Abstract
AIM
To examine the genetic profile of primary uveal melanoma (UM) as compared to UM in immune escape.
METHODS
Dendritic cells (DC) loaded with lysates of UM cells of high metastatic potential were used to stimulate CTLs(CTLs). When CTLs co-cultured with the UM cells, most UM cells could be eliminated. Survival UM cells grew slowly and were considered to be survival variants and examined by a microarray analysis. These differential genes were analyzed further with Venn Diagrams and functions related to immune escape. We additionally examined transcriptional changes of manually selected survival variants of UM cells and of clinical UM samples by quantitative real-time polymerase chain reaction (qRT-PCR), and analyzed the correlation of these expressions and patients' survival.
RESULTS
Gene expression analyses revealed a marked up-regulation of SLAMF7 and CCL22 and a significant down-regulation of KRT10, FXYD3 and ABCC2. The expression of these genes in the relapsed UM was significantly greater than those in primary UM. UM patients with overexpression of these genes had a shorter survival period as compared with those of their underexpression.
CONCLUSION
Gene expression, in particular of SLAMF7, CCL22, KRT10, FXYD3 and ABCC2, differed between primary UM cells and survival variants of UM cells.
Keywords: uveal melanoma, dendritic cells, immune escape, gene expression profile, SLAMF7
INTRODUCTION
Uveal melanoma (UM) is the most common malignant intraocular tumor in adults and the second most common type of melanoma[1]. The primary tumor can be treated by irradiation or by surgical resection, the survival of patients with UM is strongly linked to the development of distant metastases, which usually arise in the liver[2]. As a malignant intraocular tumor, UMs may benefit from the immune privilege of intraocular compartment. It has been assumed that the UM cells when metastasizing keep elements of the ocular immune privilege as protection against an attack and destruction by lymphocytes[3]. In addition, UM cells can escape the immune destruction by inhibiting the immunostimulatory function of dendritic cells (DC)[4], by impairing the T-cell function of programmed death-1 (PD-1) and programmed death-ligand 1 (PD-L1) interaction[5], and by autocrine secretion of the Fas ligand (FasL or CD95L)[6]. The development and propagation of UM cells can thus take place despite the activation of cytotoxic T-lymphocytes (CTLs) as response of the host immune system. Although the eye is an immunoprivileged site, enlarging UMs can show a dense lymphocytic infiltration[7]. Interestingly, the amount of the anti-tumor immune response characterized by a dense lymphocytic tumor infiltration including T-suppressor/CTLs[8], macrophages and regulatory T-cells has been correlated with the immune escape of the tumor cells[9]–[10].
To explain the high immunogenicity of UMs[11], previous studies analyzed the secretion of cytokines and chemokines which mediated the inflammatory responses[7], or inhibited the immune cell functions[4]–[6]. These studies revealed that these cytokines, chemokines and infiltrating immune cells could influence the inflammatory tumor microenvironment that not only helped the tumor to suppress immune reactions but also aided the growth of new blood vessels for supporting the tumor growth[12]. Immune cells involved in this process were Foxp3+ regulatory T-cells[10], myeloid-derived suppressor cells[13], and CD68+/CD163+ M2 macrophages[14]. However, these studies did not address the phenomenon of the immune escape of UM cells that can lead to extraocular metastases. They also have not examineed the changes occured in the survivial, variants of UM cells, such as alterations in gene expression or protein translation. Little was known about the gene expression profile of the immune escape variants of UM cells. As a consequence, molecular markers that would allow a selective targeting of UM cells and which could thus interrupt the metastatic cascade have not been developed yet. The development of tumor cell escape variants due to an immunoediting may also explain the failure of CTLs to eliminate tumor cells completely[15]. One of the other common mechanisms for the escape form CTLs associated immunosurveillance is the lack of surface expression of HLA class I molecules on UM cells and subsequent loss of the non-classical major histocompatibility complex (MHC) molecules MIC-A/B expression during the progression of UM cells[16]–[17]. UM cells lost HLA class I would be immunoselected under immune pressure.
MATERIALS AND METHODS
Cell Culture
The UM cell line MUM-2B was obtained from the Beijing Ophthalmology and Visual Science Key Laboratory. MUM-2B is an epithelial cell type with a high metastatic potential. It was maintained in RMPI-1640 (Gibco, Life Technologies Grand Island, NY, USA) with 10% fetal calf serum (Gibco, Life Technologies, Carlsbad, CA, USA) at 37°C in a 5% CO2 incubator.
Preparation of Human Dendritic Cells
Human immature DCs were prepared from peripheral blood mononuclear cells which were obtained from a healthy donor[18]. The DCs were prepared by a Ficoll/Paque density gradient centrifugation (Invitrogen, Life Technologies, Carlsbad, CA, USA). Peripheral blood mononuclear cells were seeded (9×106 cells/3 mL/well) into 6-well plates (Corning Costar Corp., Cambridge, MA, USA) in RPMI-1640 medium. After 1.5h of incubation at 37°C, adherent cells were used for the generation of DCs by adding granulocyte macrophage colony stimulating factor (1000 IU/mL, Peprotech, Rocky Hill, NJ, USA) and interleukin 4 (500 IU/mL, Peprotech).
At least 3 to 6 million MUM-2B cells in suspension were smashed by an ultrasonic cell disrupter (Safer Co., Nanjing, China) to prepare cells as antigens[19]. Tumor antigen protein concentrations were determined using Bio-Rad protein assay reagents (Bio-Rad Laboratories, Hercules, CA, USA)[19].
T-cell Stimulation and Cytotoxic T-cell Assays Against High Metastatic Potential Uveal Melanoma Cell in Vitro
The non-adherent lymphocytes were stimulated with autologous DCs loaded with MUM-2B lysates at a ratio of 5:1. The MUM-2B lysates and the DCs were co-cultured overnight before they were seeded with the lymphocytes. Interleukin-2 (300 IU/mL; Peprotech) was added to the cell cultures the next day and every 3d thereafter. The lymphocytes were re-stimulated with DCs every 5d for up to 2 stimulations, which consistently resulted in more than 90% of CD3+ T-cells[20]–[21]. Cytotoxic T-lymphocyte-mediated MUM-2B cytotoxicity was tested using the lactate dehydrogenase release assay (CytoTox 96 Non-Radioactive Cytotoxicity Assay, Promega Co., Madison, WI, USA) according to the manufacturer's protocol. Target cells were the cells of the high metastatic potential UM cell line MUM-2B. The ratio of effector and target cells was 2.5:1, 5:1, 10:1, 20:1 and 40:1.
Flow Cytometry Analysis
For flow cytometry analysis, one million cells were stained with a solution containing 1% pharmingen stain buffer (BSA), 2 mmol ethylenediaminetetraacetic acid (EDTA), and flow antibodies for 30min at 4°C.
We used the following antibodies: anti-CD11c (conjugated with fluorescein isothiocyanate), anti-CD83 (conjugated with phycoerythrin), HLA-DR (conjugated with peridinin-chlorophyll-protein), anti-CD86 (conjugated with phycoerythrin), anti-CD40 (conjugated with fluorescein isothiocyanate), and anti-CD80 (conjugated with phycoerythrin) (BioLegend, Inc., San Diego, CA, USA). The stimulated T-cells used anti-CD8 (conjugated with fluorescein isothiocyanate), anti-CD56 (conjugated with phycoerythrin), and CD3 (conjugated with peridinin-chlorophyll-protein), anti-CD25 (conjugated with fluorescein isothiocyanate), and anti-CD4 (conjugated with phycoerythrin) (BioLegend, Inc.). Mouse fluorescein isothiocyanate-IgG1, phycoerythrin-IgG1 and peridinin-chlorophyll-protein-IgG1 were used as isotype controls respectively. The flow cytometric analysis was carried out with a BD-FACSCalibur device (BD Biosciences, San Jose, CA, USA). The data were analyzed with the Cell Quest software (Becton Dickinson, Heidelberg, Germany). A minimum of 100 000 viable cells were measured per sample. Using forward and side scatter profile, debris and dead cells were gated out.
Cell Proliferation Assay
In the cytotoxic experiment of killing UM cells, T cells were washed by used 1×PBS (pH=7.4) because T cells were suspension growth, so we collected these survival MUM-2B cells which were adherent growth. For un-interfusing T cells, we sorted CD3+ T cells and obtained single MUM cells by using MACS method[22]–[23]. To perform a cell proliferation assay, both the survival and original MUM-2B cells were seeded in 96-well plates in a concentration of 1000 cells/well. The cell medium also included CTLs which were primed by DCs which were loaded by MUM-2B. The cell counting kit-8 (CCK8) assay (Biyuntian, Beijing, China) was used to determine the relative cell growth every 6, 12, 24 or 48h. Ten microliter of CCK8 solution was added to the media for incubation for 2h at 37°C. The absorbance at 450 nm was measured by a Microplate Reader (Bio-Rad, Hercules, CA, USA).
Preparation of RNA from Original Uveal Melanoma Cell and Its Survival Cells
Both the survival and original MUM-2B cells were collected for extracting total RNA that used for microarray analysis. Total RNA was extracted by using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) and it was purified with an RNA Isolation Kit (Ambion, Austin, TX, USA). The purity and concentration of RNA were determined from OD 260/280 (optical density at 260 nm/optical density at 280 nm) readings using a spectrophotometer (NanoDrop ND-1000, Thermo Specific Co., Wilmington, DE, USA). The RNA integrity was determined by a 1% formaldehyde denaturing gel electrophoresis. Only RNA extracts with RNA integrity number values >6 were included in the in further analysis.
Gene Expression Profiling
The gene expression profiling was performed using the Affymetrix Human Transcriptome Array 2.0 array (Affymetrix Co., Santa Clara, CA, USA). The human transcriptome array was designed with 10 per exon and 4 per junction, with approximately 4.8 million probes selected from all 98 million possible 25 mers. The array contained 7 million probes, so that the comprehensive transcriptome detected more than 0.28 million transcripts including more than 0.24 million coding transcripts and more than 0.04 million non-coding transcripts.
Microarray Data Analysis
The Affymetrix HTA 2.0 arrays were hybridized according to Affymetrix recommendations using the Ambion WT protocol (Life Technologies, Carlsbad, CA, USA) and the Affymetrix labeling and hybridization kits. One hundred ng of total RNA were processed in parallel with an external microarray quality control (MAQC) A RNA to control the robustness of data. The labeled DNA mean yield was 7.19 µg (minimum: 6.27 µg; maximum: 7.57 µg). The Affymetrix GeneChip® Human Transcriptome 2.0 microarrays (HTA2) were hybridized with 4.7 µg of labeled DNA. The raw data, transcript data and the exon data were controlled with the Expression console (Affymetrix) at the Institute Curie microarray core facility (Santa Clara, CA, USA).
The Affymetrix HTA2 dataset was analyzed by the Transcriptome Analysis Console (TAC) 2.0 (www.Affymetrix.com)[24]–[25]. We performed an unpaired Student's t-test to compare gene intensities between the primary MUM-2B cells and the survival variants of MUM-2B cells. Genes were considered to be significantly differentially expressed when the fold-change was ≥2 and when the P-value was <0.05 (unadjusted P-value). The results were considered statistically significant for unadjusted P-values <0.05 and for fold-changes of ≥2 for the expressing analysis. After bioinformatic analysis of the microarray data, a manual inspection using the Molecular Annotation System was conducted to select differential genes, its gene ontology and its pathways. The data was logarithmically (log2) transformed and the median centered by genes using the adjust data function of the CLUSTER 3.0 software was then further analyzed with hierarchical clustering with average linkage.
For screening the candidate genes, these different expressed genes were analyzed further with Venn Diagrams and functions related to tumor metastatic and immune escaping[23].
Target Genes Validation by Quantitative Real-time Polymerase Chain Reaction
We screened the candidate genes which were statistically significant for unadjusted P-values <0.05 and for fold-changes of ≥2 for the expressing analysis. i.e. SLAMF7, CCL22, AQP9, KRT10, FXYD3 and ABCC2, and validated further by quantitative real-time polymerase chain reaction (qRT-PCR). A cDNA synthesis was carried out using a PrimeScript II 1st strand cDNA Synthesis Kit (TaKaRa, Dalian, China). A qRT-PCR was performed using the ABI 7300 sequence detection system (Applied Biosystems, Foster City, CA, USA) in a volume of 20 µL containing 10 µL GoTaq® qPCR Master Mix (Promega, Madison, WI, USA) and 0.2 µm of each primer of the tested target genes. SLAMF7 fw: 5′-ATGTCTACGAGCACCTGTCAAAGCC-3′, rev: 5′-GGGCCTTCCAGGTATAAATCACATC-3′; CCL22 fw: 5′-TTCTGGGTCTTTGCCTGGGATG-3′, rev: 5′-CCAAGAATCTGCGGAGACTGTGA-3′; AQP9 fw: 5′-AGAGCAGCTTAGCGAAAGAAACCC-3′, rev: 5′-GATAGTGATGACCCCTCCAAAACG-3′; KRT10 fw: 5′-CTTGGCAGAAACAGAAGGTCGCT-3′, rev: 5′-GCAGGCTGCGGTAGGTTTGAA-3′; FXYD3 fw: 5′-AGAGTTCCTGCCGCTAAGATTTCC-3′, rev: 5′-TTCCATTTGGGGGGTTGTGC-3′; ABCC2 fw: 5′-GCTGGAAAGTCATCCCTCACAAAC-3′, rev: 5′-GTGGAGCCCAATGGAAGCAATA-3′; Actin fw: 5′-ACTTAGTTGCGTTACACCCTT-3′, rev: 5′-GTCACCTTCACCGTTCCA-3′. The reaction profile consisted of 40 cycles at 95°C for 2min, at 55°C for 30s, and at 72°C for 30s. A dissociation stage was performed at the end of the reaction consisting of 200 cycles of 7s with the temperature increased at 0.2°C /cycle to demonstrate the specificity of the amplification. The expression analysis was performed in triplicate for each sample. The housekeeping actin was used as the normalization control. The fold difference for each sample was obtained using the equation 2−dCt, where Ct is the threshold cycle (the cycle number at which the fluorescence generated within a reaction crosses the threshold) and dCt equals the mean Ct of the sample gene minus the mean Ct of actin.
Clinical Tumor Samples and Controls
Fresh tumor samples were obtained from 10 UM patients at the time of eye removal or tumor resection. There were 3 patients (“relapsing UMs”) who had undergone laser therapy and radiotherapy of the UMs prior to enucleation of the eyes. In the remaining 7 patients, the tumor had not been treated prior to the enucleation. The tissue samples were immediately frozen in liquid nitrogen and stored at -80°C until processing. The diagnosis of UM was confirmed by a surgical pathologist for all patients. The UM samples were analyzed for target genes and protein expression by performing qRT-PCR and Western blotting.
Statistical Analysis
The statistical analysis was performed using a commercially available statistical analysis program (SPSS 21.0, IBM-SPSS; Chicago, IL, USA). Data were presented as mean±standard deviation from three separate experiments. Statistical significances of difference of means throughout this study were calculated by Student's t-test in comparing data between two groups and ANOVA one-way test in comparing data from more than two groups. Correlations between the expressions of five genes were evaluated by bivariable analyses using log rank test of Kaplan-Meier methodology. Differences between groups were considered to be statistically significant with a P-value of <0.05.
Ethics Statement
All experiments and procedures were performed in accordance with the appropriate guidelines. All procedures were approved by Capital Medical University. Informed consent was obtained from all volunteers before being enrolled in the study.
RESULTS
Production of Dendritic Cells Loaded with Uveal Melanoma Cell Line (MUM-2B) Elicits Specific Cytotoxic T-lymphocytes in Vitro
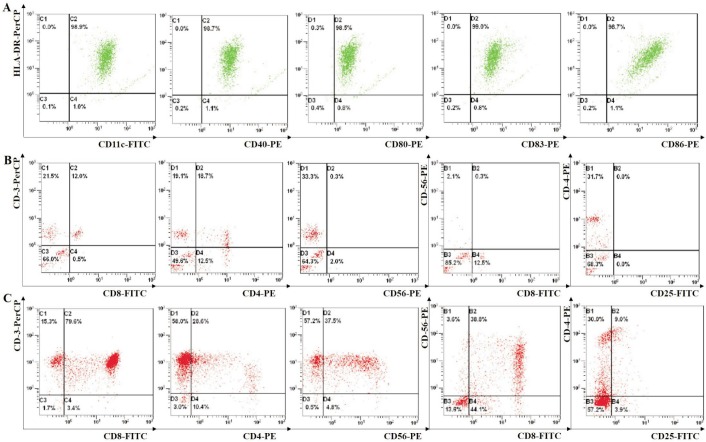
To determine the effect of the vaccination of the DCs, DCs were isolated from peripheral mononuclear blood cells of a healthy donor. They were primed in vitro using autologous DCs pulsed with MUM-2B lysates. The DCs then highly expressed the DC markers of CD11c and HLA-DR and the co-stimulatory molecules of CD80, CD86, CD83 and CD40 (Figure 1A). The lymphocytes were re-stimulated with DCs every 5d. The stimulated T cells were then tested for cells phenotypes by flow cytometry[26]. Before DCs stimulated T-lymphocytes which were derived from the same healthy donor. The phenotypes of T-lymphocytes were low level correspondingly (Figure 1B). But the stimulated T cells contained CD3+ in 94.9% of the cells, CD8+ in 79.6% of the cells, and CD3+/CD56+ in 37.5% of the natural killer T (NKT) cells, while only 28.6% of the cells expressed CD4+, and 9% of the cells were regulatory T-cells (CD4+/CD25+) (Figure 1C). It showed that DCs loaded with MUM-2B lysates stimulated the proliferation and activation of CD8+ T-lymphocytes.
Figure 1. Flow cytometric results of DCs and their activated CTLs.
A: DCs were cultured by inducing peripheral mononuclear blood cells which were derived from healthy donors. DCs were loaded with lysates of MUM-2B and matured. They then highly expressed CD11c+/HLA-DR+, and the co-stimulatory molecules CD40+, CD80+, CD83+ and CD86+. B: Before DCs stimulated T-lymphocytes which were derived from the same healthy donor. The phenotypes of T-lymphocytes had CD3+, CD8+, and CD3+/CD56+. C: DCs loaded with lysates of MUM-2B cells, stimulated proliferation and activation of T-lymphocytes which were derived from the same healthy donor. The resulting CTLs highly expressed CD3+, CD8+, and CD3+/CD56+, while in contrast CD4+ was expressed to a lower degree.
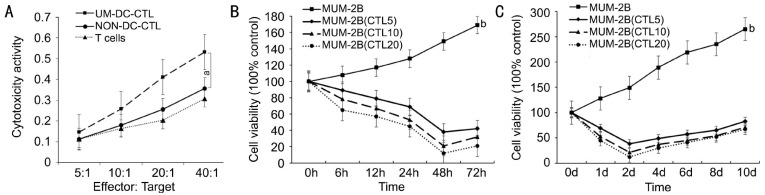
The stimulated cells were then tested for their cytotoxic ability against high metastatic potential UM cell (MUM-2B) by a cytotoxicity assay of the cytotoxic T-lymphocytes. We included CTLs which were stimulated by DCs pulsed with MUM-2B lysates (UM-DC-CTLs), CTLs stimulated by DCs without antigens (NON-DC-CTLs), and non-stimulated T-lymphocytes. With the increase of Effector/Target (E/T) ratio, the cytolysis against MUM-2B cells increased[23]–[24]. It showed that the cytotoxicity of UM-DC-CTLs was significantly higher than the cytotoxicity of NON-DC-CTLs and the cytotxicity of T-cells (Figure 2A). Moreover the cytotoxicity of NON-DC-CTLs did not differ significantly from the cytotoxicity of T-cells against the survival UM cells.
Figure 2. The cytotoxic activities against MUM-2B by T-lymphocytes activated by DCs pulsed with MUM-2B lysates.
A: The cytotoxic activities of DCs against MUM-2B by pulsing with MUM-2B lysates (UM-DC-CTL) and no antigens (NON-DC-CTL), and T-cells respectively. The cytotoxicity of UM-DC-CTL was significantly higher than of NON-DC-CTL and T-cells against UM cells (aP<0.05). B, C: The proliferation of MUM-2B cells was inhibited under the condition of immune suppression by the cytotoxic lymphocytes (bP<0.000).
Effect of Uveal Melanoma Cell Proliferation by Immune Function of Dendritic Cells Activating Cytotoxic T-lymphocytes
To test the proliferation of the UM cells under immune suppression by the DCs activated CTLs, we examined the MUM-2B viability at different time points[23]–[24]. We found that the growth of the MUM-2B cells decreased gradually. Viability was lowest at two days after co-culture with the CTLs, and then slightly increased for the next few days (Figure 2B, 2C). The proliferation of the primary MUM-2B cells differed significantly (P<0.01) from the proliferation of the MUM-2B cells under immune suppression (Figure 2B, 2C). The MUM-2B cells still survived at two days at the greatest E/T ratio, and their growth recovered by 67% at 8d later (Figure 2C).
Gene Expression Signature Discriminates Between Original Uveal Melanoma Cells and Its Survival Cells
In the gene expression analysis, samples were clustered into two groups, i.e. the group of survival variants of MUM-2B cells and the group of the original MUM-2B cells as normal controls. The survival variants of MUM-2B cells were obtained from surviving UM cells at a E/T ratio of 5/1 (group C1), a E/T ratio of 10/1 (group C2), and a E/T ratio of 20/1 (group C3). The mean expression of each gene was compared between the three survival variants of MUM-2B cell groups and the control group. With a P-value of <0.01 used as a cut-off, 824 genes were overexpressed and 69 genes were underexpressed with a ≥2.0 fold-change in merge of the C1/N, C2/N and C3/N group (P<0.01) (Figure 3).
Figure 3. Expression of mRNAs in MUM-2B cell line suppressed by CTLs activated by DCs, and in primary MUM-2B cell line.
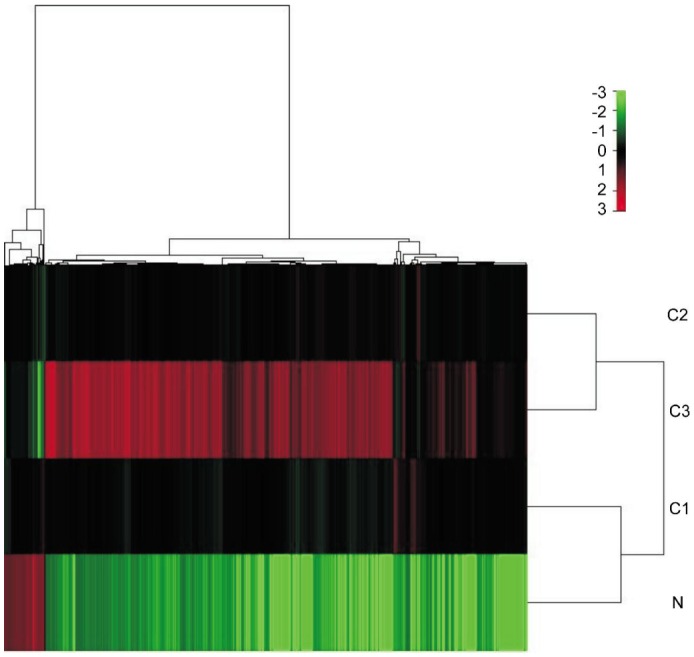
Hierarchical clustering of 893 genes showed a significantly different expression (P<0.01) in tumor cells. Row 5 represents individual genes; columns represent individual UM cells samples. The scale represents the intensity of gene expression (log2 scale ranges between 3.0 and 3.0). C1, C2 and C3 represented surviving MUM-2B cells under suppression by cytotoxic lymphocytes which activated DCs at three ratios of effectors and targets, i.e. 5/1, 10/1 and 20/1 respectively.
Gene Function Analysis and Differentially Expressed Immune Response Genes in Survival Uveal Melanoma Cells
The analysis of gene functions of differentially expressed immune response genes in the survival variants of UM cells versus the original UM cells revealed genes in the field of antigen processing and antigen presentation (109 genes, P=0.00), response to stress (226 genes, P=0.00), regulation of a defense response (70 genes, P=0.00), chemotaxis (27 genes, P=3.81×10−4), regulation of inflammatory response (40 genes, P=0.00), T-cell activation (29 genes, P=1.97×10−14), regulation of regulatory T cell differentiation (6 genes, P=1.33×10−6), regulation of interleukin-6 production (22 genes, P=1.97×10−15), regulation of interleukin-8 production (11 genes, P=3.25×10−7), regulation of interleukin-10 production (5 genes, P=2.33×10−3), and positive regulation of transforming growth factor beta production (2 genes, P=0.04).
Differential genes related to antigen processing and presentation included 109 genes which were differentially expressed (P<0.05) in merge of C1/N, C2/N and C3/N group (Table 1). These genes were up-regulated in all survival variants of MUM-2B cells. There were three genes involved in positive regulation (CD74) and negative regulation (CCR7, CD47) of DC antigen processing and presentation pathways. There were 226 differentially expressed genes which were associated with response to stress (P<0.05) (Table 1). Most of these genes are involved in the defense response, immune response and cell communication. The genes AQP9 and SLAMF7 were the genes with the most marked increase. Down-regulation of KRT10 and ABCC2 related to drug-resistance of cancer medication.
Table 1. The summary of significant overexpression and underexpression genes which were differently expressed in MUM-2B suppressed by CTLs activated by DCs.
| Significant overexpressed genes |
Significant underexpressed genes |
|||||||||
| Probe Set ID | Entrez gene | Gene symbol | Fold change | P | Probe Set ID | Entrez gene | Gene symbol | Fold change | P | |
| Antigen processing and presentation | ||||||||||
| TC01001382.hg.1 | 2207 | FCER1G | 125.13 | 0 | ||||||
| TC17001466.hg.1 | 1236 | CCR7 | 7.15 | 0 | ||||||
| TC19000174.hg.1 | 3383 | ICAM1 | 10.7 | 0 | ||||||
| TC03001629.hg.1 | 961 | CD47 | 2.32 | 0 | ||||||
| TC05001936.hg.1 | 972 | CD74 | 30.85 | 0 | ||||||
| Response to stress | ||||||||||
| TC01001372.hg.1 | 57823 | SLAMF7 | 184.97 | 0.02 | TC01002166.hg.1 | 54206 | ERRFI1 | -6.44 | 0 | |
| TC06000371.hg.1 | 7124 | TNF | 6.4 | 0 | TC01006383.hg.1 | 4582 | MUC1 | -6.59 | 0 | |
| TC09000035.hg.1 | 3717 | JAK2 | 6.86 | 0 | TC05000701.hg.1 | 1958 | EGR1 | -7.43 | 0 | |
| TC14000133.hg.1 | 4323 | MMP14 | 14.06 | 0 | TC17001472.hg.1 | 3858 | KRT10 | -36.96 | 0 | |
| TC08000305.hg.1 | 3620 | IDO1 | 5.23 | 0 | TC10000715.hg.1 | 1244 | ABCC2 | -17.81 | 0 | |
| TC15000441.hg.1 | 366 | AQP9 | 132.24 | 0 | ||||||
| Inflammatory response | ||||||||||
| TC03001355.hg.1 | 1230 | CCR1 | 20.07 | 0 | TC19000458.hg.1 | 5349 | FXYD3 | -22.76 | 0 | |
| TC16000487.hg.1 | 6367 | CCL22 | 136.23 | 0 | ||||||
| TC17000406.hg.1 | 6362 | CCL18 | 25.84 | 0 | ||||||
| TC04000408.hg.1 | 3576 | IL8 | 47.89 | 0 | ||||||
| TC02000937.hg.1 | 7130 | TNFAIP6 | 93.09 | 0 | ||||||
| TC07000687.hg.1 | 5294 | PIK3CG | 32.25 | 0 | ||||||
| TC10001559.hg.1 | 118788 | PIK3AP1 | 20.88 | 0 | ||||||
| TC12000611.hg.1 | 4069 | LYZ | 494.38 | 0 | ||||||
| T cell mediated cytotoxicity and T cell activation | ||||||||||
| TC05002050.hg.1 | 3937 | LCP2 | 14.72 | 0 | TC05000372.hg.1 | 2150 | F2RL1 | -6.55 | 0 | |
| TC13000642.hg.1 | 3936 | LCP1 | 20.3 | 0 | TC15001471.hg.1 | 4734 | NEDD4 | -6.6 | 0 | |
| TC01003006.hg.1 | 26191 | PTPN22 | 26.56 | 0 | ||||||
| TC12000955.hg.1 | 5027 | P2RX7 | 27.82 | 0 | ||||||
| TC01003407.hg.1 | 8832 | CD84 | 30.89 | 0 | ||||||
| TC01001640.hg.1 | 5788 | PTPRC | 65.05 | 0 | ||||||
| TC16000349.hg.1 | 3683 | ITGAL | 18.59 | 0 | ||||||
| TC19000174.hg.1 | 3383 | ICAM1 | 10.7 | 0 | ||||||
| Natural killer cell mediated cytotoxicity | ||||||||||
| TC01001372.hg.1 | 57823 | SLAMF7 | 184.97 | 0.02 | TC06002224.hg.1 | 79465 | ULBP3 | -8.53 | 0.02 | |
| TC12000101.hg.1 | 5777 | PTPN6 | 9.51 | 0.02 | ||||||
| TC01003996.hg.1 | 1130 | LYST | 5.46 | 0.02 | ||||||
Differential genes related to inflammation (cytokines and chemokines) were genes encoding macrophage- and T-lymphocyte-attractant chemokines and cytokine receptors (CCL3 and CCL3L3, CCR1, CCR4, CCL18, CCL22, IL8, etc), molecules related to cytokine-signaling pathways (TNFRSF1B, TNFAIP6, PIK3CG, JAK2, PIK3AP1), proteins implicated in the extravasation of leucocytes from blood to tissues (CSF1, ITGAL, ICAM1, P2RX7) and molecules that contribute to inflammation (LYZ, HCK, CXCL10,and Down-regulation of FXYD3 ) (Table 1).
Differential genes related to T-lymphocytes included those encoding for molecules that participate in the signal transduction through the T-cell receptor TCR (LCP1, LCP2, XCL1, and PTPN22); proteins involved in T-cell activation and proliferation (IL7R, CD84 and P2RX7), natural killer cell mediated cytotoxicity markers (SLAMF7, PTPN6, TGFB1 and IDO1), and T-cell mediated cytotoxicity markers (PTPRC, ITGAL, ICAM1, GNLY, CCL2) (Table 1). Genes expressed in activated immune cells that were negatively regulated by the TGF-β pathway (XCL1 and ATP6AP2), the interleukin-10 pathway (XCL1, PLA2G7, IDO1, CD48 and CD40LG), the interleukin-8 pathway (BTN3A1, CD40LG, TLR8, PPAP2B, CD80 and DOCK11), the interleukin-6 pathway (PLA2G7, TAP1, BTN3A1, TNFAIP3, HLA-B, and CD40LGl) and regulatory T-cell pathway (CTLA4, TGFB1, CD28, HLA-G, and FOXP3). These negative regulatory pathways participated in mediating tumor survival.
Validation of the Differentially Expressed Genes by Quantitative Real-time Polymerase Chain Reaction
Based on the gene ontology and pathways analysis of differential genes, we selected six candidate target genes by screening further of Venn Diagrams and gene functions. These candidate target genes were three up-regulated genes (SLAMF7, CCL22 and AQP9) and three down-regulated genes (KRT10, FXYD3 and ABCC2). Table 2 showed that SLAMF7, CCL22 and AQP9 were still highly expressed in survival variants of MUM-2B at E/T ratios of 5/1 (CTL5), 10/1 (CTL10) and 20/1 (CTL20). KRT10, FXYD3 and ABCC2 were expressed to a lower degree in the survival variants of MUM-2B cells, but only the expression of KRT10 showed a significant difference to the expression in the primary MUM-2B UM cells.
Table 2. The fold difference of target gene expression as detecting qRT-PCR in MUM-2B suppressed by CTLs activated by DCs.
| Genes | MUM-2B | MUM-2B(CTL5) | MUM-2B(CTL10) | MUM-2B(CTL20) | P |
| SLAMF7 | 1.02 | 5464.92 | 6231.42 | 4956.53 | 0.00b |
| CCL22 | 1.00 | 3539.65 | 3495.07 | 3528.87 | 0.00b |
| AQP9 | 1.00 | 2375.65 | 3542.17 | 2427.00 | 0.00b |
| KRT10 | 1.00 | 0.50 | 0.65 | 0.50 | 0.00b |
| FXYD3 | 1.00 | 0.91 | 0.89 | 0.89 | 0.23 |
| ABCC2 | 1.00 | 0.94 | 0.96 | 0.79 | 0.45 |
bP≤0.01 survival variants of MUM-2B versus original MUM-2B.
Differential Genes Validation in Clinical Uveal Melanoma Samples by Quantitative Real-time Polymerase Chain Reaction
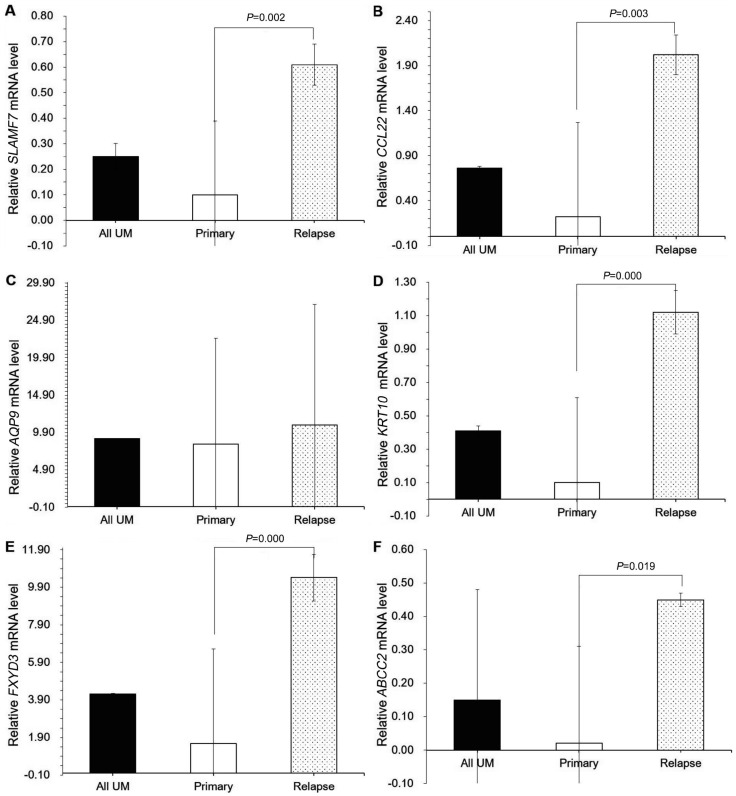
For the validation of the differential genes we collected UM cells from 10 patients with UMs samples (Table 3). We found that the expression of SLAMF7 was significantly higher in the relapsing UM cells than in the primary UM cells (Table 4; Figure 4A). The expression of CCL22 was higher in the relapsing UM cells than in the primary UM cells and in the UM tumor tissue (Table 4; Figure 4B). The expression of AQP9 did not differ significantly between the groups. The expression of KRT10 was significantly (P<0.01) lower in primary tumors than in relapsing tumors. In addition, the expression of KRT10 differed between the various subtypes of primary UMs (Table 4; Figure 4C). The expression of FXYD3 was significantly (P<0.001) higher in the relapsing UM cells than in the primary UM cells (Table 4; Figure 4E). ABCC2 was expressed significantly higher in the relapsing UM cells than in primary UMs (Table 4; Figure 4F).
Table 3. The clinical and pathological characteristics of the patients with UM.
| Variables | Values | n (%) |
| Sex | M | 11 (55) |
| F | 9 (45) | |
| Age (a) | ≤35 | 8 (40) |
| 35<Y≤60 | 12 (60) | |
| >60 | 2 (10) | |
| Location | Left choroid | 9 (45) |
| Right choroid | 11 (55) | |
| Tumor cytology | Spindle | 6 (30) |
| Epithelial | 4 (40) | |
| Mixed | 10 (60) | |
| Scleral invasion | Yes | 12 (60) |
| No | 8 (40) | |
| Recurrence | Yes | 7 (35) |
| No | 13 (65) | |
| Survival status | Dead | 11 (55) |
| Live | 9 (45) | |
| Survival time (mo) | 0-12 | 2 (10) |
| 12-24 | 4 (20) | |
| 24-36 | 5 (25) | |
| ≥36 | 9 (45) |
Table 4. The fold difference of target gene expression between uveal melanoma tumor tissue and normal choroidal tissues.
| Genes | UM tumor cytology |
1P; 2P | Tumor relapse |
P | |||
| Spindle-cell type (n=4) | Mixed cell type (n=5) | Epitheloid cell type (n=4) | Yes (n=7) | No (n=13) | |||
| SLAMF7 | 0.08±0.10 | 0.13±0.10 | 0.08±0.00 | 0.77; 0.50 | 0.61±0.30 | 0.10±0.08 | 0.00 |
| CCL22 | 0.38±0.31 | 0.13±0.09 | 0.22±0.29 | 0.16; 0.38 | 2.02±0.1.19 | 0.22±0.22 | 0.00 |
| AQP9 | 3.77±2.28 | 16.01±24.87 | 1.40±1.52 | 0.73; 0.37 | 10.89±10.86 | 8.34±16.12 | 0.81 |
| KRT10 | 0.12±0.02 | 0.03±0.03 | 0.19±0.26 | 0.01; 0.74 | 1.12±0.21 | 0.1±0.13 | 0.00 |
| FXYD3 | 0.10±0.33 | 1.45±1.12 | 2.22±2.31 | 0.71; 0.57 | 10.42±5.30 | 1.57±1.24 | 0.00 |
| ABCC2 | 0.02±0.02 | 0.02±0.02 | 0.03±0.04 | 0.99; 0.80 | 0.45±0.42 | 0.02±0.02 | 0.02 |
1P: Mixed cell type vs Spindle-cell type; 2P: Epitheloid cell type vs Spindle-cell type.
Figure 4. The fold difference of target genes obtained in UM tissues.
A: SLAMF7 was up-expressed in relapsing tissues and significantly higher than in primary UM tissues; B: CCL22 was expressed highly in relapsing tissues and significantly higher than in primary UM tissues; C: The expression of AQP9 in mixed type UM tissues was highest in these tissues. However, the expression of AQP9 hadn't significantly difference among all tissues; D: The expression of KRT10 in relapsing tissues was highest among UM tissues. KRT10 also was expressed highly in relapsing tissues and significantly higher than in primary UM tissues; E: The expression of FXYD3 was the highest in relapsing tissues. The expression of FXYD3 in relapsing UM tissues had evidently difference with in primary UM tissues. Additionally, the expression of FXYD3 in relapsing UM tissues has relevance with mixed type UM; F: ABCC2 also was up-expressed in relapsing tissues and significantly higher than in primary UM tissues.
Differential Genes Expression Correlated with Patients' Survival
Twenty UM patients were collected and their samples were detected expression of SLAMF7, CCL22, KRT10, FXYD3 and ABCC2 by qRT-PCR. The cut-off value of these genes was 2−dCt=0.23 of SLAMF7, 2−dCt=0.69 of CCL22, 2−dCt=0.45 of KRT10, 2−dCt=4.53 of FXYD3 and 2−dCt=0.07 of ABCC2, so distinguish overexpression from underexpression. There were these genes overexpression of 7 UM patients and these genes underexpression of other 13 UM patients. The survival time showed significant difference between both patients by Kaplan-Meier survival curves (log-rank test, P=0.000) (Figure 5). All patients of these genes overexpression occurred UM relapse and liver metastasis. The result elucidated that these genes could be used as targets to locate diagnostic, prognostic and therapeutic biomarkers for UM.
Figure 5. Kaplan-Meier survival curves according to the level of genes including SLAMF7, CCL22, KRT10, FXYD3 and ABCC2.
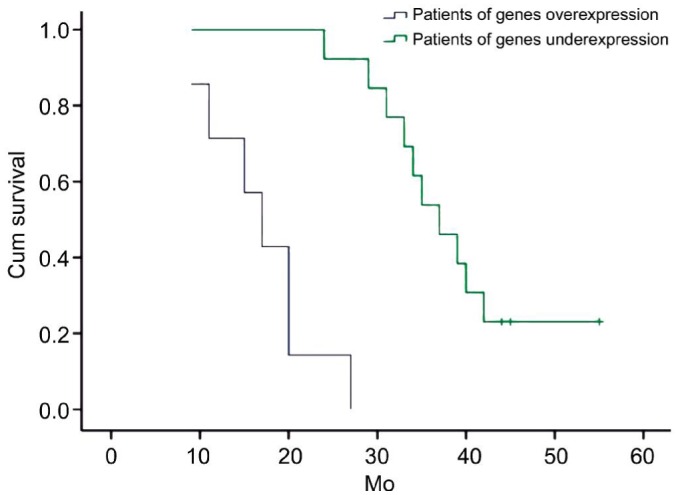
(Green: Patients of genes underexpression; Blue: genes overexpression, P=0.000) as the patients of genes overexpression show a much more relapsing clinical course (Median survival time: 72.0 vs 39.1mo, respectively).
DISCUSSION
In our experimental study, we cultured mature DCs derived from peripheral blood mononuclear cells of healthy donors. DCs were pulsed with lysates of UM cells of high metastatic potential (MUM-2B cell line) and were used to stimulate CTLs. The mature DCs highly expressed co-stimulatory molecules, and the activated CTLs highly expressed more than 90% CD3+, more than CD70% CD8+, and more than 30% CD3+/CD56+ NKT-cells (Figure1A, 1B). When mixed with the primary UM cells, the CTLs killed the majority of the MUM-2B cells within two days (Figure 2A, 2B), while about 10% of the UM cells survived and slowly proliferated under the CTLs associated immunization for 10d (Figure 2C). One may assume that the surviving UM cells escaped the immune response activated by the DCs. We then used microarray samples of these survival variants of UM cells and compared them with microarray samples of the original MUM-2B cells and analyzed differences in expressed genes and their molecular function and pathways which were closely related to the survival (Figure 3).
The survival variants of UM cells differentially expressed 109 genes associated with the antigen processing and presentation pathways which were involved in the positive regulation and negative regulation of DCs (Table 1). Important molecules were CD47 and CCR7 for the negative regulation, and MHC class II molecules that induced antigen-specific CD4+ T-cell tolerance[27]–[29]. CD47 is generally considered as an anti-phagocytotic signal expressed by cancer cells to prevent macrophages and DCs from attacking these tumor cells[27]. We found 226 genes which were differentially expressed and which were associated with the response to stress involving defense response, immune response, and cell communication (Table 1). Interestingly, we detected that AQP9 and SLAMF7 were up-regulated in the survival variants of UM cells as compared with the corresponding primary UM cells. The unique property of AQP9 is associated with an increased metabolic demand, such as spontaneous labor and inflammation[30], which plays a pivotal role in various arsenide-mediated cytocidal effects on cancer cells[31]. SLAMF7 inhibits natural killer cell activation and cytotoxicity by limiting antibody-dependent cell-mediated cytotoxicity[32]–[33]. Additionally, the down-expression level of KRT10 and ABCC2 was the first and second gene that was involved in drug-resistance of cancer[34]–[36]. PTEN improves cisplatin-resistance of human ovarian cancer cells through up-regulating KRT10 expression[34]. ABCC2 is associated with lower differentiation of cancer cells and their resistance to the cisplatin[35], moreover its overexpression is responsible for the drug-resistance of cancer stem cells[36].
The gene expression profiles revealed that the survival mechanism of UM cells was interrelated with signal transduction, such as T-cell activation and proliferation, and natural killer cell mediated cytotoxicity markers, and T-cell mediated cytotoxicity markers (Table 2). Negatively regulated pathways in activated T-cells involved in the TGF-β pathway, interleukin-10 pathway, tinterleukin-8 pathway, interleukin-6 pathway and regulatory T-cell pathway among others. In particular natural killer cells are well-developed and can contribute to the immune surveillance of tumors[37], thus SLAMF7[32]–[33], PTPN6[38], TGFB1[34],[39], and IDO1 inhibit the natural killer cell proliferation and the natural killer cell mediated cytolysis[40].
There were 29 differential cytokines and chemokines which were found to join the immune inflammation (Table 1). The presence of CTLs in UMs may be a necessary but not sufficient condition for tumor rejection. In fact, the dense infiltration of lymphocytes in UMs persists after immunoedition, probably recruited or locally expanded by the elevated expression of these specific factors in the UM microenvironment. High-expression of CCL22 and down-expression of FXYD3 in our study was of particular interest, because CCL22 belongs to macrophage-derived chemokines and regulatory T-cell cell attracting chemokines[41]–[42], which generate a microenvironment that may favor survival and growth of malignant cells, and overexpression of FXYD-3 may play an important role in the tumorgenesis and development by transforming growth factor-β signaling through ZEB1/δEF1 in human cancers[43]–[44].
Based on the gene ontology and pathways analysis of differential genes as described above, we selected three up-regulated differential genes (SLAMF7, CCL22 and AQP9) and three down-regulated differential genes (KRT10, FXYD3 and ABCC2) which were validated in the survival variants of MUM-2B cells and in the original MUM-2B cells again (Table 3). Six differential genes were detected in the UM tumor tissue samples of 10 patients (Table 4; Figure 4). In addition, the expression of both SLAMF7 and ABCC2 were significantly higher in the relapsing UM tissues than in the primary UM cells. Similarly, the expression of both CCL22 and KRT10 were higher in relapsing UM cells than that in primary UM cells (Table 4; Figure 4B, 4D). The expression of FXYD3 also was higher in relapsing tissues than in the primary UM tissues (Table 4; Figure 4E). In contrast, the expression of AQP9 did not differ significantly between the groups. We found that KRT10, FXYD3 and ABCC2 were down-regulated under immune suppression by DC stimulated CTLs, but they were up-regulated in survival variants of UM cells. Three genes are responsible for drug-resistance of cancer cells[34]–[35],[45]–[47], so that they can be up-regulated in the relapsing UM tissues, although the mechanisms have remained unclear yet. We collected relapsing UM samples of these patients who had undergone radiotherapy and laser therapy, so the expression of three genes mediated radiotherapy-resistance of UM[34]–[35],[45]–[47].
Five candidate differential genes, SLAMF7, CCL22, KRT10, FXYD3 and ABCC2 were possibly associated with the survival UM cells. These results may suggest that SLAMF7, CCL22, KRT10, FXYD3 and ABCC2 may participate in the survival variants of mechanisms of UM. Future studies may examine their molecular functions at the cell level and in vivo, to uncover their molecular mechanism in immune escaping. We found that these genes overexpression impacted the survival time of UM patients severely, moreover these patients produced UM relapse and liver metastasis.
Potential limitations of our study should be mentioned. First, as for any cell culture study, the question arises how much the results can be transferred into the clinical situation. Second, the relapsing UMs were pre-treated by radiotherapy or laser therapy, so that it was not possible to clearly distinguish between direct effects of these treatments or effects caused by the survival variants of tumor cells. Third, a main point about UM is that CTLs are most likely not the most important contributors to the killing of the tumor cells in the eye, due to the intraocular immune privileged conditions. They may, however, be relevant in the liver for the metastases[48]. Another factor of importance is the killing of the tumor cells by natural killer cells. Natural killer cells especially focus on cells that have lost HLA Class I antigens, and if cells become less lysable by CTLs, they may become more lysable by natural killer cells[48].
In conclusion, gene expression differed between primary UM cells and survival variants of UM cells. Since the present study did not provide data indicating whether the differentially expressed genes in the MUM-2B cells that survived the culture with CD8+ T Cells conferred protection from CD8+ T cells, future studies could include cytotoxicity assays with MUM-2B cells that survived a culture with CD8+ T cells, MUM-2B cells that were engineered to overexpress the genes of interest, and surviving MUM-2B cells in which these genes of interest were silenced. Future studies may also address whether the differences in the gene expression found in our study may be helpful to early detect survival variants of UM cells and may be helpful to develop targeted treatment strategies which may be more specifically focused on the survival variants of UM cells in contrast to the primary UM cells. Addressing the survival pathways which were associated with the survival of survival variants of UM cells under immunization in our study could provide targets to disrupt the survival cascade in UM.
Acknowledgments
Foundations: Supported by National Natural Science Foundation of China (No.81570891; No.81272981); the Beijing Municipal Administration of Hospitals' Ascent Plan (No.DFL20150201); Science & Technology Project of Beijing Municipal Science & Technology Commission (No.Z151100001615052); Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No.ZYLX201307); Beijing Natural Science Foundation (No.7151003); Advanced Health Care Professionals Development Project of Beijing Municipal Health Bureau (No.2014-2-003).
Conflicts of Interest: Hou F, None; Huang QM, None; Hu DN, None; Jonas JB, None; Wei WB, None.
REFERENCES
- 1.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Buder K, Gesierich A, Gelbrich G, Goebeler M. Systemic treatment of metastatic uveal melanoma: review of literature and future perspectives. Cancer Med. 2013;2(5):674–686. doi: 10.1002/cam4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niederkorn JY. Immune escape mechanisms of intraocular tumors. Prog Retin Eye Res. 2009;28(5):329–347. doi: 10.1016/j.preteyeres.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma J, Usui Y, Takeuchi M, Okunuki Y, Kezuka T, Zhang L, Mizota A, Goto H. Human uveal melanoma cells inhibit the immunostimulatory function of dendritic cells. Exp Eye Res. 2010;91(4):491–499. doi: 10.1016/j.exer.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, Chen PW, Li H, Alizadeh H, Niederkorn JY. PD-L1: PD-1 interaction contributes to the functional suppression of T-cell responses to human uveal melanoma cells in vitro. Invest Ophthalmol Vis Sci. 2008;49(6):2518–2525. doi: 10.1167/iovs.07-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallermalm K, De Geer A, Kiessling R, Levitsky V, Levitskaya J. Autocrine secretion of Fas ligand shields tumor cells from Fas-mediated killing by cytotoxic lymphocytes. Cancer Res. 2004;64(18):6775–6782. doi: 10.1158/0008-5472.CAN-04-0508. [DOI] [PubMed] [Google Scholar]
- 7.Bronkhorst IH, Jager MJ. Inflammation in uveal melanoma. Eye (Lond) 2013;27(2):217–223. doi: 10.1038/eye.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durie FH, Campbell AM, Lee WR, Damato BE. Analysis of lymphocytic infiltration in uveal melanoma. Invest Ophthalmol Vis Sci. 1990;31(10):2106–2110. [PubMed] [Google Scholar]
- 9.Woodward J, Sisley K, Reeves G, Nichols C, Parsons MA, Mudhar H, Rennie I. Evidence of macrophage and lymphocyte, but not dendritic cell, infiltration in posterior uveal melanomas, whilst cultured uveal melanomas demonstrate pluripotency by expressing CD68 and CD163. Int J Exp Pathol. 2004;85(1):35–43. doi: 10.1111/j.0959-9673.2004.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagouros E, Salomao D, Thorland E, Hodge DO, Vile R, Pulido JS. Infiltrative T regulatory cells in enucleated uveal melanomas. Trans Am Ophthalmol Soc. 2009;107:223–228. [PMC free article] [PubMed] [Google Scholar]
- 11.Rothermel LD, Sabesan AC, Stephens DJ, Chandran SS, Paria BC, Srivastava AK, Somerville R, Wunderlich JR, Lee CC, Xi L, Pham TH, Raffeld M, Jailwala P, Kasoji M, Kammula US. Identification of an immunogenic subset of metastatic uveal melanoma. Clin Cancer Res. 2016;22(9):2237–2249. doi: 10.1158/1078-0432.CCR-15-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronkhorst IH, Jager MJ. Uveal melanoma: the inflammatory microenvironment. J Innate Immun. 2012;4(5–6):454–462. doi: 10.1159/000334576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umansky V, Sevko A, Gebhardt C, Utikal J. Myeloid-derived suppressor cells in malignant melanoma. J Dtsch Dermatol Ges. 2014;12(11):1021–1027. doi: 10.1111/ddg.12411. [DOI] [PubMed] [Google Scholar]
- 14.Ly LV, Bronkhorst IH, van Beelen E, Vrolijk J, Taylor AW, Versluis M, Luyten GP, Jager MJ. Inflammatory cytokines in eyes with uveal melanoma and relation with macrophage infiltration. Invest Ophthalmol Vis Sci. 2010;51(11):5445–5451. doi: 10.1167/iovs.10-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21(2):137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Hurks HM, Metzelaar-Blok JA, Mulder A, Claas FH, Jager MJ. High frequency of allele-specific down-regulation of HLA class I expression in uveal melanoma cell lines. Int J Cancer. 2000;85(5):697–702. doi: 10.1002/(sici)1097-0215(20000301)85:5<697::aid-ijc16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Vetter CS, Lieb W, Bröcker EB, Becker JC. Loss of nonclassical MHC molecules MIC-A/B expression during progression of uveal melanoma. Br J Cancer. 2004;91(8):1495–1459. doi: 10.1038/sj.bjc.6602123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delirezh N, Moazzeni SM, Shokri F, Shokrgozar MA, Atri M, Kokhaei P. Autologous dendritic cells loaded with apoptotic tumor cells induce T cell-mediated immune responses against breast cancer in vitro. Cell Immunol. 2009;257(1–2):23–31. doi: 10.1016/j.cellimm.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Alaniz L, Rizzo MM, Mazzolini G. Pulsing dendritic cells with whole tumor cell lysates. Methods Mol Biol. 2014;1139:27–31. doi: 10.1007/978-1-4939-0345-0_3. [DOI] [PubMed] [Google Scholar]
- 20.Li YL, Wu YG, Wang YQ, Li Z, Wang RC, Wang L, Zhang YY. Bone marrow-derived dendritic cells pulsed with tumor lysates induce anti-tumor immunity against gastric cancer ex vivo. World J Gastroenterol. 2008;14(46):7127–7132. doi: 10.3748/wjg.14.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JJ, Choi BH, Kang HK, Park MS, Park JS, Kim SK, Pham TN, Cho D, Nam JH, Kim YJ, Rhee JH, Yang DH, Kim YK, Kim HJ, Chung IJ. Induction of multiple myeloma-specific cytotoxic T lymphocyte stimulation by dendritic cell pulsing with purified and optimized myeloma cell lysates. Leuk Lymphoma. 2007;48(10):2022–2031. doi: 10.1080/10428190701583975. [DOI] [PubMed] [Google Scholar]
- 22.Yu P, Lee Y, Wang Y, Liu X, Auh S, Gajewski TF, Schreiber H, You Z, Kaynor C, Wang X, Fu YX. Targeting the primary tumor to generate CTL for the effective eradication of spontaneous metastases. J Immunol. 2007;179(3):1960–1968. doi: 10.4049/jimmunol.179.3.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal M, García-Alcalde F, Concha A, Cano C, Blanco A, Garrido F, Ruiz-Cabello F. Genome-wide differential genetic profiling characterizes colorectal cancers with genetic instability and specific routes to HLA class I loss and immune escape. Cancer Immunol Immunother. 2012;61(6):803–816. doi: 10.1007/s00262-011-1147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dadkhah E, Naseh H, Farshchian M, Memar B, Sankian M, Bagheri R, Forghanifard MM, Montazer M, Kazemi Noughabi M, Hashemi M, Abbaszadegan MR. A cancer-array approach elucidates the immune escape mechanism and defects in the DNA repair system in esophageal squamous cell carcinoma. Arch Iran Med. 2013;16(8):463–470. [PubMed] [Google Scholar]
- 25.Rice J, Buchan S, Stevenson FK. Critical components of a DNA fusion vaccine able to induce protective cytotoxic T cells against a single epitope of a tumor antigen. J Immunol. 2002;169(7):3908–3913. doi: 10.4049/jimmunol.169.7.3908. [DOI] [PubMed] [Google Scholar]
- 26.Popescu I, Pipeling M, Akulian J, McDyer J. Phenotypic and functional characterization of cytotoxic T lymphocytes by flow cytometry. Methods Mol Biol. 2014;1186:21–47. doi: 10.1007/978-1-4939-1158-5_3. [DOI] [PubMed] [Google Scholar]
- 27.Saban DR. The chemokine receptor CCR7 expressed by dendritic cells: a key player in corneal and ocular surface inflammation. Ocul Surf. 2014;12(2):87–99. doi: 10.1016/j.jtos.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubrot J, Duraes FV, Potin L, Capotosti F, Brighouse D, Suter T, LeibundGut-Landmann S, Garbi N, Reith W, Swartz MA, Hugues S. Lymph node stromal cells acquire peptide-MHCII complexes from dendritic cells and induce antigen-specific CD4+ T cell tolerance. J Exp Med. 2014;211(6):1153–1166. doi: 10.1084/jem.20132000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal P, Romero R, Mazaki-Tovi S, Tromp G, Tarca AL, Kim YM, Chaiworapongsa T, Kusanovic JP, Erez O, Than NG, Hassan SS. Fetal membranes as an interface between inflammation and metabolism: increased aquaporin 9 expression in the presence of spontaneous labor at term and chorioamnionitis. J Matern Fetal Neonatal Med. 2009;22(12):1167–1175. doi: 10.3109/14767050903019692. [DOI] [PubMed] [Google Scholar]
- 30.Iriyama N, Yuan B, Yoshino Y, Hatta Y, Horikoshi A, Aizawa S, Takeuchi J, Toyoda H. Aquaporin 9, a promising predictor for the cytocidal effects of arsenic trioxide in acute promyelocytic leukemia cell lines and primary blasts. Oncol Rep. 2013;29(6):2362–2368. doi: 10.3892/or.2013.2388. [DOI] [PubMed] [Google Scholar]
- 31.Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, Starling GC, Ciarlariello D, Bhaskar S, Briercheck EL, Hughes T, Yu J, Rice A, Benson DM., Jr Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC. Cancer Immunol Immunother. 2013;62(12):1841–1849. doi: 10.1007/s00262-013-1493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veillette A, Guo H. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit Rev Oncol Hematol. 2013;88(1):168–177. doi: 10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Balasa B, Yun R, Belmar NA, Fox M, Chao DT, Robbins MD, Starling GC, Rice AG. Elotuzumab enhances natural killer cell activation and myeloma cell killing through interleukin-2 and TNF-α pathways. Cancer Immunol Immunother. 2015;64(1):61–73. doi: 10.1007/s00262-014-1610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Wang K, Liu W, Hao Q. PTEN overexpression improves cisplatin-resistance of human ovarian cancer cells through upregulating KRT10 expression. Biochem Biophys Res Commun. 2014;444(2):141–146. doi: 10.1016/j.bbrc.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Halon A, Materna V, Donizy P, Matkowski R, Rabczynski J, Lage H, Surowiak P. MRP2 (ABCC2, cMOAT) expression in nuclear envelope of primary fallopian tube cancer cells is a new unfavorable prognostic factor. Arch Gynecol Obstet. 2013;287(3):563–570. doi: 10.1007/s00404-012-2589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma X, Zhou J, Zhang CX, Li XY, Li N, Ju RJ, Shi JF, Sun MG, Zhao WY, Mu LM, Yan Y, Lu WL. Modulation of drug-resistant membrane and apoptosis proteins of breast cancer stem cells by targeting berberine liposomes. Biomaterials. 2013;34(18):4452–4465. doi: 10.1016/j.biomaterials.2013.02.066. [DOI] [PubMed] [Google Scholar]
- 37.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6(11):836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 38.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11(4):321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3(11):999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu YH, Kim JC. Expression of indoleamine 2,3-dioxygenase in human corneal cells as a local immunosuppressive factor. Invest Ophthalmol Vis Sci. 2007;48(9):4148–4152. doi: 10.1167/iovs.05-1336. [DOI] [PubMed] [Google Scholar]
- 41.Shiels MS, Pfeiffer RM, Hildesheim A, Engels EA, Kemp TJ, Park JH, Katki HA, Koshiol J, Shelton G, Caporaso NE, Pinto LA, Chaturvedi AK. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst. 2013;105(24):1871–1880. doi: 10.1093/jnci/djt309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas J, Schopp L, Storch-Hagenlocher B, Fritzsching B, Jacobi C, Milkova L, Fritz B, Schwarz A, Suri-Payer E, Hensel M, Wildemann B. Specific recruitment of regulatory T cells into the CSF in lymphomatous and carcinomatous meningitis. Blood. 2008;111(2):761–766. doi: 10.1182/blood-2007-08-104877. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto H, Mukaisho K, Sugihara H, Hattori T, Asano S. Down-regulation of FXYD3 is induced by transforming growth factor-β signaling via ZEB1/δEF1 in human mammary epithelial cells. Biol Pharm Bull. 2011;34(3):324–329. doi: 10.1248/bpb.34.324. [DOI] [PubMed] [Google Scholar]
- 44.Zhu ZL, Yan BY, Zhang Y, Yang YH, Wang MW, Zentgraf H, Zhang XH, Sun XF. Overexpression of FXYD-3 is involved in the tumorigenesis and development of esophageal squamous cell carcinoma. Dis Markers. 2013;35(3):195–202. doi: 10.1155/2013/740201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meding S, Balluff B, Elsner M, Schöne C, Rauser S, Nitsche U, Maak M, Schäfer A, Hauck SM, Ueffing M, Langer R, Höfler H, Friess H, Rosenberg R, Walch A. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J Pathol. 2012;228(4):459–470. doi: 10.1002/path.4021. [DOI] [PubMed] [Google Scholar]
- 46.Chen KG, Valencia JC, Gillet JP, Hearing VJ, Gottesman MM. Involvement of ABC transporters in melanogenesis and the development of multidrug resistance of melanoma. Pigment Cell Melanoma Res. 2009;22(6):740–749. doi: 10.1111/j.1755-148X.2009.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bibert S, Aebischer D, Desgranges F, Roy S, Schaer D, Kharoubi-Hess S, Horisberger JD, Geering K. A link between FXYD3 (Mat-8)-mediated Na,K-ATPase regulation and differentiation of Caco-2 intestinal epithelial cells. Mol Biol Cell. 2009;20(4):1132–1140. doi: 10.1091/mbc.E08-10-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jager MJ, Hurks HM, Levitskaya J, Kiessling R. HLA expression in uveal melanoma: there is no rule without some exception. Hum Immunol. 2002;63(6):444–451. doi: 10.1016/s0198-8859(02)00389-0. [DOI] [PubMed] [Google Scholar]