Abstract
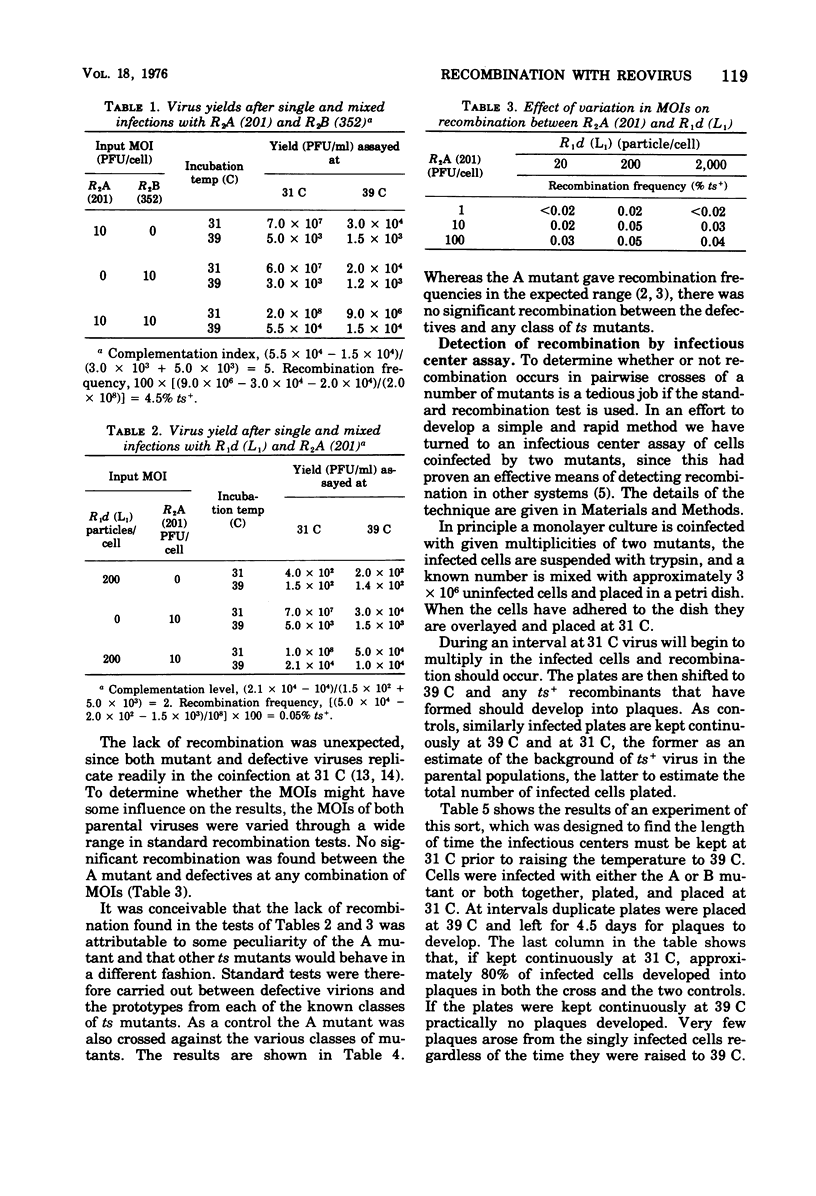
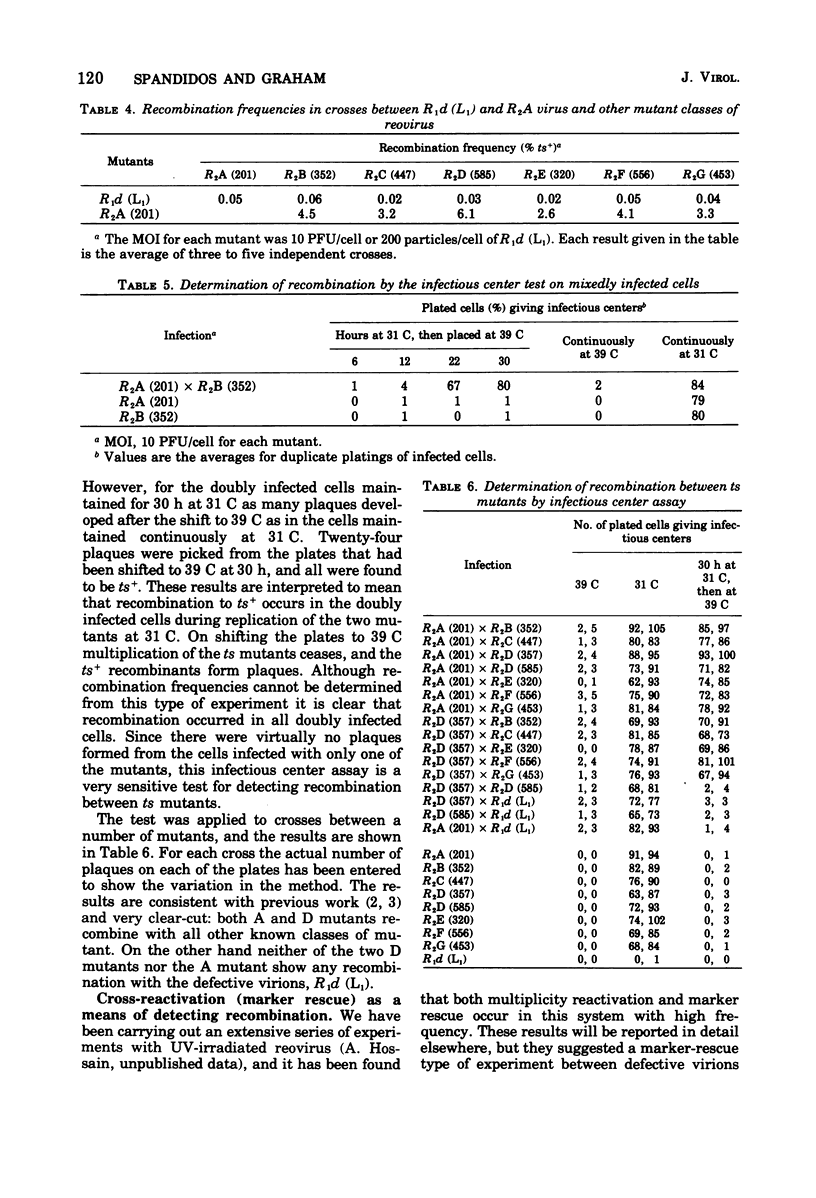
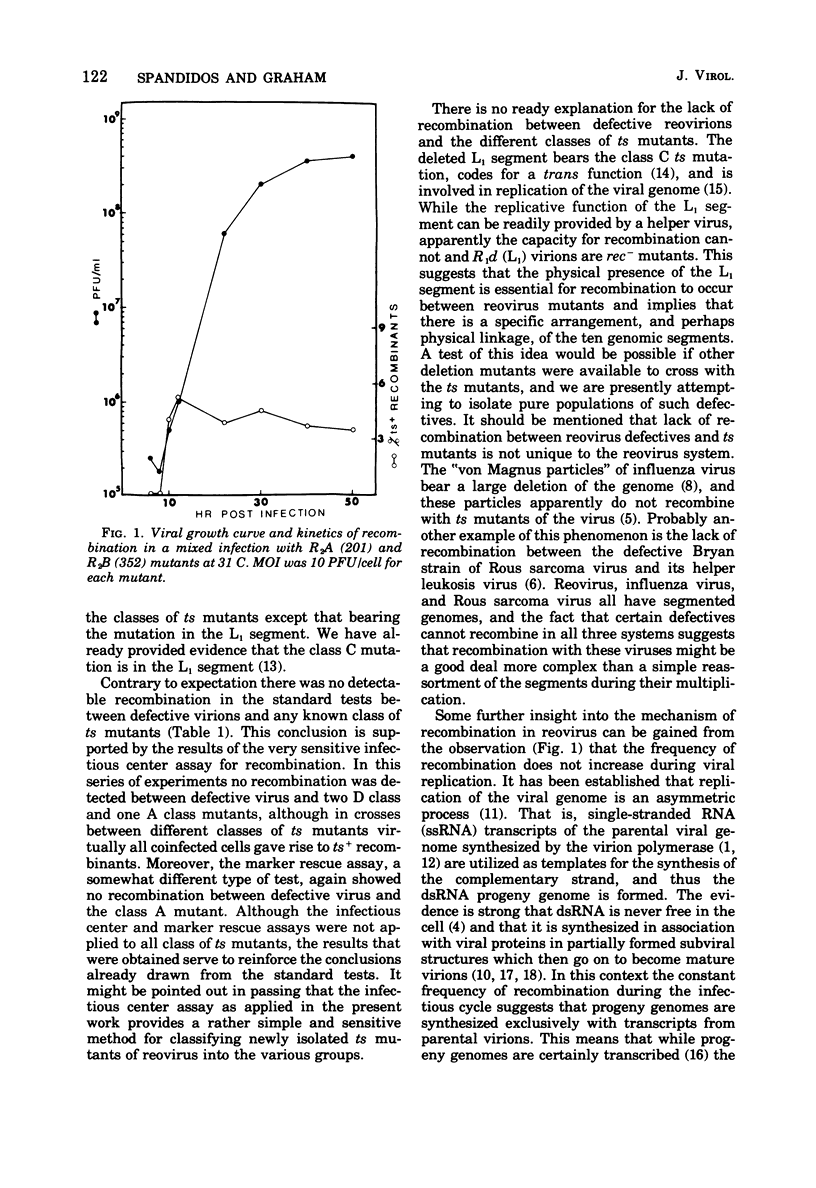
In standard pairwise crosses there was no detectable recombination between defective reovirus lacking the largest genomic segment and prototypes of the seven known classes of ts mutants. However, in such crosses between R2A (201) and the various prototypes frequencies of ts+ recombinants between 2.6 and 6.1% were observed, as others have found (Fields, 1971; Fields and Joklik, 1969). An infectious center assay was devised to measure recombination in this system, and it was found that all mixedly infected cells gave rise to ts+ recombinants in crosses between prototype ts mutants, but no recombination was detectable when the defective virus was crossed with three different ts mutants. The ts mutation of mutant R2A (201) was efficiently rescued when crossed with UV-inactivated wild-type virus but not when crossed with UV-inactivated defective virus. It is concluded from these various experiments that if there is any recombination between these defective reovirions and any known class of ts mutants it is too low to be measured by methods presently available. The kinetics of recombination were measured in cells mixedly infected with R2A (201) and R2B (352) mutants. At the earliest time progeny virus could be found in the cells the frequency of ts+ recombinants was 4.5%, and this frequency remained unchanged despite a subsequent 1,000-fold increase in progeny virus.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borsa J., Graham A. F. Reovirus: RNA polymerase activity in purified virions. Biochem Biophys Res Commun. 1968 Dec 30;33(6):895–901. doi: 10.1016/0006-291x(68)90396-3. [DOI] [PubMed] [Google Scholar]
- Fields B. N., Joklik W. K. Isolation and preliminary genetic and biochemical characterization of temperature-sensitive mutants of reovirus. Virology. 1969 Mar;37(3):335–342. doi: 10.1016/0042-6822(69)90217-7. [DOI] [PubMed] [Google Scholar]
- Fields B. N. Temperature-sensitive mutants of reovirus type 3 features of genetic recombination. Virology. 1971 Oct;46(1):142–148. doi: 10.1016/0042-6822(71)90013-4. [DOI] [PubMed] [Google Scholar]
- Gomatos P. J. RNA synthesis in reovirus-infected L929 mouse fibroblasts. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1798–1805. doi: 10.1073/pnas.58.4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst G. K. Mechanism of influenza recombination. I. Factors influencing recombination rates between temperature-sensitive mutants of strain WSN and the classification of mutants into complementation--recombination groups. Virology. 1973 Sep;55(1):81–93. doi: 10.1016/s0042-6822(73)81010-4. [DOI] [PubMed] [Google Scholar]
- Kawai S., Hanafusa H. Genetic recombination with avian tumor virus. Virology. 1972 Jul;49(1):37–44. doi: 10.1016/s0042-6822(72)80005-9. [DOI] [PubMed] [Google Scholar]
- Nonoyama M., Watanabe Y., Graham A. F. Defective virions of reovirus. J Virol. 1970 Aug;6(2):226–236. doi: 10.1128/jvi.6.2.226-236.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons M., Hirst G. K. The single- and double-stranded RNA's and the proteins of incomplete influenza virus. Virology. 1969 May;38(1):68–72. doi: 10.1016/0042-6822(69)90128-7. [DOI] [PubMed] [Google Scholar]
- Prevec L., Watanabe Y., Gauntt C. J., Graham A. F. Transcription of the genomes of type 1 and type 3 reoviruses. J Virol. 1968 Apr;2(4):289–297. doi: 10.1128/jvi.2.4.289-297.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S., Watanabe Y. Incorporation of in vitro synthesized reovirus double-stranded ribonucleic acid into virus corelike particles. J Virol. 1972 Nov;10(5):943–950. doi: 10.1128/jvi.10.5.943-950.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg M., Silverstein S. C., Levin D. H., Acs G. Asynchronous synthesis of the complementary strands of the reovirus genome. Proc Natl Acad Sci U S A. 1971 Feb;68(2):505–508. doi: 10.1073/pnas.68.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin A. J., Sipe J. D. RNA polymerase activity in purified reoviruses. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1462–1469. doi: 10.1073/pnas.61.4.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos D. A., Graham A. F. Complementation between temperature-sensitive and deletion mutants of reovirus. J Virol. 1975 Dec;16(6):1444–1452. doi: 10.1128/jvi.16.6.1444-1452.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos D. A., Graham A. F. Complementation of defective reovirus by ts mutants. J Virol. 1975 Apr;15(4):954–963. doi: 10.1128/jvi.15.4.954-963.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos D. A., Krystal G., Graham A. F. Regulated transcription of the genomes of defective virions and temperature-sensitive mutants of reovirus. J Virol. 1976 Apr;18(1):7–19. doi: 10.1128/jvi.18.1.7-19.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Kudo H., Graham A. F. Selective inhibition of reovirus ribonucleic acid synthesis by cycloheximide. J Virol. 1967 Feb;1(1):36–44. doi: 10.1128/jvi.1.1.36-44.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweerink H. J., Ito Y., Matsuhisa T. Synthesis of reovirus double-stranded RNA within virionlike particles. Virology. 1972 Nov;50(2):349–358. doi: 10.1016/0042-6822(72)90386-8. [DOI] [PubMed] [Google Scholar]
- Zweerink H. J. Multiple forms of SS leads to DS RNA polymerase activity in reovirus-infected cells. Nature. 1974 Feb 1;247(5439):313–315. doi: 10.1038/247313a0. [DOI] [PubMed] [Google Scholar]



