Abstract
X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is a cytokine-regulated, tumor necrosis factor (TNF) receptor-associated factor (TRAF) domain-containing protein that has a poorly defined cellular function. Here, we show that ectopically-expressed XAF1 inhibits TNF-α-induced NF-κB activation, whereas shRNA silencing of endogenous XAF1 augments it. Our data suggest that XAF1 may inhibit TNF-α-induced NF-κB activation by disrupting assembly of the TRADD/TRAF2/RIP1 complex (complex I) downstream of TNF receptor activation. XAF1 interacts with TRAF2 and inhibits TRAF2-dependent NF-κB activation, in part, by blocking TRAF2 poly-ubiquitination. Our findings also indicate that although XAF1 does not directly inhibit RIP1-dependent NF-κB activation, it binds RIP1 and disrupts RIP1/TRADD association. Our data suggest that XAF1 acts as a feedback regulator of the TNF receptor signaling pathway to suppress NF-κB activation.
Keywords: XAF1, TRAF2, NF-κB, RIP1, TNF, inflammatory response
Introduction
Tumor necrosis factor alpha (TNF-α) is a dual function cytokine that mediates pro-survival or pro-apoptotic responses, depending upon the physiological or biochemical context of the activated signaling [1–3]. Under most conditions, TNF-α binding to TNF receptor (TNFR) 1 leads to receptor subunit multimerization and recruitment of the adaptor protein TNFR1-associated death domain (TRADD) through death domain-mediated interactions between the receptor cytoplasmic tail and TRADD. TRADD associates directly with TNF receptor associated factor (TRAF) 2 and receptor interacting protein (RIP) 1, which in turn recruit additional proteins to initiate complex I formation [4–8] including proteins involved in regulating protein phosphorylation, ubiquitination and deubiquitination, culminating in activation of nuclear factor-kappa B (NF-κB). A critical step in TNF-α signaling is the lysine 63 (K63)-linked poly-ubiquitination of RIP1, which is thought to serve as a platform for transforming growth factor beta (TGF-β) activated kinase (TAK) 1/TAK1 binding protein (TAB) 1/TAB2 complex docking and activation of TAK1 [9]. The inhibitor of NF-κB kinase (IKK) alpha/beta subunit IKKα/IKKβ/NF-κB essential modulator (NEMO) enzyme complex is also recruited to K63-linked poly-ubiquitinated RIP1 through NEMO’s ubiquitin binding domain. Activated TAK1 phosphorylates IKKβ promoting a structural change in the enzyme [10–12]. The activated IKK complex phosphorylates inhibitor of NF-κB (IκB) on serine 32 and 36, leading to its K48-linked ubiquitination and proteasomal degradation [13]. As a consequence, free NF-κB translocates into the nucleus and induces gene expression that promotes cell survival and other responses (inflammation in particular). Under certain circumstances, instead of triggering cell survival signaling TNF-α/TNFR1/TRADD recruits Fas-associated death domain protein (FADD) and caspase-8 to form the complex II leading to caspase-3-dependent cell death [5,14]. In addition, c-jun N-terminal kinases (JNKs), p38s and extracellular signal-regulated kinases (ERKs) are also potently activated in response to TNF-α through the mitogen-activated protein kinase (MAPK) pathway leading to stimulation of activator protein AP-1 [15,16].
TRAF proteins contribute to a variety of biological responses, including those that regulate immunity, inflammation, embryonic development and bone metabolism through regulating signaling pathways that impact cell survival, differentiation and death [17,18]. TRAF family members are characterized by a highly conserved C-terminal TRAF domain that is required for interactions with other TRAF proteins, receptors and downstream signaling molecules [13]. Some family members also contain RING finger/zinc finger domains that exhibit E3 ubiquitin ligase activity [13]. As such, many TRAFs regulate signaling by participating directly or indirectly in ubiquitination processes that regulate or mediate signal propagation [19,20]. TRAF2, in particular, activates NF-κB via NF-κB-inducing kinase (NIK) or IKK activation [21–23]. TRAF2 deficiency leads to early lethality in mice and sensitizes cells to TNF-α-induced apoptosis [24,25]. TRAF2 functions as a scaffold protein by interacting directly or indirectly with receptors in the TNFR family, kinases in MAPK kinase kinase (MAP3K) family and inhibitor of apoptosis proteins (cIAP)-1 and -2. TRAF2 also catalyzes nonproteolytic K63-dependent ubiquitination of RIP1, in conjunction with the E2 ubiquitin-conjugating enzyme Ubc13, which serves as a recruitment signal that ultimately leads to NF-κB activation [26–29]. TRAF2 can also target itself for degradation through K48-linked ubiquitination [30] and can induce 26S-dependent proteolysis of TRAF3 [31] and perhaps other proteins. TRAF2’s effects are regulated by other ubiquitin-editing enzymes, including A20 and CYLD deubiquitinases [29,32]. A20, in particular, is a potent inhibitor of several key components of the TNF-α signaling pathway and is itself transcriptionally upregulated by TNF-α-induced NF-κB [29,33,34]. Other negative feedback regulators such as IκB, IL-10 and optineurin [35–37] are also induced by TNF-α to regulate the duration of inflammatory response.
X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is a cytoplasmic protein that is widely expressed in normal tissues, albeit at low basal levels [38]. XAF1 is upregulated by interferons (IFN) [39] and other cytokines like TNF-α [39–41]. The physiological role of XAF1 remains somewhat obscure, although several groups have implicated a role in pro-apoptotic responses. Early studies suggested that XAF1 associated with XIAP to redirect XIAP from the cytosol to the nucleus and inhibit XIAP’s anti-caspase activity, thereby inducing apoptosis [38]. However, not all subsequent work has identified an association with XIAP, and although other IAP family members can interact with XAF1 [42], the consequences of these associations remain unclear. Regardless, overexpression of XAF1 leads to suppression of cancer cell growth and sensitizes cancer cells to death induced by various stimuli [39,40,43–45]. Structurally, XAF1 contains an N-terminal zinc-finger TRAF-like domain [39,46] suggesting a probable function in regulating protein-protein interactions and perhaps in regulating TRAF-dependent responses. We have identified XAF1 as a TNF-α-induced gene, and in this report we investigate the role for XAF1 in TNF-α/TNFR1 signaling. Our data show that XAF1 inhibits TNF-α-induced NF-κB activation and TRAF2 ubiquitination and activity. Formation of TNFR complex I was disrupted by XAF1, perhaps providing insight into its previously identified pro-apoptotic activity. Together, these results implicate XAF1 as a negative feedback regulator of TNF-α signaling to NF-κB.
Materials and Methods
Cell lines, plasmids and reagents
Human embryonic kidney (HEK) 293 cells and human renal carcinoma cells ACHN (both from ATCC, Manassas, VA, USA) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Expression plasmids for XAF1 and its truncated mutants were described previously [39,46], except for the untagged XAF1 cDNA, which was generated by moving the full length XAF1 cDNA from the HA-XAF1 plasmid into the EcoRI/KpnI sites in pcDNA3.1. Human TRAF2 was cloned into pcDNA3.1 in frame with a C-terminal myc/HIS tag for ectopic expression analysis. p3XFLAG-RIP1 and pcDNA3.1-HA-TRADD were gifts from G. Nuñez (University of Michigan, MI, USA), and pcDNA3.1-HA-Ubiquitin was a gift from D. Chadee (University of Toledo, OH, USA). The rabbit anti-XAF1 polyclonal antiserum was generated by immunization with full-length GST-tagged XAF1, and XAF1-specific immunoglobulin affinity purified with the antigen. Human recombinant TNF-α was purchased from Invitrogen (Carlsbad, CA, USA), MG132, cycloheximide (CHX), anti-Actin monoclonal antibody and anti-FLAG monoclonal antibody from Sigma Aldrich (St. Louis, MO, USA), anti-phospho-IκBα monoclonal antibody, anti-HA monoclonal antibody and anti-XAF1 monoclonal antibody from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-capase-3 polyclonal antibody, anti-TRAF2 polyclonal antibody and anti-myc monoclonal antibody from Cell Signaling Technologies (Boston, MA, USA), and anti-RIP1 monoclonal antibody and anti-TRADD monoclonal antibodies from BD Biosciences (San Jose, CA, USA). All transfection studies were performed using Lipofectamine reagents (Invitrogen, Carlsbad, CA, USA).
XAF1 inducible cells
Plasmids of pUbiq.irtTA, encoding the glucocorticoid/tetracycline-inducible transactivator [47], and pTRE2hyg were gifts of F. Dong (University of Toledo, OH, USA). HEK293 cells were co-transfected with pUbiq.irtTA along with pBabe-puro plasmid, stably transfected clones selected in 2 µg/ml puromycin and then stably re-transfected with XAF1- pTRE2hyg vector, followed by stable clone selection in hygromycin/puromycin. XAF1 induction in individual clones was assessed by immunoblotting for XAF1 protein in cell lysates after 24 h treatment of cells with 100 nM dexamethasone (Dex) and 1 µg/ml doxycycline (Dox). A single clone with tight regulation (low basal and strongly induced XAF1 protein expression) was selected for use in the studies reported herein. A HEK293 clone stably expressing pUbiq.irtTA and empty pTRE2hyg plasmids was used as a control in all induced expression studies.
RNA interference
A XAF1-directed shRNA (NM_199139, 5'-AAACCCAGGGCTGCCTTGGAAAAG-3') in lentiviral vector pLKO.1-puro (Open biosystems, Huntsville, AL, USA) was co-transfected with the packaging vectors psPAX2 and pMD2.G (Addgene, Cambridge, MA, USA) into HEK293 cells. A vector of scrambled shRNA was transfected in parallel to generate negative control virus. After 48 and 72 h, virus-containing supernatants were harvested and used to infect ACHN cells in the presence of 4 µg/ml polybrene for 24 h. After that time, cells were cultured in 2 µg/ml puromycin for another 72 h to generate a stably infected cell population. XAF1 protein expression was examined by immunoblot analysis in surviving cells.
Luciferase reporter assays
NF-κB transcriptional activity was assessed using a 3XκB-Luc reporter plasmid (B. Ashburner, University of Toledo, OH, USA), which contained three NF-κB binding sites, derived from the major histocompatibility complex class I gene, inserted ahead of the luciferase gene in vector pGL3-basic [48]. HEK293 or ACHN cells were co-transfected with 3XκB-Luc, an SV40 β-galactosidase-expressing plasmid that served as an internal control for transfection efficiency and plasmids expressing proteins of interest. After 24 h, cells were lysed in Reporter Lysis Buffer (Promega, Madison, WI, USA) and luciferase activity measured as described [49]. Where indicated, cells were exposed to 20 ng/ml TNF-α for 8 h prior to cell lysis. Relative light unit readings were normalized to β-galactosidase activity and expressed as a percentage of appropriate control values.
Reverse transcription-coupled polymerase chain reaction (RT-PCR)
Cellular RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed to cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA). PCR amplification was performed in a Bio-Rad PCR thermocycler using a three cycle program with an optimized annealing temperature and Taq Polymerase MasterMix (5 PRIME, Gaithersburg, MD, USA). For semi-quantitative PCR studies, amplification products were visualized on a 1% agarose gel with ethidium bromide staining under UV light. Quantitative real-time RT-PCR utilized SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) and a 2-step amplification program (30 second at 95°C and 60 second at 62°C) performed on an Eppendorf Realplex thermal cycler. Quantitation was performed using the Δ(ΔCt) method with values normalized to GAPDH in each experiment. PCR primer sequences were as follows (all 5’-to-3)’: XAF1 (For: GAGCACCAGCAGGTTGGGTG, Rev: AATCATTTGGTTGCAATAAT), IL-8 (For: CAGTTTTGCCAAGGAGTGCT, Rev: ACTTCTCCACAACCCTCTGC) and GAPDH (For: AAATCCCATCACCTTCC, Rev: CTCTCCCATGACGAACATGG).
Immunoblot and immunoprecipitation analyses
Cell lysates were prepared by passive lysis on ice (lysis buffer: 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% TRITON X-100 and proteasome inhibitor cocktail) followed by centrifugation at 12,000 rpm for 15 min at 4°C to remove cell debris. Immunoblot analyses followed standard protocols [39]. Where indicated, FLAG or HA tagged proteins were immunoprecipiated using anti-Flag affinity gel (Sigma Aldrich, St. Louis, MO, USA) or anti-HA antibody-conjugated protein G agarose (GE health care, Stockholm, Sweden) respectively. HIS-tagged proteins were captured by Ni-NTA Agarose resin (Qiagen, Valencia, CA, USA). For immunoprecipitation of endogenous TRAF2, anti-TRAF2 agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used.
Statistical Analysis
For quantitative data presented graphically, the mean ± standard deviations were calculated based on at least three separate experiments under the same conditions. Student's t-tests were used to compare data pairs and the degree of significance (p-value) indicated in the figure legends.
Results and Discussion
XAF1 inhibited TNF-α-induced NF-κB activation
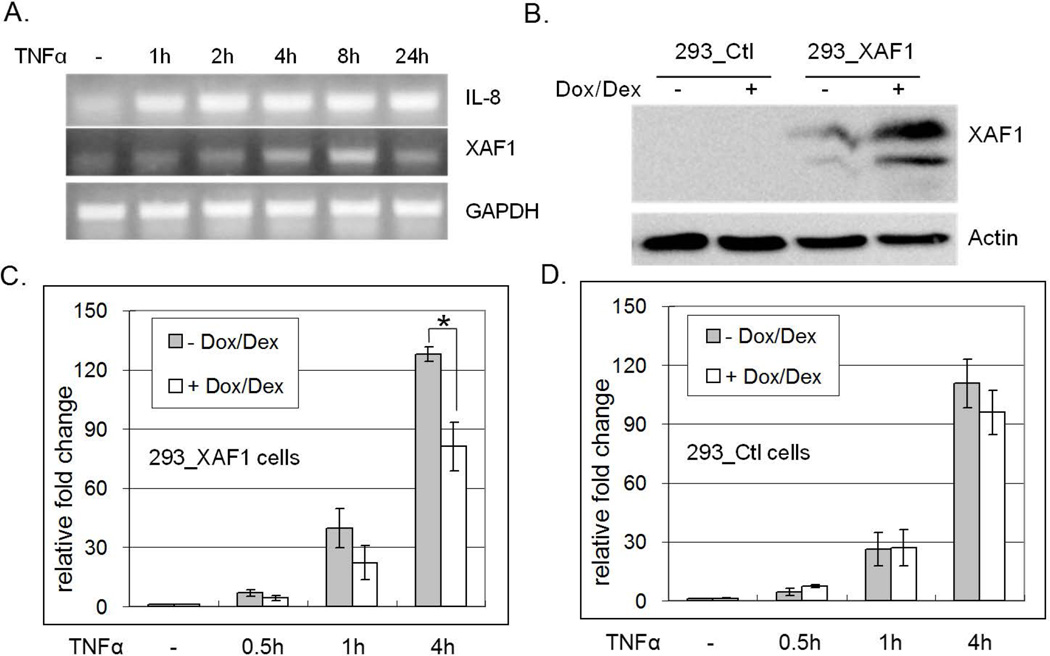
We had previously implicated XAF1 as a regulator of IFN and TRAIL induced apoptosis [39]. To extend those observations, time course analysis of TNF-α target gene induction was conducted in the renal carcinoma ACHN cell line. IL-8 and XAF1 mRNA levels were measured by RT PCR with and without TNF-α treatment (1–24 h). Whereas TNF-α induced sustained IL-8 mRNA expression within 1 h through 24 h, induced XAF1 mRNA levels were not observed until 8 h post-treatment, and returned to baseline by 24 h (Figure 1A). These data suggested that the regulation of XAF1 by TNF-α was distinct from that of IL-8, both in terms of initiation and duration.
Figure 1. XAF1 inhibits TNF-α-induced IL-8 expression.
(A) ACHN cells were treated with 20 ng/ml TNF-α, RNA isolated at the indicated times and reverse transcribed. IL-8, XAF1 and GAPDH mRNAs were amplified by RT PCR. (B) Dox/Dex regulated 293 cells were generated as described in Materials and Methods. XAF1 induction by Dox/Dex in the 293_XAF1 cells, but not empty vector control cells, was confirmed by immunoblot using anti-XAF1 antibody. XAF1-inducible cells (C) or empty vector control cells (D) were treated with Dox/Dex overnight or left untreated and then treated with 20 ng/ml TNF-α for the indicated times. IL8 transcript was quantified by real-time RT PCR using GAPDH as an internal control. Relative IL8 mRNA levels are compared to untreated samples. The mean and SEM were calculated based on data from three independent experiments (* p<0.05).
To assess the impact of XAF1 on TNF-α-induced signal transduction, we used a Tet-on system to inducibly express ectopic XAF1 in HEK293 cells. Cells expressing the Dox/Dex-regulated pUbiq.irtTA plasmid were stably transfected with empty pTREhyg vector or pTREhygXAF1. Stable transfectants were selected in hygromycin and assessed for Dox/Dex dependent regulation of XAF1 protein (Figure 1B). Stable lines displaying tight XAF1 regulation were then treated with Dox/Dex for 16 h prior to TNF-α treatment (0–4 h), followed by RNA isolation to assess gene induction. Real-time RT PCR analysis revealed that TNF-α-induced IL-8 mRNA was reduced in the presence of ectopic XAF1 expression (Figure 1C). Parallel experiments in pTREhyg vector control cells confirmed that the inhibition was not due to Dox/Dex (Figure 1D). These data implicated XAF1 as a putative negative regulator of TNF-α responsiveness.
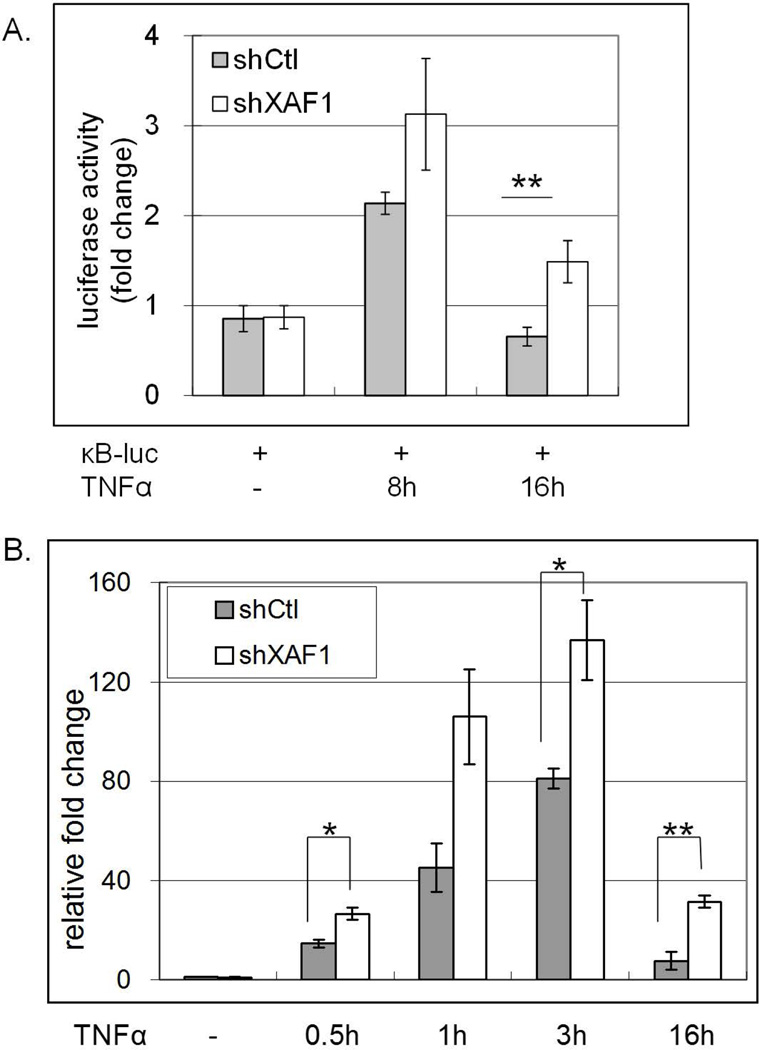
To extend these studies, we assessed a role for endogenous XAF1 in TNF-α responsiveness by performing transient knockdown of XAF1 in ACHN cells by using shRNA. NF-κB activation was measured by luciferase assays after TNF-α treatment, and showed that NF-κB activation was significantly enhanced in XAF1 knockdown cells compared to control cells after 16 h TNF-α treatment (Figure 2A). Down-regulating XAF1 also enhanced TNF-α induced IL-8 expression (Figure 2B). These data suggest that endogenous XAF1 was a physiologically relevant regulator of TNF-α responsiveness.
Figure 2. XAF1 knockdown aμgments TNF-α response.
(A) Transient knockdown of XAF1 in ACHN cells was achieved by infection with XAF1 shRNA lentivirus as described in Materials and Methods. XAF1-knockdown cells or cells receiving control virus were cotransfected for 24 h with an NF-κB responsive luciferase plasmid (0.4 µg) and a β-galactosidase-expression plasmid (0.4µg), followed by 20 ng/ml TNF-α treatment for another 8 h or 16 h. Luciferase activities were measured, normalized to β-galactosidase values and results presented as fold change relative to controls without TNF-α treatment. The mean and SEM were calculated based on data from three independent experiments (**p<0.01). (B) XAF1-knockdown cells or control cells were treated with 20 ng/ml TNF-α and RNA isolated at the indicated times and reverse transcribed. Quantitative real-time RT PCR was used to quantify IL-8 mRNA levels, which were normalized to GAPDH mRNA levels. Relative IL-8 mRNA levels were calculated by comparing to untreated controls. The mean and SEM were based on three independent experiments (**p<0.01; * p<0.05).
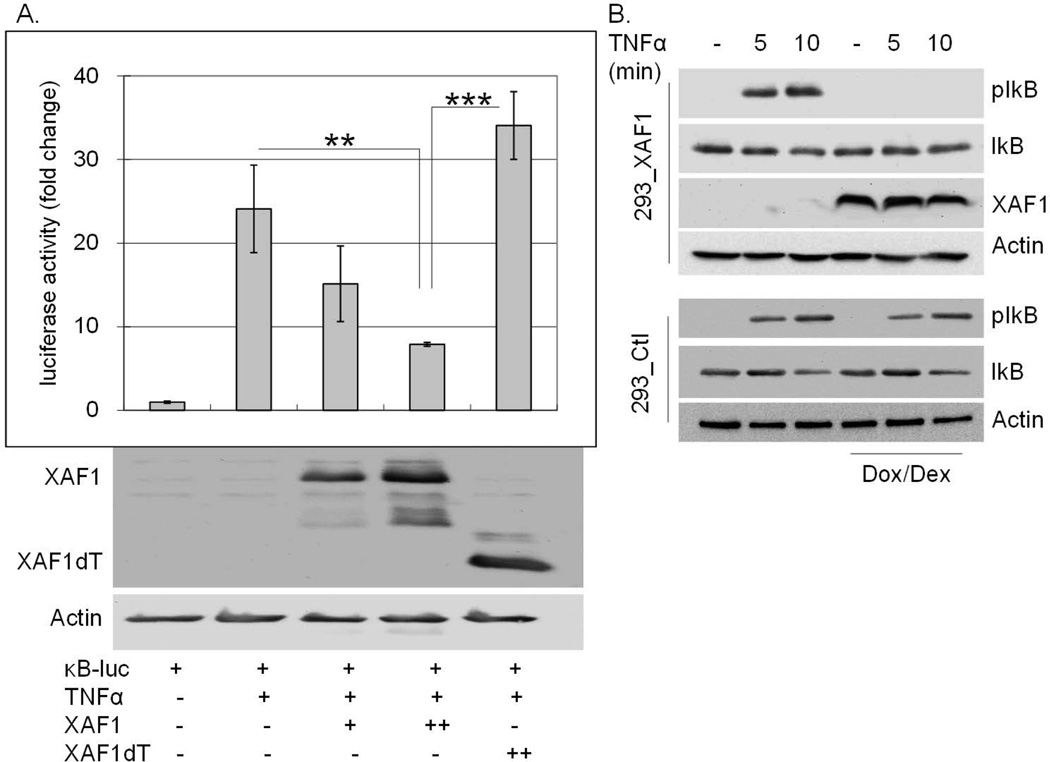
To investigate the impact of XAF1 on TNF-α-induced NF-κB activation, HEK293 cells were co-transfected with a NF-κB response element regulated luciferase reporter construct and either wild type or TRAF-domain mutated XAF1 (XAF1dT) expression plasmids. After 24 h, cells were treated with TNF-α for 8 h and luciferase activity measured in cell lysates. XAF1 co-transfection blocked TNF-α-induced NF-κB activation in a dose-dependent manner whereas the TRAF domain deleted XAF1 did not impact NF-κB activity (Figure 3A). XAF1 induction in the Dox/Dex cells similarly blocked IκB phosphorylation by TNF-α (Figure 3B). Together, these data implicated XAF1 as a negative regulator of NF-κB activation by TNF-α and suggested that this TNF-α induced protein functioned as a negative feedback regulator of TNF-α signaling to NF-κB.
Figure 3. XAF1 inhibits TNF-α-induced NF-κB activation.
(A) 293 cells were cotransfected for 24 h with an NF-κB responsive luciferase plasmid (0.1 µg) and a β-galactosidase-expression plasmid (0.1 µg) with or without plasmids expressing HA-XAF1 (0.2 [+] or 0.4 [++] µg) or HA-XAF1dT (0.4 [++] µg), followed by 20 ng/ml TNF-α treatment for another 8 h. Luciferase activities were measured, normalized to β-galactosidase values and results presented as fold change relative to empty vector controls without TNF-α treatment. The mean and SEM were calculated based on data from three independent experiments (*** p<0.001; ** p<0.01). These cell lysates were also subjected to SDS PAGE separation followed by immunoblot using an anti-HA antibody to confirm the protein expression of HA-XAF1 or HA-XAF1dT. (B) XAF1-inducible (293_XAF1) or control (293_Ctl) cells were left untreated or treated with Dox/Dex overnight followed by 20 ng/ml of TNF-α for 0, 5 or 10 min. Cell lysates were separated by SDS PAGE and immunoblotted with anti-phospho-IκB, XAF1 and Actin antibodies. Data are representative of at least three independent experiments.
XAF1 inhibited TRAF2-mediated NF-κB activation
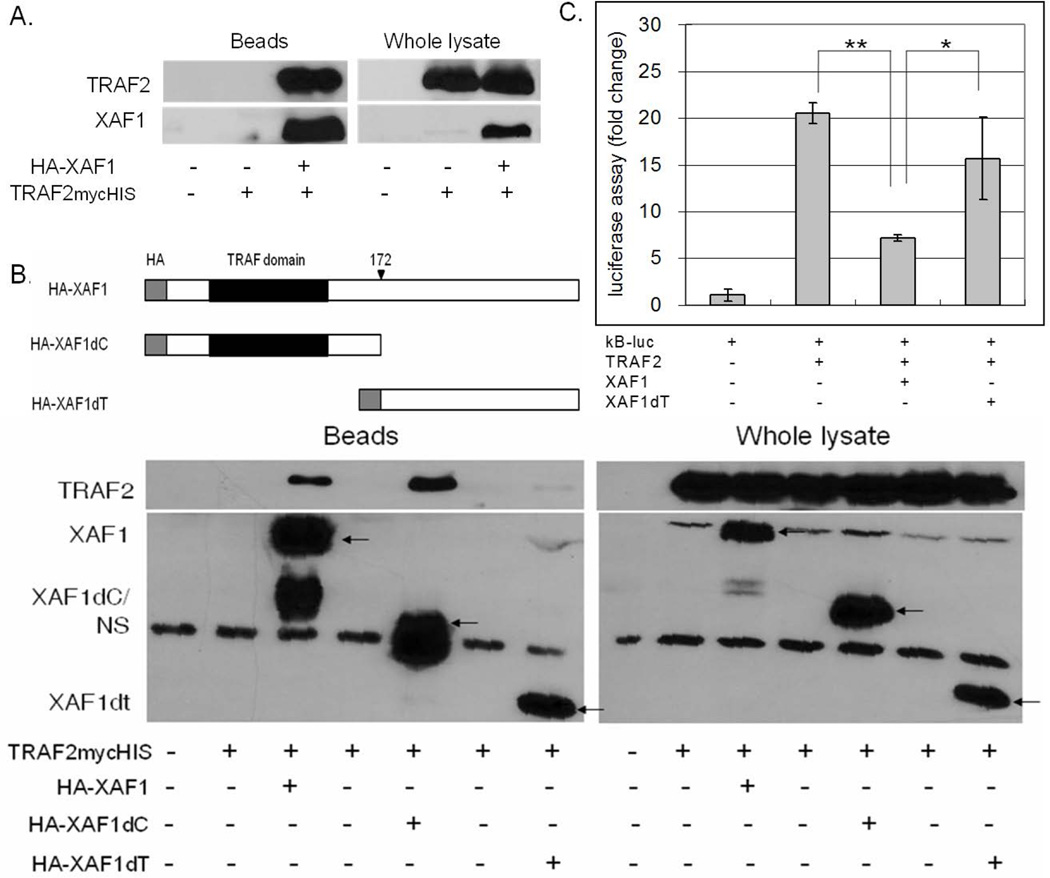
To investigate the mechanism of XAF1 inhibition, the direct association and impact of XAF1 on various TNF-α signaling intermediates were assessed, starting with TRAF2. Co-immunoprecipitation assays demonstrated that HA-tagged XAF1 interacted with mycHIS-tagged TRAF2 when ectopically expressed in HEK293 cells (Figure 4A). By using N-terminal or C-terminal deletions of XAF1, it was found that the TRAF domain was required for XAF1/TRAF2 interaction (Figure 4B). Since TRAF2 plays an important role in TNF-α-induced NF-κB activation, we assessed a role for XAF1 in regulating this response by using luciferase reporter experiments in HEK293 cells. Cells were co-transfected with an NF-κB luciferase reporter in the presence or absence of TRAF2 with or without full length XAF1 or the XAF1 TRAF domain deletion mutant. TRAF2-mediated NF-κB activation was potently inhibited by full length XAF1 but not the TRAF domain deleted XAF1, suggesting that the TRAF domain was essential for its activity (Figure 4C).
Figure 4. XAF1 interacts with TRAF2.
(A) 293 cells were transiently transfected with plasmids expressing TRAF2mycHis and/or HA-XAF1. 24 h after transfection, XAF1 was captured on anti-HA-agarose and then eluted proteins assessed by immunoblot. Input cell lysates were immunoblotted in parallel for comparison. (B) 293 cells were transfected with plasmids expressing TRAF2mycHis and HA-XAF1, C-terminus deleted XAF1 (HA-XAF1dC) or TRAF domain deleted XAF1 (HA-XAF1dT). After 24 h XAF1 variants were captured on anti-HA-agarose, and proteins in the whole cell lysate and eluted complexes assessed by immunoblot analysis. (C) 293 cells were cotransfected with an NF-κB responsive luciferase plasmid (0.1 µg), a β-galactosidase-expressing plasmid (0.1 µg) and plasmids expressing TRAF2mycHis (0.2 µg), HA-XAF1 (0.2 µg) or HA-XAF1dT (0.2 µg). After 24 h, luciferase activities were measured and normalized to β-galactosidase activity. Results are presented as a fold change in normalized luciferase activity as compared to cells transfected with the empty vector. The mean and SEM were calculated based on data from three independent experiments (** p<0.01; * p<0.05).
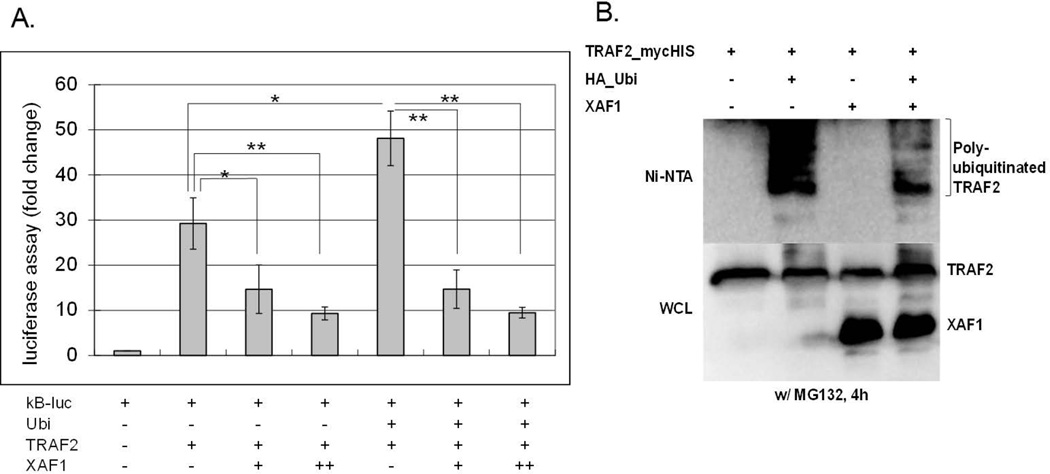
NF-κB signal transduction requires both K48- and K63-linked ubiquitination of multiple proteins, many of which are mediated by the E3 ligase activities of TRAF proteins [13]. We therefore assessed XAF1 impact on TRAF2 ubiquitination. Co-transfection of TRAF2 with ubiquitin augmented TRAF2-mediated NF-κB activation, an effect that was inhibited by co-transfection with XAF1 (Figure 5A). HEK293 cells ectopically expressing TRAF2, XAF1 and/or ubiquitin were subject to Ni-NTA pull-down followed by immunoblot analysis. Poly-ubiquitination of TRAF2 was blocked by XAF1 (Figure 5B). These data suggest that XAF1 inhibition of TRAF2 activity and/or ubiquitination may regulate TNF-α signaling.
Figure 5. XAF1 inhibited ubiquitin-enhanced TRAF2 activity.
(A) 293 cells were cotransfected with a NF-κB responsive luciferase plasmid (0.1 µg), a β-galactosidaseexpressing plasmid (0.1 µg) and plasmids expressing XAF1 (0.2 [+] or 0.4 [++] µg) or TRAF2 (0.2 µg) or ubiquitin (0.2 µg). After 24 h, Luciferase activities were measured and normalized to β-galactosidase values. Results are presented as fold change relative to empty vector controls without TNF-α treatment. The mean and SEM were calculated based on data from three independent experiments (** p<0.01; * p<0.05). (B) 293 cells were transiently transfected with TRAF2mycHis, HA-ubiquitin, and/or untagged XAF1 expression plasmids. 24 h after transfection, TRAF2mycHis was pulled down by Ni-NTA beads followed by SDS PAGE and immunoblotting with an anti-HA antibody to detect ubiquitinated TRAF2 (top panel). Total XAF1 and TRAF2 were detected in immunoblots of the total cell lysates with antibodies for XAF1 and myc, respectively. Data are representative of at least three independent experiments.
XAF1 regulated complex I formation
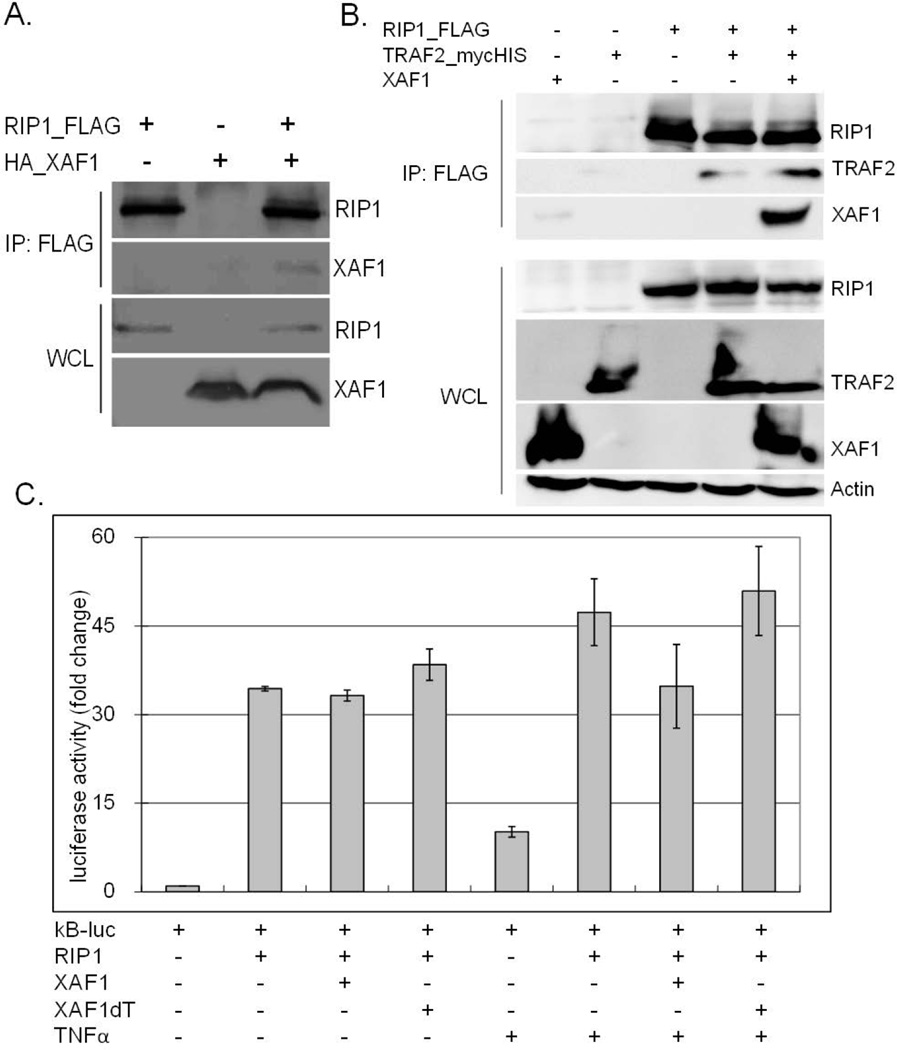
In addition to TRAF2, TNF-α-induced complex I also contains TRADD and RIP1. In co-transfection experiments, XAF1 did not associate with TRADD (data not shown) but did co-immunoprecipitate with RIP1, with or without TRAF2 co-expression (Figure 6A and 6B). Interestingly, ectopic XAF1 did not affect RIP1-induced NF-κB promoter activation although it did reduce TNF-α-augmented NF-κB luciferase induction (Figure 6C). These data suggest that its impact on signaling occurred prior to RIP1 activation, or in a parallel regulatory pathway.
Figure 6. XAF1 interacted with RIP1.
(A) 293 cells were transiently transfected with plasmids expressing FLAG tagged RIP1 with or without HA tagged XAF1 or an empty vector. After 24 h, RIP1 was precipitated by anti-FLAG affinity gel, and eluted proteins separated by SDS PAGE and assessed by immunoblot with the indicated antibodies. Whole cell lysates were immunoblotted in parallel to verify XAF1 and RIP1 expression. (B) 293 cells ectopically expressing FLAG-RIP1, TRAF2mycHis, and/or untagged XAF1 were lysed and FLAG-RIP1 immunoprecipitated using anti-FLAG affinity gel. The proteins in the immune complex and whole cell lysates were separated by SDS PAGE and immunoblotted with epitope tag-specific, XAF1-specific or actin-specific antibodies. Data are representative of at least three independent experiments. (C) 293 cells were cotransfected with a NF-κB response element-regulated luciferase plasmid (0.1 µg), along with a β-galactosidase-expressing plasmid (0.1 µg) and plasmid expressing FLAG-RIP1 (0.2 µg), HA-XAF1 (0.2 µg) or HA-XAF1dT (0.4 µg). After 24 h, cells were left untreated or treated with TNF-α for 8 h, luciferase activities measured and normalized to β-galactosidase values. Results are presented as fold change relative to empty vector controls without TNF-α treatment. The mean and SEM were calculated based on data from three independent experiments.
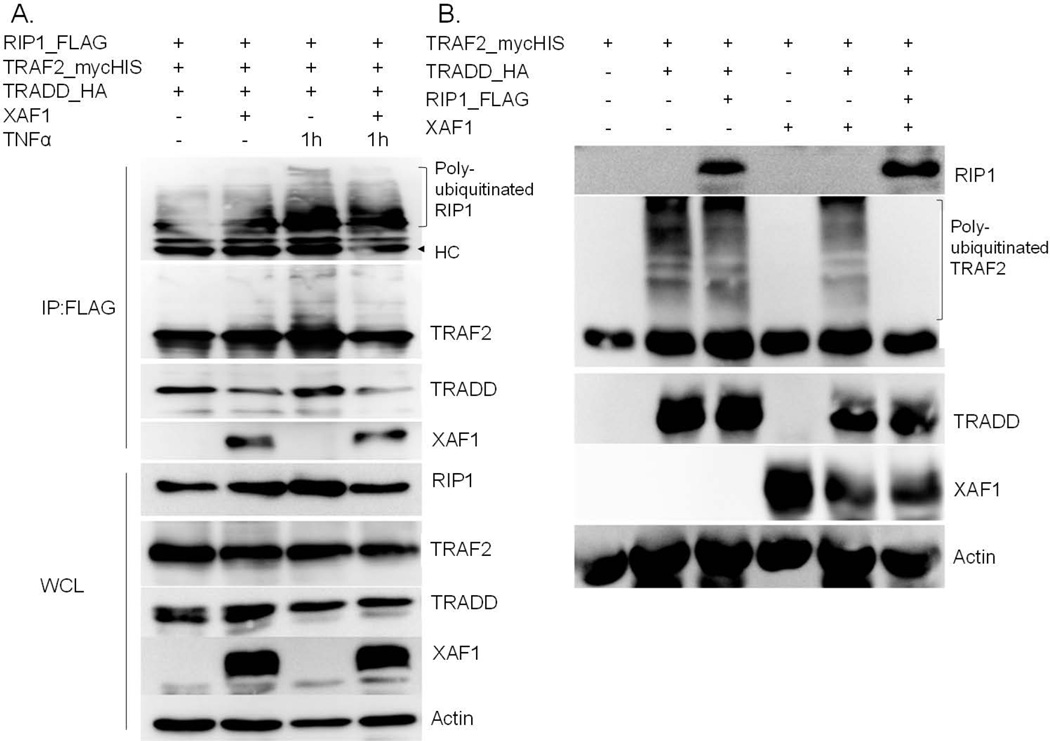
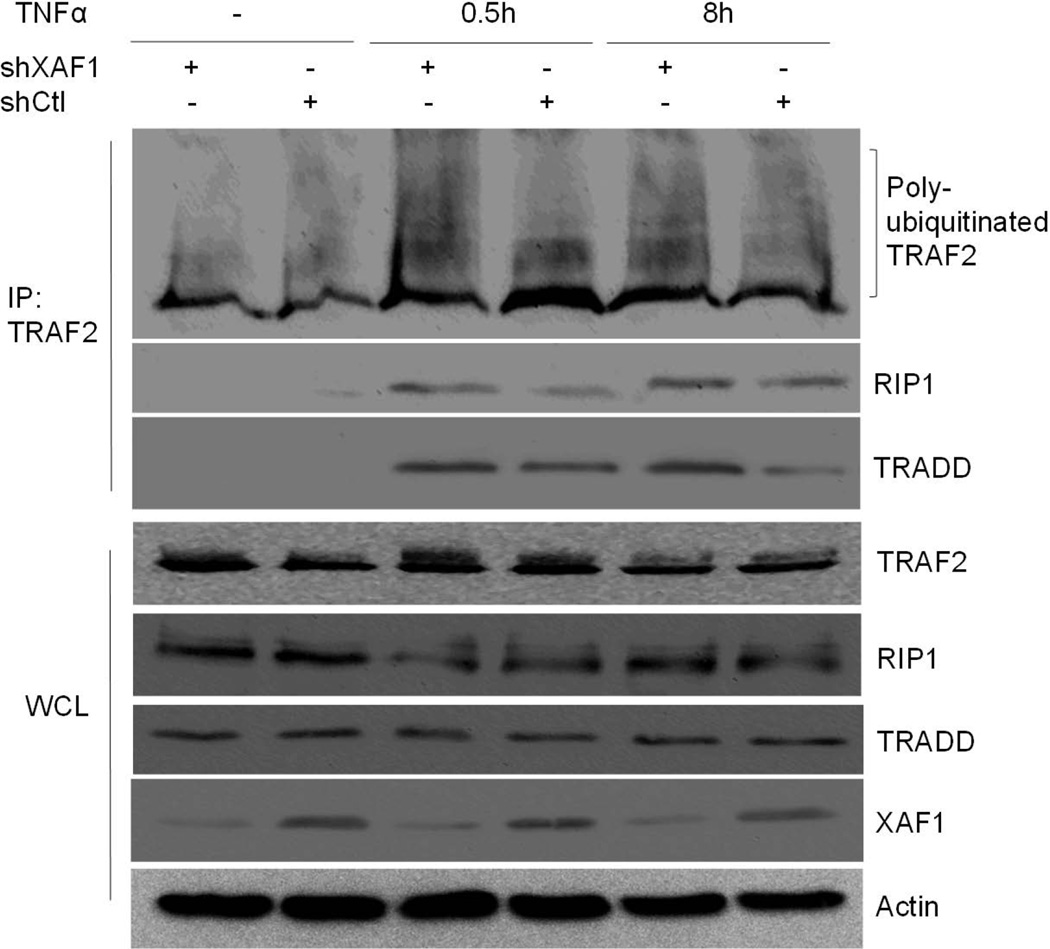
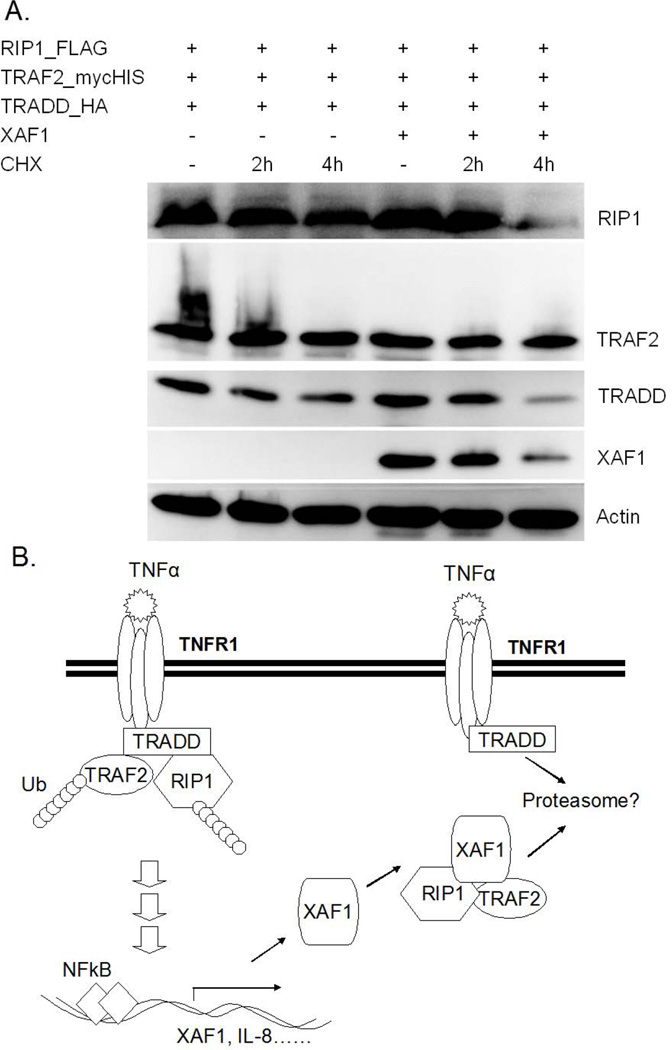
To elucidate whether XAF1 impacted complex I formation, key components of this complex (TRADD, TRAF2 and RIP1) were transiently expressed in HEK293 with or without XAF1 co-expression. After 24 h, FLAG-tagged RIP1 was captured on FLAG beads, and co-immunoprecipitated proteins assessed by immunoblot analysis. Our data suggested that interaction of RIP1 and TRADD was disrupted by XAF1 (Figure 7A), and that this led to reduced TNF-α-induced TRAF2 ubiquitination (Figure 7A). Interestingly, RIP1 plus XAF1 blocked TRAF2 ubiquitination by TRADD more than either alone (Figure 7B). These data suggested that RIP1 may be a critical determinant of XAF1 action. The effect of XAF1 on TRAF2/TRADD/RIP1 complex formation was further confirmed by immunoprecipitation of endogenous TRAF2 in XAF1 shRNA knockdown cells or control shRNA infection cells. The results showed that TRAF2 underwent rapid ubiquitination following TNF-α treatment and exhibited strong interaction with RIP1 and TRADD, both effects were augmented by XAF1 down-regulation (Figure 8). It is worth noting that the shRNA suppression of XAF1 impacted levels of proteins associated with TRAF2, but not steady state total cellular levels of RIP1 and TRADD (Fig. 8). These data suggested that XAF1 was not a global regulator of protein levels, but impacted just those within the complex. Further evidence for this was obtained by inducing association of RIP1, TRADD and TRAF2 by ectopically overexpressing them in the presence or absence of XAF1 with or without CHX treatment (Fig. 9). XAF1 enhanced TRADD and RIP1 degradation upon addition of the protein synthesis inhibitor, enforcing the idea that XAF1 impacted their stabilities or half-life only when co-expressed or otherwise in association with these proteins.
Figure 7. XAF1 interfered with RIP1-TRADD association.
(A) 293 cells were transfected with FLAG-RIP1, TRAF2mycHis and HA-TRADD with or without untagged XAF1 cotransfection for 24 h and then left untreated or treated with TNF-α for 1h before lysate preparation. RIP1- associated proteins were captured on anti-FLAG beads and proteins in the immune complexes or whole cell lysates separated by SDS PAGE and transfected proteins detected by immunoblotting with epitope tagspecific antibodies. XAF1 and actin were detected by anti-XAF1 and anti-actin antibodies, respectively. Data are representative of at least three independent experiments. (B) 293 cells were transfected as in (A) with the indicated expression plasmids. After 24 h, whole cell lysates were collected for SDS PAGE and immunoblot analysis with the same antibodies as (A). Data are representative of at least three independent experiments.
Figure 8.
XAF1 knockdown prolonged TNF-α-induced TRAF2 ubiquitination and TRAF2/RIP1/TRADD complex. XAF1-knockdown cells or control cells (generated as described in Materials and Methods) were treated with 20 ng/ml TNF-α and cell lysates collected after 0.5 h or 8 h. TRAF2-associated proteins were captured on anti-TRAF2 beads. The proteins in the immune complexes or whole cell lysates were separated by SDS PAGE and detected by immunoblotting with antibodies against the proteins of interest. Data are representative of at least three independent experiments.
Figure 9. XAF1 regulates TRADD and RIP1 half-life.
(A) 293 cells were transfected with FLAG-RIP1, TRAF2mycHis, and HA-TRADD with or without untagged XAF1. After 24 h, some cells received 100 μgmL −1 of cycloheximide for 2 or 4 h to block protein synthesis, after which time cell lysates were collected and subjected to SDS/PAGE and immunoblot analysis using epitope tag-specific antibodies or anti-XAF1 and anti-actin antibodies. Data are representative of at least three independent experiments. (B) A model of XAF1 action based on the collected data. XAF1 is induced by TNF-a and feeds back to inhibit the signaling complex. By associating with both TRAF2 and RIP1, XAF1 appears to disrupt interaction between these proteins and TRADD, thereby disrupting the TNFR signaling complex and leading to suppression of NF- jB signaling. Data in (A) suggested that components of the disrupted complex may be targeted for degradation.
Overall, our data suggest that XAF1 expression inhibits TNF-α-induced NF-κB activation via disruption of TRADD/TRAF2/RIP1 complex function (Figure 9B). It is possible that XAF1 physically blocks interaction between the components of the complex, preventing them from receiving or retaining K63-ubiquitination that is essential for complex formation. This could lead to premature proteasomal degradation of TRADD or RIP1. Although XAF1 has not previously been implicated in TNF signal suppression, it may figure prominently only in certain cell types or under certain physiological conditions, such as inflammatory cells primed by other cytokines. Regardless, the impact of XAF1 on complex I has the potential to enhance cellular sensitivity to pro-apoptotic signals, thereby providing a possible mechanism behind the long-recognized pro-apoptotic function of XAF-1 [50]. As such, these observations provide important new insight into a previously unidentified role for XAF-1 in cytokine cross-talk and inflammatory responses.
Acknowledgments
We thank Drs. Haiying Li, Qi Ke and Shaun Rosebeck for helpful comments and discussions. This work was supported by the National Institutes of Health Grants R01AI068133 (to DWL) and R21AI063014 (to DWL).
References
- 1.Gruss HJ, Dower SK. Tumor necrosis factor ligand superfamily: involvement in the pathology of malignant lymphomas. Blood. 1995;85:3378–3404. [PubMed] [Google Scholar]
- 2.Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 3.Tracey KJ, Cerami A. Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu Rev Med. 1994;45:491–503. doi: 10.1146/annurev.med.45.1.491. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 5.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 6.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 7.Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 8.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 9.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 10.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 11.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [corrected] [DOI] [PubMed] [Google Scholar]
- 13.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 15.Derijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 16.Westwick JK, Weitzel C, Minden A, Karin M, Brenner DA. Tumor necrosis factor alpha stimulates AP-1 activity through prolonged activation of the c-Jun kinase. J Biol Chem. 1994;269:26396–26401. [PubMed] [Google Scholar]
- 17.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 18.Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 19.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 20.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 21.Mercurio F, et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 22.Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 23.Song HY, Regnier CH, Kirschning CJ, Goeddel DV, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-kappaB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci U S A. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SY, Kaufman DR, Mora AL, Santana A, Boothby M, Choi Y. Stimulus-dependent synergism of the antiapoptotic tumor necrosis factor receptor-associated factor 2 (TRAF2) and nuclear factor kappaB pathways. J Exp Med. 1998;188:1381–1384. doi: 10.1084/jem.188.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh WC, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 29.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 30.Brown KD, Hostager BS, Bishop GA. Regulation of TRAF2 signaling by self-induced degradation. J Biol Chem. 2002;277:19433–19438. doi: 10.1074/jbc.M111522200. [DOI] [PubMed] [Google Scholar]
- 31.Hostager BS, Haxhinasto SA, Rowland SL, Bishop GA. Tumor necrosis factor receptor-associated factor 2 (TRAF2)-deficient B lymphocytes reveal novel roles for TRAF2 in CD40 signaling. J Biol Chem. 2003;278:45382–45390. doi: 10.1074/jbc.M306708200. [DOI] [PubMed] [Google Scholar]
- 32.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 33.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci U S A. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc Natl Acad Sci U S A. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platzer C, Meisel C, Vogt K, Platzer M, Volk HD. Up-regulation of monocytic IL-10 by tumor necrosis factor-alpha and cAMP elevating drugs. Int Immunol. 1995;7:517–523. doi: 10.1093/intimm/7.4.517. [DOI] [PubMed] [Google Scholar]
- 37.Sudhakar C, Nagabhushana A, Jain N, Swarup G. NF-kappaB mediates tumor necrosis factor alpha-induced expression of optineurin, a negative regulator of NF-kappaB. PLoS One. 2009;4:e5114. doi: 10.1371/journal.pone.0005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liston P, et al. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 39.Leaman DW, Chawla-Sarkar M, Vyas K, Reheman M, Tamai K, Toji S, Borden EC. Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J Biol Chem. 2002;277:28504–28511. doi: 10.1074/jbc.M204851200. [DOI] [PubMed] [Google Scholar]
- 40.Straszewski-Chavez SL, Visintin IP, Karassina N, Los G, Liston P, Halaban R, Fadiel A, Mor G. XAF1 mediates tumor necrosis factor-alpha-induced apoptosis and X-linked inhibitor of apoptosis cleavage by acting through the mitochondrial pathway. J Biol Chem. 2007;282:13059–13072. doi: 10.1074/jbc.M609038200. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, et al. c-Jun N-terminal kinase (JNK1) upregulates XIAP-associated factor 1 (XAF1) through interferon regulatory factor 1 (IRF-1) in gastrointestinal cancer. Carcinogenesis. 2009;30:222–229. doi: 10.1093/carcin/bgn271. [DOI] [PubMed] [Google Scholar]
- 42.Arora V, Cheung HH, Plenchette S, Micali OC, Liston P, Korneluk RG. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J Biol Chem. 2007;282:26202–26209. doi: 10.1074/jbc.M700776200. [DOI] [PubMed] [Google Scholar]
- 43.Chung SK, et al. Frequent alteration of XAF1 in human colorectal cancers: implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132:2459–2477. doi: 10.1053/j.gastro.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, MacLeod AR, Borden EC. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 45.Yu LF, et al. XAF1 mediates apoptosis through an extracellular signal-regulated kinase pathway in colon cancer. Cancer. 2007;109:1996–2003. doi: 10.1002/cncr.22624. [DOI] [PubMed] [Google Scholar]
- 46.Yin W, Cheepala S, Clifford JL. Identification of a novel splice variant of X-linked inhibitor of apoptosis-associated factor 1. Biochem Biophys Res Commun. 2006;339:1148–1154. doi: 10.1016/j.bbrc.2005.11.128. [DOI] [PubMed] [Google Scholar]
- 47.Anastassiadis K, Kim J, Daigle N, Sprengel R, Scholer HR, Stewart AF. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene. 2002;298:159–172. doi: 10.1016/s0378-1119(02)00979-4. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell T, Sugden B. Stimulation of NF-kappa B-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]