electroporated twice at 48-h interval with 400 nM siRNA using the Gene Pulser Xcell system (Bio-Rad Laboratories) at 300 V and 500 µF. Silencing of WASP/N-WASP expression was optimal 24 h after the second electroporation. HeLa cells were transfected with 10 nM siRNA using Lipofectamine RNAiMAX (Invitrogen). AT2R silencing was optimal 48 h after transfection.
Mouse studies
8-wk-old female mice (C57BL/6NCrl; Charles River) or congenic CD45.1 mice (B6.SJL-Ptprca Pepcb/BoyCrlPas; from our animal facilities) were housed under pathogen-free conditions with food and water ad libitum. The described experiments received the approval of the French Ministry of Higher Education and Research. They were performed in compliance with national guidelines and regulations.
Statistical analysis
Two group comparisons used the Mann-Whitney rank test. Statistical analyses were performed with StatView 5 software (SAS Institute, Inc.), and values of P ≤ 0.05 were considered significant. Prism software (5.0d; GraphPad Software) was used for graphical representation.
Online supplemental material
Fig. S1 shows chemical structures of all mycolactone and cotransin analogues used in this study, mapping of mycolactone-resistance mutations in a three-dimensional model of Sec61α structure, and IVT controls. Fig. S2 shows cytotoxicity of mycolactone and CT8 in human fibroblasts and T lymphocytes and effects of WASP/N-WASP and AT2R silencing on mycolactone-mediated inhibition of secretory protein production. Table S1 shows mycolactone-susceptible proteins in Jurkat T cells, as detected by our SILAC analysis.
Supplementary Material
Acknowledgments
We thank T. Chaze and M. Matondo-Bouzanda from the Pasteur Proteomics platform for assistance with proteome analyses. D. Tranter is acknowledged for valuable technical assistance. We are also grateful to P. Cossart and A. Echard for critical reading of the manuscript.
This work was supported by the Fondation pour la Recherche Médicale (FRM 2012 DEQ20120323704 to C. Demangel), the Association Raoul Follereau (C. Demangel), the Région Ile de France (dim130027 to C. Demangel), the Academy of Finland (289737 to V.O. Paavilainen), the Sigrid Juselius Foundation (V.O. Paavilainen), and Biocentrum Helsinki (V.O. Paavilainen). F. Impens received financial support from a Pasteur-Roux Fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BiP
- binding immunoglobulin protein
- BU
- Buruli ulcer
- CRM
- canine rough microsome
- iNOS
- inducible nitric oxide synthase
- IO
- ionomycin
- IRES
- internal ribosome entry site
- IVT
- in vitro translation
- LC-MS/MS
- liquid chromatography–tandem MS
- MS
- mass spectrometry
- PLN
- peripheral LN
- SILAC
- stable-isotope labeling with amino acids in cell culture
- SRP
- signal recognition particle
- WASP
- Wiskott–Aldrich syndrome protein
References
- Besemer J., Harant H., Wang S., Oberhauser B., Marquardt K., Foster C.A., Schreiner E.P., de Vries J.E., Dascher-Nadel C., and Lindley I.J.. 2005. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 436:290–293. 10.1038/nature03670 [DOI] [PubMed] [Google Scholar]
- Bieri R., Bolz M., Ruf M.T., and Pluschke G.. 2016. Interferon-γ is a crucial activator of early host immune defense against Mycobacterium ulcerans infection in mice. PLoS Negl. Trop. Dis. 10:e0004450 10.1371/journal.pntd.0004450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulkroun S., Guenin-Macé L., Thoulouze M.I., Monot M., Merckx A., Langsley G., Bismuth G., Di Bartolo V., and Demangel C.. 2010. Mycolactone suppresses T cell responsiveness by altering both early signaling and posttranslational events. J. Immunol. 184:1436–1444. 10.4049/jimmunol.0902854 [DOI] [PubMed] [Google Scholar]
- Chany A.C., Casarotto V., Schmitt M., Tarnus C., Guenin-Macé L., Demangel C., Mirguet O., Eustache J., and Blanchard N.. 2011. A diverted total synthesis of mycolactone analogues: an insight into Buruli ulcer toxins. Chemistry. 17:14413–14419. 10.1002/chem.201102542 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Demangel C., Stinear T.P., and Cole S.T.. 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7:50–60. 10.1038/nrmicro2077 [DOI] [PubMed] [Google Scholar]
- Flynn J.L., and Chan J.. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93–129. 10.1146/annurev.immunol.19.1.93 [DOI] [PubMed] [Google Scholar]
- Garrison J.L., Kunkel E.J., Hegde R.S., and Taunton J.. 2005. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 436:285–289. 10.1038/nature03821 [DOI] [PubMed] [Google Scholar]
- George K.M., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., and Small P.L.. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 283:854–857. 10.1126/science.283.5403.854 [DOI] [PubMed] [Google Scholar]
- Guarner J., Bartlett J., Whitney E.A., Raghunathan P.L., Stienstra Y., Asamoa K., Etuaful S., Klutse E., Quarshie E., van der Werf T.S., et al. 2003. Histopathologic features of Mycobacterium ulcerans infection. Emerg. Infect. Dis. 9:651–656. 10.3201/eid0906.020485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin-Macé L., Carrette F., Asperti-Boursin F., Le Bon A., Caleechurn L., Di Bartolo V., Fontanet A., Bismuth G., and Demangel C.. 2011. Mycolactone impairs T cell homing by suppressing microRNA control of L-selectin expression. Proc. Natl. Acad. Sci. USA. 108:12833–12838. 10.1073/pnas.1016496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin-Macé L., Veyron-Churlet R., Thoulouze M.I., Romet-Lemonne G., Hong H., Leadlay P.F., Danckaert A., Ruf M.T., Mostowy S., Zurzolo C., et al. 2013. Mycolactone activation of Wiskott-Aldrich syndrome proteins underpins Buruli ulcer formation. J. Clin. Invest. 123:1501–1512. 10.1172/JCI66576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin-Macé L., Baron L., Chany A.C., Tresse C., Saint-Auret S., Jönsson F., Le Chevalier F., Bruhns P., Bismuth G., Hidalgo-Lucas S., et al. 2015. Shaping mycolactone for therapeutic use against inflammatory disorders. Sci. Transl. Med. 7:289ra85 10.1126/scitranslmed.aab0458 [DOI] [PubMed] [Google Scholar]
- Hall B., and Simmonds R.. 2014. Pleiotropic molecular effects of the Mycobacterium ulcerans virulence factor mycolactone underlying the cell death and immunosuppression seen in Buruli ulcer. Biochem. Soc. Trans. 42:177–183. 10.1042/BST20130133 [DOI] [PubMed] [Google Scholar]
- Hall B.S., Hill K., McKenna M., Ogbechi J., High S., Willis A.E., and Simmonds R.E.. 2014. The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog. 10:e1004061 10.1371/journal.ppat.1004061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S., Zhao N., Manganaro L., Hultquist J.F., Noel J., et al. 2016. Targeting viral proteostasis limits influenza virus, HIV, and Dengue virus infection. Immunity. 44:46–58. 10.1016/j.immuni.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Coutanceau E., Leclerc M., Caleechurn L., Leadlay P.F., and Demangel C.. 2008. Mycolactone diffuses from Mycobacterium ulcerans–infected tissues and targets mononuclear cells in peripheral blood and lymphoid organs. PLoS Negl. Trop. Dis. 2:e325 10.1371/journal.pntd.0000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Junne T., Wong J., Studer C., Aust T., Bauer B.W., Beibel M., Bhullar B., Bruccoleri R., Eichenberger J., Estoppey D., et al. 2015. Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J. Cell Sci. 128:1217–1229. 10.1242/jcs.165746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C., Kim H., Nilsson I., White S.H., and von Heijne G.. 2008. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for Nin-Cout and Nout-Cin transmembrane helices. Proc. Natl. Acad. Sci. USA. 105:15702–15707. 10.1073/pnas.0804842105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon A.L., Garrison J.L., Hegde R.S., and Taunton J.. 2007. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J. Am. Chem. Soc. 129:14560–14561. 10.1021/ja076250y [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon A.L., Paavilainen V.O., Sharma A., Hegde R.S., and Taunton J.. 2014. An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. eLife. 3:e01483 10.7554/eLife.01483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maifeld S.V., MacKinnon A.L., Garrison J.L., Sharma A., Kunkel E.J., Hegde R.S., and Taunton J.. 2011. Secretory protein profiling reveals TNF-α inactivation by selective and promiscuous Sec61 modulators. Chem. Biol. 18:1082–1088. 10.1016/j.chembiol.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion E., Song O.R., Christophe T., Babonneau J., Fenistein D., Eyer J., Letournel F., Henrion D., Clere N., Paille V., et al. 2014. Mycobacterial toxin induces analgesia in buruli ulcer by targeting the angiotensin pathways. Cell. 157:1565–1576. 10.1016/j.cell.2014.04.040 [DOI] [PubMed] [Google Scholar]
- McKenna M., Simmonds R.E., and High S.. 2016. Mechanistic insights into the inhibition of Sec61-dependent co- and post-translational translocation by mycolactone. J. Cell Sci. 129:1404–1415. 10.1242/jcs.182352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monné M., Gafvelin G., Nilsson R., and von Heijne G.. 1999. N-tail translocation in a eukaryotic polytopic membrane protein: synergy between neighboring transmembrane segments. Eur. J. Biochem. 263:264–269. 10.1046/j.1432-1327.1999.00498.x [DOI] [PubMed] [Google Scholar]
- Paatero A.O., Kellosalo J., Dunyak B.M., Almaliti J., Gestwicki J.E., Gerwick W.H., Taunton J., and Paavilainen V.O.. 2016. Apratoxin kills cells by direct blockade of the Sec61 protein translocation channel. Cell Chem Biol. 23:561–566. 10.1016/j.chembiol.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Park E., and Rapoport T.A.. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41:21–40. 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- Phillips R., Sarfo F.S., Guenin-Macé L., Decalf J., Wansbrough-Jones M., Albert M.L., and Demangel C.. 2009. Immunosuppressive signature of cutaneous Mycobacterium ulcerans infection in the peripheral blood of patients with buruli ulcer disease. J. Infect. Dis. 200:1675–1684. 10.1086/646615 [DOI] [PubMed] [Google Scholar]
- Sarfo F.S., Le Chevalier F., Aka N., Phillips R.O., Amoako Y., Boneca I.G., Lenormand P., Dosso M., Wansbrough-Jones M., Veyron-Churlet R., et al. 2011. Mycolactone diffuses into the peripheral blood of Buruli ulcer patients - implications for diagnosis and disease monitoring. PLoS Negl. Trop. Dis. 5:e1237 10.1371/journal.pntd.0001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Mariappan M., Appathurai S., and Hegde R.S.. 2010. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol. Biol. 619:339–363. 10.1007/978-1-60327-412-8_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg T., and Kishi Y.. 2010. Highly sensitive, operationally simple, cost/time effective detection of the mycolactones from the human pathogen Mycobacterium ulcerans. Chem. Commun. (Camb.). 46:1410–1412. 10.1039/b924896j [DOI] [PubMed] [Google Scholar]
- Walter P., and Blobel G.. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96:84–93. 10.1016/S0076-6879(83)96010-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.