Abstract
Previously, we have shown that flies under-expressing the two mitochondrial peroxiredoxins (Prxs), dPrx3 and dPrx5, display increases in tissue-specific apoptosis and dramatically shortened life span, associated with a redox crisis, manifested as changes in GSH:GSSG and accumulation of protein mixed disulfides. To identify specific pathways responsible for the observed biological effects, we performed a transcriptome analysis. Functional clustering revealed a prominent group enriched for immunity-related genes, including a considerable number of NF-kB-dependent antimicrobial peptides (AMP) that are up-regulated in the Prx double mutant. Using qRT-PCR analysis we determined that the age-dependent changes in AMP levels in mutant flies were similar to those observed in controls when scaled to percentage of life span. To further clarify the role of Prx-dependent mitochondrial signaling, we expressed different forms of dPrx5, which unlike the uniquely mitochondrial dPrx3 is found in multiple subcellular compartments, including mitochondrion, nucleus and cytosol. Ectopic expression of dPrx5 in mitochondria but not nucleus or cytosol partially extended longevity under normal or oxidative stress conditions while complete restoration of life span occurred when all three forms of dPrx5 were expressed from the wild type dPrx5 transgene. When dPrx5 was expressed in mitochondria or in all three compartments, it substantially delayed the development of hyperactive immunity while expression of cytosolic or nuclear forms had no effect on the immune phenotype. The data suggest a critical role of mitochondria in development of chronic activation of the immune response triggered by impaired redox control.
Keywords: Peroxiredoxin, mitochondria, immunity, aging, redox, Drosophila
1. Introduction
Peroxiredoxins (Prxs) constitute a family of enzymes that catalyze the degradation of H2O2 and other peroxides in multiple cellular compartments. Consequently they help reduce oxidative stress and are well positioned to serve as sensors whereby fluctuations in redox state can be transmitted via the Prxs to redox-sensitive targets (1–6). Based on their ability to act as oxidative stress-reducing agents and also as mediators of redox-sensitive signaling, Prxs are implicated in a variety of cellular processes, including metabolism, immunity and aging (7–13).
Like their mammalian orthologs, Drosophila Prxs reside in different subcellular compartments, including mitochondria, a major generator of cellular reactive oxygen species (ROS) (11,14). Proper functioning of mitochondria is critical for control of fundamental cellular processes and regulation of pathways that determine cell life or death, while malfunctioning is associated with a number of disorders, such as chronic inflammation and premature senescence (15,16). Normal functioning of mitochondria requires peroxidase activity in the form of peroxiredoxins and/or glutathione peroxidases (17). In Drosophila peroxidase activity in the mitochondria is provided solely by two Prxs, the mitochondrial-specific dPrx3 and dPrx5, which in addition to its presence in the mitochondria has also been localized to cytosolic and nuclear compartments (11,14). Overexpression of peroxiredoxins and other H2O2-degrading enzymes has been used to suppress mitochondrial H2O2 levels in a series of transgenic studies in both mice and flies (14,18–21), giving rise to an array of physiological outcomes, both beneficial and detrimental. The beneficial effects, such as greater resistance to oxidative stress, may be ascribed to improved antioxidant function, whereas the negative effects presumably derive from the impact of lowered H2O2 levels on redox-sensitive signaling (14,18,21–24).
Recently, we reported that the peroxiredoxins residing in mitochondria, dPrx3 and dPrx5, play a central role in maintaining cellular redox homeostasis and cell viability (11). Under-expression of dPrx3 and dPrx5 together, but not separately, had a broad impact on the redox milieu resulting in proapoptotic changes and a dramatic shortening of life span to approximately 20% (11). Interestingly the progression of changes in redox, manifested as the accumulation of GSSG and protein mixed disulfides, as well as the patterns of apoptosis largely paralleled those seen during normal aging, albeit at an accelerated pace (25–28), suggesting that mitochondrial Prxs may impact longevity pathways and are responsible for the shortened life span phenotype observed in flies underexpressing dPrx3 and dPrx5, named double mutant (DM) hereafter.
In an attempt to identify the longevity pathways modulated by Prxs, we conducted genomewide analysis of gene expression in flies, mutant for both dPrx3 and dPrx5 as well as mutants for the individual Prxs, and found significant overlap with transcriptome responses typically observed in normal aging. Furthermore, we ascertained that it is the mitochondrial form of dPrx5 rather than the nuclear or cytosolic forms that plays the more predominant role in immunity and aging.
2. Material and methods
2.1. Generation of UAS-mit dPrx5, UAS-cyt dPrx5, and UAS-nucl dPrx5 transgenic flies
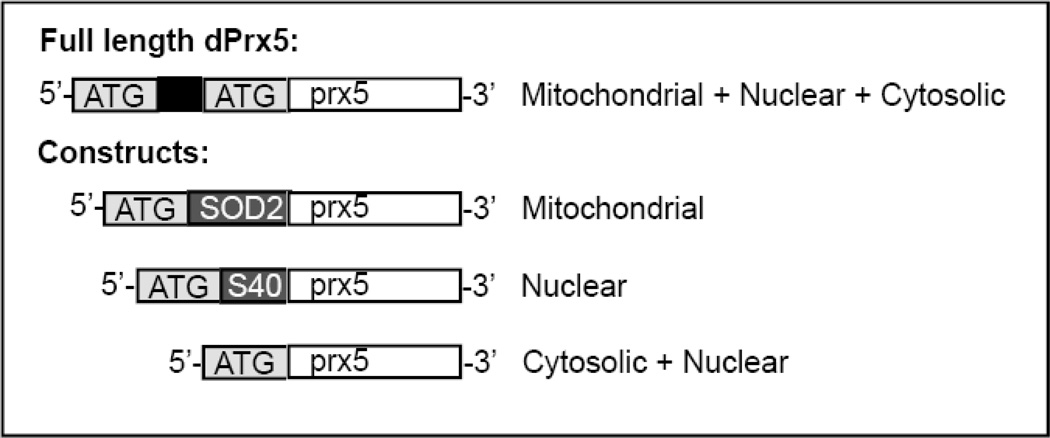
As reported earlier (29), Drosophila dPrx5 is expressed from the full-length dPrx5 gene, containing mitochondrial pre-sequence and an alternate AUG codon, from which the shorter form, found in cytosolic and nuclear compartments, is presumably translated (14). To target dPrx5 to a specific compartment, three different transgenes were generated (Fig. 1).
Figure 1. Structure of the dPrx5 transgenic constructs.
The UAS-dPrx5 construct coding for wild type full length dPrx5 polypeptide was made previously (14). Briefly, the construct included a mitochondrial pre-sequence (black), which is subsequently cleaved at a second in-frame methionine upon entry into the mitochondria. This second in-frame methionine also serves as an alternative translation site giving rise to the short dPrx5 form (in white), found in cytosol and nucleus (14). The SOD2 mitochondrial leader peptide (in dark grey) and nuclear-targeting SV40 NLS sequence (in dark grey) were used to target dPrx5 to the mitochondrial and nuclear compartments respectively while removal of the mitochondrial targeting domain restricted expression of dPrx5 largely to the cytosolic compartment, and secondarily to the nucleus.
A dPrx5 transgene construct lacking the mitochondrial targeting sequence was made, such that translation was initiated from the second in-frame methionine (Fig. 1, Cytosolic+nuclear variant). This polypeptide form of dPrx5 is localized largely to the cytosol and secondarily to the nucleus, as previously reported (14). For construction, the shorter ORF of dPrx5 without its mitochondrial pre- sequence was amplified using dPrx5 cDNA as a template and primers, forward 5’-tgc aca gaa ttc aaa ATG GTG AAA GTA GGA GAC TCC C-3’ and reverse 5’-act act tct aga TTA CTT CTT GCC AAT GTT GTT G-3’. Non-coding sequences are shown in lowercase and coding sequences in uppercase; EcoRI and XbaI sites are underlined; start and stop codons are in italic.
To enhance the presence of dPrx5 in nucleus (Fig. 1, Nuclear), a nuclear-targeting construct was generated by fusing the nuclear localization signal (NLS) of SV40 large-T antigen (30,31) to a short form of dPrx5. The NLS coding sequence (shown by bold), and the N terminus of the short dPrx5 ORF (wave underlined) were fused using the forward primer 5’- tac aca gaa ttc aaa ATG GGG CCA CCA AAG AAG AAG CGA AAG GTC GGC GTG AAA GTA GGA GAC TCC C-3’ together with the reverse primer used for the cytosolic+nuclear construct indicated above.
To allow specific targeting to mitochondria, the second methionine was removed and a mitochondrial targeting sequence from the Drosophila sod2 gene was fused to the ORF of the shorter form of dPrx5, as indicated in Fig. 1. In this case, the cleavage site was preserved but the removal of a second methionine prevented translation of the cytosolic+nuclear short form. For construction, dPrx5 cDNA was amplified using a forward primer, composed of sod2 mitochondrial targeting sequence (shown in bold) fused to the N terminus of the shorter dPrx5 form, 5’-tgc aca gaa ttc aaa ATG TTC GTG GCC CGT AAA ATT TCG CAA ACT GCA AGC CTG GCG GTG CGT GGC AAG CAC GTG AAA GTA GGA GAC TCC C-3’ together with the reverse primer used for the cytosolic+nuclear construct indicated above.
These compartment-specific dPrx5 transgenes were subsequently inserted into the pUASTattb vector via the EcoRI and XbaI sites (32) and injected into D. melanogaster strain attP40 using the services of BestGene, Inc. (Chino Hills, CA) (33).
2.2. Fly Strains and Procedures
All transgenic, mutant and enhancer fly lines were backcrossed into the reference y w strain background. Flies under-expressing dPrx3 by RNAi were made by crossing UAS RNAi-dprx3 transgenes (11) to the high-level global daughterless (Da-GAL4) driver. The dprx5 mutant and double mutant (DM) flies, under-expressing both dPrx3 and dPrx5, were generated as described previously (11,14,29). Ectopic expression of the compartment-specific dPrx5 forms from their corresponding transgenes in a DM background was achieved by generation of fly lines indicated in Table 1. Controls were heterozygous flies carrying transgene and driver alleles. Since no differences in effects on longevity and other fly characteristics were observed between transgene and driver controls (11) and not shown), we only present here the data for one of them, Da-GAL4, dprx5/+.
Table 1.
Genotypes of fly lines expressing different forms of dPrx5.
| Line name | genotype | abbreviation |
|---|---|---|
| Cytosolic dPrx5 | UAS-cyt dPrx5/+; RNAi-dprx3, dprx5/Da-GAL4, dprx5 | Cyt |
| Mitochondrial dPrx5 | UAS-mit dPrx5/+; RNAi-dprx3, dprx5/Da-GAL4, dprx5 | Mit |
| Nuclear dPrx5 | UAS-nucl dPrx5/+; RNAi-dprx3, dprx5/Da-GAL4, dprx5 | Nucl |
| Full-length dPrx5 | UAS-dPrx5/+; RNAi-dprx3, dprx5/Da-GAL4, dprx5 | dPrx5 |
In all experimental studies, flies were maintained on standard sucrose-cornmeal fly food at 25°C. Age-synchronized cohorts of flies were generated by collecting newly-eclosed flies over a period of 48 hours. Approximately 25 flies were placed in each vial and transferred to fresh food on a daily basis. Survivorship studies under normal and oxidative stress conditions were conducted as described previously (11). Briefly, flies were maintained on regular food or fed 1% sucrose solutions containing ROS-generating agents, paraquat (PQ) and hydrogen peroxide at concentrations indicated in figure legends. Fly deaths were recorded approximately every 12 hours.
2.3. Transcriptome analysis by RNA-Sequencing
Analysis has been performed with two independent cohorts of flies for each genotype. Total RNA was isolated from 25 whole bodies of 13-day old males using Trizol reagent (Thermo Fisher Scientific). Samples were treated with Promega RQ1 DNAse (~1u/ug RNA) and submitted for global transcriptome gene expression analysis conducted by the University of Rochester Genomics Resource Center (URGRC). RNA-seq library preparations were done using Illumina TruSeq mRNAseq and 100nt single-end reads were obtained by using Illumina HiSeq 2500 RNAseq. Sequenced reads were cleaned according to a rigorous pre-processing workflow (Trimmomatic-0.32) and mapped to the D. melanogaster genome (ncbi 5.3) with SHRiMP2.2.3. Differential expression analysis was performed with cufflinks2.0.2 (34) with an FDR cutoff of 0.05 (95% confidence interval). Genes that displayed statistically significant variations in expression levels between experimental groups and controls were selected for further functional classification and functional annotation clustering carried out by DAVID bioinformatics resources (35,36). Gene interaction analysis, coexpression and co-localization clustering has been done by using GeneMania analysis (www.genemania.com).
2.4. qRT-PCR
Quantitative RT-PCR was performed as described (10); primers are listed in the Supplementary Content (Suppl. Table 1). The signals obtained for each gene were standardized against signals obtained for the rp49 housekeeping gene and expressed as arbitrary mRNA units.
2.5. Subcellular fractionation and immunoblotting
The expression of dPrx5 in different subcellular compartments was analyzed after fractionation of cellular organelles in dprx5 null flies, expressing different dPrx5 forms from the transgenes. Subcellular fractionation was performed using gradient centrifugation essentially as described in Radyuk et al. (14). Briefly, a nuclear pellet produced after centrifugation at 1,000 g was deposited on three layers of iodixanol (Sigma D1556) (35%, 30%, 25%) and centrifuged at 10,000 g for 40 min, resulting in a nuclei-containing band at the 30%–35% interface. For mitochondrial and cytosol fractionation, thoraces from a group of 50 flies were dissected and placed in a mortar containing 500 µL of pre-chilled mitochondria isolation buffer (20 mM Tris/HCl, pH 7; 0.32 M sucrose, 1 mM EDTA, 1mM EGTA and 1% BSA). Thoraces were then gently pounded and filtered through 10 microns mesh Nylon filter to remove the tissue particles. Mitochondria were pelleted by centrifuging at 500 g at 4°C for 3 min and washed twice with the isolation buffer without BSA. The supernatant contained the cytosolic fraction.
For immunoblot analysis, proteins were extracted from whole fly tissues or separated organelles using protein extraction buffer containing protease inhibitors (Roche). Samples for loading were made essentially as described (37) after determining protein concentrations by the Bio-Rad Protein Assay reagent (Bio-Rad). Ten µg of protein extract for each sample were resolved by SDS/10% PAGE, transferred to PVDF membrane (Millipore), and immunoblots were processed as described (11).
Levels of dPrx3 and dPrx5 proteins in the whole body lysates extracted from DM flies expressing different transgene-derived dPrx5 forms were evaluated using anti-dPrx3 and anti-dPrx5 antibodies, as well as anti-actin antibodies (MP Biomedicals) to control for loading. Levels of dPrx5 proteins in separated subcellular fractions were determined using anti-dPrx5 antibody. Antibodies for dPrx3 (mitochondrial (11)), GCLm (modulatory subunit of glutamate-cysteine ligase; cytosolic (38)) and Histone 3 HDAC (histone deacetylase, nuclear, BD Biosciences) were used to assess the purity of the subcellular fractions and control for loading.
2.6. Statistical analysis
Differences in mRNA and protein levels were compared between groups by analysis of variance by using Prism for Macintosh version 4.0a software (GraphPad Software, Inc.). Statistical significance of the age-specific variations in mRNA levels between fly lines was determined by comparing the slopes and intercepts among regression lines. The mean and median survivorship time and statistical significance of differences between survival curves were assessed using the logrank test.
3. Results
3.1. Transcriptome profiling of mitochondrial peroxiredoxin mutants
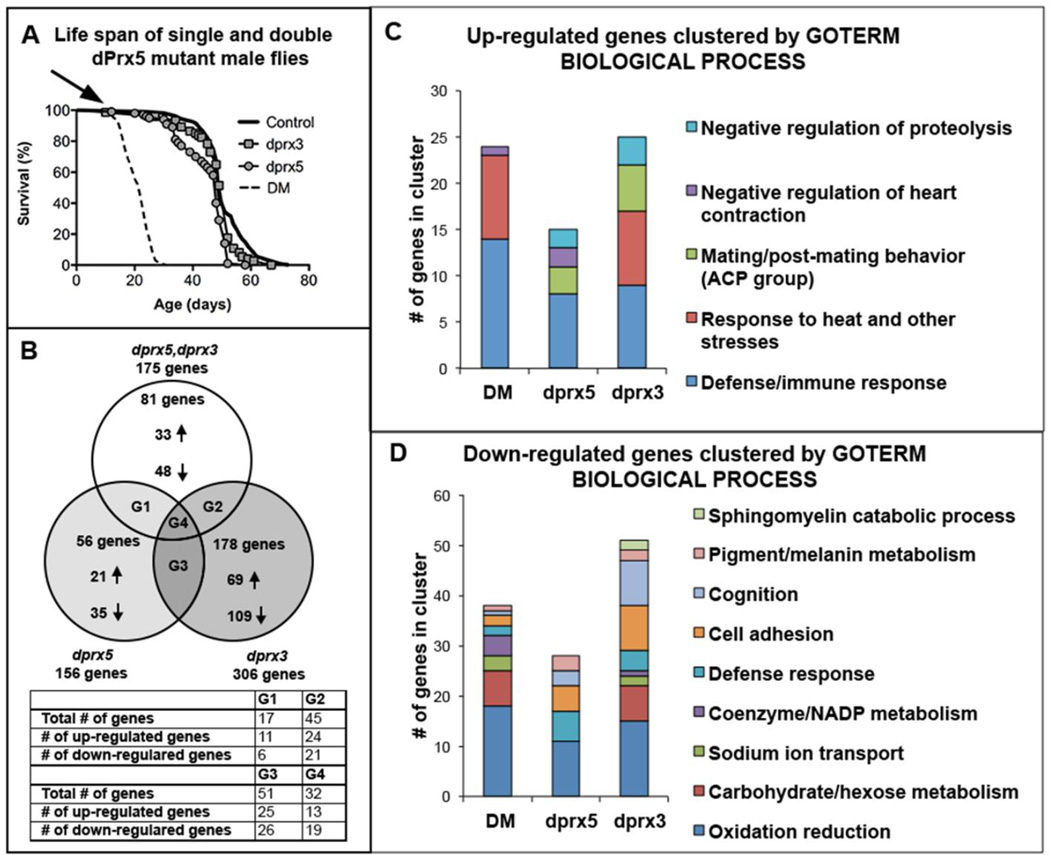
In previous studies, we found that flies under-expressing both dPrx3 and dPrx5 (DM) acquired a phenotype, which was distinct from that in the single mutants (11). On the basis of changes in longevity, redox state and apoptosis, we suggested that the DM might represent a model of accelerated aging. To identify specific pathways by which the removal of peroxidase activity from Drosophila mitochondria elicits these effects, we have performed total transcriptome profiling of flies under-expressing dPrx3 and dPrx5 individually and together. We compared changes in gene expression in individual and double mutants relative to control in flies of the same chronological age (13 days old), corresponding to the age when the double mutant population has declined by approximately 10% (Fig. 2A).
Figure 2. Transcriptome analysis of Prx mutants.
The genotypes of flies subjected to analysis were Da-GAL4, dprx5/+ (control); Da-GAL4, dprx5/dprx5 (dprx5 mutant); Da-GAL4, dprx5/RNAidprx3 (dprx3) and Da-GAL4, dprx5/RNAi-dprx3, dprx5 (DM). A, Survivorship of Prx single and double mutants along with control. RNA was extracted for RNA-seq analysis before the onset of the rapid death observed in the DM at 13 day, indicated by the arrow. B, Venn diagram representing the overlapping and non-overlapping genes, whose expression was altered in the double and single mutants compared to control. The diagram shows the total number of up-regulated and downregulated genes unique for each group. G1, G2, G3 and G4 – groups of overlapping up- and downregulated genes, shown in the table. C, Functional clustering of 131, 70 and 81 genes up-regulated in dprx3, dprx5 and double mutants respectively. D, Functional clustering of 175, 86 and 94 genes down-regulated in dprx3, dprx5 and double mutants respectively. Biological processes GO with an enrichment score >1 are depicted. Suppl. Table 1 contains lists of the genes included in B–D.
The decrease in dPrx activity in double and single mutants significantly altered expression of 1–2% of the ~ 14,000 genes probed by RNA-seq, and ~ 1/2 of these displayed common expression patterns in either pair-wise comparisons among the mutants (G1 + G2 + G3) or in the comparison between all three (G4) (Fig. 2B and Suppl. Table 2). On the other hand, a considerable number of genes changes were unique for each mutant background (58% for dprx3, 36% for dprx5 and 46% for the DM), indicating some mutant-specific effects.
3.1.1. Functional clustering
Functional annotation clustering of the altered genes was performed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov). Three gene ontology (GO) terms (biological processes (BP), cellular compartment (CC) and molecular function (MF) were used to analyze the functional significance of the sets of up- and down-regulated genes, affected by each treatment. The analysis revealed that under-expression of dPrx3 specifically contributed to the DM phenotype by affecting response to abiotic stimuli, primarily via up-regulation of heat shock 70 proteins (Hsp). Both dprx3 and dprx5 mutants exhibited a strong up-regulation of the immune defense genes, which is clearly manifest in the DM (Fig. 2C and Suppl. Table 2), This gene cluster included a number of antimicrobial peptides (AMPs) and other components of the immune pathways, as well as the TotC gene, which is responsive to many abiotic and microbial stresses (39).
In both dprx3 and dprx5 single mutants and the DM, down-regulation of genes was observed prominently amongst the oxidation-reduction GO, comprising genes involved in regulation of mitochondrial function and detoxification (Fig. 2D and Suppl. Table 2). In addition, under-expression of dPrx3 but not dPrx5 resulted in down-regulation of genes related to glucose and mannose metabolism (Fig. 2D and Suppl. Table 2), which was noted in the DM as well and has been associated with models of mitochondrial disease (40).
3.1.2. DM unique and differentially expressed genes
Approximately 46% (Fig. 2B) of the genes differentially expressed in the DM are unique, indicating the existence of specific targets/pathways that require the activity of both Prxs. Functional classification of the 33 uniquely affected genes, up-regulated in response to the absence of dPrx3 and dPrx5, revealed a prominent GO cluster comprised of the immune/defense-related genes (Suppl. Table 3). Of the 48 genes down-regulated in the DM, 16 of them were clustered to oxidation reduction processes and carbohydrate metabolism, and included a number of hydrolases and cytochrome P450s (Cyp) (Suppl. Table 3).
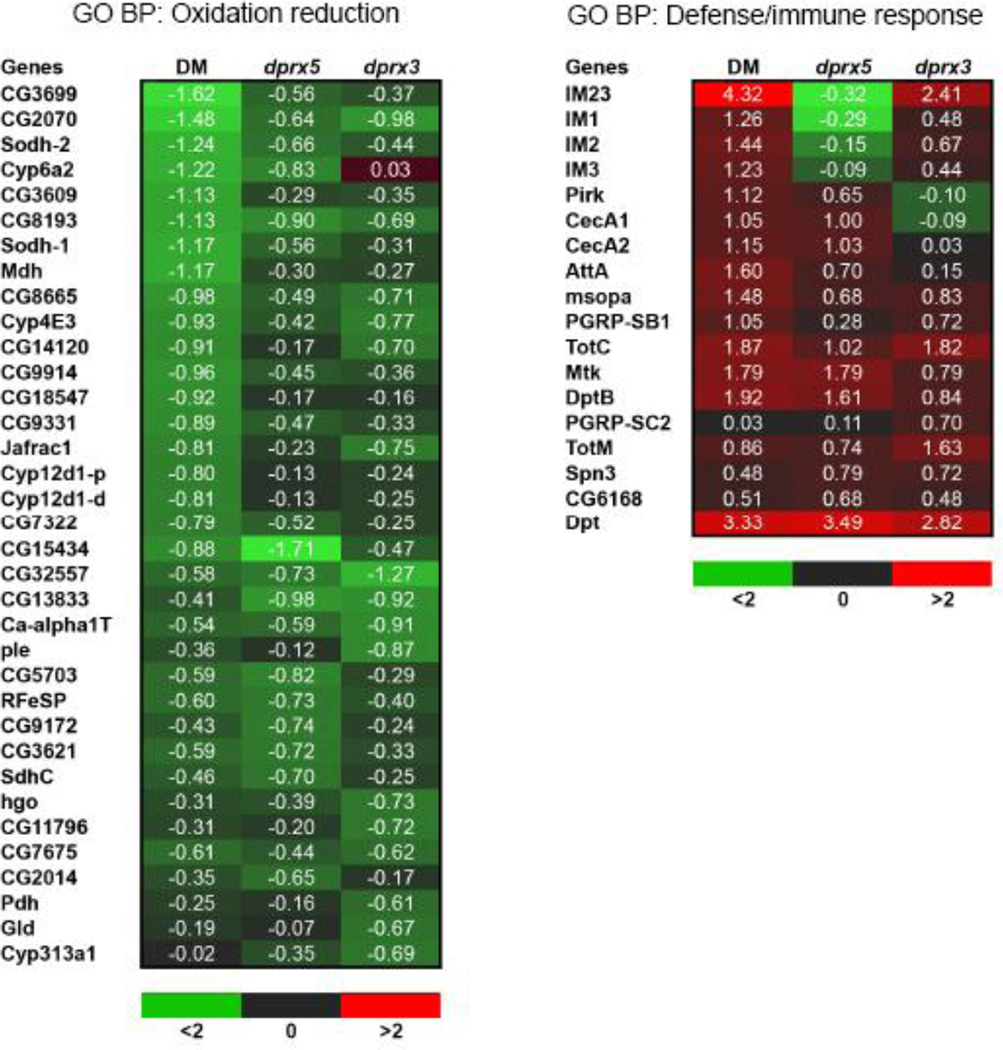
The significant overlap in genes whose expression is affected by the individual dprx3 and dprx5 mutations and the double mutation (Fig 2B, groups G1, G2 and G4) suggests that the individual Prx mutations contribute to the DM phenotype in an additive or synergistic mode. The analysis showed that, in general, DM enhances up-regulation of the immunity genes and suppresses a number of metabolism and detoxification-related genes (Fig. 3). The list of up-regulated genes also included peptidoglycan recognition proteins (PGRPs) that were ascribed to GO aminoglycan metabolism but are also known components of immune signaling in Drosophila (Fig. 3) (41).
Figure 3. Heat map demonstrating changes in expression of the defense/immune-related (right) and oxidation/reduction-related genes (left).
The results are expressed as fold ratio of the log2 gene expression values in the following comparisons: DM vs control (DM), dprx3 vs control (dprx3) and dprx5 vs control (dprx5). The red color depicts an increase in expression level of the mutants relative to control, and green represents a decrease. Shown is GO Biological Process (BP).
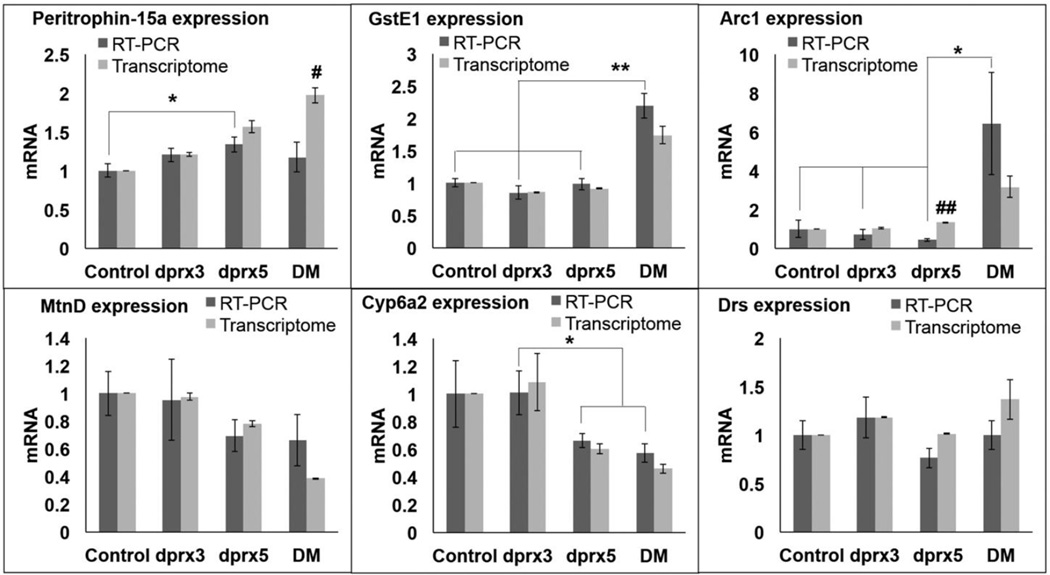
The list of genes uniquely affected by the DM included genes unclassified by functional clustering but whose changes in expression levels were considerably greater than in the single mutants. Differential expression of these genes, including Arc1 and GstE1, as well as other selected representative genes was validated by qRT-PCR and were generally consistent with the data obtained by RNA-seq analysis (Fig. 4). Since the analyzed material was collected from different cohorts of flies of the same chronological age, slight variations in physiological aging could account for the observed differences in the expression of age-sensitive genes.
Figure 4. Validation of transcriptome analysis by qRT-PCR.
Analysis of gene expression in control and the double (DM) and single mutants (dprx5 and dprx3) has been done in flies of the same chronological age (12–13 days). Primers for gene amplification are listed in Suppl. Table 1. Averages and SEM from at least tree independent samples are shown. Asterisks denote statistically significant differences obtained in RT-PCR analysis (*P<0.05; **P<0.01). Differences between values obtained by RT-PCR and transcriptome data are shown by # (P<0.05) and ## (P<0.01).
3.2. Age-dependent changes in gene expression in double and single dprx mutants
Since the material for RNA-seq was taken from flies of the same chronological age but at widely different points in their life span trajectory, with the DM flies entering the rapid death phase at ~10% mortality while controls were still in their early adult phase (Fig. 2A), it was plausible that some of the transcriptional fluctuations may derive from the effects of physiological aging. In an attempt to distinguish between possible aging effects and toxic effects, we analyzed mRNA levels in experimental and control flies at ages that reflected equivalent points of progression through life trajectory (Table 2). Thus, young flies were collected approximately 16% into their respective life spans, which corresponded to approximately 12 days for control and single mutants and 3 days for the DM (Fig. 2A, Table 2). Physiologically older flies were collected at the onset of initial death (~ 10% mortality), which corresponded to ~ 30 days for single mutants and control, and ~12 days for the DM (Fig. 2A, Table 2). Our primary focus was on the GO cluster with the highest enrichment score, the immunity-related genes/AMPs, but we also analyzed genes that displayed particularly dramatic differences in expression between the DM and single mutant flies, shown in Fig. 4.
Table 2.
Ages of flies collected for the gene expression analysis
| Line | Age | % of Life Span | % of dead flies |
|---|---|---|---|
| Control | 3 | 4 | 0 |
| 12 | 16 | 0 | |
| 30 | 45 | 10 | |
| 50 | 70 | 40 | |
| dprx3 | 3 | 4 | 0 |
| 12 | 16 | 0 | |
| 30 | 45 | 10 | |
| 50 | 70 | 40 | |
| dprx5 | 3 | 4 | 0 |
| 12 | 16 | 0 | |
| 30 | 45 | 10 | |
| 50 | 65 | 40 | |
| DM | 3 | 16 | 0 |
| 6 | 25 | 0 | |
| 12 | 45 | 10 | |
| 16 | 65 | 40 | |
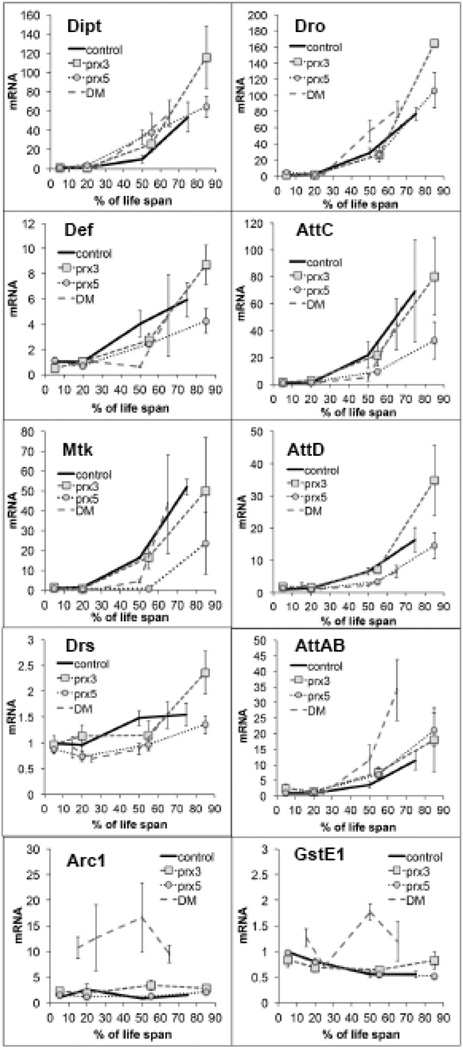
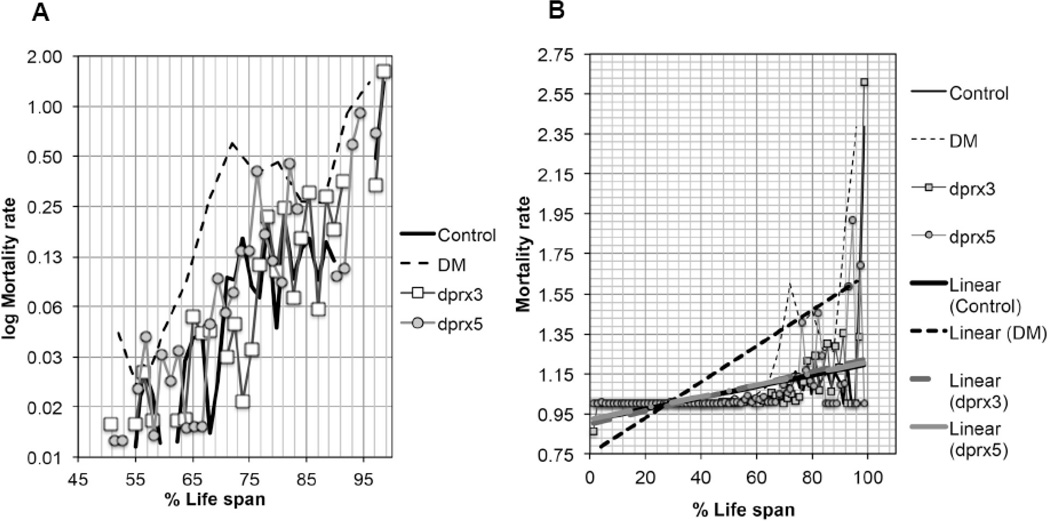
In most cases, mRNA levels of AMPs in single and DM flies, when scaled to life span, exhibited changes similar to those observed in control flies (Fig. 5), as well as in our long-lived y w reference strain (data not shown). One exception to this was the accelerated accumulation of AttAB in the DM, suggesting that the effects cannot be attributed solely to accelerated physiological aging (Fig. 5). Indeed, changes in mortality rate differed between the DM and the single mutants and control, indicating the contribution of the true aging and pathology (Fig. 6).
Figure 5. Effects of dPrx3 and dPrx5 under-expression on gene expression during aging.
Genotypes of flies were described in Table 1. Control flies are heterozygotes carrying the dprx5 allele along with the Da-GAL4 driver (Da-GAL4, dprx5/+). All groups of flies were collected at different ages, as indicated in Table 2. RNA was isolated from at least 10 flies in each group. Primers for gene amplification are listed in the Suppl. Table 1. Results are means ± SEM of two replicates performed with three independent cohorts of flies (total n=6). There were no statically significant differences in age-specific changes in the levels of Dipt, Def, AttC, and Drs between the DM and single mutants and Control, as determined by analysis of the slopes of corresponding regression lines. Statistically significant differences were observed between DM and control (P=0.0433) and dprx3 mutant and control (P=0.021) for Dro; between Control and dprx5 (P=0.0347) for Mtk; between DM and Control (P=0.0138), and DM and dprx3 (P=0.0092) for AttD. Age-specific increase in AttA&B expression was significantly higher in DM compared to single dprx3 and dprx5 mutants and Control (P=0.0018, P=0.0141, P=0.0115). Extremely significant were differences in the expression of Arc1 between the DM and other fly lines, as determined by analysis of the intercepts (P<0.0001). Analysis of GstE1 expression, relied on comparison of the slopes and intercepts, showed statistical significance between the DM and dprx3 (P=0.0002), dprx5 (0.0458), and Control (P=0.0354).
Figure 6. Changes in mortality rate in the double (DM), the single mutants (dprx5, dprx3) and control.
Mortality rate was calculated as reported (67), and plotted on a log2 scale (A) as a function of physiological age (% of life span). Approximately 100–125 flies were used for each fly line. B, The linear trend lines defined for mortality rates for each group of flies, are shown to demonstrate the divergence between the DM and the single mutants and controls.
3.3. Effects of mitochondrial peroxiredoxins on fly physiology and immune response
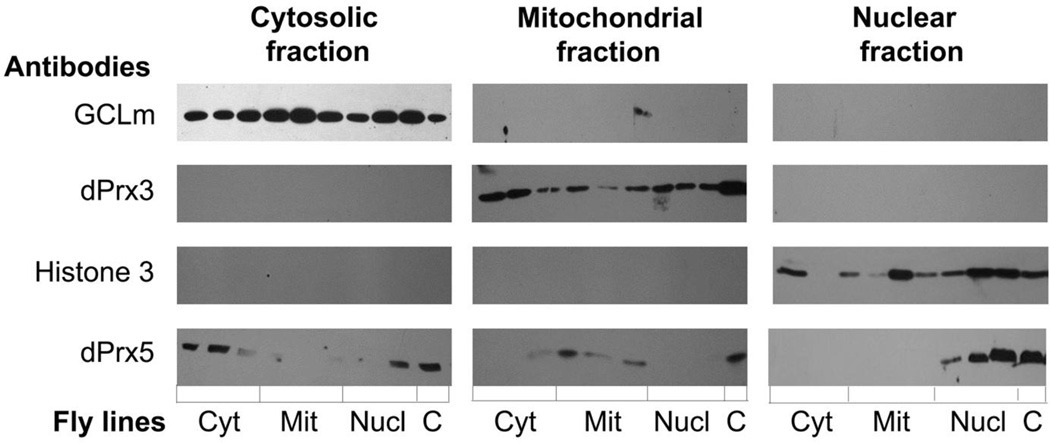
Although the depletion of both dPrx3 and dPrx5 in the double mutant flies had a significant lifespan shortening effect, the mean and median life spans of the single mutants were comparable to the controls indicating that a single wild type copy of either dPrx3 or dPrx5 was sufficient to support normal longevity (11,14). While the dPrx3 protein resides solely in the mitochondria, dPrx5 has multiple subcellular localizations, including mitochondrion (mit), nucleus (nucl) and cytosol (cyt) (14). To define a role for the different dPrx5 forms in the observed effects on aging and the temporal profiles of the genes affected by the double mutation, and to further explore pathways that determine the observed phenotypes, we generated flies ectopically expressing dPrx5 in the different subcellular compartments (Material and Methods, Fig. 1). Organelle-specific expression of dPrx5 was verified by subjecting the nuclear, mitochondrial and cytosolic subcellular fractions to immunoblot analysis (Fig. 7). As expected, localization of dPrx5 that lacks the mitochondrial pre-sequence was mainly cytosolic with some proportion of the protein distributed to the nucleus but not the mitochondrion. Removal of an alternate methionine resulted in effective targeting of dPrx5 to mitochondria while it was not detectable in cytosolic and nuclear fractions, and incorporation of an SV40 nuclear localization sequence drove expression into the nucleus.
Figure 7. Immunoblot analysis of subcellular fractions isolated from fly lines expressing different dPrx5 forms.
dPrx5 protein expression from different UAS transgenes was assessed in the dprx5 null background. Genotypes of fly lines were as follows: cytosolic – UAS-cyt dPrx5/+; Da-GAL4, dprx5/dprx5; nuclear – UAS-nucl dPrx5/+; Da-GAL4, dprx5/dprx5; mitochondrial – UAS-mit dPrx5/+; Da-GAL4, dprx5/dprx5. For each form, three independent fly lines were generated and immunoblot analysis was performed in triplicate. Flies expressing the endogenous dPrx5 protein served as a positive control (C). Antibodies raised to compartment-specific proteins (GCLm, Histone H3 and dPrx3) were used to control for loading and purity of subcellular fractions.
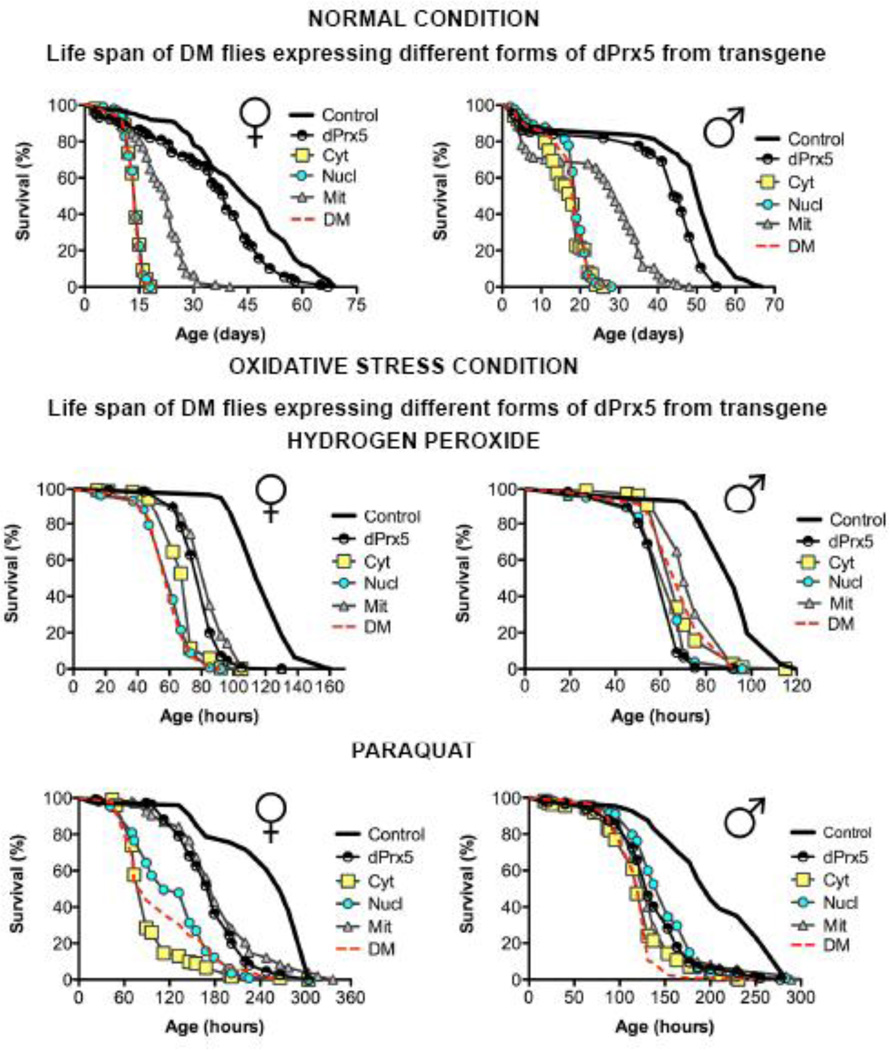
Since the effects of dPrx5 on survivorship and other traits could be masked in the presence of dPrx3, the different dPrx5 forms were investigated in a double mutant background. The ectopic expression of nuclear and/or cytosolic forms of dPrx5 by global high-level Da-GAL4 driver did not improve the survivorship of DM under normal or oxidative stress conditions (Fig. 8). In contrast, expression of dPrx5 solely in the mitochondria conferred a significant rescue effect on longevity, although production of all three dPrx5 forms was still required for complete restoration of life span (Fig. 8, top).
Figure 8. Effects of different dPrx5 forms on fly survivorship under normal (top) and oxidative stress conditions (bottom).
Shown are representative data of two experiments with independent cohorts. In each experiment, approximately 100–125 flies were used for each line. Oxidative stress was caused by feeding sucrose solutions supplemented with 0.125M H2O2 for males and 0.25M for females. PQ was added at the concentration of 5mM for both males and females. Statistically significant differences (P<0.05) between survivorship curves were determined by the log rank test. All statistical data are shown in Tables 3 and 4. The genotypes of flies are described in Table 1.
dPrx5 expression from the wild type (dPrx5) or from mitochondria-targeting (Mit) transgenes partially protected flies from oxidative stress induced by hydrogen peroxide or paraquat, and these effects were seen only in females (Fig. 8 bottom). Since flies expressing dPrx5 in all three compartments and those expressing dPrx5 in the mitochondria exhibited comparable protective effects, it may be inferred that Prx expression in the mitochondrion is critical in conferring protection against exogenous oxidative stress, at least in females. The observed sex bias in sensitivity of different fly lines to oxidative stress could be related to mitochondrial function, which is subject to genetic and physiological differences between the sexes (reviewed in (42), (43). Alternatively, a role for fat body function cannot be excluded (44).
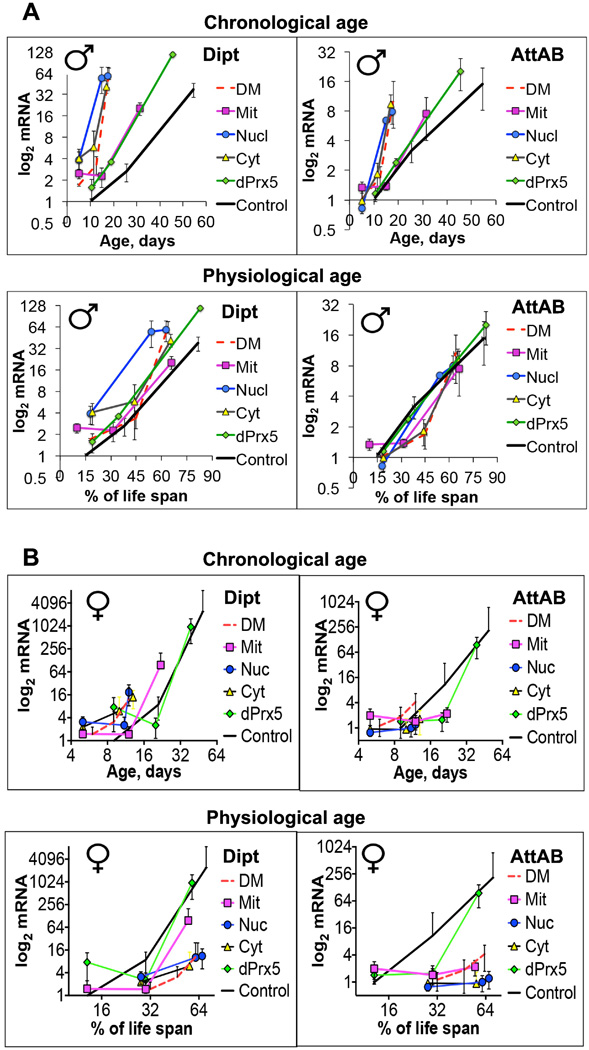
Given the clear impact of dPrx3 and dPrx5 on immunity-related genes, we also investigated a role for the different dPrx5 forms on immune response. The expression of the cytosolic or nuclear forms had little if any influence on the expression of AMPs in flies with a DM background (Fig. 9). However, when dPrx5 was expressed in mitochondria, it substantially delayed the development of the hyperactive immunity phenotype (Fig. 9). Surprisingly, dPrx5 expressed from the full-length transgene was less effective in suppressing the hyper-activation of AMP expression (Fig. 9). When we plotted AMP expression against % of life span, all survivorship curves followed similar trajectories (Fig. 9 A), suggesting a strong link between physiological aging and the state of Drosophila immunity.
Figure 9. Effects of different dPrx5 forms on activation of the immune response genes in males (A) and females (B).
The genotypes of flies are described in Table 1. Flies were collected at the ages indicated in Table 5. RNA was isolated from at least 10 flies for each group. qRT-PCR was performed as indicated in Material and Methods and Fig. 5. Results are means ± SEM of three replicates performed with three independent cohorts (total n=9). There were no statically significant differences in age-specific changes in the levels of Dipt and AttA&B between the DM and flies expressing cytosolic or nuclear forms of dPrx5. The differences were significant between the DM and Control (P=0.025 for AttA&B and P=0.0022 for Dipt), the DM and flies expressing mitochondrial dPrx5 (P=0.0399 for AttA&B and P=0.0061 for Dipt) as determined by analysis of the slopes of corresponding regression lines. Differences between flies expressing mitochondrial, full length dPrx5 and Control were not significant. When Dipt or AttA&B mRNA levels were plotted against % of life span, no significant differences in the regression curves slopes were determined (bottom graphs). B, Both chronological and physiological aging conferred differential effects on diptericin expression between flies expressing mitochondrial (Mit) or predominantly mitochondrial (dPrx5) forms and the DM (P=0.0048 and P<0.0001) while differences between the DM and flies expressing the cytosolic or nuclear forms of dPrx5 were not statistically significant. Expression of attacin differs significantly between the DM and all other fly lines at 11–13 days of age, at a time when short-lived DM as well as the cytotolic and nuclear expressors are undergoing rapid death (P <0.001).
As in the case of resistance to oxidative stress, we observed immune-related dichotomy between the sexes (Fig. 9). Unlike males, where comparable levels of AMP were observed in different fly lines to track with physiological age, females were characterized by greater activation of diptericin and attacin genes in longer-lived fly lines, including controls and flies expressing dPrx5 in mitochondrion and in all three compartments. Thus, the activation of AMP in females tracked with chronological age, and it may be surmised that the short-lived DM flies and flies expressing dPrx5 in the nuclear and cytosolic compartments died prior to development of a full-scale hyperactive immune response. In addition, the effects of mitochondrially-expressed dPrx5 on activation of AttA and B were different from the effects on Dipt, which could be attributed to differential effects of the steroid hormone ecdysone and its receptor on the regulation of Imd pathway-related genes (45). On the other hand, there were also some similarities in immune response in both male and female flies. As in males, the mitochondrial form of dPrx5 expressed separately or as a major component derived from the full-length dPrx5 transgene substantially delayed the increase in diptericin levels compared to those observed in DM flies as well as flies expressing the nuclear and cytosolic forms of dPrx5. Changes in AttA and B levels showed a similar trend, albeit not statistically significant (Fig. 9 B).
4. Discussion
Mitochondria play a central role in regulation of a variety of cell functions and processes, including energy conversion, cellular metabolism, and apoptosis. An essential component in the regulation of these processes is H2O2, which is produced as a by-product of mitochondrial respiration and whose levels in this organelle are maintained by residential peroxidases. The downstream reactions of H2O2 include oxidation of susceptible cysteine residues, which may impart functional consequences on redox-sensitive signaling pathways (46). Here, by performing transcriptome analysis, we determined that the major outcome of impaired redox homeostasis resulting from the absence of mitochondrial peroxidase activity is a prominent hyperactivation of the immunity and stress response genes and simultaneous decrease in the expression of genes involved in detoxification and mitochondrial function maintenance (Fig. 2 C–D, Fig. 3, Suppl. Table 2, and Suppl. Table 3).
Given that the absence of mitochondrial peroxidase activity led to enhanced apoptosis and a shift in redox state, it was somewhat unexpected that no notable shifts in the expression of gene clusters related to either apoptosis or oxidative stress were detected (Fig. 2 C–D and Suppl. Table 2, and Suppl. Table 3). In prior studies measuring gene expression response to oxidative stress conditions in flies, antioxidant gene clusters were commonly affected (47). One might infer then that the redox state changes in the absence of peroxidase activity are qualitatively distinct from the effects of manipulations previously described and might indeed interfere with elements of NRF2-like signaling.
The gene cluster induced most strongly in response to the reduction in peroxidase activity in both single and double mutant flies fell into the category of the immune response genes (IRG) (Fig. 2C). These included a number of known components of the Imd and Toll immune pathways, as well as genes whose expression is triggered via JAK/STAT signaling in response to multiple stresses (Fig. 2C, Fig. 3 and Suppl. Table 2, (39,48)).
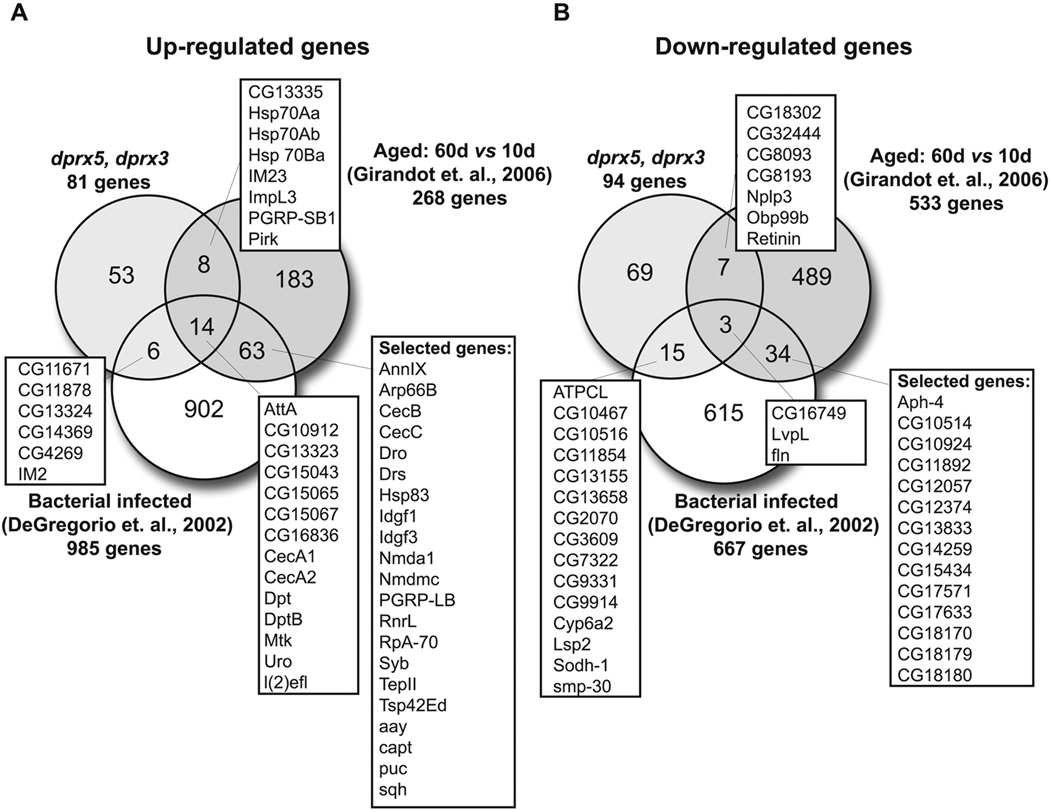
Induction of the stress response genes may simply reflect activation of the adaptive response to the increasingly pro-oxidizing environment in the single and particularly the double Prx mutants. In the case of the double mutants such changes might also reflect in part physiological aging, given that these flies were collected when the mortality had reached 10% in the life span trajectory (Fig 2A). Indeed, numerous genome-wide studies conducted by different labs identified the immune response genes as those most commonly affected with age (49). When we compared our data with aging and infection-related transcriptome databases, we noted a significant overlap of affected genes related to humoral immunity that were present in both the DM and also in old flies (50,51) as well as flies infected with bacteria (48,52), as shown in the Venn diagram (Fig. 10).
Figure 10. Comparison of transcriptional responses to depletion of mitochondrial dPrxs, infection and aging.
Venn diagram of genes differentially expressed in dprx3, dprx5 double mutant flies, aged flies (51), and bacteria-infected flies (48). A, Genes with increased expression, and B, genes with decreased expression.
Considering the well-established correlations between aging and changes in endogenous oxidative stress due to altered mitochondrial function and a shift in cellular redox to a more prooxidative state (27,53,54), the up-regulation of the IRG, including AMPs, in older flies may reflect responses to this altered cellular redox status. The similar changes that occur in the DM may also represent a response to analogous changes in redox state that manifest at a significantly earlier age in these flies (11). Recently, it was reported that the key immunity components, together with other genes involved in oxidative stress response and detoxification, such as GstE1 were implicated in response to hyperoxia (55). Moreover, experimental overexpression of AMPs, such as Dipt and Att was shown not only to increase resistance to bacterial infection, which is their primary function, but also play a significant role in tolerance to exogenous hyperoxia (55). Together, these data suggest that in addition to their antimicrobial function, some AMPs may act as free radical scavengers because of the cationic nature of the surface charge. In this regard, it is interesting to note that a number of the AMP genes (Dipt, CecD, AttA and Mtk) contain ARE consensus sequences in their promoters as revealed by the Alibaba program (gene-regulation.com) (data not shown).
On the other hand, AMP over-expression may also cause adverse effects on cell physiology, witness the neurodegeneration observed when AMPs were overexpressed in Drosophila brain tissues (56). Recently, we found that high-level constitutive over-expression of AMPs in transgenic flies had cytotoxic effects and resulted in increased tissue-specific cell death and shortened longevity. These effects were most prominent in flies overexpressing AttA (unpublished data). It is plausible then that the apoptosis observed in the DM (11), could be a secondary event caused by cytotoxicity due to high levels of AMPs.
While the immune response was upregulated in the DM, there was simultaneous decrease in the expression of cytochrome P450s (Fig. 3 and Suppl. Table 2). Recent studies have established a regulatory role for cytochrome P450 epoxygenases and hydroxylases in inflammation via the ability to metabolize multiple substrates related to the regulation of inflammation and lipid homeostasis (57). Perhaps, downregulation of cytochromes may exacerbate the hyperactive immunity phenotype, observed in the DM. In general the genes specifically affected by the DM were highly related and tended to cluster within the Drosophila genome as if to facilitate co-regulation. For instance, the immune-induced molecules (IM1, IM3 and CGs that have Drosophila immune-induced molecule (Dim) domain) form a tight cluster spanning 10 KB at cytogenetic position 55C4 on the right arm of the second chromosome. Protein network analysis also revealed considerable evidence for physical interaction/co-localization amongst these co-regulated genes (GeneMania analysis, data not shown) (58). Other genes, as those that have Cyp P450 domain or heat shock proteins, were found physically interacting (GeneMania analysis, data not shown). There was also a significant degree of co-expression and co-localization, as in case of JAK/STAT regulated Turandot proteins and IRG (GeneMania analysis, data not shown), which may indicate their involvement in the same biological processes or pathways.
The biological processes affected in the individual dprx3 and dprx5 mutants include significant overlap with each other as well as with those affected in the double mutant. Moreover these changes in transcription profiles paralleled to some extent those observed in other animal models as well as those that accompany aging and stress response in Drosophila. Thus, like Drosophila, absence of Prx3 in the mouse resulted in a significant down-regulation of genes involved in carbohydrate metabolism (18,22,59) (Fig. 2D, Suppl. Table 2). Similar groups of genes related to sugar/mannose metabolism were down-regulated in old flies and flies exposed to different stressors, such as hydrogen peroxide and heat (49). Other genes down-regulated in both older flies and flies lacking mitochondrial Prxs included a group of membrane-bound proteins associated with the mitochondrial respiratory electron transport chain, ATP synthesis, and complex 1 activity as well as genes associated with detoxification, mainly represented by cytochromes. Cytochrome P450s are also essential for synthesis and degradation of signaling molecules, and suppression of Cyp expression in C. elegans resulted in failed detoxification and increased mortality rates (60). The importance of Prx3 in detoxification and mitigating hepatotoxic damage was also shown in KO mice (61). The effects that were specific for the absence of dprx3 were related to cellular responses to environmental stresses, such as heat, hypoxia, and UV (Fig. 2C, Suppl. Table 2), and included a number of heat stress proteins and chaperones, which are normally induced in response to the accumulation of unfolded proteins formed under different stress conditions (62). Overall, it seems that Prx3 plays a defining role in mitochondrial functioning, and this notion is supported by studies conducted in mammals (63,64). Nevertheless, there is a considerable degree of redundancy with Prx5, based on the overlap of gene expression patterns of the single mutants and the additive nature of transcript levels in the DM (Fig. 2 C–D, Fig. 3, Suppl. Table 2).
Given the overlap between dPrx3 and dPrx5 transcription profiles, it remains possible that dPrx5 has functional effects that are distinct from those of dPrx3, particularly in view of the distribution of dPrx5 in different subcellular compartments, including mitochondrial, cytosolic and nuclear (14). Consequently we investigated which form of dPrx5 was responsible for the DM phenotype. Unexpectedly, the ectopic expression of compartment-targeted dPrx5 (Cyt, Nucl and Mit) or the wild type form (dPrx5), which targets all three compartments, conferred little protection from oxidative stress caused by H2O2 or PQ. Only a partial and sex-specific increase in resistance was observed in flies expressing dPrx5 from mitochondrial (Mit) and wild type (dPrx5) transgenes using the global Da11 GAL4 driver (Fig. 1, Table. 1). Since dprx3 mutant flies exhibit no loss in susceptibility to oxidative stress relative to wild type, it would appear that sufficient expression of the ectopically expressed dPrx5 transgenes was not achieved in critical cells/ tissues, resulting in only partial rescue (11). Nevertheless, the data obtained here are consistent with mammalian studies showing a more important protective role of the mitochondrial form of Prx5 (65,66).
Although mitochondria-localized dPrx5 had only moderate and sex-specific effects on resistance to acute exogenous oxidative stress, the beneficial effects on fly life span were more pronounced (Fig. 8). Thus, the localization of dPrx5 to the mitochondria appears to be particularly important in mitigating the redox crisis of the DM. However, it was only the wild type form of dPrx5 that completely restored survivorship of flies to the levels observed in controls (Fig. 8), indicating that distribution of dPrx5 in the cytosol and/or the nucleus is also required to achieve full rescue. For instance, nuclear dPrx5 might contribute to long-term protecting effects by preventing the formation of DNA damage (65).
The observed differences in the effects of mitochondria dPrx5 expression on survivorship under normal and oxidative stress conditions suggest that the effects on longevity proceed not only through a mechanism that necessitates increased stress resistance and management of chronic lowlevel oxidative stress, but also involve other processes. Considering the major changes observed in IRG transcription profiles, we also investigated the effects of compartment-specific dPrx5 expression on the state of humoral immunity. It was only the mitochondrially-localized dPrx5 that significantly attenuated over-activation of the immune responses in the DM flies (Fig. 9), and had a beneficial effect on longevity (Fig. 8). To the best of our knowledge, this is a first report showing that the infection-irrelevant immune response depends at least in part on the activity of redox-regulating enzymes in the mitochondria. It is yet to be determined whether immune signaling originated in the mitochondria proceeds through the same cascades that are activated in response to microbereceptor interactions and utilize components of the Toll and Imd immune pathways. Furthermore, given the recent finding that the endoplasmic reticulum (ER)-localized dPrx4 regulates abiotic inflammatory responses in Drosophila (12), it will be critical to assess potential cross-talk between the ER and mitochondrial-localized Prxs. To conclude, we have determined that mitochondrial pathways modulated by peroxiredoxin activity play a significant role in pro-inflammatory response and aging.
Supplementary Material
Table 3.
Mean life span of DM female flies expressing different forms of dPrx5 from transgene
| Normal conditions | |||||
|---|---|---|---|---|---|
| Fly Lines | Mean, days | % vs Control | p value | % vs DM | p value |
| Control | 44.5 | +68.54 | <0.0001 | ||
| 55 | +76.36 | <0.0001 | |||
| dPrx5 | 39 | −12.36 | <0.0001 | +64.10 | <0.0001 |
| 35 | −36.36 | <0.0001 | +62.86 | <0.0001 | |
| Cyt | 14 | −68.54 | <0.0001 | 0 | 0.3707 |
| 14 | −74.55 | <0.0001 | +7.14 | <0.0001 | |
| Nucl | 14 | −68.54 | <0.0001 | 0 | 0.4957 |
| 14 | −74.55 | <0.0001 | +7.14 | <0.0001 | |
| Mit | 23 | −48.31 | <0.0001 | +39.13 | <0.0001 |
| 23 | −58.18 | <0.0001 | +43.48 | <0.0001 | |
| DM | 14 | −68.54 | <0.0001 | ||
| 13 | −76.36 | <0.0001 | |||
|
Hydrogen peroxide | |||||
| Genotype | Mean, hours | % vs Control | P value | % vs DM | P value |
| Control | 114 | +45.61 | <0.0001 | ||
| 141 | +47.52 | <0.0001 | |||
| dPrx5 | 85 | −25.44 | <0.0001 | +27.06 | <0.0001 |
| 92 | −34.75 | <0.0001 | +19.57 | <0.0001 | |
| Cyt | 73 | −35.96 | <0.0001 | +15.07 | <0.0001 |
| 83 | −41.13 | <0.0001 | +10.84 | 0.5718 | |
| Nucl | 62 | −45.61 | <0.0001 | 0 | 0.6329 |
| 67 | −52.48 | <0.0001 | −9.46 | 0.0085 | |
| Mit | 85 | −25.44 | <0.0001 | +27.06 | <0.0001 |
| 97 | −31.21 | <0.0001 | +23.71 | <0.0001 | |
| DM | 62 | −45.61 | <0.0001 | ||
| 74 | −47.52 | <0.0001 | |||
|
Paraquat | |||||
| Control | 266 | <0.0001 | +66.54 | <0.0001 | |
| 249 | <0.0001 | +54.22 | <0.0001 | ||
| dPrx5 | 179 | −32.71 | <0.0001 | +50.28 | <0.0001 |
| 163 | −34.54 | <0.0001 | +30.06 | <0.0001 | |
| Cyt | 89 | −66.54 | <0.0001 | 0 | 0.0643 |
| 63 | −74.70 | <0.0001 | −44.74 | <0.0001 | |
| Nucl | 112 | −57.89 | <0.0001 | +20.54 | 0.2107 |
| 114 | −54.22 | <0.0001 | 0 | 0.6203 | |
| Mit | 179 | −32.71 | <0.0001 | +50.28 | <0.0001 |
| 131 | −47.39 | <0.0001 | +12.98 | 0.0119 | |
| DM | 89 | −66.54 | <0.0001 | ||
| 114 | −54.22 | <0.0001 | |||
Life span data in column 2 are mean life span of the groups shown in Fig.8. Column 3 and 4 indicate the percent changes in experimental groups vs control (Da-GAL4, dprx5/+), and the significance probabilities (p values) of the associated log rank tests. Similarly, columns 5 and 6 indicate percent changes and p values in relation to DM (Da-GAL4, dprx5/ RNAi-dprx3, dprx5). Genotypes of fly lines are shown in Table 1.
Table 4.
Mean life span of DM male flies expressing different forms of dPrx5 from transgene
| Normal conditions | |||||
|---|---|---|---|---|---|
| Fly Lines | Mean, days | % vs Control | p value | % vs DM | p value |
| Control | 51.00 | +62.72 | <0.0001 | ||
| 49.00 | +55.10 | <0.0001 | |||
| dPrx5 | 44.50 | −12.75 | <0.0001 | +57.30 | <0.0001 |
| 47.00 | −4.08 | <0.0001 | +53.19 | <0.0001 | |
| Cyt | 18.00 | −64.71 | <0.0001 | −5.26 | 0.3664 |
| 23.00 | −53.06 | <0.0001 | +4.35 | 0.2983 | |
| Nucl | 19.00 | −62.75 | <0.0001 | 0 | 0.4218 |
| 20.00 | −59.18 | <0.0001 | −9.09 | 0.0063 | |
| Mit | 29.00 | −43.14 | <0.0001 | +34.48 | <0.0001 |
| 27.00 | −44.89 | <0.0001 | +18.52 | <0.0001 | |
| DM | 19.00 | −62.75 | <0.0001 | ||
| 22.00 | −55.10 | <0.0001 | |||
|
Hydrogen peroxide | |||||
| Genotype | Mean, hours | % vs Control | P value | % vs DM | P value |
| Control | 92.00 | +27.17 | <0.0001 | ||
| 85.00 | +27.06 | <0.0001 | |||
| dPrx5 | 67.00 | −27.17 | <0.0001 | 0 | <0.0001 |
| 62.00 | −27.06 | <0.0001 | 0 | 0.3336 | |
| Cyt | 67.00 | −27.17 | <0.0001 | 0 | 0.4578 |
| 62.00 | −27.06 | <0.0001 | 0 | 0.0124 | |
| Nucl | 67.00 | −27.17 | <0.0001 | 0 | <0.0001 |
| 62.00 | −27.06 | <0.0001 | 0 | 0.0040 | |
| Mit | 70.00 | −23.91 | <0.0001 | +4.29 | 0.0254 |
| 67.00 | −21.18 | <0.0001 | +7.46 | 0.0007 | |
| DM | 67.00 | −27.17 | <0.0001 | ||
| 62.00 | −27.06 | <0.0001 | |||
|
Paraquat | |||||
| Control | 200 | +34.50 | <0.0001 | ||
| 199 | +26.63 | <0.0001 | |||
| dPrx5 | 131 | −34.50 | <0.0001 | 0 | <0.0001 |
| 146 | −26.63 | <0.0001 | 0 | 0.1438 | |
| Cyt | 119 | −40.50 | <0.0001 | −9.16 | 0.2122 |
| 146 | −26.63 | <0.0001 | 0 | 0.8483 | |
| Nucl | 153 | −23.50 | <0.0001 | +14.38 | <0.0001 |
| 112 | −43.71 | <0.0001 | −23.29 | <0.0001 | |
| Mit | 131 | −34.50 | <0.0001 | 0 | <0.0001 |
| 168 | −15.58 | <0.0001 | +13.10 | <0.0001 | |
| DM | 131 | −34.50 | <0.0001 | ||
| 146 | −26.63 | <0.0001 | |||
Life span data in column 2 are mean life span of the groups shown in Fig.8. Column 3 and 4 indicate the percent changes in experimental groups vs control (Da-GAL4, dprx5/+), and the significance probabilities (p values) of the associated log rank tests. Similarly, columns 5 and 6 indicate percent changes and p values in relation to DM (Da-GAL4, dprx5/ RNAi-dprx3, dprx5). Genotypes of fly lines are shown in Table 1.
Table 5.
Ages of flies collected for the gene expression analysis
| MALES | FEMALES | ||||
|---|---|---|---|---|---|
| Line | Age | % of Life Span | Line | Age | % of Life Span |
| Control | 11 | 16 | Control | 9 | 13 |
| 26 | 38 | 21 | 30 | ||
| 55 | 82 | 49 | 71 | ||
| Mit | 5 | 10 | Mit | 5 | 13 |
| 15 | 32 | 12 | 30 | ||
| 32 | 66 | 22 | 55 | ||
| Nucl | 5 | 18 | Nucl | 5 | 28 |
| 15 | 54 | 11 | 61 | ||
| 18 | 63 | 12 | 67 | ||
| Cyt | 5 | 19 | Cyt | 5 | 28 |
| 12 | 44 | 10 | 56 | ||
| 17 | 66 | 13 | 72 | ||
| dPrx5 | 11 | 19 | dPrx5 | 9 | 13 |
| 19 | 35 | 20 | 30 | ||
| 46 | 83 | 39 | 58 | ||
| DM | 5 | 18 | DM | 6 | 32 |
| 13 | 45 | 9 | 47 | ||
| 18 | 64 | 12 | 63 | ||
Genotypes of fly lines are shown in Table 1.
Highlights.
Underexpression of mitochondrial peroxiredoxins induces the immune response
Mitochondrial peroxiredoxin 5 delays the onset of hyperactive immunity
Mitochondrial peroxiredoxin 5 partially protects from oxidative stress
Shortened longevity of the double dprx3&5 mutant is rescued by mitochondrial dPrx5
Acknowledgments
This work was supported by the grant R01 AG032342 from the National Institute on Aging/National Institutes of Health. We would like to thank Judith Benes, SMU, for the technical assistance in fly lab. We also thank the University of Rochester Genomics Research Center (URGRC) for conducting the RNA-seq procedure and their senior bioinformatics analyst/programmer Jason R. Myers for the data analysis. We are also grateful to Pawel Michalak (Virginia Tech) for invaluable advice on transcriptome analyses.
List of Abbreviations
- Prx
Peroxiredoxin
- AMP
antimicrobial peptide
- ROS
reactive oxygen species
- DM
double mutant
- NLS
nuclear localization signal
- Da
daughterless
- PQ
paraquat
- Cyp
cytochrome P450
- Cyt
cytosolic
- Mit
mitochondrial
- Nucl
nuclear
- IRG
immune response genes
- Hsp
heat shock protein
- PGRP
peptidoglycan recognition proteins
- BP
biological process
- ER
endoplasmic reticulum.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid Redox Signal. 2011;15:781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 2.Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flohe L. The impact of thiol peroxidases on redox regulation. Free radical research. 2016;50:126–142. doi: 10.3109/10715762.2015.1046858. [DOI] [PubMed] [Google Scholar]
- 4.Fourquet S, Huang ME, D'Autreaux B, Toledano MB. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid Redox Signal. 2008;10:1565–1576. doi: 10.1089/ars.2008.2049. [DOI] [PubMed] [Google Scholar]
- 5.Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 6.Netto LE, Antunes F. The Roles of Peroxiredoxin and Thioredoxin in Hydrogen Peroxide Sensing and in Signal Transduction. Molecules and cells. 2016;39:65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee SG. Overview on Peroxiredoxin. Molecules and cells. 2016;39:1–5. doi: 10.14348/molcells.2016.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoops B, Argyropoulou V, Becker S, Ferte L, Kuznetsova O. Multiple Roles of Peroxiredoxins in Inflammation. Molecules and cells. 2016;39:60–64. doi: 10.14348/molcells.2016.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampton MB, O'Connor KM. Peroxiredoxins and the Regulation of Cell Death. Molecules and cells. 2016;39:72–76. doi: 10.14348/molcells.2016.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radyuk SN, Michalak K, Klichko VI, Benes J, Orr WC. Peroxiredoxin 5 modulates immune response in Drosophila. Biochim Biophys Acta. 2010;1800:1153–1163. doi: 10.1016/j.bbagen.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radyuk SN, Rebrin I, Klichko VI, Sohal BH, Michalak K, Benes J, Sohal RS, Orr WC. Mitochondrial peroxiredoxins are critical for the maintenance of redox state and the survival of adult Drosophila. Free Radic Biol Med. 2010;49:1892–1902. doi: 10.1016/j.freeradbiomed.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klichko VI, Orr WC, Radyuk SN. The role of peroxiredoxin 4 in inflammatory response and aging. Biochimica et biophysica acta. 2016;1862:265–273. doi: 10.1016/j.bbadis.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledano MB, Delaunay-Moisan A. Keeping Oxidative Metabolism on Time: Mitochondria as an Autonomous Redox Pacemaker Animated by H2O2 and Peroxiredoxin. Molecular cell. 2015;59:517–519. doi: 10.1016/j.molcel.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. The Biochemical journal. 2009;419:437–445. doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeles DC, Ho P, Chua LL, Wang C, Yap YW, Ng C, Zhou Z, Lim KL, Wszolek ZK, Wang HY, Tan EK. Thiol peroxidases ameliorate LRRK2 mutant-induced mitochondrial and dopaminergic neuronal degeneration in Drosophila. Human molecular genetics. 2014;23:3157–3165. doi: 10.1093/hmg/ddu026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace DC. Mouse models for mitochondrial disease. American journal of medical genetics. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- 17.De Simoni S, Goemaere J, Knoops B. Silencing of peroxiredoxin 3 and peroxiredoxin 5 reveals the role of mitochondrial peroxiredoxins in the protection of human neuroblastoma SH-SY5Y cells toward MPP+ Neurosci Lett. 2008;433:219–224. doi: 10.1016/j.neulet.2007.12.068. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Na R, Gu M, Salmon AB, Liu Y, Liang H, Qi W, Van Remmen H, Richardson A, Ran Q. Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell. 2008;7:866–878. doi: 10.1111/j.1474-9726.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong LK, Mockett RJ, Bayne AC, Orr WC, Sohal RS. Decreased mitochondrial hydrogen peroxide release in transgenic Drosophila melanogaster expressing intramitochondrial catalase. Arch Biochem Biophys. 2000;383:303–308. doi: 10.1006/abbi.2000.2093. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay SS, Leung KS, Hicks MJ, Hastings PJ, Youssoufian H, Plon SE. Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J Cell Biol. 2006;175:225–235. doi: 10.1083/jcb.200607061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang HH, Arscott LD, Ballou DP, Williams CH., Jr Acid-base catalysis in the mechanism of thioredoxin reductase from Drosophila melanogaster. Biochemistry. 2008;47:1721–1731. doi: 10.1021/bi702040u. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Shoji W, Takano H, Nishimura N, Aoki Y, Takahashi R, Goto S, Kaifu T, Takai T, Obinata M. Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPSinduced oxidative stress. Biochem Biophys Res Commun. 2007;355:715–721. doi: 10.1016/j.bbrc.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 23.Lee KP, Shin YJ, Cho SC, Lee SM, Bahn YJ, Kim JY, Kwon ES, Jeong DY, Park SC, Rhee SG, Woo HA, Kwon KS. Peroxiredoxin 3 has a crucial role in the contractile function of skeletal muscle by regulating mitochondrial homeostasis. Free Radic Biol Med. 2014;77C:298–306. doi: 10.1016/j.freeradbiomed.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Radyuk SN, Klichko VI, Michalak K, Orr WC. The effect of peroxiredoxin 4 on fly physiology is a complex interplay of antioxidant and signaling functions. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:1426–1438. doi: 10.1096/fj.12-214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebrin I, Sohal RS. Pro-oxidant shift in glutathione redox state during aging. Adv Drug Deliv Rev. 2008;60:1545–1552. doi: 10.1016/j.addr.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc Natl Acad Sci U S A. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebrin I, Bayne AC, Mockett RJ, Orr WC, Sohal RS. Free aminothiols, glutathione redox state and protein mixed disulphides in aging Drosophila melanogaster. Biochem J. 2004;382:131–136. doi: 10.1042/BJ20040506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebrin I, Sohal RS. Comparison of thiol redox state of mitochondria and homogenates of various tissues between two strains of mice with different longevities. Exp Gerontol. 2004;39:1513–1519. doi: 10.1016/j.exger.2004.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem Biophys Res Commun. 2008;368:273–278. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 31.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 32.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radyuk SN, Klichko VI, Spinola B, Sohal RS, Orr WC. The peroxiredoxin gene family in Drosophila melanogaster. Free Radic Biol Med. 2001;31:1090–1100. doi: 10.1016/s0891-5849(01)00692-x. [DOI] [PubMed] [Google Scholar]
- 38.Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J Biol Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- 39.Ekengren S, Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem Biophys Res Commun. 2001;284:998–1003. doi: 10.1006/bbrc.2001.5067. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Ayala DJ, Chen S, Kemppainen E, O'Dell KM, Jacobs HT. Gene expression in a Drosophila model of mitochondrial disease. PloS one. 2010;5:e8549. doi: 10.1371/journal.pone.0008549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tower J. Mitochondrial maintenance failure in aging and role of sexual dimorphism. Arch Biochem Biophys. 2015;576:17–31. doi: 10.1016/j.abb.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burger JM, Promislow DE. Sex-specific effects of interventions that extend fly life span. Sci Aging Knowledge Environ. 2004;2004:pe30. doi: 10.1126/sageke.2004.28.pe30. [DOI] [PubMed] [Google Scholar]
- 44.Argue KJ, Neckameyer WS. Altering the sex determination pathway in Drosophila fat body modifies sex-specific stress responses. Am J Physiol Regul Integr Comp Physiol. 2014;307:R82–R92. doi: 10.1152/ajpregu.00003.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, Yates E, Tatar M, Silverman N. Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J. 2013;32:1626–1638. doi: 10.1038/emboj.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 47.Wilson RH, Lai CQ, Lyman RF, Mackay TF. Genomic response to selection for postponed senescence in Drosophila. Mech Ageing Dev. 2013;134:79–88. doi: 10.1016/j.mad.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landis G, Shen J, Tower J. Gene expression changes in response to aging compared to heat stress, oxidative stress and ionizing radiation in Drosophila melanogaster. Aging (Albany NY) 2012;4:768–789. doi: 10.18632/aging.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girardot F, Lasbleiz C, Monnier V, Tricoire H. Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girardot F, Monnier V, Tricoire H. Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics. 2004;5:74. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill S, Van Remmen H. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol. 2014;2:936–944. doi: 10.1016/j.redox.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Droge W. Oxidative stress and aging. Adv Exp Med Biol. 2003;543:191–200. doi: 10.1007/978-1-4419-8997-0_14. [DOI] [PubMed] [Google Scholar]
- 55.Zhao HW, Zhou D, Haddad GG. Antimicrobial peptides increase tolerance to oxidant stress in Drosophila melanogaster. The Journal of biological chemistry. 2011;286:6211–6218. doi: 10.1074/jbc.M110.181206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci U S A. 2013;110:E1752–E1760. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christmas P. Role of Cytochrome P450s in Inflammation. Adv Pharmacol. 2015;74:163–192. doi: 10.1016/bs.apha.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, Gygi SP, Celniker SE, Obar RA, Artavanis-Tsakonas S. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huh JY, Kim Y, Jeong J, Park J, Kim I, Huh KH, Kim YS, Woo HA, Rhee SG, Lee KJ, Ha H. Peroxiredoxin 3 is a key molecule regulating adipocyte oxidative stress, mitochondrial biogenesis, and adipokine expression. Antioxid Redox Signal. 2012;16:229–243. doi: 10.1089/ars.2010.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pakharukova MY, Vavilin VA, Sripa B, Laha T, Brindley PJ, Mordvinov VA. Functional Analysis of the Unique Cytochrome P450 of the Liver Fluke Opisthorchis felineus. PLoS Negl Trop Dis. 2015;9:e0004258. doi: 10.1371/journal.pntd.0004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bae SH, Sung SH, Lee HE, Kang HT, Lee SK, Oh SY, Woo HA, Kil IS, Rhee SG. Peroxiredoxin III and sulfiredoxin together protect mice from pyrazole-induced oxidative liver injury. Antioxid Redox Signal. 2012;17:1351–1361. doi: 10.1089/ars.2011.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta. 2015;1847:1448–1456. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee KP, Shin YJ, Cho SC, Lee SM, Bahn YJ, Kim JY, Kwon ES, Jeong do Y, Park SC, Rhee SG, Woo HA, Kwon KS. Peroxiredoxin 3 has a crucial role in the contractile function of skeletal muscle by regulating mitochondrial homeostasis. Free Radic Biol Med. 2014;77:298–306. doi: 10.1016/j.freeradbiomed.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Li L, Yu AQ. The functional role of peroxiredoxin 3 in reactive oxygen species, apoptosis, and chemoresistance of cancer cells. J Cancer Res Clin Oncol. 2015;141:2071–2077. doi: 10.1007/s00432-015-1916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banmeyer I, Marchand C, Verhaeghe C, Vucic B, Rees JF, Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of Chinese hamster ovary cells: effects on cytotoxicity and DNA damage caused by peroxides. Free Radic Biol Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 66.Sabharwal SS, Waypa GB, Marks JD, Schumacker PT. Peroxiredoxin-5 targeted to the mitochondrial intermembrane space attenuates hypoxia-induced reactive oxygen species signalling. Biochem J. 2013;456:337–346. doi: 10.1042/BJ20130740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Promislow DE, Tatar M, Khazaeli AA, Curtsinger JW. Age-specific patterns of genetic variance in Drosophila melanogaster. I. Mortality. Genetics. 1996;143:839–848. doi: 10.1093/genetics/143.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.