Escalante et al. show that a highly prevalent mouse intestinal protozoa, Tritrichomonas muris, was found to be a confounding factor in murine colitis. Mice infected with this parasite had elevated baseline levels of Th1 cytokines and developed exacerbated Th1-mediated disease.
Abstract
The mammalian gastrointestinal tract hosts a diverse community of microbes including bacteria, fungi, protozoa, helminths, and viruses. Through coevolution, mammals and these microbes have developed a symbiosis that is sustained through the host’s continuous sensing of microbial factors and the generation of a tolerant or pro-inflammatory response. While analyzing T cell–driven colitis in nonlittermate mouse strains, we serendipitously identified that a nongenetic transmissible factor dramatically increased disease susceptibility. We identified the protozoan Tritrichomonas muris as the disease-exacerbating element. Furthermore, experimental colonization with T. muris induced an elevated Th1 response in the cecum of naive wild-type mice and accelerated colitis in Rag1−/− mice after T cell transfer. Overall, we describe a novel cross-kingdom interaction within the murine gut that alters immune cell homeostasis and disease susceptibility. This example of unpredicted microbial priming of the immune response highlights the importance of studying trans-kingdom interactions and serves as a stark reminder of the importance of using littermate controls in all mouse research.
Introduction
Microorganisms within the mammalian gastrointestinal tract have developed complex relationships with the host, such that the host provides a nutrient-rich microbial niche while the microbes provide vitamins, metabolites, and other nutrients otherwise inaccessible to the host (Hooper et al., 2012). To maintain this symbiosis, the host has evolved many microbial sensors, such as Toll-like receptors and Nod-like receptors, that stimulate the immune system to secrete factors such as mucous, antimicrobial peptides, and IgA, to keep microbes in check (Philpott et al., 2014; Caballero and Pamer, 2015). In turn, these microbial signals are necessary for the proper development of the mucosal immune system (Honda and Littman, 2012; Hooper et al., 2012).
Although it is clear that the presence of certain bacteria (Ivanov et al., 2009; Atarashi et al., 2013), viruses (Pfeiffer and Virgin, 2016), and helminths (Elliott and Weinstock, 2012) within the microbiota can impact intestinal homeostasis, the potential immune-shaping role of other kingdoms, such as Protista, has been less well studied. Protozoa are unicellular eukaryotes known to cause human diseases such as malaria, giardiasis, and trichomoniasis (Lindsay et al., 2008). In wild and laboratory mice, several protozoa have been documented to be disease causing, whereas others, such as Entamoeba muris and Tritrichomonas muris, are considered nonpathogenic members of the murine microbiome (Baker, 2008). However, recent studies suggest that T. muris and a related organism, Tritrichomonas musculis, have the potential to be pathogenic as the presence of these protists in the gut microbiota promote type 2 immune responses (Howitt et al., 2016) and Th1 inflammation (Chudnovskiy et al., 2016), respectively. These observations suggest that even though murine health is not overtly altered, protozoa colonization has the potential to modify the immune response in disease models.
While analyzing colitis induced using a T cell transfer model in two nonlittermate mouse lines, we unexpectedly observed that the exacerbated disease susceptibility of one of the lines was transmissible by cohousing and was dominant in littermates. We further identified the protozoan parasite T. muris to be the pro-colitogenic confounding factor in this T cell transfer model of colitis. Indeed, mice infected with T. muris developed an increased IFN-γ+ CD4 T cell response and accelerated epithelial damage within the colon. We have also found that colonizing mice chronically with isolated T. muris altered the baseline number of Th1 T cells in the mouse intestine. This work reveals an unexpected critical role of a murine intestinal protozoan parasite in exacerbating colitis. This also highlights the need for monitoring parasite infections in specific pathogen–free animal facilities and argues for the standard use of littermate controls in all murine research.
Results and discussion
Rip2−/−Rag1−/− mice have accelerated T cell transfer colitis pathology
With the aim of studying the role of NOD1 and NOD2 signaling in regulating non–T/B cells during colitis, we generated a Rip2−/−Rag1−/− mouse line for use in the T cell transfer model of colitis. Indeed, RIP2 is an essential signaling adapter molecule downstream of both NOD1 and NOD2 (Magalhaes et al., 2011). After transfer of wild-type CD4+CD45RBHigh naive T cells, Rip2−/−Rag1−/− mice rapidly lost weight compared with nonlittermate Rag1−/− mice and were euthanized at week 4 because of their poor health (Fig. 1 A). Upon examination, Rip2−/−Rag1−/− mice had larger spleens and short, thick colons, consistent with an increase in cellularity and significant colon pathology (Fig. 1, B–E). Preparation of colon lamina propria (LP) cells revealed a decreased frequency but a significant absolute increase in the number of IFN-γ+CD4+ T cells (Fig. 1, F and G). There was also an increased number of TNFα+CD4+ T cells but no increase in IL-17A+CD4+ T cells (Fig. 1, F and G). Analysis of colon explant cultures revealed an elevated protein level of IFN-γ and IL-12/IL-23p40 (Fig. 1 H). These results demonstrated that, in comparison with nonlittermate Rag1−/− mice, Rip2−/−Rag1−/− mice had an increased pro-inflammatory response and exacerbated colon pathology after naive T cell transfer.
Figure 1.
Nonlittermate Rip2−/−Rag1−/− mice develop accelerated colitis. Nonlittermate Rip2−/−Rag1−/− and Rag1−/− mice were injected i.p. with 0.5 × 106 CD45RBHigh CD4 T cells. (A) Mice were weighed weekly, and the percentage of initial body weight was calculated. Data from four experiments were pooled, n = 15 mice per group. (B) Mice were sacrificed at week 4. Colons were fixed, H&E stained, and scored for colitis severity. Data from three experiments were pooled. (C) Representative spleens and colons were imaged. (D and E) Spleen, colon LP, and MLNs were harvested at week 4, and isolated cells were quantified by flow cytometry. (F and G) Colon cells were restimulated with plate-bound anti-CD3/anti-CD28 antibody and analyzed by flow cytometry to determine the total (F) and frequency (G) of cytokine-positive cells. Data from three experiments were pooled with 5–15 mice per group. (H) Proximal colon explants were cultured for 18 h, and supernatants were analyzed by ELISA for cytokine production. Data from two experiments were pooled with four to seven samples per group. (A, B, and D–H) Mean ± SEM is shown with *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 using an unpaired Student’s t test.
Rip2−/−Rag1−/− mice are not protected by regulatory cells
Co-transfer of regulatory CD4+CD45RBLow T cells in Rag1−/− mice has been shown to protect mice from CD4+CD45RBHigh-mediated colon pathology (Ostanin et al., 2009). Co-transfer of CD4+CD45RBLow T cells into Rip2−/−Rag1−/− mice, however, did not protect the mice from colitis (Fig. 2 A). These mice still developed increased cellularity in the spleen and increased CD4+ T cell infiltration of the colon LP and mesenteric lymph node (MLN; Fig. S1, A and B). Increased numbers of IFN-γ+CD4+ T cells and TNFα+CD4+ T cells were also still observed in the colon, along with an increase in colon IFN-γ and IL-12/IL-23p40 protein (Fig. 2, B–D; and Fig. S1, C and D). Various other cytokines, such as IL-1β, IL-6, IL-10, IL-12p70, IL-13, KC, MIP-1α, and MIP-2 were also increased in Rip2−/−Rag1−/− colons, regardless of the T cells transferred (Fig. 2 E). This lack of protection was not caused by a lack of CD4+Foxp3+ regulatory T cells (T reg cells) as there was a similar ratio of CD4+Foxp3+ to CD4+Foxp3− T cells in both Rag1−/− and Rip2−/−Rag1−/− mice when CD4+CD45RBLow T cells were co-transferred (Fig. 2 C). These results suggest that the early increase in pro-inflammatory mediators in the Rip2−/−Rag1−/− mice may be blocking the ability of the CD4+CD45RBLow T cells to suppress inflammation.
Figure 2.
Nonlittermate Rip2−/−Rag1−/− mice are not protected from pathology by regulatory CD45RBLow T cells. Nonlittermate Rip2−/−Rag1−/− and Rag1−/− mice were injected i.p. with 0.5 × 106 CD45RBHigh with or without 0.25 × 106 CD45RBLow CD4 T cells and sacrificed at week 4. (A) Colons were fixed, H&E stained, and scored for colitis severity. Data from three experiments were pooled. (B and C) Spleen, colon LP, and MLNs were harvested, and isolated cells were counted, restimulated with plate bound anti-CD3/anti-CD28 antibody, and analyzed by flow cytometry to determine tissue Foxp3+ (C) and colon IFN-γ+ T cell numbers (B). Data from two experiments were pooled with 4–12 mice per group. (D) Proximal colon explants were cultured for 18 h, and supernatants were analyzed by ELISA for cytokine production. Data from two experiments were pooled with three to seven samples per group. (A–D) Mean ± SEM is shown with *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 using a one-way ANOVA and Tukey’s post-hoc analysis. (E) Proximal colon explants were cultured for 18 h, and supernatants were analyzed by Bio-Plex assay for cytokine production. Data are one representative of three experiments with two samples per group. *, samples below minimum level of detection; #, samples above maximal level of detection.
Exacerbated Rip2−/−Rag1−/− pathology is transmissible
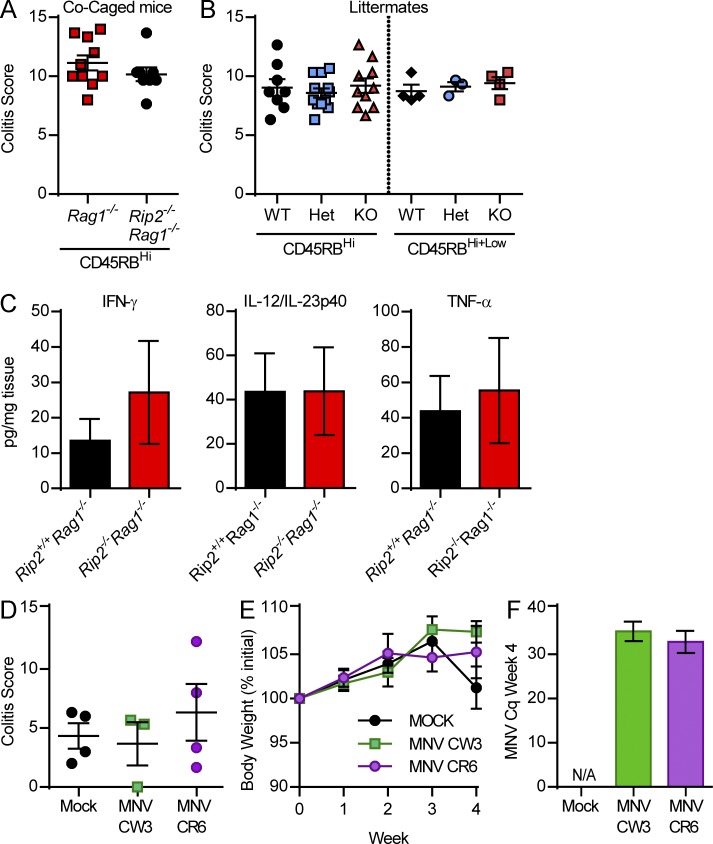
To determine whether the accelerated colitis seen in Rip2−/−Rag1−/− mice was driven by Rip2 deficiency or was caused by other factors such as a dysbiotic microbiota, Rip2−/−Rag1−/− and Rag1−/− mice were co-caged at weaning and 5 wk later injected with CD4+CD45RBHigh T cells. Strikingly, 4 wk after transfer, co-caged Rag1−/− mice developed equally accelerated pathology as the Rip2−/−Rag1−/− mice (Fig. 3 A), suggesting that the disease-inducing factor could be horizontally transferred. Littermate mice were then generated, and the resulting Rip2+/+Rag1−/− mice also developed accelerated colitis and had similar pro-inflammatory cytokine levels as their Rip2−/−Rag1−/− littermates (Fig. 3, B and C). These results indicate that microbial factors, rather than Rip2 genotype, were likely driving the exacerbated colitis in Rip2−/−Rag1−/− mice.
Figure 3.
Co-caged and littermate mice develop equally exacerbated T cell–induced colitis. (A and B) Co-caged (A) and littermate (B) Rip2−/−Rag1−/− and Rip2+/+Rag1−/− mice were injected i.p. with 0.5 × 106 CD45RBHigh CD4 T cells and sacrificed at week 4. Colons were fixed, H&E stained, and scored for colitis severity. Data from three experiments were pooled. Mean ± SEM is shown, and an unpaired Student’s t test was performed. (C) Proximal colon explants from week 4 after transfer littermate mice were cultured for 18 h, and supernatants were analyzed by ELISA for cytokine production. Data from two experiments were pooled with five to six samples per group. Mean ± SEM is shown, and a Student’s t test was performed. (D–F) Rag1−/− mice were orally gavaged with 106 PFU MNV CR6 or MNV CW3 and 2 wk later injected i.p. with 0.5 × 106 CD45RBHigh CD4 T cells. (D) Body weight was monitored, and mice were sacrificed at week 4. (E) Colons were fixed, H&E stained, and scored for colitis severity. (F) Fecal pellets were confirmed positive by qPCR. Data are from one experiment with three to four mice per group. Mean ± SEM is shown, and a one-way ANOVA was performed.
To determine the potential pro-colitogenic microbial factors, Rip2−/−Rag1−/− and Rag1−/− nonlittermate and littermate mice were screened for viral infections. Interestingly, Rip2−/−Rag1−/− nonlittermate and all littermates, regardless of genotype, were positive for murine norovirus (MNV) infection (IDEXX testing by quantitative real-time PCR [qPCR]), whereas nonlittermate Rag1−/− mice were negative. MNV is commonly found in the gastrointestinal tract of laboratory mice, and Rag1−/− mice have been reported to be persistently infected after exposure (Karst et al., 2003). Because previous studies had shown that MNV infection may exacerbate both chemical- and bacterial-induced colitis in mice (Lencioni et al., 2008; Cadwell et al., 2010), we investigated the hypothesis that MNV may exacerbate colitis in Rag1−/− mice during T cell transfer colitis. To test this, we infected Rag1−/− mice with MNV CW3 or MNV CR6 and 2 wk later compared them with their noninfected littermates during T cell transfer colitis. Although MNV levels were still detectable by qPCR 4 wk after CD4+CD45RBHigh T cell transfer, no differences were observed in pathology or weight loss, suggesting that MNV infection is not a contributing factor to T cell transfer colitis (Fig. 3, D–F).
T. muris infection exacerbates T cell transfer colitis
Rip2−/−Rag1−/− and Rag1−/− nonlittermate mice were also screened for parasitic infection. All mice were found to be negative for E. muris, Giardia muris, and Spironucleus muris. Notably, however, the original Rip2−/−Rag1−/− mouse line and littermate mice were all found to be positive for the protozoa T. muris, whereas Rag1−/− nonlittermates were not. Indeed, large numbers of these protozoa could be observed by microscope in the cecal contents of Rip2−/−Rag1−/− but not Rag1−/− nonlittermate mice (Fig. 4 A). Giemsa staining revealed the characteristic three anterior flagella of T. muris (Fig. 4 B). qPCR screening of our colony revealed that 18 out of 24 separate mouse strains were T. muris colonized. T. muris is transferred between mice by ingestion of infected fecal pellets containing pseudocysts, but transmission between cages is limited when sterile technique is strictly followed (Baker, 2008). Co-caging of mice led to transmission of the parasitic infection by 2 wk (not depicted). We next tested the hypothesis that T. muris infection could be the confounding factor that accelerated colitis in our T cell transfer experiments.
Figure 4.
T. muris accelerates T cell–induced colitis. (A) Fresh cecal contents were diluted in phosphate-buffered saline and visualized under a microscope. (B) T. muris from the cecal contents of Rip2−/−Rag1−/− mice were stained with Giemsa and visualized. (C–G) T. muris was isolated from the cecum of Rip2−/−Rag1−/− mice, and 106 protozoans were orally gavaged into Rag1−/− mice. 2 wk later, mice were injected i.p. with 0.5 × 106 CD45RBHigh CD4 T cells. (C) Body weight was measured, and mice were sacrificed at week 4. (D–F) Colons were fixed, H&E stained, and scored for colitis severity. Data from three experiments were pooled with 12 mice per group. Bars: (A) 100 μm; (B) 10 μm; (F) 200 μm. (G) Proximal colon explants from week 4 after T cell transfer T. muris–infected and control mice were cultured for 18 h, and supernatants were analyzed by ELISA for cytokine production. Data from two experiments were pooled with 9–10 samples per group. (C–E and G) Mean ± SEM is shown with *, P ≤ 0.05; **, P ≤ 0.01 using an unpaired Student’s t test.
T. muris was isolated from the cecal contents of Rip2−/−Rag1−/− mice (Fig. S2 A) and orally gavaged into Rag1−/− mice. 2 wk later, CD4+CD45RBHigh T cells were transferred into the T. muris–infected Rag1−/− mice and their noninfected littermate controls. By 4 wk after T cell transfer, the T. muris–infected mice lost significantly more weight than their mock-challenged siblings (Fig. 4 C). The T. muris–infected mice had significantly increased loss of crypt structure, epithelial damage (P = 0.0545), and lymphocyte infiltration (chronic inflammation score; Fig. 4, D–F). IFN-γ and IL-12/IL-23p40 protein levels were also elevated in the colons of the infected mice, whereas IL-18 was not significantly increased (Fig. 4 G). Together, these data show that T. muris infection contributed to an elevated Th1 pro-inflammatory environment in the colon that resulted in increased colon pathology during T cell transfer colitis.
Chronic T. muris infection alters baseline T cell homeostasis
To determine whether T. muris could be altering the steady-state level of immune activation in the gut of immunocompetent mice during natural infection, we created a line of chronically infected C57BL/6 mice by infecting a pair of C57BL/6 breeders. Baseline T cell numbers and cytokine production in the offspring of these infected mice were compared with pups from noninfected breeders (littermates to the infected breeders). Interestingly, infected mice had increased frequencies and a tendency for numbers (P = 0.0617) of IFN-γ+ CD4 T cells in their cecal LP (Fig. 5 A). In contrast, frequencies of IL-17A+ CD4 T cells were decreased in the colon LP (Fig. 5 B). Rorγt−Foxp3+ T reg cell frequencies and numbers were similar between infected and noninfected mice, but Rorγt+Foxp3+ T reg cell frequencies were decreased (Fig. 5, A and B). IFN-γ protein levels were below the assay level of detection in cultured colon explant supernatants, and IL-18 was not significantly different, but interestingly, IL-12/IL-23p40 protein was elevated in T. muris colonized mice (Fig. 5 C). This baseline increase in IL-12/IL-23p40 protein and IFN-γ–producing CD4 T cells, accompanied by the decrease in T reg cells, indicated a shift to a more pro-inflammatory rather than tolerant mucosal environment in the presence of T. muris.
Figure 5.
T. muris infection chronically increases cecal IFN-γ+ T cells. (A–D) C57BL/6 breeders were orally gavaged with 106 isolated protozoans. (A and B) Ceca (A) and colons (B) of infected pups and control pups from noninfected breeders were harvested. Cells were isolated and restimulated for 4 h with PMA and ionomycin, in the presence of protein transport inhibitor cocktail, before flow cytometry analysis. Data from two experiments were pooled with three to six mice per group. (C) Proximal colon explants were cultured for 24 h, and supernatants were analyzed by ELISA for IL-12/IL-23p40 and IL-18 protein. Data from two experiments were pooled with six to seven samples per group. (D) Small intestines were fixed, stained for tufts cells (DCLK1+DAPI+), and quantified. Data from two experiments were pooled with six mice per group. (A–D) Mean ± SEM is shown with *, P ≤ 0.05 using an unpaired Student’s t test.
Although we focused on a Th1 immune response in the large intestine, the wide array of cytokines found to be increased in the Rip2−/−Rag1−/− colons during T cell transfer colitis suggested that T. muris may be acting as a general stimulator of the mucosal immune system and not specifically favoring a Th1 response. This would agree with the study from Howitt et al. (2016) that outlined the induction of a type 2 response in the small intestine upon T. muris colonization. Interestingly, we also observed a significant increase in type 2–inducing tuft cells in the small intestines of our chronically infected wild-type mice, suggesting T. muris responses may differ in distinct gut regions (Fig. 5 D). Recently, Chudnovskiy et al. (2016) reported that acute infection of mice with a related protozoan T. musculis induced IL-18–driven Th1 and Th17 responses that altered the outcome of bacterial-induced colitis, T cell–driven colitis, and colorectal tumor formation. Although our study also demonstrates increased Th1 responses after protozoa infection, IL-18 and IL-17 levels were not significantly increased. This difference may be the result of chronic versus acute infection or the result of different protozoa. Indeed, the 28s rRNA primers used in our study cannot differentiate between T. muris and T. musculis. Collectively, these studies are further evidence that T. muris and related intestinal parasites could alter disease outcomes in many other mouse models.
Because of the presence of T. muris in our Rip2−/−Rag1−/− mice, we were unable to determine whether NOD1 and 2 signaling could alter colitis onset in the T cell transfer model of colitis. A previous study has described a pro-colitogenic bacterial composition in Nod2−/− and Rip2−/− mice; however, no details about the colonization of these mice with protozoa were given (Couturier-Maillard et al., 2013). Although the relationship between T. muris and bacteria within the gut is unknown, the large number of protozoa present in the cecal contents of T. muris colonized mice suggests a likely impact on bacterial composition. Thus, along with searching for potential T. muris–specific antigens, the potential of a T. muris–induced bacterial dysbiosis could also be explored.
T. muris screening is not standard in animal facilities; therefore, the current prevalence of T. muris in laboratory mice is unknown. Treatment with metronidazole has been reported to rid mice of T. muris (Roach et al., 1988; Howitt et al., 2016); however, antibiotic treatment also alters bacterial composition, which could have other confounding effects on a given phenotype. Our experience suggests that cross-fostering newborn pups with T. muris–free dams is a potential option. Although T. muris may now be flagged as a potential confounding factor in intestinal disease models, whether it impacts systemic disease remains to be determined. Moreover, there still remains many other unidentified protozoans or microbes that may also play immunomodulatory roles in mouse models, underscoring the importance of proper littermate controlled experiments.
Pentatrichomonas hominis and Dientamoeba fragilis are two species of trichomonads known to sporadically colonize the human gastrointestinal tract (Maritz et al., 2014). Although these trichomonads were originally described to be commensal protists, the correlation between the presence of these organisms in patients presenting gastrointestinal symptoms and the work presented here calls for a reevaluation of their pathogenicity (Meloni et al., 2011; Stark et al., 2016). Overall, our findings highlight the need for a better understanding of cross-kingdom interactions between host and protozoa within the gastrointestinal tract and emphasize the importance of controlling for microbial factors in mouse models of disease using littermate controls.
Materials and methods
Animals
All animals were housed in specific pathogen–free conditions at the University of Toronto, Division of Comparative Medicine. Rag1−/− and C57BL/6 mice were purchased from The Jackson Laboratory and subsequently bred in house. Rip2−/− mice were obtained from R.A. Flavell (Yale University School of Medicine, New Haven, CT; Kobayashi et al., 2002). Rip2−/−Rag1−/− mice were generated by crossing Rag1−/− mice with in-house Rip2−/− mice. All animal experiments were approved by the animal care committee, University of Toronto.
T cell transfer colitis
CD4+CD45RBHigh and CD4+CD45RBLow T cells were sorted from T. muris–negative C57BL/6 spleens. 0.5 × 106 CD4+CD45RBHigh with or without 0.25 × 106 CD4+CD45RBLow T cells were injected i.p. into 8-wk-old Rag1−/− or Rip2−/−Rag1−/− mice. Mice were weighed twice a week to obtain a mean weekly weight. Mice were sacrificed at 4 wk after T cell transfer, and colons were fixed with 10% formalin and hematoxylin and eosin (H&E) stained for pathological scoring or used for LP preps and flow cytometry. Colon pathology was scored by a blinded pathologist as previously described (Ostanin et al., 2009).
Tissue preparations
Colon LP cells were isolated as previously described (Goodyear et al., 2014). In brief, colons were washed in pre-stripping buffer (HBSS with 5 mM DTT, penicillin, streptomycin, and FBS) to remove mucous and fecal contents before incubation in epithelial stripping buffer (HBSS with 5 mM EDTA, penicillin, streptomycin, and FBS). Colons were then washed in HBSS + 10 mM HBSS, minced, and digested in digestion buffer (HBSS with 10 mM Hepes, penicillin, streptomycin, 20 µg/ml DNase, and 0.2U/ml Liberase TM). Colon pieces were titurated with an 18GA needle, filtered, and washed. Spleen and MLNs were mashed through a filter, and cells were isolated by centrifugation. Red blood cells were lysed using ammonium chloride.
Flow cytometry
For intracellular cytokine staining, cells were restimulated for 18 h on plates coated with anti-CD3 and anti-CD28 antibody or for 4 h with PMA and ionomycin. Brefeldin A and Monensin (Protein transport inhibitor cocktail; eBioscience) were added during the last 4 h of restimulation. Dead cells were marked using the LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Molecular Probes), and Fc receptors were blocked using mouse CD16/CD32 antibody (eBioscience). Surface staining was performed with anti–mouse CD4 (GK1.5), CD45RB (C363-16A), and TCRβ (H57-597) from eBioscience. Cells were fixed and permeabilized using Foxp3/Transcription Factor staining buffer set (eBioscience) before staining with anti–mouse IFN-γ (XMG1.2), TNFα (MP6-XT22), IL-17A (eBio17B7), and Foxp3 (FJK-16s) from eBioscience. Samples were analyzed on a FACSCanto II or LSR Fortessa X20 (BD).
Colon explants, ELISAs, and Bio-Plex assay
Proximal colon explants were weighed, rinsed in RPMI, and cultured in 500 µl complete RPMI for 18–24 h as previously described (Hue et al., 2006). Supernatants were centrifuged and used for cytokine measurement by IL-12/IL-23p40 (eBioscience; sensitivity 2 pg/ml), IFN-γ (eBioscience; sensitivity 15 pg/ml), and IL-18 (MBL; sensitivity 12.5 pg/ml) ELISA or Bio-Plex Mouse Cytokine 23-plex Assay (Bio-Rad Laboratories) according to the manufacturer’s instructions. Results were normalized to tissue weight.
Immunofluorescence staining
Small intestines were collected, formalin fixed, and paraffin embedded. Paraffin sections (5 µm) were deparaffinized in xylene and rehydrated in an ethanol gradient. Antigen retrieval was performed in 10 mM sodium citrate buffer, pH 6.0, at 95°C for 20 min. Sections were stained with rabbit polyclonal anti-DCLK1 (1:1,000; Abcam) overnight at 4°C, followed by staining with Alexa Fluor 647–conjugated anti–rabbit secondary antibody (1:4,000; Molecular Probes), and counterstaining with DAPI (1:10,000; Sigma-Aldrich). Images were acquired with the Axio Scan Z.1 Slide Scanner (ZEISS). Quantification was performed blind by counting the total number of tuft cells per crypt-villus with the Zen Blue 2 software (ZEISS).
MNV infection
MNV CR6 and CW3 stocks and plasmids were provided by H.W. Virgin (Washington University School of Medicine, St. Louis, MO). MNV was propagated, quantified, and detected by qPCR as previously described (Hwang et al., 2014). Mice were mock infected or infected with 106 PFU MNV and confirmed to be positive by qPCR (IDEXX Bioresearch or in house as previously described).
T. muris isolation and infection
Cecal contents of Rip2−/−Rag1−/− mice were resuspended in PBS and filtered twice through a 70-µm cell strainer. Flow through was centrifuged, resuspended, and refiltered twice. Protozoa were double sorted based on size, granularity, and violet autofluorescence on an Influx (BD; Fig. S2 A) and visualized under a microscope (BX43; Olympus) to confirm protozoa isolation. Eubacteria 16s rDNA levels in sort samples were determined by qPCR as previously described (Robertson et al., 2013) to measure the reduction of bacterial contamination (Fig. S2 B). 106 protozoa in PBS or protozoa-free PBS was orally gavaged into Rag1−/− or C57BL/6 mice. 2 wk after infection, mice were confirmed to be T. muris positive and used for T cell transfer colitis or as breeders. T. muris testing was performed by IDEXX Bioresearch, cecal content wet smear, and/or by qPCR for the 28s rRNA as previously described (Howitt et al., 2016). For qPCR, fecal DNA was isolated using a NucleoSpin Soil kit (MACHEREY-NAGEL) and amplified using Power SYBR green master mix (Invitrogen). For microscopic visualization, T. muris were fixed with paraformaldehyde, adhered to slides by cytospin, and stained with Giemsa.
Statistical analysis
Unpaired Student’s t tests were performed when two groups were compared. A one-way ANOVA was used for comparison of more than two groups, followed by a post-hoc Tukey’s multiple comparisons test if needed.
Online supplemental material
Supporting data for Fig. 2 (CD4+CD45RBHigh+Low T cells into Rip2−/−Rag1−/− vs. Rag1−/− mice), T. muris sorting strategy, and measurement of bacterial contamination are provided. Fig. S1 shows Rip2−/−Rag1−/− are not protected from pathology by regulatory CD45RBLow T cells. Fig. S2 shows the sorting strategy for T. muris.
Supplementary Material
Acknowledgments
The authors of this paper would like to thank Dionne White and Joanna Warzyszynska for their expert FACS assistance, Elisabeth Foerster for her microscopy expertise, and the University of Toronto Department of Comparative Medicine animal facility for their assistance.
This work was supported by grants from the Canadian Institutes of Health Research (to D.J. Philpott) and the Crohn’s and Colitis Foundation of Canada (to T. Mallevaey). N.K. Escalante was supported by a Vanier Canada Graduate Scholarship.
The authors declare no competing financial interests.
Author contributions: Conceptualization, N.K. Escalante, D.J. Philpott, T. Mallevaey, and S.E. Girardin; methodology, N.K. Escalante, A. Mortha, D.J. Philpott, T. Mallevaey, and S.E. Girardin; formal analysis, N.K. Escalante; investigation, N.K. Escalante, P. Lemire, M. Cruz Tleugabulova, D. Prescott, and C.J. Streutker; writing—original draft, N.K. Escalante; writing—review and editing, D.J. Philpott and S.E. Girardin; visualization, N.K. Escalante; supervision, D.J. Philpott, T. Mallevaey, and S.E. Girardin; project administration, N.K. Escalante, D.J. Philpott, and T. Mallevaey; funding acquisition, D.J. Philpott and T. Mallevaey.
Footnotes
Abbreviations used:
- LP
- lamina propria
- MLN
- mesenteric lymph node
- MNV
- murine norovirus
- qPCR
- quantitative real-time PCR
References
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., et al. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 500:232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Baker D.G. 2008. Parasites of rats and mice. In Flynn’s Parasites of Laboratory Animals. Second edition. D.G. Baker, editor. Blackwell Publishing Ltd., Ames, IA. 303–397. [Google Scholar]
- Caballero S., and Pamer E.G.. 2015. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 33:227–256. 10.1146/annurev-immunol-032713-120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C.Y., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., and Virgin H.W.. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141:1135–1145. 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A., Mortha A., Kana V., Kennard A., Ramirez J.D., Rahman A., Remark R., Mogno I., Ng R., Gnjatic S., et al. 2016. Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell. 167:444–456.e14. 10.1016/j.cell.2016.08.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier-Maillard A., Secher T., Rehman A., Normand S., De Arcangelis A., Haesler R., Huot L., Grandjean T., Bressenot A., Delanoye-Crespin A., et al. 2013. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123:700–711. 10.1172/JCI62236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.E., and Weinstock J.V.. 2012. Helminth-host immunological interactions: prevention and control of immune-mediated diseases. Ann. N. Y. Acad. Sci. 1247:83–96. 10.1111/j.1749-6632.2011.06292.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear A.W., Kumar A., Dow S., and Ryan E.P.. 2014. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods. 405:97–108. 10.1016/j.jim.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Honda K., and Littman D.R.. 2012. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30:759–795. 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., and Macpherson A.J.. 2012. Interactions between the microbiota and the immune system. Science. 336:1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt M.R., Lavoie S., Michaud M., Blum A.M., Tran S.V., Weinstock J.V., Gallini C.A., Redding K., Margolskee R.F., Osborne L.C., et al. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 351:1329–1333. 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S., Ahern P., Buonocore S., Kullberg M.C., Cua D.J., McKenzie B.S., Powrie F., and Maloy K.J.. 2006. Interleukin-23 drives innate and T cell–mediated intestinal inflammation. J. Exp. Med. 203:2473–2483. 10.1084/jem.20061099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Alhatlani B., Arias A., Caddy S.L., Christodoulou C., Cunha J.B., Emmott E., Gonzalez-Hernandez M., Kolawole A., Lu J., et al. 2014. Murine norovirus: propagation, quantification, and genetic manipulation. Curr. Protoc. Microbiol. 33:1–61: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Lay M., Davidson J., and Virgin H.W. IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 299:1575–1578. 10.1126/science.1077905 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Inohara N., Hernandez L.D., Galán J.E., Núñez G., Janeway C.A., Medzhitov R., and Flavell R.A.. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 416:194–199. 10.1038/416194a [DOI] [PubMed] [Google Scholar]
- Lencioni K.C., Seamons A., Treuting P.M., Maggio-Price L., and Brabb T.. 2008. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp. Med. 58:522–533. [PMC free article] [PubMed] [Google Scholar]
- Lindsay D.S., Sundermann C.A., and Baker D.G.. 2008. Biology of the protozoa. In Flynn’s Parasites of Laboratory Animals. Second edition. D.G. Baker, editor. Blackwell Publishing Ltd., Ames, IA. 15–25. [Google Scholar]
- Magalhaes J.G., Lee J., Geddes K., Rubino S., Philpott D.J., and Girardin S.E.. 2011. Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands. Eur. J. Immunol. 41:1445–1455. 10.1002/eji.201040827 [DOI] [PubMed] [Google Scholar]
- Maritz J.M., Land K.M., Carlton J.M., and Hirt R.P.. 2014. What is the importance of zoonotic trichomonads for human health? Trends Parasitol. 30:333–341. 10.1016/j.pt.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni D., Mantini C., Goustille J., Desoubeaux G., Maakaroun-Vermesse Z., Chandenier J., Gantois N., Duboucher C., Fiori P.L., Dei-Cas E., et al. 2011. Molecular identification of Pentatrichomonas hominis in two patients with gastrointestinal symptoms. J. Clin. Pathol. 64:933–935. 10.1136/jcp.2011.089326 [DOI] [PubMed] [Google Scholar]
- Ostanin D.V., Bao J., Koboziev I., Gray L., Robinson-Jackson S.A., Kosloski-Davidson M., Price V.H., and Grisham M.B.. 2009. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G135–G146. 10.1152/ajpgi.90462.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J.K., and Virgin H.W.. 2016. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 351:aad5872 10.1126/science.aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott D.J., Sorbara M.T., Robertson S.J., Croitoru K., and Girardin S.E.. 2014. NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol. 14:9–23. 10.1038/nri3565 [DOI] [PubMed] [Google Scholar]
- Roach P.D., Wallis P.M., and Olson M.E.. 1988. The use of metronidazole, tinidazole and dimetridazole in eliminating trichomonads from laboratory mice. Lab. Anim. 22:361–364. 10.1258/002367788780746287 [DOI] [PubMed] [Google Scholar]
- Robertson S.J., Zhou J.Y., Geddes K., Rubino S.J., Cho J.H., Girardin S.E., and Philpott D.J.. 2013. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 4:222–231. 10.4161/gmic.24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark D., Barratt J., Chan D., and Ellis J.T.. 2016. Dientamoeba fragilis, the neglected trichomonad of the human bowel. Clin. Microbiol. Rev. 29:553–580. 10.1128/CMR.00076-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.