Barrott et al. show that PI3′-lipid signaling potentiates metastasis in a genetically engineered mouse model of synovial sarcomagenesis and drives cancer cells to express CSF1, recruiting macrophages to the tumor microenvironment.
Abstract
Solid tumor metastasis is a complex biology, impinged upon by a variety of dysregulated signaling pathways. PI3′-lipid signaling has been associated with metastasis and inflammation in many cancers, but the relationship between tumor cell–intrinsic PI3′-lipid signaling and inflammatory cell recruitment has remained enigmatic. Elevated PI3′-lipid signaling associates with progression of synovial sarcoma, a deadly soft tissue malignancy initiated by a t(X;18) chromosomal translocation that generates an SS18-SSX fusion oncoprotein. Here, we show in genetically engineered mouse models of locally induced expression of SS18-SSX1 or SS18-SSX2 that Pten silencing dramatically accelerated and enhanced sarcomagenesis without compromising synovial sarcoma characteristics. PTEN deficiency increased tumor angiogenesis, promoted inflammatory gene expression, and enabled highly penetrant spontaneous pulmonary metastasis. PTEN-deficient sarcomas revealed infiltrating myeloid-derived hematopoietic cells, particularly macrophages and neutrophils, recruited via PI3′-lipid–induced CSF1 expression in tumor cells. Moreover, in a large panel of human synovial sarcomas, enhanced PI3′-lipid signaling also correlated with increased inflammatory cell recruitment and CSF1R signal transduction in both macrophages and endothelial cells. Thus, both in the mouse model and in human synovial sarcomas, PI3′-lipid signaling drives CSF1 expression and associates with increased infiltration of the monocyte/macrophage lineage as well as neutrophils.
Introduction
Synovial sarcoma (SS) is the most common soft-tissue sarcoma in adolescents and young adults (Herzog, 2005). Once metastatic, it is usually incurable. Characterized by a balanced t(X;18) chromosomal translocation that produces an SS18-SSX fusion oncogene (Turc-Carel et al., 1987; Clark et al., 1994; de Leeuw et al., 1995), SS has been shown to require few other genetic derangements beyond the fusion (Barretina et al., 2010; Przybyl et al., 2014; Mikami et al., 2015; Vlenterie et al., 2015; Jones et al., 2016).
Expression of either SS18-SSX1 or SS18-SSX2 has proven sufficient to drive synovial sarcomagenesis in the mouse (Haldar et al., 2007, 2009; Jones et al., 2016). These models, and most genetic models of fusion oncogene–driven sarcomagenesis, have fallen slightly short of displaying the full biology of metastasis (Haldar et al., 2007, 2009; Straessler et al., 2013; Goodwin et al., 2014; Jones et al., 2016). This has raised the question of what additional genetic or epigenetic changes are necessary for progression to metastasis, as distinct from initiation of sarcomagenesis (Przybyl et al., 2014; Jones et al., 2016).
One of the most frequent secondary genetic changes demonstrated in human SS is loss of the phosphatase and tensin homologue on chromosome 10 (PTEN) gene (Barretina et al., 2010). The negative regulator of phosphatidyl inositol 3′-lipid (PI3′-lipid) signaling, PTEN is a tumor suppressor gene that is frequently silenced or lost in many cancer types (Ali et al., 1999; Vivanco and Sawyers, 2002; Oda et al., 2005; Zbuk and Eng, 2007; Chalhoub and Baker, 2009). The resultant increase in PI3′-lipid signaling leads, among many effects, to phosphorylation and activation of protein kinase B (termed pAKT), which results in a variety of downstream signaling that encourages proliferation and prevents apoptosis (Tamura et al., 1998; Patel et al., 2001; Chow and Baker, 2006).
Loss of PTEN associates with tumor aggressiveness and progression to metastasis in some cancers (Depowski et al., 2001; Deocampo et al., 2003; Vasudevan et al., 2009; Kim et al., 2015). Such a relationship has not been shown specifically in SS, but increased PI3′-lipid signaling correlates with a worse prognosis in this disease (Friedrichs et al., 2011; Setsu et al., 2013). As a result of the many and pleiotropic effects of loss of PTEN, especially when interwoven into genetically complex cancers, such as melanoma and carcinomas of the lung, prostate, and breast, the precise contribution of PTEN loss to oncogenesis can be difficult to decipher (Oda et al., 2005; Stemke-Hale et al., 2008; Chalhoub and Baker, 2009). We therefore determined to test the impact of Pten silencing in the genetic mouse model of SS, in the hope that it could provide tractable information with regard to the biology of PI3′-lipid signaling in the progression of solid tumor malignancies.
Results
Pten silencing enhances synovial sarcomagenesis in the mouse
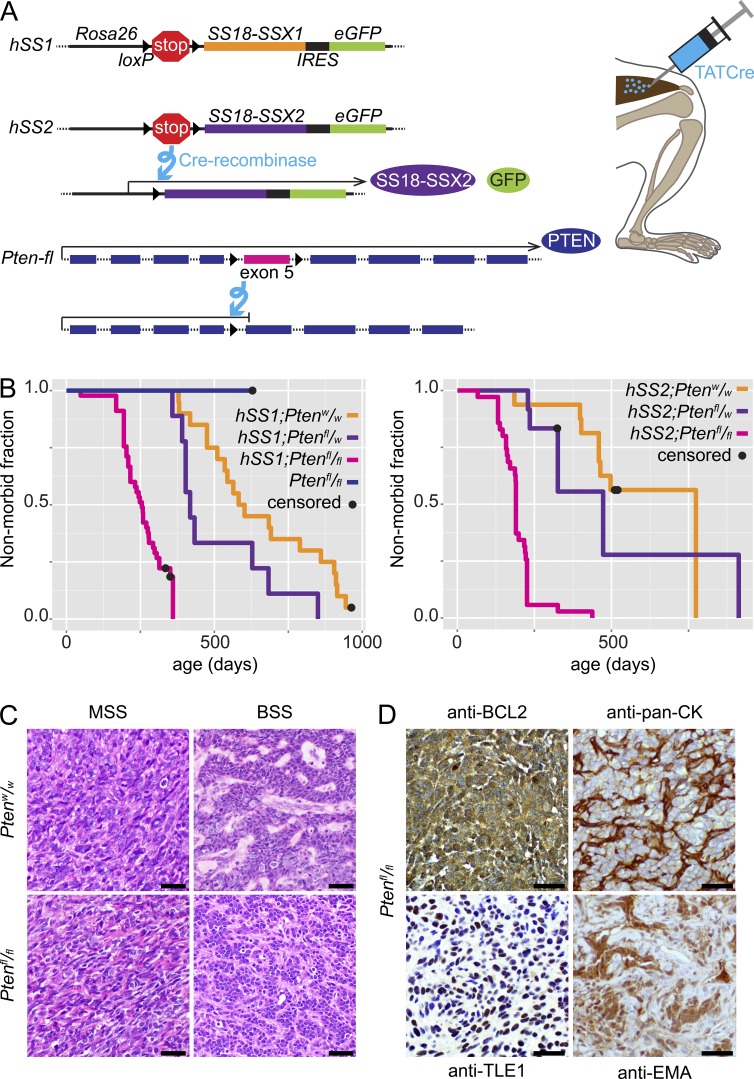
To test the impact of Pten silencing in SS, mice bearing conditional expression alleles of SS18-SSX1 (hSS1) or SS18-SSX2 (hSS2) were bred to mice bearing a conditional disruption allele of Pten (Fig. 1 A). Tumorigenesis was initiated in mice of each genotype by the injection of TATCre, which was previously shown to induce synovial sarcomagenesis from activation of hSS1 or hSS2 in a majority of mice, but only after a long latency (Barrott et al., 2015; Jones et al., 2016).
Figure 1.
Pten silencing enhances synovial sarcomagenesis. (A) Schematic of alleles, recombination products, and TATCre injection technique (IRES, internal ribosomal entry site). (B) Kaplan-Meier plots of the nonmorbid fraction of hSS1 (left) or hSS2 (right) mice with Pten genotypes of homozygous wild-type (n = 20 and 16 for hSS1 and hSS2, respectively), heterozygous (n = 9 and 12), or homozygous-floxed (n = 45 and 35), injected at age 1 mo with TATCre in the hindlimb. Pten homozygous floxed mice with wild-type Rosa26, also injected at 1 mo with TATCre, are presented on the hSS1 plot (n = 6). Statistical difference (Log-rank test) between Ptenw/w and Ptenfl/fl for hSS1, P = 0.0004, and for hSS2, P = 0.0173. (C) Photomicrographs of H&E histology examples of monophasic (MSS) and biphasic (BSS) SS18-SSX-induced synovial sarcomas from each Pten genotype in TATCre-injected mice. (D) Photomicrographs after immunohistochemistry on hSS;Ptenfl/fl tumor tissue sections with noted primary antibodies, demonstrating characteristic SS staining patterns. Bars, 25 µm.
The addition of homozygous conditional silencing of Pten was sufficient to decrease the latency to tumorigenesis and increase the prevalence of tumorigenesis to full penetrance when combined with expression of either SS fusion gene (Fig. 1 B). A control group with TATCre-induced homozygous silencing of Pten alone developed no tumors by 600 d after injection. The tumors that developed from activation of hSS1 or hSS2 and Pten silencing were compared with mouse tumors from TATCre-activated hSS1 or hSS2 alone, as well as to human SSs. Pten silencing did not change the development of the classic histological features of SS in the mouse tumors (Fig. 1 C and Fig. S1 A). These features included biphasic histology, wherein cells of both mesenchymal and epithelial characteristics and morphology intermingle and the latter even coalesce into glandular structures. As the development of these features requires a full reprogramming of germ-layer-derived cellular identity, it is considered a hallmark of SS and is pathognomonic for the disease. The ratio of biphasic to monophasic (only mesenchymal-character cells) histology did not differ between Pten-proficient and -deficient sarcomas. Immunohistochemical stains, used to make the histological diagnosis of SS clinically, also demonstrated classic patterns, regardless of Pten silencing (Fig. 1 D and Fig. S1 B). Thus, PTEN deficiency accelerated and enhanced fusion-initiated synovial sarcomagenesis in the mouse.
Pten silencing promotes pulmonary metastasis from mouse synovial sarcoma
In contrast to virally delivered Cre-recombinase expression vectors, which have potential for systemic escape from the localized injection, distribution to a distant site, cell infection, and Cre-mediated recombination, the injection of TATCre is more confidently localized in its Cre-mediated recombination effect. Because multiple Cre-recombinase proteins are required in each cell to mediate the recombination events, the likelihood that systemic escape of a few injected protein molecules will result in distant recombination events is vanishingly small. TATCre induction also differs meaningfully from Cre-expression from any promoter, either transgenic or targeted in the mouse genome, where multiple anatomical sites of disease are initiated simultaneously or metachronously, reducing the opportunity to clarify primary versus metastatic sites of sarcoma. TATCre induced tumorigenesis therefore offered a unique opportunity to test for true metastasis in this model.
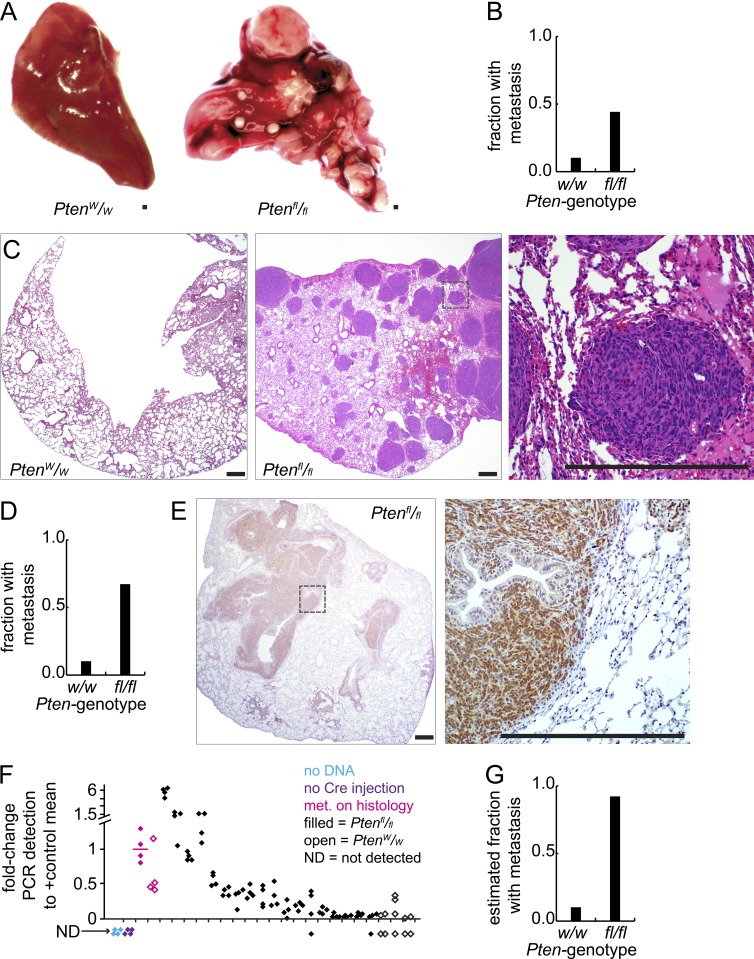
Mice of each Pten genotype were assessed for the development of metastatic foci of SS, initially by gross dissection of organs. Approximately 44% of the hSS1;Ptenfl/fl and hSS2;Ptenfl/fl mice had grossly apparent pulmonary metastatic disease, but only 1 out of 10 of the Pten-wild-type comparators had apparent metastasis (Fig. 2, A and B), that in a mouse aged to nearly 2 yr. Histological sectioning of the pulmonary parenchyma confirmed the same (Fig. 2 C). Histology expanded the number of metastatic foci identified per mouse, as well as the prevalence of hSS;Ptenfl/fl mice (but neither hSS1 nor hSS2 alone) with any metastasis, which approached 70% (Fig. 2 D). Immunohistochemical staining against GFP, which is expressed with the fusion gene from an internal ribosomal entry site that is present on both the hSS1 and hSS2 alleles, enabled definitive confirmation not only of the tumor-identity of the metastatic cells, but also their expression of the fusion oncogene (Fig. 2 E).
Figure 2.
Pten silencing promotes synovial sarcoma metastasis. (A) Gross photomicrographs of pulmonary lobes from hSS;Ptenw/w and hSS;Ptenfl/fl mice that were injected with TATCre at age 1 mo. (B) Graph depicts the fraction of mice in each group with metastasis detectable by gross inspection alone (hSS;Ptenw/w, n = 10, hSS;Ptenfl/fl, n = 88). (C) Low power H&E photomicrographs of lungs from mice bearing control or Pten-silenced tumors, as well as higher power H&E (right, from box in center image). (D) Graph depicts the fraction of mice in each group with metastasis detectable by histology. (E) Photomicrographs of lungs from tumor bearing hSS;Ptenfl/fl mice, demonstrating immunohistochemistry against GFP (indicates SS18-SSX expression). (F) Quantitative PCR results from lung tissue derived from tumor-bearing mice for the recombined Rosa26 locus as a marker of disseminated tumor cells. Two positive control samples that demonstrated metastasis (met.) on histology (one of each Pten genotype are noted in magenta), followed by testing for additional samples of each genotype without histologically detectable metastasis. Data points represent four technical replicates for each biological sample. (G) Graph depicts the estimated combined fraction of mice in each group with disseminated tumor cells detected by gross dissection, histology, or qPCR. Bars, 250 µm.
To determine whether Pten silencing enabled greater tumor cell dissemination to the lungs or simply greater growth after dissemination, we developed a PCR assay to detect a recombined genomic DNA segment in the Rosa26 locus that would only be present in tumor cells disseminated to the lung. This assay was used to test lung tissue for disseminated tumor cells in mice of each Pten genotype that had developed tumors, but no grossly or histologically detectable metastasis. Many of the lungs of Pten-silenced hSS1 or hSS2 mice without ostensible metastatic foci (detectable by gross dissection or histological assessment) demonstrated the presence of disseminated tumor cells by PCR (Fig. 2 F). Lungs of mice with PTEN-proficient sarcomas did not. This increased the estimated fraction of hSS;Ptenfl/fl mice with evidence of disseminated tumor cells in the pulmonary parenchyma by the time of sacrifice as a result of morbidity to >90% (Fig. 2 G). This suggests that Pten silencing specifically promoted sarcoma cell dissemination.
Metastatic synovial sarcomas were more vascular but otherwise indistinct
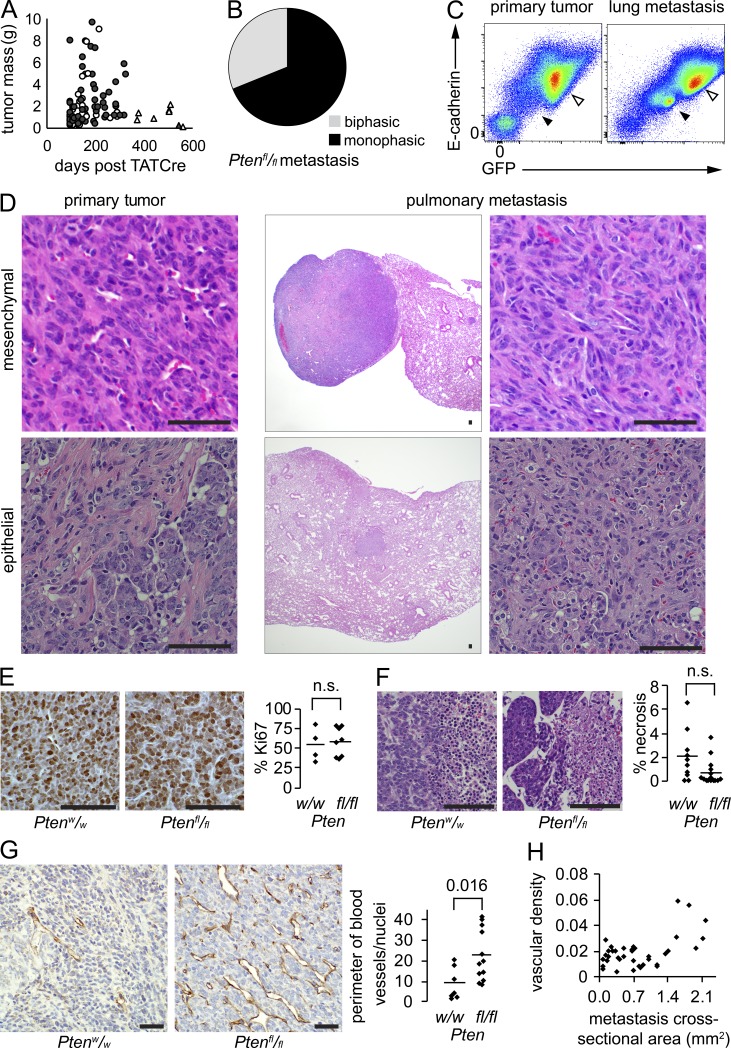
Clinically, risk of sarcoma metastasis correlates with primary tumor size, as well as loosely with time between tumor development and treatment initiation. Primary tumor size did not differ significantly between tumors of each genotype at the time of the morbidity-driven host euthanasia and tumor harvest (P = 0.12). Primary tumor size also did not correlate with detectable metastasis in Pten-silenced tumors (P = 0.51). Age at sacrifice only trended toward older ages for mice of the Ptenfl/fl;hSS1 and Ptenfl/fl;hSS2 genotypes that developed grossly or histologically detectable metastasis compared with those that did not (P = 0.14; Fig. 3 A).
Figure 3.
Pten silencing drives increased vascularity in synovial sarcoma. (A) Plot of tumor mass at harvest against time elapsed after TATCre injection into mice bearing conditional SS18-SSX and either Ptenw/w (triangles) or Ptenfl/fl (circles). Filled shapes indicate tumors with associated histologically detected metastasis. (B) Pie chart demonstrating histological subtype fractions for metastatic tumors in hSS;Ptenfl/fl mice (n = 59). (C) Flow cytometry of a primary tumor and lung metastasis stained with E-cadherin and analyzed for intrinsic GFP expression. Tumor population (open arrow) and endothelial cells (filled arrow) both express higher levels of E-cadherin than the immune infiltrates (lower left). (D) Example photomicrographs of tumors with retained histological appearance between primary tumor and metastasis. (E) Example anti–Ki-67 immunohistochemistry photomicrographs and chart of proliferative indices for SS18-SSX-induced tumors in each Pten genotype (student’s t test). (F) Example H&E photomicrographs of necrosis (right side of each) identified in tumors and graph of the mean necrotic area per tumor cross section by histology (Student’s t test). (G) Example anti-CD31 immunohistochemistry photomicrographs demonstrating vascular density in hSS tumors arising with wild-type or silenced Pten. Graph presents mean vascular density in primary tumors of each genotype (Student’s t test). (H) Plot of vascular density indicated as vessel perimeter per cross-sectional area against cross-sectional area of each metastasis in hSS;Ptenfl/fl mice. Bars, 50 µm.
Theoretical models for the metastasis of solid tumor cells to distant sites typically involve strengthening of the cancer hallmarks of proliferation, angiogenesis, metabolism, and invasion, often characterized in epithelial-derived carcinomas by a transition to a mesenchymal state or the development of necrosis. We therefore compared each of these parameters between primary tumors with wild-type Pten, primary tumors with silenced Pten, and metastatic tumors of the latter genotype. Ratios of monophasic and biphasic (more epithelial) histology did not differ between the tumor groups. Additionally, the epithelial histology was retained in many of the metastatic foci from epithelial primary tumors (Fig. 3, B–D; and Fig. S2 A).
Ki-67 proliferative indices did not differ between PTEN-proficient versus -deficient sarcomas (Fig. 3 E). To answer the question whether increased necrosis correlated with PTEN deficiency, and thus could contribute to metastatic potential, we measured percent necrosis between the two Pten genotypes and found no significant difference (Fig. 3 F). Although SS is considered a strongly angiogenic tumor, with characteristic hemangiopericytomatous vascular patterns as one of the diagnostic features, Pten silencing increased the vascularity of primary tumors significantly (Fig. 3 G). This increased density of vessels was less pronounced in small metastatic foci, but increased relative to cross-sectional area in larger metastases (Fig. 3 H and Fig. S2 B), suggesting that environmental signals or oxygen tension can yet impact angiogenesis, rather than an exclusive cell-intrinsic impact of Pten silencing alone.
Pten silencing drives an inflammatory transcriptome in mouse synovial sarcomas
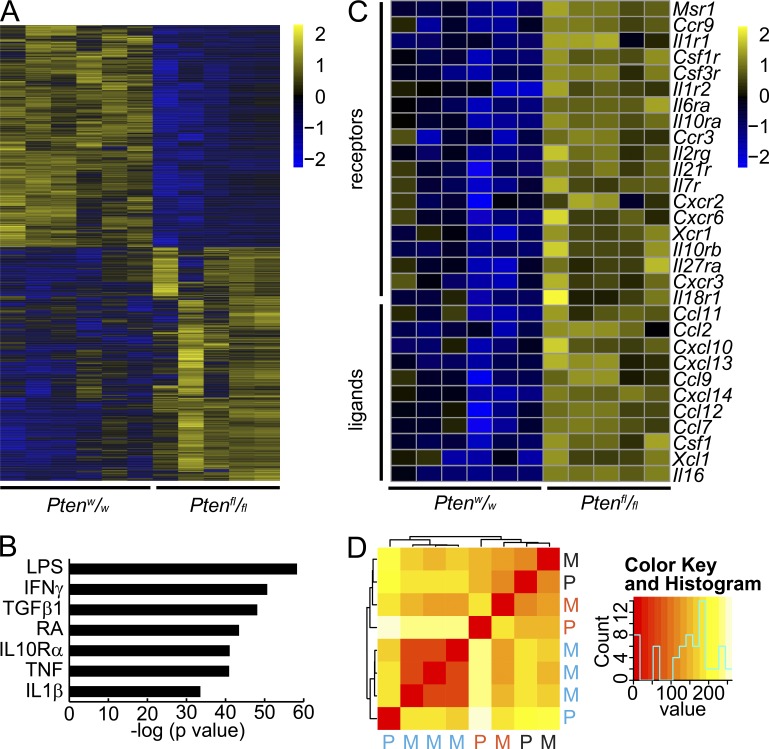
To compare the transcriptional impact of PTEN deficiency on SS, we sequenced total RNA from tumors initiated by TATCre-induced hSS1 or hSS2 and either wild-type or homozygous floxed Pten. The transcriptomes of tumors grouped by Pten genotype demonstrated differential expression of 4,051 genes with an absolute log2 ratio > 1 and adjusted P < 0.05 significance level (Fig. 4 A and Table S1). Unbiased ingenuity pathway analysis of these differentially expressed genes identified inflammatory pathways and inflammatory upstream regulators as the top hits (Fig. 4 B and Table S2). In detail, PTEN-deficient sarcomas demonstrated increased inflammation-related gene transcription. Especially increased was the expression of inflammatory recruiting signals and their receptors: Ccl7 and Ccl11 with Ccr3, Xcl1 with Xcr1, and CSF1 with CSF1R (Fig. 4 C).
Figure 4.
Pten silencing in synovial sarcoma drives an inflammatory transcriptome. (A) Heat map demonstrating the expression of 4,051 genes that are twofold and significantly differentially expressed between TATCre induced SS18-SSX–expressing tumors of either homozygous wild-type (w/w) or homozygous floxed (fl/fl) Pten genotype. (B) Plot of the P-values of the most significant Ingenuity Pathway Analysis results for likely upstream regulators of the differential expression between tumors of the two Pten genotypes (RA, retinoic acid; others are gene symbols; right tailed Fisher Exact Test and Benjamini-Hochberg correction for multiple testing). (C) Heat map of inflammatory receptors and ligands that can lead to recruitment of myeloid cells into tissues. (D) Nonhierarchical heat map clustering of primary (P) and metastatic (M) tumors color coded by individual mouse hosts.
We also compared transcriptomes between primary tumors and their associated metastases. Each metastasis clustered tightly with its primary tumor (Fig. 4 D), suggesting that SS tumors harbor minimal intratumoral heterogeneity from which sub-clones metastasize, and that even very distinct environments (the limb versus the lung) minimally impact the whole-tumor transcriptome. Metastatic tumors also demonstrated inflammatory transcriptomes, but not as pronounced as their related primary tumors (Tables S3 and S4).
Pten silencing increases myeloid-derived cell infiltration into synovial sarcomas
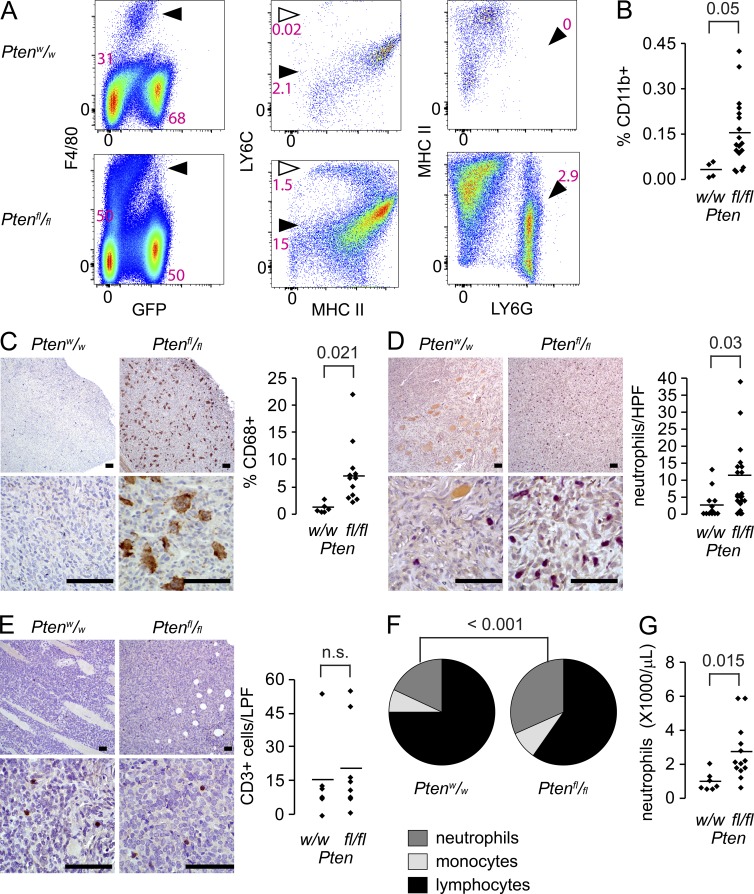
Suspecting that tumor cells were not independently responsible for the inflammatory gene expression patterns, we analyzed tumors of each genotype by flow cytometry. The three cell subtypes that were most dramatically increased were two subsets of F4/80+ cells, Ly6Cmid;MHC-II+ cells and Ly6Chi;MHC-II+ cells, and Ly6C+;Ly6G+ cells, representing macrophages, monocytes, and neutrophils, respectively (Fig. 5 A and Fig. S3 A). Specifically within the non-GFP population, the myeloid cell fraction (CD11b+) increased fivefold in PTEN-deficient sarcomas (Fig. 5 B and Fig. S3 B).
Figure 5.
Pten silencing associates with increased infiltration of myeloid-derived cells. (A) Example flow cytometry comparing an hSS;Ptenw/w tumor (top) to an hSS;Ptenfl/fl tumor (bottom) for F4/80+/GFP− monocytes/macrophages (arrowhead, left plot), their MHCII+/Ly6Chigh newly recruited monocyte (open arrow, middle plot) and MHCII+/Ly6Cmid tissue macrophage (filled arrowhead, middle plot) subpopulations, and Ly6C+/Ly6G+ neutrophils (arrowhead, right plot). (PE, phycoerythrin; APC, allophycocyanin; PerCP, perdinin chlorophyll protein). (B) Graph comparing the percent of infiltrating CD11B+ cells in hSS tumors of each Pten genotype (Student’s t test). (C) Representative low and high power photomicrographs and quantitation of CD68+ monocytes/macrophages (brown) by immunohistochemistry in hSS;Ptenw/w and hSS;Ptenfl/fl tumors (Student’s t test). (D) Representative low and high power photomicrographs and quantitation per high power field (HPF) of Leder cytochemically stained neutrophils (magenta) in hSS;Ptenw/w and hSS;Ptenfl/fl tumors (Student’s t test). (E) Representative photomicrographs of CD3 immunohistochemistry in hSS;Ptenw/w and hSS;Ptenfl/fl primary tumors and an associated graph demonstrating low and equivalent numbers of CD3+ lymphocytes (Student’s t test). (F) Pie charts presenting the mean fractions of neutrophils, monocytes, and lymphocytes in the peripheral blood of morbid tumor-bearing hSS;Ptenw/w and hSS;Ptenfl/fl mice (n = 7 and 13, respectively; Chi-squared test). (G) Graph of peripheral blood neutrophil counts in morbid tumor-bearing hSS;Ptenw/w and hSS;Ptenfl/fl mice (Student’s t test). Bars, 50 µm. Graph points present individual tumors; bars present means.
Tissue sections from tumors of each genotype were also assessed by anti-CD68 immunohistochemistry. This confirmed increased macrophages/monocytes in tumors of the Ptenfl/fl;hSS genotypes (Fig. 5 C). Neutrophils were assessed using Leder cytochemical staining for the granulocytic lineage, which demonstrated higher prevalence in PTEN-deficient tumors (Fig. 5 D). To evaluate the presence of lymphocytic infiltrates as a contributing factor to the inflammatory phenotype, tumors from each genotype were immunohistochemically stained with anti-CD3, which resulted in low (0.5 ± 0.1%; mean ± SEM) yet similar numbers of CD3+ cells in Pten-silenced and Pten-wild-type tumors (Fig. 5 E).
Evaluation of peripheral blood counts demonstrated elevated neutrophil fractions of leukocytes (Fig. 5 F), as well as increased neutrophil counts in mice growing PTEN-deficient tumors (Fig. 5 G).
Mouse synovial sarcoma cells with added Pten silencing express Csf1
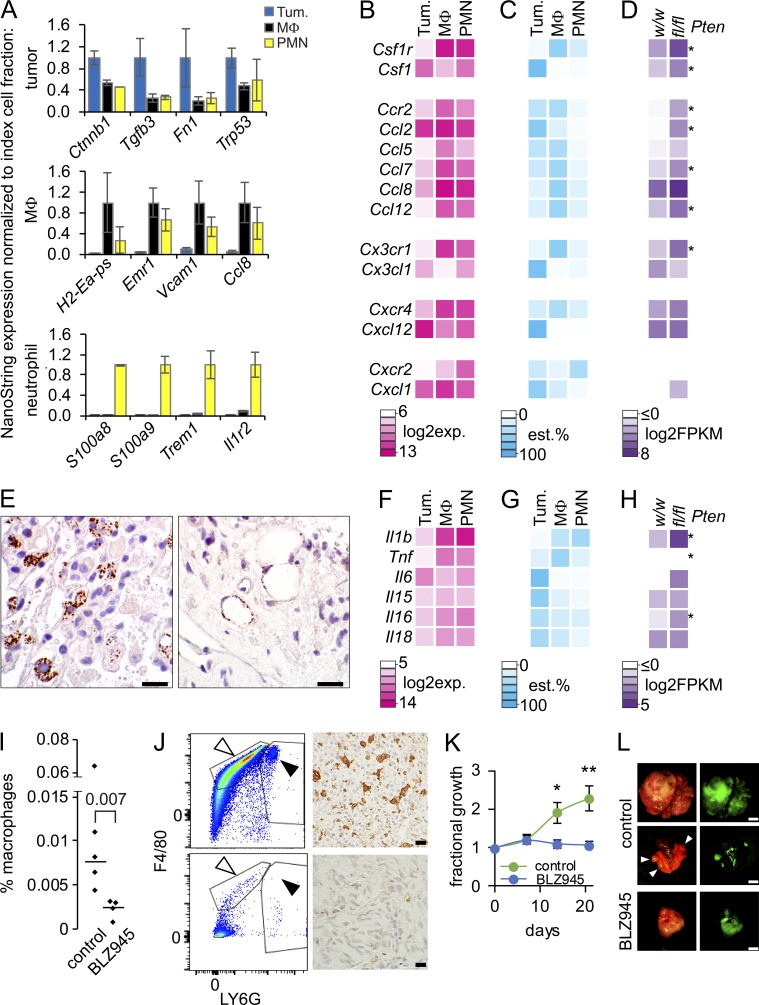
To determine the specific cellular sources of the abundant myeloid recruiting signals and receptors apparent in the whole-tumor transcriptomes of Pten-silenced and SS18-SSX–expressing mouse sarcomas, freshly harvested tumors were dissociated and sorted by flow cytometry into GFP+ (and therefore SS18-SSX-expressing) tumor cells, macrophages, and neutrophils (Fig. 5 A and Fig. S3 A). Because FACS preparation of fresh tumor samples requires multiple hours of dissociation and labeling, we did not anticipate that it would render accurate quantitative transcriptional levels of genes, but merely expression relative to the other cell fractions to enable identification of the principal source cell fraction for each gene’s transcripts. Total RNA isolated from each cell fraction was subjected to NanoString analysis, using the inflammatory mediator panel. As proof of the assay’s validity as a test of cell fraction relative expression, four genes considered markers of each cell fraction (i.e., SS tumor cells, macrophages, and neutrophils) demonstrated elevated expression in the index cell fraction (Fig. 6 A). The normalized expression data for myeloid recruitment ligand/receptor pairs (Fig. 6 B) were then multiplied by the numerical fraction of each cell type by flow analysis to render an estimated percentage of the overall transcripts in the whole tumor that came from each cell type (Fig. 6 C and Table S5).
Figure 6.
Pten silencing enhanced synovial sarcoma cells express Csf1. (A) Nanostring expression graphs of marker genes for each cellular fraction from whole hSS;Ptenfl/fl tumors, with expression normalized to that in each index cell type (MΦ, macrophage; PMN, polymorphonuclear cell or neutrophil). (B) NanoString expression levels heat map of macrophage recruitment receptors and their corresponding ligands in each source cell fraction of tumors. (C) Heat map of the estimated (est.) percent contribution to each gene’s overall expression in the tumor from each source cell fraction, generated by normalizing the NanoString expression by the mean numerical fraction of cells. (D) Heat map of the mean log2-transformed FPKM expression level for each gene in hSS;Ptenw/w and hSS;Ptenfl/fl tumors. *, P < 0.05. (E) Photomicrograph of immunohistochemistry with an anti-pCSF1R antibody, demonstrating macrophages (left) and endothelial cells (right) in a tumor section actively receiving and transducing CSF1 signal. Bar, 10 µm. (F–H) Heat maps as in B–D, but of neutrophil stimulants. Notably, G-CSF (Csf3) and GM-CSF (Csf2) were not significantly expressed by tumors in whole transcriptome profiling, with or without Pten silencing. (I) Graph comparing the percent macrophage population in control tumors or tumors treated with BLZ945. Statistical test did not include outlier in control with high percentage of macrophage (Student’s t test). (J) Representative flow cytometry demonstrating a decreased population of macrophages in the BLZ945-treated tumors (bottom) compared with control (top). Photomicrograph of immunohistochemistry with an anti-CD68 antibody, showing a decrease in macrophages in BLZ945 treated tumors (bottom) compared with control (top). (K) Graph of percent tumor growth over baseline for 21 d of treatment with or without 200 mg/kg of BLZ945. *, P < 0.05; **, P < 0.01. n = 5 per group, Student’s t test. (L) Examples of photomicrographs of control tumors and BLZ945-treated tumors with corresponding GFP expression demonstrating the twofold increase in control tumor volume. Middle panel is control lungs with grossly visible metastases (arrowheads). Bars: (black) 10 µm; (white) 5 mm.
Of five potential macrophage chemoattractant receptors, Csf1r, Ccr2, Cx3cr1, Cxcr4, and Cxcr2, the estimated expression percentages of all but the last were highest in macrophages. The largest percentage of Cxcr2 expression came from neutrophils. Tumor cells were the most prominent source of the matched, recruiting ligands Csf1, Ccl2, Cx3cl1, Cxcl2, and Cxcl1. However, in our whole-tumor RNAseq data (Fig. 6 D), only the first two were significantly up-regulated in tumors with silenced Pten compared with wild-type Pten. This suggests that Pten-silenced tumor cells expressing Csf1 and/or Ccl2 recruit macrophages expressing Csf1r and Ccr2. Although tumor cells also expressed high levels of Cxcl12, which can recruit Cxcr4-expressing macrophages, this expression cannot explain the increased infiltrating macrophages associated with Pten-silencing, because Pten-wild-type tumors also expressed high levels of this recruiting ligand (Fig. 6 D). Macrophages were the greatest source cell fraction for the other potential ligands for Ccr2, suggesting the presence of auto-recruitment signals in addition to the tumor-expressed recruitment by Ccl2 and Csf1 (Fig. 6, B–D).
Immunohistochemistry demonstrated macrophages with phosphorylated CSF1R, which indicates ligand activation, in PTEN-deficient tumor sections (Fig. 6 E). These pCSF1R+ cells were typically in close proximity to tumor cells, not neutrophils, further suggesting that tumor cells drive monocytic lineage recruitment to the tumor microenvironment. However, macrophages were not the only cell fraction affected by tumor secreted CSF1, as activated pCSF1R was detected in endothelial cells as well (Fig. 6 E). Macrophages were the dominant, nonautocrine, source cell fraction for the expression of the systemic neutrophil stimulant Il1b (Fig. 6, F–H).
To evaluate if inhibition of CSF1 signaling reduces the recruitment of macrophages in PTEN-deficient SSs, we treated mice for 3 wk with a potent and selective inhibitor of CSF1R, BLZ945 (Pyonteck et al., 2013). As indicated by flow cytometry analysis for F4/80+ macrophages and immunohistochemical analysis for CD68+ macrophages, inhibition of CSF1R reduced their presence in dissociated tumors and FFPE tumors (Fig. 6, I and J). BLZ945 at 200 mg/kg also limited tumor volume growth compared with a twofold increase in the control-treated mice (Fig. 6, K and L).
PI3′-lipid signaling in synovial sarcomas drives CSF1 expression
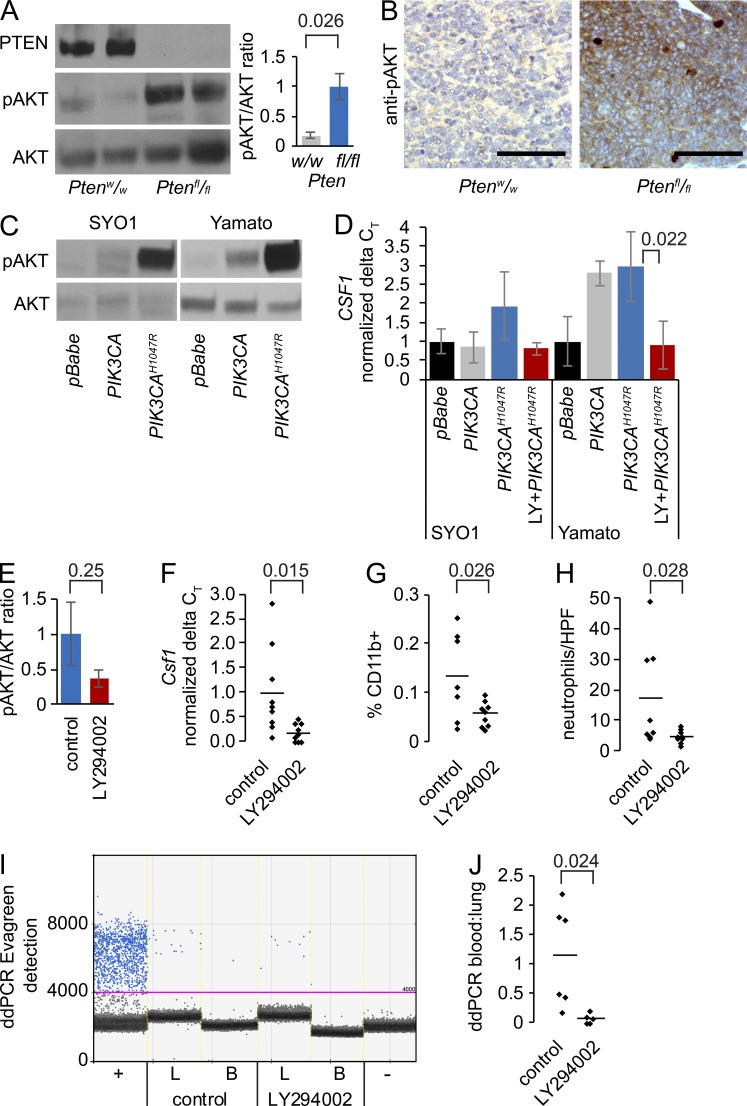
To confirm Pten silencing in the tumors that arose after TATCre injection into hSS;Ptenfl/fl mice, we isolated whole tumor protein lysates and performed Western blots. These demonstrated the absence of PTEN and strong increases in pAKT, but not total AKT, compared with Pten-wild-type tumors (Fig. 7 A). Immunohistochemistry against pAKT verified this result at the tissue level (Fig. 7 B and Fig. S4 A).
Figure 7.
PI3′-lipid signaling drives CSF1 expression in synovial sarcoma. (A) Representative Westerns for PTEN, pAKT, and AKT in hSS;Ptenw/w and hSS;Ptenfl/fl tumors, with quantitation from four tumors from each. Performed independently four times with eight different hSS;Ptenfl/fl tumors and four different hSS;Ptenw/w (Student’s t test). (B) Representative photomicrographs of pAKT immunohistochemistry in hSS;Ptenw/w and hSS;Ptenfl/fl tumors. (C) Westerns for pAKT and AKT from two human SS cell lines transfected with empty vector (pBabe), wild-type PIK3CA, or constitutively active mutant PIK3CA (H1047R). Performed independently three times. (D) Graph of RT-qPCR expression of CSF1 in cell lines transfected with empty vector, wild-type PIK3CA, or constitutively active mutant PIK3CA (H1047R) with or without PI3′-lipid kinase inhibition with LY294002 (noted as LY). Performed independently at least three times (Student’s t test). (E) Graph of the mean ± SE of the pAKT suppression (not significant) in hSS;Ptenfl/fl tumors treated in vivo for 1 wk with 70 mg/kg body mass LY294002 or vehicle control (Student’s t test). (F) Graph of the reduced Csf1 expression by RT-qPCR in hSS;Ptenfl/f tumors treated in vivo for 1 wk with LY294002 or control (Student’s t test). (G) Graph of the percent CD11B+ cells by flow cytometry in hSS;Ptenfl/fl tumors treated in vivo for 1 wk with LY294002 or control (Student’s t test). (H) Graph of the neutrophil counts by Leder stain in tissue sections from hSS;Ptenfl/fl tumors treated in vivo for 1 wk with LY294002 or control (Student’s t test). (I) Digital droplet PCR from lung tissue (L) and blood (B) derived from tumor-bearing mice for the recombined Rosa26 locus as a marker of disseminated tumor cells after tumors were treated in vivo for 1 wk with LY294002 or control. Primary tumor was used as a positive control (+) and hSS;Ptenfl/fl mouse blood without TATCre was used as a negative control (-). (J) Graph of the ratio of tumor DNA detected in the blood to lungs of hSS;Ptenfl/fl mice treated in vivo for 1 wk with LY294002 or control (Student’s t test).
To assess the broader relationship between increased PI3′-lipid signaling and myeloid chemoattractant expression, we transfected an active mutant (H1047R) of PIK3CA into two human SS cell lines. SYO1 expresses SS18-SSX2. Yamato-SS expresses SS18-SSX1. Compared with empty vector transfections, each cell line increased pAKT levels and increased CSF1 expression by RT-qPCR. This effect was blocked by the application of 5 µM LY294002, a pan-PI3′-lipid kinase inhibitor (Fig. 7, C and D).
Although it lacks the superior pharmacokinetics of developed drugs, LY294002 has also demonstrated PI3′-lipid kinase inhibition in vivo (Semba et al., 2002). To test the dependence of chemoattractant signal expression on PI3′-lipid signaling in vivo, Ptenfl/fl;hSS mice with large tumors were randomized to daily oral gavage administration of LY294002 or vehicle control for 1 wk before tumor harvest. Westerns and immunohistochemistry demonstrated only modest reductions in pAKT (Fig. 7 E and Fig. S4 B), confirming our anticipation that LY294002 was not likely to demonstrate serious tumor-impacting PI3′-lipid inhibition. The hypothesized impact on Csf1 expression; however, was confirmed (Fig. 7 F). Further, tumor infiltrating CD11b+ by flow cytometry and Leder-stained neutrophil counts on histological sections were also reduced after LY294002 administration compared with vehicle control treatment (Fig. 7, G and H). As a result of the short duration of treatment, there was no expectation that treatment would significantly affect the primary tumors and large lung metastases. Nevertheless, it was expedient to assay the disseminated tumor cells in the circulation to see if LY294002 treatments decreased the presence of these cells in the blood. Indeed, we observed by digital droplet PCR (ddPCR) the presence of the recombined Rosa26 locus in the lungs and blood of control-treated animals. The ratio of lung/blood tumor DNA was reduced significantly in the LY294002-treated samples (Fig. 7, I and J).
Tumor-associated neutrophils and monocytes correlate with PI3′-lipid signaling in SS
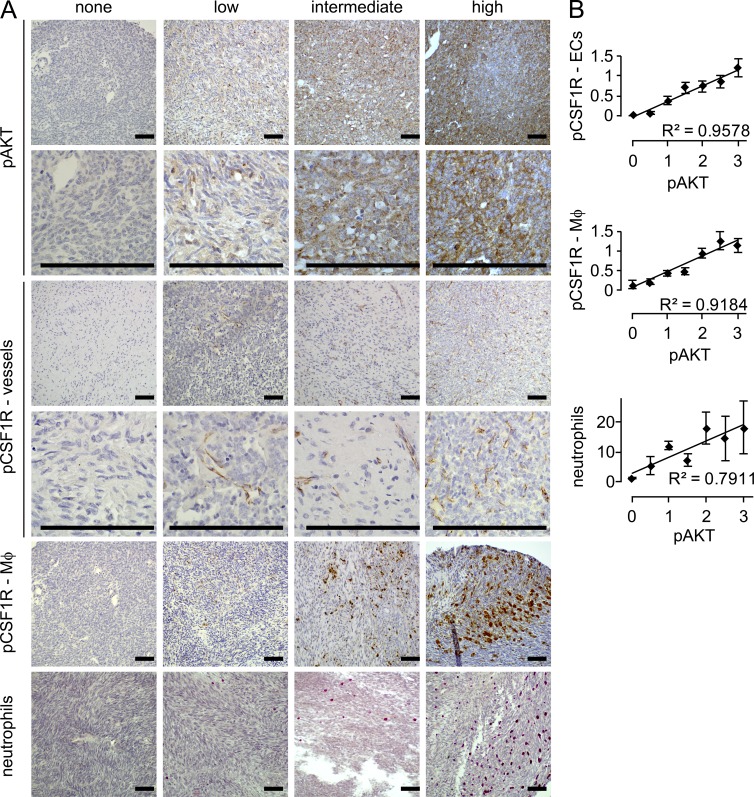
Silencing of Pten in the mouse tumors primarily represents an experimental in vivo boost to PI3′-lipid signaling. PI3′-lipid signaling may be increased in human SS through a variety of other means. We therefore tested the more general relationship between pAKT presence, as a common marker of PI3′-lipid signaling, and infiltrating myeloid-derived cells in a large panel of human SS on a tissue microarray (TMA). With 382 samples from 191 human SSs, we screened cases for pAKT by immunohistochemistry, identifying 4.7% negative, 34% low, 36.6% intermediate, and 24.6% high for expression of pAKT (Fig. 8 A). Human SSs were grouped into the different levels of pAKT and correlative stains of pCSF1R and Leder cytochemical stains were performed in blinded fashion. Mean scores per human tumor were plotted against pAKT. All three comparisons demonstrated correlation coefficients >0.79 and ANOVA showed a statistically significant difference across samples in the pAKT and pCSF1R comparison (P < 3 × 10−6; Fig. 8 B).
Figure 8.
Tumor-infiltrating myeloid cells correlate with pAKT in human synovial sarcoma. (A) Representative photomicrographs of blinded scoring of a human tissue microarray with 191 cases of human synovial sarcoma for immunohistochemistry against tumor cell staining against pAKT, endothelial cell staining for pCSF1R, macrophage (MΦ) staining for pCSF1R, and cytochemical Leder staining for neutrophils. (B) Plots of correlation between categorical pAKT staining (mean between two TMA samples per tumor) and the endothelial pCSF1R staining, macrophage pCSF1R staining, and Leder-stained neutrophil counts. Linear regression correlation was calculated with the R2 values being provided. A single-factor ANOVA was also performed on pCSF1R values between the different pAKT bins.
Discussion
Others have previously demonstrated diverse facets of PI3′-lipid signaling in SS. By transcriptome profiling, many human SSs express receptor tyrosine kinases and their corresponding ligands, such as PDGFRα and PDGFα (Tamborini et al., 2004), FGFRs and FGFs (Ishibe et al., 2005), or EGFR and EGF (Nielsen et al., 2002; Blay et al., 2004; Thomas et al., 2005; Teng et al., 2011). There is evidence for epigenetic down-regulation of PTEN in many human SS cell lines (Su et al., 2010), which have also shown PI3′-lipid activity by pAKT levels and the inhibition of pAKT and proliferation with the PI3′-lipid kinase inhibitor LY294002 (Friedrichs et al., 2011). Immunohistochemistry on human tumor samples has also shown a correlation between pAKT and poor prognosis, including development of metastatic disease (Setsu et al., 2013). Although PTEN loss, PIK3CA-activating mutations, RAS-activating mutations, and other genetic means of increasing PI3′-lipid signaling have each been identified in only a small minority of human SSs (Barretina et al., 2010), we used one of these to effect a material change in the metastatic potential of SS18-SSX–driven tumors in mice. Alternatively, increased PI3′-lipid signaling might be achieved through the full range of upstream signaling, in many human SSs that progress. Importantly, the PI3′-lipid alterations in this model were tumor cell–intrinsic, distinct from any myeloid cell PI3′-lipid signaling.
Several other mouse genetic models of cancer have demonstrated increased metastasis upon the addition of Pten silencing (Stambolic et al., 2000; Abate-Shen et al., 2003; Wang et al., 2003; Knobbe et al., 2008). In most of these, an already apparent metastatic phenotype was enhanced by the silencing of Pten. The clarity of the phenotype in the genetically simple model of synovial sarcomagenesis certainly presents an opportunity for deeper study of the means by which PI3′-lipid signaling enables metastasis in future investigation. We have shown already that Pten silencing–mediated proliferation alone has not enabled this phenotype by mere outgrowth of already disseminated cells. Other than the one metastatic tumor in a single, 2-yr-old mouse, Pten-wild-type tumors had no evidence of even disseminated cells by PCR. Further, another model with the addition of β-catenin stabilization to SS18-SSX2 expression grew even faster than tumors enhanced by Pten silencing, but did not exhibit the prevalent metastatic phenotype (Barrott et al., 2015). The effects of PI3′-lipid signaling in a tumor cell are pleiotropic, including growth, survival, proliferation, apoptosis, cell migration, inflammation, and metabolism (Chalhoub and Baker, 2009). The contribution of each may participate in metastasis.
Among these, inflammation has particularly been linked to PTEN absence. Increased inflammation has been identified in models of Pb-Cre+;Ptenlox/lox prostate cancer (Maxwell et al., 2013; Garcia et al., 2014) Pten−/−;p53−/− breast cancer (Kim et al., 2015), and Dct::TVA;BrafCA;Cdkn2alox/lox melanoma (Cho et al., 2015), as well as in human samples of breast cancer (Lin et al., 2002) and squamous cell carcinomas of the head and neck (Young and Lathers, 1999) associated with PTEN loss. The mechanism for this PI3′-lipid–related inflammation and metastasis has been somewhat elusive, and generally attributed to increased growth rates, increased necrosis, or genetic instability driving the creation of neoantigens. One study linked xenograft breast metastasis to IL-6–mediated inflammatory signaling in a Pten/Trp53-null background. However, there was no connection to inflammatory cell recruitment as a result of the NOD/SCID mouse background (Kim et al., 2015). A study of UV-induced melanoma showed that an inflammatory response promoted angiogenesis, melanocyte migration, and neutrophil recruitment (Bald et al., 2014), but the inflammatory response was attributed to cell-extrinsic factors rather than the tumor cells themselves. In the SS mice and in human cell lines and tumor samples, we have linked Csf1/CSF1 expression to increased PI3′-lipid signaling and found this to be a manipulable program for myeloid recruitment.
The role of these inflammatory cells in either suppressing or supporting metastasis will also be the subject of intensive future investigation. Both relationships have been demonstrated in other model systems. Studies in human leiomyosarcoma have associated macrophage recruitment to high CSF1 expression in the sarcoma, which correlates to a worse prognosis (Lee et al., 2008; Espinosa et al., 2009). In the inflammation-associated melanoma model, neutrophil recruitment promotes tumor–endothelial interactions in the lungs to promote seeding (Huh et al., 2010; Cools-Lartigue et al., 2013; Hiratsuka et al., 2013). We observed in the SS model the presence of some aspects of described prometastatic roles for myeloid inflammatory cells (Gabrilovich, 2015). The mouse tumor cells express Ccl2, which has been described as a means of communication between tumor cells and monocytes that enables metastasis (Bonapace et al., 2014). Also, the SS tumor–associated macrophages demonstrated high expression of IL-1β, the systemic effects of which were evident in peripheral blood neutrophil counts. This signaling, through γδT cells at distant sites, has been shown to enable neutrophil-mediated metastasis (Coffelt et al., 2015). The complexity of these relationships may be optimally tested in this new model, especially considering that most other models depend on engrafted tumor cell lines or dissemination of tumor cell lines by embolization in the pulmonary parenchyma in mice with altered immune systems.
Herein, we have not demonstrated any specific, driving role for tumor-associated inflammatory cells in the aggressiveness of this mouse model of SS. Nevertheless, the association between inflammation and SS metastasis encourages the prospect of pharmacologically targeting inflammatory cells to impact the tumor cells indirectly. Already, strategies are being investigated in other disease models with cautious enthusiasm. Vaccines that specifically target tumor-associated macrophages in colon, breast, and lung carcinoma metastases have reduced both tumor volumes and macrophage numbers (Luo et al., 2006). Likewise clodronate liposomal depletion showed promising results in models of lung carcinoma (Fritz et al., 2014). Targeting CSF1R and CCR2 with small molecule inhibitors or monoclonal antibodies in pancreatic and breast carcinomas depleted macrophages, which resulted in deleterious effects on the tumors (DeNardo et al., 2011; Mitchem et al., 2013; Ries et al., 2014). Most of the tumor cytoreduction using these strategies was attributed to loss of modulation of the lymphocytic T cell response. SS presents an ideal opportunity to interrogate potentially targetable roles for inflammation distinct from T cell modulation.
As a result of the genetic simplicity of the SS mouse model with Pten conditional ablation alleles and working with a fully competent immune system, we may begin to unravel the complexities of PI3′-lipid signaling that result in tumor inflammation and metastasis. With a more precise knowledge of the recruitment signals required for inflammatory cell tumor infiltration or the reciprocal secreted factors that may stimulate metastasis, we can rigorously test and exploit these vulnerabilities to reduce metastatic burden.
Materials and methods
Mice
Mouse experiments were conducted with the approval of the University of Utah institutional animal care committee and use committee in accordance with legal and ethical standards. The previously described hSS1, hSS2, and Ptenfl/fl mice were maintained on a mixed strain background, C57BL/6 and SvJ (Groszer et al., 2001; Haldar et al., 2007; Jones et al., 2016). TATCre injections were 10 µl vol of 42 µM into the quadriceps. BLZ945 (MedChem Express) was dosed at 200 mg/kg by daily oral gavage in 20% β-cyclodextrin. LY294002 (MedChem Express) was dosed at 70 mg/kg by daily oral gavage in 0.5% hydroxypropyl methylcellulose/0.1% Tween 20. All experimental mouse treatments were conducted with littermates as controls. Complete blood counts were performed at the start of treatment or upon necropsy on a HemaTrue (Heska).
Imaging
Brightfield and fluorescent images were obtained with a AF6000 dissecting microscope and software (Leica). A 3.5×–90× LED low heat zoom stereo microscope equipped with 5 MP digital camera was used to image gross lung metastases (Amscope). All imaging was performed at room temperature in air medium. Light photomicrographs were obtained with an Olympus BX43 microscope, a DP26 camera and cellSensEntry software (Olympus). For Figs. 2 (C and E) and 3 D, a Plan N 2× objective with 0.06 numerical aperture was used. For Figs. 2 (C and E), 5 (C and D), and S2 (A and B), a Plan N 20× objective with 0.40 numerical aperture was used. For Figs. 1 (C and E), 3 (D–F), 5 (C and D), 7 B, 8 A, S1 (A and B), and S4 (A and B), a Plan N 40× objective with 0.65 numerical aperture was used. For Figs. 5 E, 6 E, and 8 A, a PlanC N 60× objective with 0.80 numerical aperture was used. Images were cropped for panels with Adobe Photoshop software and magnification bars placed directly in Adobe Illustrator, during figure generation. No gamma adjustments were performed.
Transcriptome analyses
Total RNA was isolated with the RNeasy mini kit (QIAGEN). Complementary DNAs were generated using Superscript III (Invitrogen). Quantitative PCR used the qPCR 2X GREEN Master Mix (Genesee Scientific) and a CFX Connect (Bio-Rad Laboratories) for detection. Primers were generated using software PrimerBot!. Primer sequences can be found in Table S6.
For transcriptome sequencing of TATCre tumors, RNA was prepared using the Illumina TruSeq RNA kit (Illumina), checked with the Bioanalyzer RNA 6000 chip (Agilent Technologies), captured using the RiboZero method (Illumina), and 50-cycle end-read sequenced on an Illumina HiSeq 2000. Reference fasta files were generated by combining the chromosome sequences from mm10 with splice junction sequences generated by USeq (v8.8.8) MakeTranscriptome using Ensembl transcript models (build 74). Reads were aligned with Novoalign (v2.08.01), allowing up to 50 alignments per read. USeq’s SamTranscriptomeParser selected the best alignment for each and converted the coordinates of reads aligning to splices back to genomic space. Differential gene expression was measured using USeq DefinedRegionDifferentialSeq, which counts the number of reads aligned to each gene and then call DESeq2 (v1.4.5) using default settings. R package pheatmap (v1.0.2) or heatmap.2 generated heat maps. Log2 (FPKM) values were centered and scaled by gene.
For NanoString gene expression analysis, RNA was isolated from three different tumor cell populations: GFP+, CD11b+/Ly6C+/Ly6G+, and CD11b+/Ly6Cmid/F4-80+/MHC IIhigh. 10 ng of RNA was combined with the mouse immunology panel of 545 gene probes and analyzed on the nCounter (NanoString).
Histology
Mouse tissues were fixed in 4% paraformaldehyde overnight, and embedded in paraffin. The TMA was created by taking two 0.8-mm punches from 191 independent donor human synovial sarcoma tumor blocks from pathology archives and arraying them into recipient blocks using a Beecher MTA-1 manual TMA apparatus as previously described (Lahat et al., 2010). Paraffin-embedded tissues were stained by immunohistochemistry by rehydrating slides through a citrosolv and ethanol dilution wash. Antigen-retrieval was performed in 10 mM sodium citrate (pH 6.0). Sections were immunostained with primary antibodies followed by horseradish peroxidase detection methods and counterstained with hematoxylin. The complete list of antibodies can be found in Table S7. Leder staining with Naphthol AS-D chloroacetate esterase (Sigma-Aldrich) was performed per manufacturer recommendations.
Flow cytometry
Specimens were minced, enzymatically digested (Tumor Dissociation kit; Miltenyi Biotec), and mechanically dissociated (GentleMACS Tissue Homogenizer; Miltenyi Biotec). Tissue debris was removed using 70-µm MACS Smart strainers (Miltenyi Biotec). Cells from all tissues were washed with PBS containing 0.5 mg/ml bovine serum albumin, and then stained using a cocktail of rat anti–mouse antibodies from BD: Ly6C, CD11b, I-A/I-E, F4/80, Ly-6G, and DAPI (Invitrogen). Cell expression was determined by multicolor flow cytometry on a FACSCanto flow cytometer (BD) and cell sorting was performed using a FACSAria (BD) and FlowJo 8.7.1 (Tree Star) for analysis.
Cell lines
The human SYO-1 (Kawai et al., 2004) and Yamato-SS (Naka et al., 2010) cell lines were maintained in DMEM with 10% FBS, and validated every 3 mo by expression of and dependence (siRNA) on SS18-SSX2 or 1, respectively. Transfection with pBabe-puro-HA, pBabe-puro-HA PIK3CA, or pBabe-puro-HA PIK3CA (H1047R; Addgene) used Lipofectamine 3000 (Invitrogen). LY294002 (5 µM) treatments began 24 h after transfection and lasted for 48 h.
Immunoblotting
Protein was isolated using a mild lysis buffer (10 mM Tris-HCl, pH 8.1, 10 mM NaCl, 0.5% NP-40, and proteinase inhibitors), clarified by centrifugation, loaded at 25 µg/sample for gel electrophoresis, and then transferred to PVDF membranes. Western blots were detected with goat anti–rabbit secondary antibodies and horseradish peroxidase.
Digital droplet PCR
DNA was isolated from tumor, lung, and blood tissues using the DNeasy kit (QIAGEN). 25 ng of DNA was analyzed using primers that amplified the recombined Rosa26 locus, following the manufacturer’s recommendations for emulsification in a QX200 droplet generator, thermocycling on a C1000, and analysis using QuantaSoft software (all from Bio-Rad Laboratories).
Statistical methods
For most comparisons between PTEN-proficient and -deficient sarcomas, a two-tailed Student’s t test was performed. All sample sizes are stated in the figure legends unless each data point is plotted in the figure. Statistical significance for the Kaplan Meier plots was determined by using a log-rank test. Ingenuity pathway analyses used a right tailed Fisher Exact Test and Benjamini-Hochberg correction for multiple testing. Pie chart percentage comparisons were tested using a χ2 test. In the tissue microarray, the mean values of pCSF1R IHC and Leder stain were plotted against pAKT IHC and linear regression correlation was calculated with the R2 values being provided. A single-factor ANOVA was also performed on pCSF1R values between the different pAKT bins. All counting and scoring was performed under reader blinding.
Accession no.
Raw RNA sequencing data are available from NCBI GEO under accession no. GSE81476.
Online supplemental material
Fig. S1 shows Pten disruption enhances synovial sarcomagenesis. Fig. S2 shows lung metastases retain primary tumor histology and E-cadherin expression and increases in vascularity with size. Fig. S3 shows Pten disruption associates with increased infiltration of myeloid-derived cells. Fig. S4 shows PI3′-lipid signaling drives phosphorylation of AKT and downstream expression of CSF1. Table S1 lists differential gene expression between Ptenw/w and Ptenfl/fl synovial sarcomas. Table S2 lists ingenuity pathway analysis of potential upstream regulators between Ptenw/w and Ptenfl/fl. Table S3 lists differential gene expression between primary and lung metastases Ptenfl/fl synovial sarcomas. Table S4 lists Ingenuity pathway analysis for potential upstream regulators between primary and lung metastases Ptenfl/fl synovial sarcomas. Table S5 Nanostring gene expression on sorted cell poplulations with normalized values based on prevelance in flow cytometry. Tables S1–S5 are available as Excel files. Tables S6 lists specific antibodies used, and Table S7 lists specific primers used.
Supplementary Material
Acknowledgments
The authors thank Sheri Holmen of Huntsman Cancer Institute for sharing wisdom and the Pten-floxed mouse strain. The authors gratefully received histology technical assistance from Sheryl Tripp at ARUP laboratories and Blake Anderson at Huntsman Cancer Institute Histology Core, RNA sequencing and NanoString assistance from Brian Dalley and John O’Shea at the Huntsman Cancer Institute Genomics core, digital droplet PCR assistance from Brian Whitmire and Hector Macias-Saldivar at Bio-Rad Laboratories, Heska training from Tony Pomicter of the Deininger laboratory at Hunstman Cancer Institute, and flow cytometry assistance from James Marvin and Chris Leukel at the University of Utah Flow Cytometry core.
The Paul Nabil Bustany Memorial Fund for Synovial Sarcoma Research, the Sarcoma Cancer Foundation of Canada (Beth England’s Sarcoma Research Fund), and the Damon Runyon Cancer Research Foundation provided funding for this project. This work was also partly supported by National Cancer Institute award P30CA042014. Work in the University of Utah Flow Cytometry Facility was also supported by the National Center for Research Resources of National Institutes of Health, 1S10RR026802-01.
The authors declare no competing financial interests.
Author contributions: J.J. Barrott and K.B. Jones conceived the idea, designed experiments, and wrote the manuscript. J.J. Barrott, L.A. Kafchinski, H. Jin, S.D. Kannan, and R. Kennedy carried out experiments. T. Mosbruger performed bioinformatics on RNAseq and NanoString data and generated heat maps and clustering figures. A.J. Lazar and T. Liu provided the histopathological review, and A.J. Lazar, W.-L. Wang, J.-W. Tsai, and D.M. Araujo generated the human tissue microarray. All authors reviewed and edited the manuscript.
Footnotes
Abbreviations used:
- PI3′
- phosphatidyl inositol 3′
- SS
- synovial sarcoma
- TMA
- tissue microarray
References
- Abate-Shen C., Banach-Petrosky W.A., Sun X., Economides K.D., Desai N., Gregg J.P., Borowsky A.D., Cardiff R.D., and Shen M.M.. 2003. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 63:3886–3890. [PubMed] [Google Scholar]
- Ali I.U., Schriml L.M., and Dean M.. 1999. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 91:1922–1932. 10.1093/jnci/91.22.1922 [DOI] [PubMed] [Google Scholar]
- Bald T., Quast T., Landsberg J., Rogava M., Glodde N., Lopez-Ramos D., Kohlmeyer J., Riesenberg S., van den Boorn-Konijnenberg D., Hömig-Hölzel C., et al. 2014. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature. 507:109–113. 10.1038/nature13111 [DOI] [PubMed] [Google Scholar]
- Barretina J., Taylor B.S., Banerji S., Ramos A.H., Lagos-Quintana M., Decarolis P.L., Shah K., Socci N.D., Weir B.A., Ho A., et al. 2010. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 42:715–721. 10.1038/ng.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrott J.J., Illum B.E., Jin H., Zhu J.F., Mosbruger T., Monument M.J., Smith-Fry K., Cable M.G., Wang Y., Grossmann A.H., et al. 2015. β-catenin stabilization enhances SS18-SSX2-driven synovial sarcomagenesis and blocks the mesenchymal to epithelial transition. Oncotarget. 6:22758–22766. 10.18632/oncotarget.4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay J.Y., Ray-Coquard I., Alberti L., and Ranchère D.. 2004. Targeting other abnormal signaling pathways in sarcoma: EGFR in synovial sarcomas, PPAR-gamma in liposarcomas. Cancer Treat. Res. 120:151–167. 10.1007/1-4020-7856-0_9 [DOI] [PubMed] [Google Scholar]
- Bonapace L., Coissieux M.M., Wyckoff J., Mertz K.D., Varga Z., Junt T., and Bentires-Alj M.. 2014. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 515:130–133. 10.1038/nature13862 [DOI] [PubMed] [Google Scholar]
- Chalhoub N., and Baker S.J.. 2009. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 4:127–150. 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.H., Robinson J.P., Arave R.A., Burnett W.J., Kircher D.A., Chen G., Davies M.A., Grossmann A.H., VanBrocklin M.W., McMahon M., and Holmen S.L.. 2015. AKT1 activation promotes development of melanoma metastases. Cell Reports. 13:898–905. 10.1016/j.celrep.2015.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L.M., and Baker S.J.. 2006. PTEN function in normal and neoplastic growth. Cancer Lett. 241:184–196. 10.1016/j.canlet.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Clark J., Rocques P.J., Crew A.J., Gill S., Shipley J., Chan A.M., Gusterson B.A., and Cooper C.S.. 1994. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 7:502–508. 10.1038/ng0894-502 [DOI] [PubMed] [Google Scholar]
- Coffelt S.B., Kersten K., Doornebal C.W., Weiden J., Vrijland K., Hau C.S., Verstegen N.J., Ciampricotti M., Hawinkels L.J., Jonkers J., and de Visser K.E.. 2015. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 522:345–348. 10.1038/nature14282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools-Lartigue J., Spicer J., McDonald B., Gowing S., Chow S., Giannias B., Bourdeau F., Kubes P., and Ferri L.. 2013. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Invest. 67484 10.1172/JCI67484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw B., Balemans M., Olde Weghuis D., and Geurts van Kessel A.. 1995. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum. Mol. Genet. 4:1097–1099. 10.1093/hmg/4.6.1097 [DOI] [PubMed] [Google Scholar]
- DeNardo D.G., Brennan D.J., Rexhepaj E., Ruffell B., Shiao S.L., Madden S.F., Gallagher W.M., Wadhwani N., Keil S.D., Junaid S.A., et al. 2011. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1:54–67. 10.1158/2159-8274.CD-10-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deocampo N.D., Huang H., and Tindall D.J.. 2003. The role of PTEN in the progression and survival of prostate cancer. Minerva Endocrinol. 28:145–153. [PubMed] [Google Scholar]
- Depowski P.L., Rosenthal S.I., and Ross J.S.. 2001. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod. Pathol. 14:672–676. 10.1038/modpathol.3880371 [DOI] [PubMed] [Google Scholar]
- Espinosa I., Beck A.H., Lee C.H., Zhu S., Montgomery K.D., Marinelli R.J., Ganjoo K.N., Nielsen T.O., Gilks C.B., West R.B., and van de Rijn M.. 2009. Coordinate expression of colony-stimulating factor-1 and colony-stimulating factor-1-related proteins is associated with poor prognosis in gynecological and nongynecological leiomyosarcoma. Am. J. Pathol. 174:2347–2356. 10.2353/ajpath.2009.081037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs N., Trautmann M., Endl E., Sievers E., Kindler D., Wurst P., Czerwitzki J., Steiner S., Renner M., Penzel R., et al. 2011. Phosphatidylinositol-3′-kinase/AKT signaling is essential in synovial sarcoma. Int. J. Cancer. 129:1564–1575. 10.1002/ijc.25829 [DOI] [PubMed] [Google Scholar]
- Fritz J.M., Tennis M.A., Orlicky D.J., Lin H., Ju C., Redente E.F., Choo K.S., Staab T.A., Bouchard R.J., Merrick D.T., et al. 2014. Depletion of tumor-associated macrophages slows the growth of chemically induced mouse lung adenocarcinomas. Front. Immunol. 5:587 10.3389/fimmu.2014.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D. 2015. Fatal attraction: How macrophages participate in tumor metastases. J Exp Med. 212:976. 10.1084/jem.2127insight1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A.J., Ruscetti M., Arenzana T.L., Tran L.M., Bianci-Frias D., Sybert E., Priceman S.J., Wu L., Nelson P.S., Smale S.T., and Wu H.. 2014. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol. Cell. Biol. 34:2017–2028. 10.1128/MCB.00090-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin M.L., Jin H., Straessler K., Smith-Fry K., Zhu J.F., Monument M.J., Grossmann A., Randall R.L., Capecchi M.R., and Jones K.B.. 2014. Modeling alveolar soft part sarcomagenesis in the mouse: a role for lactate in the tumor microenvironment. Cancer Cell. 26:851–862. 10.1016/j.ccell.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M., Erickson R., Scripture-Adams D.D., Lesche R., Trumpp A., Zack J.A., Kornblum H.I., Liu X., and Wu H.. 2001. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 294:2186–2189. 10.1126/science.1065518 [DOI] [PubMed] [Google Scholar]
- Haldar M., Hancock J.D., Coffin C.M., Lessnick S.L., and Capecchi M.R.. 2007. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 11:375–388. 10.1016/j.ccr.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Haldar M., Hedberg M.L., Hockin M.F., and Capecchi M.R.. 2009. A CreER-based random induction strategy for modeling translocation-associated sarcomas in mice. Cancer Res. 69:3657–3664. 10.1158/0008-5472.CAN-08-4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog C.E. 2005. Overview of sarcomas in the adolescent and young adult population. J. Pediatr. Hematol. Oncol. 27:215–218. 10.1097/01.mph.0000161762.53175.e4 [DOI] [PubMed] [Google Scholar]
- Hiratsuka S., Ishibashi S., Tomita T., Watanabe A., Akashi-Takamura S., Murakami M., Kijima H., Miyake K., Aburatani H., and Maru Y.. 2013. Primary tumours modulate innate immune signalling to create pre-metastatic vascular hyperpermeability foci. Nat. Commun. 4:1853 10.1038/ncomms2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.J., Liang S., Sharma A., Dong C., and Robertson G.P.. 2010. Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res. 70:6071–6082. 10.1158/0008-5472.CAN-09-4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibe T., Nakayama T., Okamoto T., Aoyama T., Nishijo K., Shibata K.R., Shima Y., Nagayama S., Katagiri T., Nakamura Y., et al. 2005. Disruption of fibroblast growth factor signal pathway inhibits the growth of synovial sarcomas: potential application of signal inhibitors to molecular target therapy. Clin. Cancer Res. 11:2702–2712. 10.1158/1078-0432.CCR-04-2057 [DOI] [PubMed] [Google Scholar]
- Jones K.B., Barrott J.J., Xie M., Haldar M., Jin H., Zhu J.F., Monument M.J., Mosbruger T.L., Langer E.M., Randall R.L., et al. 2016. The impact of chromosomal translocation locus and fusion oncogene coding sequence in synovial sarcomagenesis. Oncogene. 35:5021–5032. 10.1038/onc.2016.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A., Naito N., Yoshida A., Morimoto Y., Ouchida M., Shimizu K., and Beppu Y.. 2004. Establishment and characterization of a biphasic synovial sarcoma cell line, SYO-1. Cancer Lett. 204:105–113. 10.1016/j.canlet.2003.09.031 [DOI] [PubMed] [Google Scholar]
- Kim G., Ouzounova M., Quraishi A.A., Davis A., Tawakkol N., Clouthier S.G., Malik F., Paulson A.K., D’Angelo R.C., Korkaya S., et al. 2015. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene. 34:671–680. 10.1038/onc.2014.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobbe C.B., Lapin V., Suzuki A., and Mak T.W.. 2008. The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene. 27:5398–5415. 10.1038/onc.2008.238 [DOI] [PubMed] [Google Scholar]
- Lahat G., Tuvin D., Wei C., Wang W.L., Pollock R.E., Anaya D.A., Bekele B.N., Corely L., Lazar A.J., Pisters P.W., and Lev D.. 2010. Molecular prognosticators of complex karyotype soft tissue sarcoma outcome: a tissue microarray-based study. Ann. Oncol. 21:1112–1120. 10.1093/annonc/mdp459 [DOI] [PubMed] [Google Scholar]
- Lee C.H., Espinosa I., Vrijaldenhoven S., Subramanian S., Montgomery K.D., Zhu S., Marinelli R.J., Peterse J.L., Poulin N., Nielsen T.O., et al. 2008. Prognostic significance of macrophage infiltration in leiomyosarcomas. Clin. Cancer Res. 14:1423–1430. 10.1158/1078-0432.CCR-07-1712 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Gouon-Evans V., Nguyen A.V., and Pollard J.W.. 2002. The macrophage growth factor CSF-1 in mammary gland development and tumor progression. J. Mammary Gland Biol. Neoplasia. 7:147–162. 10.1023/A:1020399802795 [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhou H., Krueger J., Kaplan C., Lee S.H., Dolman C., Markowitz D., Wu W., Liu C., Reisfeld R.A., and Xiang R.. 2006. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J. Clin. Invest. 116:2132–2141. 10.1172/JCI27648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.J., Coulter J., Walker S.M., McKechnie M., Neisen J., McCabe N., Kennedy R.D., Salto-Tellez M., Albanese C., and Waugh D.J.. 2013. Potentiation of inflammatory CXCL8 signalling sustains cell survival in PTEN-deficient prostate carcinoma. Eur. Urol. 64:177–188. 10.1016/j.eururo.2012.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami T., Kurose A., Javed F., and Takeda Y.. 2015. Detection of rare variant of SS18-SSX1 fusion gene and mutations of important cancer-related genes in synovial sarcoma of the lip: gene analyses of a case and literature review. J. Oral Maxillofac. Surg. 73:1505–1515. 10.1016/j.joms.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Mitchem J.B., Brennan D.J., Knolhoff B.L., Belt B.A., Zhu Y., Sanford D.E., Belaygorod L., Carpenter D., Collins L., Piwnica-Worms D., et al. 2013. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 73:1128–1141. 10.1158/0008-5472.CAN-12-2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka N., Takenaka S., Araki N., Miwa T., Hashimoto N., Yoshioka K., Joyama S., Hamada K., Tsukamoto Y., Tomita Y., et al. 2010. Synovial sarcoma is a stem cell malignancy. Stem Cells. 28:1119–1131. 10.1002/stem.452 [DOI] [PubMed] [Google Scholar]
- Nielsen T.O., West R.B., Linn S.C., Alter O., Knowling M.A., O’Connell J.X., Zhu S., Fero M., Sherlock G., Pollack J.R., et al. 2002. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 359:1301–1307. 10.1016/S0140-6736(02)08270-3 [DOI] [PubMed] [Google Scholar]
- Oda K., Stokoe D., Taketani Y., and McCormick F.. 2005. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 65:10669–10673. 10.1158/0008-5472.CAN-05-2620 [DOI] [PubMed] [Google Scholar]
- Patel L., Pass I., Coxon P., Downes C.P., Smith S.A., and Macphee C.H.. 2001. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr. Biol. 11:764–768. 10.1016/S0960-9822(01)00225-1 [DOI] [PubMed] [Google Scholar]
- Przybyl J., Sciot R., Wozniak A., Schöffski P., Vanspauwen V., Samson I., Siedlecki J.A., Rutkowski P., and Debiec-Rychter M.. 2014. Metastatic potential is determined early in synovial sarcoma development and reflected by tumor molecular features. Int. J. Biochem. Cell Biol. 53:505–513. 10.1016/j.biocel.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F., Olson O.C., Quick M.L., Huse J.T., Teijeiro V., et al. 2013. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19:1264–1272. 10.1038/nm.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries C.H., Cannarile M.A., Hoves S., Benz J., Wartha K., Runza V., Rey-Giraud F., Pradel L.P., Feuerhake F., Klaman I., et al. 2014. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 25:846–859. 10.1016/j.ccr.2014.05.016 [DOI] [PubMed] [Google Scholar]
- Semba S., Itoh N., Ito M., Harada M., and Yamakawa M.. 2002. The in vitro and in vivo effects of 2-(4-morpholinyl)-8-phenyl-chromone (LY294002), a specific inhibitor of phosphatidylinositol 3′-kinase, in human colon cancer cells. Clin. Cancer Res. 8:1957–1963. [PubMed] [Google Scholar]
- Setsu N., Kohashi K., Fushimi F., Endo M., Yamamoto H., Takahashi Y., Yamada Y., Ishii T., Yokoyama K., Iwamoto Y., and Oda Y.. 2013. Prognostic impact of the activation status of the Akt/mTOR pathway in synovial sarcoma. Cancer. 119:3504–3513. 10.1002/cncr.28255 [DOI] [PubMed] [Google Scholar]
- Stambolic V., Tsao M.S., Macpherson D., Suzuki A., Chapman W.B., and Mak T.W.. 2000. High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/- mice. Cancer Res. 60:3605–3611. [PubMed] [Google Scholar]
- Stemke-Hale K., Gonzalez-Angulo A.M., Lluch A., Neve R.M., Kuo W.L., Davies M., Carey M., Hu Z., Guan Y., Sahin A., et al. 2008. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68:6084–6091. 10.1158/0008-5472.CAN-07-6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straessler K.M., Jones K.B., Hu H., Jin H., van de Rijn M., and Capecchi M.R.. 2013. Modeling clear cell sarcomagenesis in the mouse: cell of origin differentiation state impacts tumor characteristics. Cancer Cell. 23:215–227. 10.1016/j.ccr.2012.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Cheng H., Sampaio A.V., Nielsen T.O., and Underhill T.M.. 2010. EGR1 reactivation by histone deacetylase inhibitors promotes synovial sarcoma cell death through the PTEN tumor suppressor. Oncogene. 29:4352–4361. 10.1038/onc.2010.204 [DOI] [PubMed] [Google Scholar]
- Tamborini E., Bonadiman L., Greco A., Gronchi A., Riva C., Bertulli R., Casali P.G., Pierotti M.A., and Pilotti S.. 2004. Expression of ligand-activated KIT and platelet-derived growth factor receptor beta tyrosine kinase receptors in synovial sarcoma. Clin. Cancer Res. 10:938–943. 10.1158/1078-0432.CCR-03-0059 [DOI] [PubMed] [Google Scholar]
- Tamura M., Gu J., Matsumoto K., Aota S., Parsons R., and Yamada K.M.. 1998. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 280:1614–1617. 10.1126/science.280.5369.1614 [DOI] [PubMed] [Google Scholar]
- Teng H.W., Wang H.W., Chen W.M., Chao T.C., Hsieh Y.Y., Hsih C.H., Tzeng C.H., Chen P.C., and Yen C.C.. 2011. Prevalence and prognostic influence of genomic changes of EGFR pathway markers in synovial sarcoma. J. Surg. Oncol. 103:773–781. 10.1002/jso.21852 [DOI] [PubMed] [Google Scholar]
- Thomas D.G., Giordano T.J., Sanders D., Biermann S., Sondak V.K., Trent J.C., Yu D., Pollock R.E., and Baker L.. 2005. Expression of receptor tyrosine kinases epidermal growth factor receptor and HER-2/neu in synovial sarcoma. Cancer. 103:830–838. 10.1002/cncr.20847 [DOI] [PubMed] [Google Scholar]
- Turc-Carel C., Dal Cin P., Limon J., Rao U., Li F.P., Corson J.M., Zimmerman R., Parry D.M., Cowan J.M., and Sandberg A.A.. 1987. Involvement of chromosome X in primary cytogenetic change in human neoplasia: nonrandom translocation in synovial sarcoma. Proc. Natl. Acad. Sci. USA. 84:1981–1985. 10.1073/pnas.84.7.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan K.M., Barbie D.A., Davies M.A., Rabinovsky R., McNear C.J., Kim J.J., Hennessy B.T., Tseng H., Pochanard P., Kim S.Y., et al. 2009. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 16:21–32. 10.1016/j.ccr.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I., and Sawyers C.L.. 2002. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2:489–501. 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- Vlenterie M., Hillebrandt-Roeffen M.H., Flucke U.E., Groenen P.J., Tops B.B., Kamping E.J., Pfundt R., de Bruijn D.R., Geurts van Kessel A.H., van Krieken H.J., et al. 2015. Next generation sequencing in synovial sarcoma reveals novel gene mutations. Oncotarget. 6:34680–34690. 10.18632/oncotarget.5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G.V., Li G., Roy-Burman P., Nelson P.S., et al. 2003. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 4:209–221. 10.1016/S1535-6108(03)00215-0 [DOI] [PubMed] [Google Scholar]
- Young M.R., and Lathers D.M.. 1999. Myeloid progenitor cells mediate immune suppression in patients with head and neck cancers. Int. J. Immunopharmacol. 21:241–252. 10.1016/S0192-0561(99)00008-9 [DOI] [PubMed] [Google Scholar]
- Zbuk K.M., and Eng C.. 2007. Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer. 7:35–45. 10.1038/nrc2037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.