Abstract
Gut microbes are essential for the degradation of dietary oxalate, and this function may play a role in decreasing the incidence of kidney stones. However, many oxalate-degrading bacteria are susceptible to antibiotics and the use of oxalate-degrading probiotics has only led to an ephemeral reduction in urinary oxalate. The objective of the current study was to determine the efficacy of using whole-community microbial transplants from a wild mammalian herbivore, Neotoma albigula, to increase oxalate degradation over the long term in the laboratory rat, Rattus norvegicus. We quantified the change in total oxalate degradation in lab rats immediately after microbial transplants and at 2- and 9-month intervals following microbial transplants. Additionally, we tracked the fecal microbiota of the lab rats, with and without microbial transplants, using high-throughput Illumina sequencing of a hyper-variable region of the 16S rRNA gene. Microbial transplants resulted in a significant increase in oxalate degradation, an effect that persisted 9 months after the initial transplants. Functional persistence was corroborated by the transfer, and persistence of a group of bacteria previously correlated with oxalate consumption in N. albigula, including an anaerobic bacterium from the genus Oxalobacter known for its ability to use oxalate as a sole carbon source. The results of this study indicate that whole-community microbial transplants are an effective means for the persistent colonization of oxalate-degrading bacteria in the mammalian gut.
Keywords: Gut microbiota, Dietary oxalate, Microbial transplant, Urinary oxalate
Introduction
Oxalate, a simple organic acid, is an important nephrotoxin that can have a considerable impact on humans and mammalian herbivores [1–3]. Oxalate is widely produced as a secondary compound by plants to deter herbivory and is found in many of the foods consumed by humans [4–6]. To a lesser extent, mammals will also produce oxalate endogenously in the liver, as a terminal metabolite derived from several dietary precursors such as glyoxalate, glycine, and hydroxyproline, among others [2, 7]. If the concentration of calcium oxalate in the urine exceeds the metastable limit, crystallization of the mineral can occur and eventually calcium oxalate deposits can aggregate into kidney stones [8–13]. About 80% of all kidney stones in humans are composed of calcium oxalate, and in severe cases, end-stage renal disease can develop [4, 6, 13–15]. Despite its toxicity, oxalate cannot be metabolized by mammals but rather is metabolized by oxalate-degrading bacteria common in the mammalian gut [16–20].
A diversity of oxalate-degrading bacteria have been isolated from humans, sheep, rats, dairy, and other mammalian sources [18, 21–23]. Oxalobacter formigenes is an obligate oxalate consumer, as it requires oxalate as a carbon and energy source, and the presence of this species in the human gut is negatively associated with kidney stone formation [18, 24, 25]. Facultative oxalate-degrading bacteria include species from the Lactobacillus, Enterococcus, and Bifidobacterium genera among others, and these bacteria can also have a significant impact on the balance of oxalate in mammals [19, 22, 26–28]. However, repeated use of antibiotics may be reducing the incidence of oxalate-degrading bacteria in the human gut [25, 29–31]. Given that these bacteria may play an important role in preventing kidney stone formation, their loss represents a considerable public health issue [32, 33].
Previous attempts to introduce oxalate-degrading microbes into the human or rat gut have resulted in an ephemeral decrease in urinary oxalate excretion. These probiotic formulations include O. formigenes alone or different combinations of Lactobacillus, Bifidobacterium, Enterococcus, and other facultative oxalate degraders. With all formulations, the probiotics initially lead to a reduction in urinary oxalate excretion in both humans and rodents, but the bacteria and their oxalate-degrading function are typically lost in as little as 5 days after oxalate is removed from the diet [26, 28, 34–36]. The loss of the probiotic oxalate-degrading bacteria after the removal of dietary oxalate is in contrast to other mammals, such as fat sand rats (Psammomys obesus), sheep, and some humans, that harbor oxalate-degrading bacteria natively. In these mammals, the oxalate-degrading bacterial populations are maintained across generations, even when oxalate is removed from the diet [37–39].
The microbial community of Sprague–Dawley rats (SDR), like humans, is dominated by the Bacteroidetes phylum, followed by the Firmicutes, Proteobacteria, and others [40]. While SDR will exhibit some oxalate degradation with initial consumption of oxalate, they do not typically harbor O. formigenes, and their level of oxalate degradation decreases with continued exposure to oxalate, indicative of maladaptation of their native gut microbiota to oxalate consumption [34]. In contrast, Neotoma albigula is capable of degrading >90% of the dietary oxalate ingested by N. albigula at levels up to 9% dietary oxalate by dry weight [41]. Moreover, N. albigula maintains this highly effective oxalate-degrading microbiota even after 6 months on a low (0.2 %) oxalate diet [42]. Thus, this pair of species makes an excellent model to examine the effect of fecal transplants on oxalate degradation.
The purpose of the current study was to determine the efficacy of using whole-community microbial transplants to confer persistent oxalate degradation across species. We had three primary objectives. The first objective was to determine the efficacy of using whole-community microbial transplants from N. albigula in conferring the oxalate-degrading function to another rodent, Rattus norvegicus (SDR). The second objective was to determine the persistence of the transferred function. Finally, the gut microbiota was tracked to determine the differential response of oxalate on the gut microbiota between animals receiving a microbial transplant and those with their native microbiota.
Materials and Methods
Location, Collection, and Diet of Animals
Three N. albigula collected with Sherman live traps from Castle Valley, Utah (38.63° N, 109.41° W), in October 2012 served as the microbial community donors. Donors included one male and two female animals. After trapping, N. albigula were transported to the University of Utah Department of Biology Animal Facility and housed in individual cages (48 × 27 × 20 cm) under a 12:12-h light/dark cycle, at 28 °C and 20% humidity. Animals were fed high-fiber rabbit chow (Harlan Teklad formula 2031, Denver, CO, USA; 0.2% oxalate) for 10 months prior to experimentation. Additionally, nine male Sprague–Dawley laboratory rats (20–21 days old) were purchased from Harlan Laboratories (Denver, CO, USA). Sprague–Dawley rats (SDR) were fed standard rat chow (Harlan Teklad formula 2018) for 2 weeks prior to experimentation. All methods were approved by the IACUC under protocol no. 12-12010.
To determine the efficacy of transferring the function of oxalate degradation across species, all animals were placed in the following diet trial. Initially, all donor animals received a 0.05% oxalate diet and all SDR received a 0% oxalate diet for 5 days. This protocol allowed for the quantification of endogenous oxalate excretion. The SDR diet consisted of a custom purified rat chow with no quantifiable oxalate (Table S1). N. albigula received the same custom rat chow with a high-fiber rabbit chow mixed in at a 3:1 ratio. The addition of the high-fiber rabbit chow was necessary because N. albigula would not consume the rat chow without it. After 5 days on these no oxalate diets, all animals were fed a 1.5% oxalate diet (by dry weight) for 3 days, prepared by mixing sodium oxalate (Fisher Scientific, Pittsburgh, PA, USA) into the purified rat chow. This diet regimen permitted the quantification of dietary oxalate excretion by SDR with their native microbiota and the acclimation of the gut microbiota of N. albigula for oxalate degradation. After 3 days on 1.5% oxalate, fresh feces (<6 h old) were collected from donor animals from the top of a fecal collection tube attached to a metabolic cage. This approach was taken to minimize time that microbes were exposed to aerobic conditions. Feces were ground with a sterilized pestle and mortar and homogenized into the purified rat chow of six of the SDRs (2.9 g woodrat feces per lab rat) similar to previous studies [43]. Three control SDR did not receive any feces. Following the single fecal transplant, both the transplant and no-transplant SDRs were maintained on the 1.5% oxalate diet for an additional 3 days to quantify oxalate excretion. To determine the persistence of the transferred microbial oxalate function, after the initial diet trial, all SDR were returned to the 0% oxalate diet and the diet trial was repeated 2 and 9 months after the microbial transplant, with no additional transplants in subsequent trials. During the course of all trials, both food and water were given ad libitum.
During the diet trial, metabolic cages were used to separate urine and feces, which were collected daily in 50-ml conical tubes. The urine and feces were used for the quantification of oxalate excretion and fecal microbial communities (discussed below). Additionally, we collected daily data on body mass and food and water intake, along with fecal and urinary output. Using the food intake and fecal output data, we estimated dry matter digestibility (DMD) as 1 − (fecal output ÷ food consumed by dry weight). Data were evaluated with repeated measures ANOVA and post hoc Tukey’s analyses.
Oxalate Assays
Quantification of urinary and fecal oxalate has been described previously [41]. Briefly, urine samples were collected daily from each animal and pooled together across days for each treatment period for a total of nine samples per treatment period. Urine samples were acidified to a pH of <3 with H2SO4 to solubilize oxalate crystals, and remaining precipitates were removed by centrifugation. The pH of the supernatant was brought up to 7 with NaOH. At pH 7, CaCl2 was added to precipitate oxalate, and the resulting calcium oxalate was isolated with centrifugation. A volume of distilled water matching the total urinary volume was added to calcium oxalate precipitate. Samples were then titrated as described below.
For fecal oxalate assays, feces were collected daily for each animal and dried overnight at 45 °C and then pooled across days by animal at the end of each treatment period. Dried feces were ground and added to 5 ml 6 N H2SO4 for 15 min to extract oxalate. Then, 25 ml of distilled water was added and filtered to remove particulates. The filtrate was brought up to a pH of 7 with NaOH. CaCl2 was added to precipitate oxalate. Calcium oxalate was isolated from the filtrate with centrifugation. Then, a volume of distilled water equal to that recovered after filtration was added, and the samples were titrated.
The urine and fecal extracts containing calcium oxalate were titrated with KMnO4 in triplicate. Aliquots were acidified with H2SO4 and heated to 70–90 °C. The KMnO4 was added until a pink color persisted for 30 s, and the volume of KMnO4 added was recorded. These volumes were then compared to titrations of a standard series of oxalate solutions to determine the concentration of oxalate in the sample. Standard curves were made by adding 0, 5, 10, 15, or 20 mM of sodium oxalate to the urine or feces of SDR consuming 0% oxalate. After extraction and titration of standard solutions, the titration volume for samples with no oxalate added was subtracted from all other samples to account for endogenous oxalate production. Through these methods, we recovered 103.36 ± 10.61% of the oxalate from urine and 96.4 ± 5.11% of the oxalate from feces. Both titration curves were linear with r2 values >0.9. To estimate total oxalate degradation, we subtracted oxalate excreted from oxalate consumed. With this metric, the excretion of endogenous oxalate in some cases led to negative values of oxalate degradation. However, when endogenous oxalate excretion was accounted for based on total excretion on a 0% oxalate diet, all animals exhibited some level of oxalate degradation. Accounting for endogenous excretion did not change the significance of the data.
Microbial Inventories
Fresh feces were collected for microbial inventories on the last day of each diet treatment (nine time points). Feces were frozen at −80 °C until DNA extraction. DNA extraction was completed with the QIAamp DNA Stool Mini Kit (Qiagen, Germantown, MD, USA), from 180 to 220 mg of feces. Microbial inventories were generated from 84 fecal samples by amplifying the V4 region of the 16S ribosomal RNA (rRNA) gene with the primers 515F and 806R [44]. Primers for the initial PCR step contained a 12-base barcode sequence, and PCR products were multiplexed into a single-lane sequencing run on an Illumina MiSeq (Illumina, Sand Diego, CA, USA) with paired-end sequencing of 150 base pairs per paired end, as previously described [45].
Resulting sequences were analyzed in QIIME software [44]. Standard quality control [46] was conducted in QIIME, and sequences were demultiplexed with default parameters. A de novo picking strategy was utilized to classify operational taxonomic units (OTUs) with UCLUST [47] at a minimum sequence identity of 97 %. This strategy resulted in an OTU table and phylogenetic tree, which were used in downstream analyses. Sequences identified by UCLUST as chloroplasts or mitochondria or that had fewer than ten representations across the dataset were removed. Additionally, samples of microbial communities with fewer than 3000 sequence reads total were removed from further data analysis.
For comparative analyses, samples were rarified to an equal sampling depth of 33,472, which was the highest number that included all samples remaining after quality control. To compare the microbial communities across individuals and treatments, unweighted and weighted UniFrac analyses of the OTU table were preformed generating metrics on community membership and structure, respectively [48]. Statistical differences between treatment groups were calculated with an analysis of similarity (ANOSIM) after 999 permutations. Additionally, a log-likelihood ratio analysis was conducted comparing the average frequency of an OTU for each group after feeding on 0% oxalate for 5 days and after 6 days on a 1.5% oxalate diet 2 and 9 months after the transplant. To determine which OTUs transferred to SDR, which were common and which were ephemeral, a Venn diagram was generated depicting the shared and unique OTUs from all three groups immediately after the transplant period. Significance for each metric was set at a P value <0.05 after a false discovery rate (FDR) correction.
Results
The Effect of Whole Microbial Community Transplants on SDR and Oxalate Degradation
The SDR group receiving the fecal transplant exhibited a 16% increase in food intake and a 14% increase in oxalate intake compared to before the transplant (Table 1). The transplant group produced 13.3 ± 11% more feces; however, there was no significant difference in fecal output (P = 0.06) compared to controls. There was no significant difference between treatment groups with respect to body mass, DMD, water intake, or urine output (Table 1).
Table 1.
Host metrics between the transplant (n = 5) nd no transplant (n = 3) groups after 6 days on 1.5% oxalate for each of the three replicate diet trials, which corresponds to 3, 69, and 279 days after the microbial transplant
| Metric | Mean (g) ± SE |
F value | P value | |||||
|---|---|---|---|---|---|---|---|---|
| Time point | 3 Days |
69 Days |
279 Days |
|||||
| Treatment | Transplant | No transplant | Transplant | No transplant | Transplant | No transplant | ||
| Body mass | 173.7 ± 2.3 | 173.7 ± 0.7 | 441.3 ± 1 | 444.1 ± 6.3 | 563.8 ± 16.9 | 541.6 ± 10.3 | 0.0 | 0.94 |
| Food intake | 17.2 ± 0.29 | 15.58 ± 0.25 | 18.16 ± 0.48 | 17.6 ± 0.17 | 17.93 ± 1.01 | 12.87 ± 0.07 | 16.4 | <0.001 |
| Oxalate intake | 0.26 ± 0.004 | 0.23 ± 0.007 | 0.27 ± 0.007 | 0.26 ± 0.003 | 0.26 ± 0.19 | 0.19 ± 0.007 | 13.6 | 0.001 |
| Fecal output | 3.5 ± 0.1 | 3.3 ± 0.1 | 3.3 ± 0.2 | 3.3 ± 0.3 | 3.2 ± 0.3 | 2.3 ± 0.2 | 4.1 | 0.06 |
| DMD | 0.80 ± 0.007 | 0.79 ± 0.004 | 0.82 ± 0.006 | 0.81 ± 0.01 | 0.82 ± 0.009 | 0.82 ± 0.02 | 0.9 | 0.37 |
| Water intake | 27.1 ± 1.7 | 26.9 ± 0.8 | 27.9 ± 3.1 | 23.5 ± 1.2 | 26.9 ± 2 | 31.6 ± 2.0 | 0.0 | 0.99 |
| Urine output | 9.6 ± 1.5 | 8.8 ± 0.9 | 10.5 ± 1.9 | 6.4 ± 1.3 | 11.5 ± 1.3 | 13.5 ± 1.3 | 0.4 | 0.55 |
Data were analyzed with a repeated measures ANOVA (df = 1.8). The F and P values presented are for the contrast between transplant and no transplant
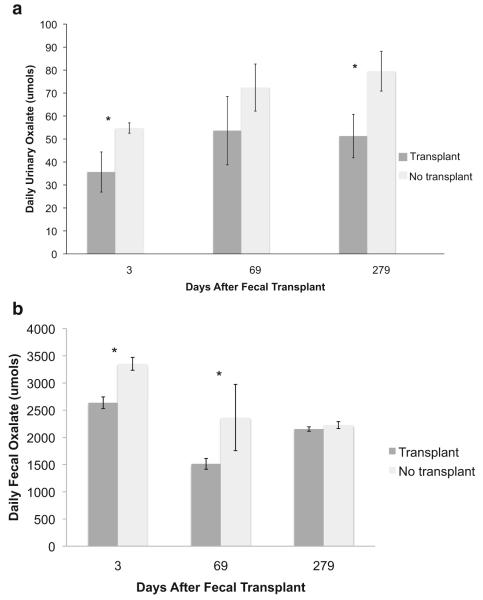
Microbial community transplants altered oxalate degradation in SDRs. There was a 323% increase in total microbial oxalate degradation of the SDRs with the transplant versus the group without the transplant (Fig. 1). The increase in oxalate degradation was apparent in the reduction of both urinary and fecal oxalate excretions. Transplant SDRs excreted 48% less urinary oxalate and 32.5% less fecal oxalate compared with the no-transplant SDR (Fig. 2). The oxalate-degrading function persisted over 9 months in the transplant SDRs. However, a significant time effect was observed in both groups with the greatest degree of degradation occurring at approximately 2 months post microbial transplant (Fig. 1).
Fig. 1.
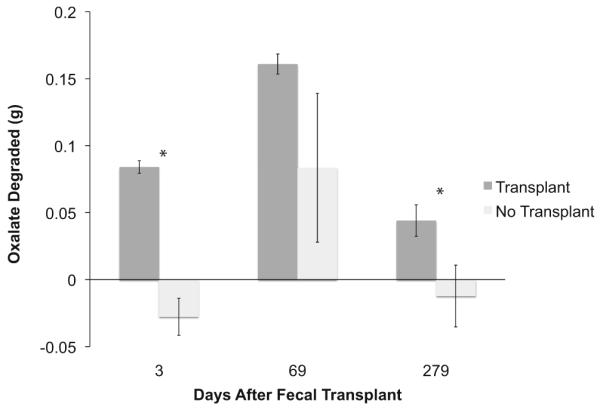
Daily oxalate degradation between the transplant and no-transplant groups 3, 69, and 279 days after the transplant. Data were analyzed with a repeated measures ANOVA (treatment (1,8) P < 0.001; time (2,18) P < 0.001). *Significant differences within each time point as assessed with a Holm’s corrected, Tukey’s post hoc analysis
Fig. 2.
Oxalate excretion between the transplant and no-transplant groups 3, 69, and 279 days after the fecal transplant. a Urinary oxalate excretion. Data were analyzed with a repeated measures ANOVA (treatment (1,8) P = 0.03). *Significant differences within each time point as assessed with a Tukey’s post hoc analysis. b Fecal oxalate excretion. Data were analyzed with a repeated measures ANOVA (treatment (1,8) P < 0.001, time (2,18) P < 0.001, time × treatment (2,32) P = 0.008). *Significant differences within each time point as assessed with a Tukey’s post hoc analysis
Microbial Transplants, Time, and Oxalate Affect the Gut Microbiota
We obtained 9,443,135 paired-end, high-quality 16S V4 region sequence reads of a median read length of 302 and an average of 145,279 ± 11,089 reads per sample. The dataset from one transplant SDR was removed because one of the inventories had <3000 sequence reads, thereby reducing the sample size of this group to five animals. A total of 21,403 OTUs were defined at 97% similarity across all fecal samples, with 739,460 OTUS removed for having <10 sequences, two mitochondrial OTUs (46 sequences) removed, and three chloroplast OTUs (110 sequences) removed across the entire dataset. Based on rarefaction analysis, 33,472 sequence reads provided a good estimate of true diversity (Fig. S1) but removed an additional 862 OTUs from the dataset. Of the remaining 20,541 OTUs, 98.9% were assignable to 13 phyla and 52.8% of OTUs were assignable to 90 genera.
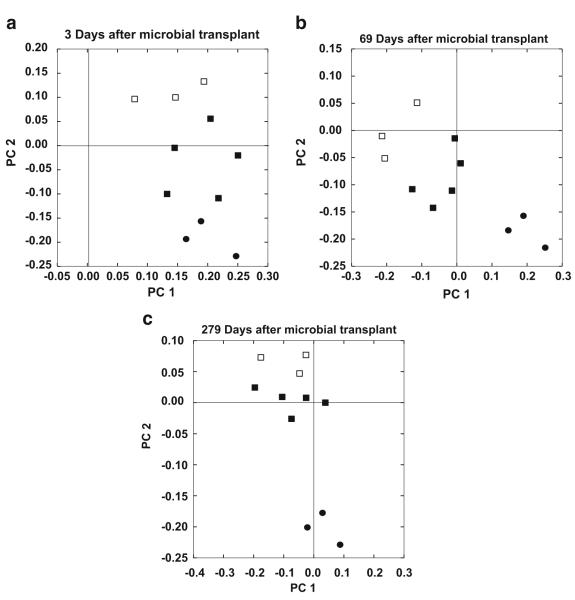
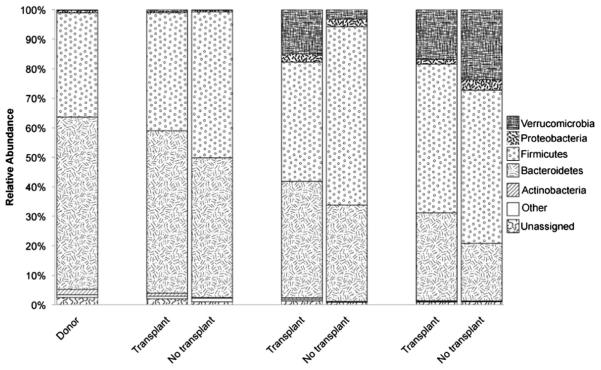
Microbial transplants resulted in a gut microbiota that was a mix of N. albigula and SDR bacteria (Figs. 3 and S2). After the transplant, there were 607 OTUs that were unique to the donor and transplant groups, with 552 OTUs present in all three groups. The two SDR groups shared 1776 OTUs in addition to the 552 OTUs shared by all groups. Each group also exhibited the presence of unique OTUs not found in the other groups (Fig. S2, Table S3). The gut microbiota of the SDRs was most similar to N. albigula 3 days after the fecal transplant compared to the other time points (Fig. 3a), and over time, the gut microbiota of the transplant group drifted to be more SDR-like (Fig. 3b, c). This drift corresponded with the proliferation of the Verrucomicrobia and Proteobacteria phyla over time in both the transplant and no-transplant groups (Fig. 4). Overall, there was a significant effect of microbial transplant on community membership in the SDRs as assessed by an analysis of similarity (ANOSIM) on the unweighted UniFrac analysis (Fig. 3). A similar transplant effect was seen in the structure of the microbial community when assessed by ANOSIM on the weighted UniFrac analysis (P = 0.01).
Fig. 3.
a–c PCoA plot based upon an unweighted UniFrac analysis depicting the effect of a fecal transplant on the microbial community at three time points a 3, b 69, and c 279 days. Symbols indicate woodrat donors (circles), transplant SDRs (closed squares), and no-transplant SDRs (open squares). There was a significant difference in community membership between transplant and no-transplant groups across all time points (ANOSIM P = 0.01). For PCoA plots (a–c), PC1 explains 8.06% of variation; PC2 explains 4.01 %
Fig. 4.
The relative abundance of the dominant phyla in the feces of the donor woodrats and the transplant and no-transplant groups 3, 69, and 279 days after the microbial transplant. Phyla in columns are stacked in the same order as the legend
The addition of dietary oxalate resulted in a net positive effect on OTUs in the transplant group, in terms of the number of OTUs exhibiting a significant increase in frequency, and anet negative effect on OTUs for the no-transplant group (Fig. 5). For the transplant group, 227 OTUs exhibited a significant increase in frequency, which included a species of Oxalobacter transferred from the donors and persisted for the duration of the experiment. For no-transplant group, only 80 OTUs exhibited a significant increase, with no Oxalobacter present. The transplant group had 160 OTUs that significantly decreased in frequency after 6 days on oxalate, whereas the no-transplant group had 205 OTUs that decreased in frequency (Table S2).
Fig. 5.
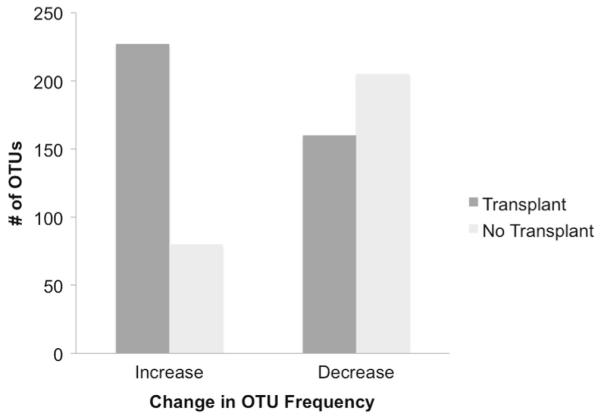
The number of OTUs exhibiting a significant change in frequency (increase or decrease) from 0 to 6 days of exposure to 1.5% dietary oxalate in the transplant and no-transplant groups (log-likelihood ratio analysis)
Discussion
Mammalian herbivores offer a vast and untapped resource of microbial communities attuned to the degradation of toxins that hold considerable relevance to public health. Oxalate, a simple organic acid that is a component in 80% of kidney stones, is metabolized exclusively by the gut microbiota, with no contribution from mammalian enzymes [16]. Thus, oxalate-degrading probiotics offer a promising target for therapeutic intervention. The objective of the current study was to determine if microbial oxalate degradation could be transferred across host species using fecal microbial communities from a mammalian herbivore that regularly consumes high amounts of oxalate. Our results reveal that the transplant of fecal microbial communities increased total oxalate degradation by more than threefold with a corresponding decrease in urinary oxalate excretion by 48 %. Our results are on par with other transplant studies involving O. formigenes, a species that requires oxalate as a carbon and energy source, which have yielded a reduction in urinary oxalate of 49–70% [36, 37].
Few studies have attempted to quantify total microbial oxalate degradation in the gut, and our study is the first to quantify total oxalate degradation after bacteriotherapy. While urinary oxalate is indicative of the probability of kidney stone formation and has been quantified in all of the probiotic studies (e.g., [26, 28, 34–36]), total oxalate degradation is more reflective of microbial oxalate metabolism. When using bacteriotherapy to reduce oxalate loads, it is important to quantify the total effect of the treatment. Previous studies did not quantify total oxalate degradation, which includes a quantification of both fecal oxalate excretion and oxalate consumption in addition to urinary oxalate excretion.
Our results suggest that the differences in oxalate degradation between the two SDR groups over the course of the 9-month experiment were multi-factorial. Donor animals had a relatively low number of unique OTUs compared to the SDR (Fig. S2) and to freshly caught animals but maintained a high capacity for oxalate degradation (unpublished). Changes in the gut microbiota composition of animals, including the same species as the donors, brought into captivity have been documented previously [49]. Microbial transplants involving the N. albigula microbiota resulted in the persistent colonization of bacteria, including O. formigenes, to the SDR and the concomitant retention of oxalate-degrading function 9 months after the transplant, indicating the robustness of the community to captive conditions. In contrast, humans and SDRs inoculated with O. formigenes alone typically exhibit a loss of the bacteria and their associated function within 5 days to 5 weeks [34–36]. Some individuals can retain O. formigenes 9 months after inoculation, indicative of potentially important differences among individuals that may facilitate colonization [32]. The use of fecal microbial transplants often results in a more persistent shift of the recipient’s microbiota for several months to up to a year or more after the initial transplant [50–53]. In the current study, the number of shared OTUs between the donor and transplant groups indicates a high degree of transferability of bacteria (Fig. S2). However, many OTUs appear to be common to both N. albigula and the SDR. Previous studies have shown that the recipients’ microbiota drifts away from the initial donor microbiota over time. In the current study, the shift in microbiota composition was dominated by an increase in the relative abundance of Verrucomicrobia and Proteobacteria. This result is similar to age-related shifts in the gut microbiota of rodents, which may stem from initially rare taxa that proliferate with the changing conditions of the host and may facilitate the changes in oxalate degradation in both SDR groups over the course of the experiment [54]. The connection between gut microbiota and oxalate degradation is further evidenced by the fact that even in the control group, several OTUs exhibited a significant increase in relative abundance with oxalate consumption (Fig. 5).
In a previous study, 116 OTUs from the gut microbiota of N. albigula exhibited a significant increase in relative abundance with oxalate consumption, whereas only one OTU significantly decreased in relative abundance [42]. For SDR in the current study, dietary oxalate likewise affected specific taxa within the gut microbiota in both the transplant and no-transplant groups. In the transplant group, this was manifested as an increase in frequency of several donor taxa including 22 that exhibited a significant increase in the Miller et al. [42] experiment. For the no-transplant group, only two of the OTUs that exhibited a significant increase in frequency were found in the group of 116 OTUs from the previous study (Fig. 5). Overall, there was a net positive effect of oxalate consumption for the transplant group and a net negative effect for the no-transplant group in terms of change in frequency. These results may represent co-adaptation over time between the gut microbiota, mammalian host, and the consumption of a diet high in oxalate.
Treatments to alleviate recurrent kidney stones, such as increasing fluid intake, decreasing oxalate consumption, and increasing calcium intake, have not led to a significant decrease in the prevalence of kidney stones [55]. Attempts at using oxalate-degrading probiotics to reduce oxalate excretion do not provide a lasting effect. The current study shows that the whole-community microbial transplants from a mammalian herbivore can produce a persistent reduction in oxalate levels and particularly in urinary oxalate, which is an important indicator for the probability of kidney stone formation [56]. While the current study focused on compositional changes to the gut microbiota with transplant and addition of oxalate, future studies will focus on the metabolic changes that occur beyond simple oxalate degradation. The gut microbiota of N. albigula represents a novel source of microbial communities that can be used as a resource to develop community-based probiotics that are more effective and persistent than what is currently available.
Supplementary Material
Acknowledgments
This project was funded by NSF (grant DEB-1342615 to M. Denise Dearing and Colin Dale) and NIH (grant 1F32DK102277-01A1 to Aaron W. Miller). We thank Kevin Kohl for help in collecting animals; Adam Schmidt, Caleb Felicetti, and Ky-Phuong Luong for help with diet trials; and Bob Weiss for feedback on the experiment.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00248-016-0800-2) contains supplementary material, which is available to authorized users.
References
- 1.James L, Butcher J. Halogeton poisoning of sheep: effect of high level oxalate intake. J Anim Sci. 1972;35:1233–1238. doi: 10.2527/jas1972.3561233x. [DOI] [PubMed] [Google Scholar]
- 2.Conyers RA, Bais R, Rofe AM. The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin Chem. 1990;36:1717–1730. [PubMed] [Google Scholar]
- 3.Massey L, Roman-Smith H, Sutton R. Effect of dietary oxalate and calcium on urinary oxalate and risk of formation of calcium oxalate kidney stones. J Am Diet Assoc. 1993;93:901–906. doi: 10.1016/0002-8223(93)91530-4. [DOI] [PubMed] [Google Scholar]
- 4.Coe F, Evan A, Worcester E. Kidney stone disease. J Clin Invest. 2005;115:2598–2608. doi: 10.1172/JCI26662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franceschi VR, Nakata PA. Calcium oxalate in plants: formation and function. Annu Rev Plant Biol. 2005;56:41–71. doi: 10.1146/annurev.arplant.56.032604.144106. [DOI] [PubMed] [Google Scholar]
- 6.Taylor E, Curhan G. Determinants of 24-hour urinary oxalate excretion. Clin J Am Soc Nephrol. 2008;3:1453–1460. doi: 10.2215/CJN.01410308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes RP, Goodman HO, Assimos DG. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 2001;59:270–276. doi: 10.1046/j.1523-1755.2001.00488.x. [DOI] [PubMed] [Google Scholar]
- 8.Dussol B, Berlan Y. Urinary kidney stone inhibitors. what is the new? Urol Int. 1998;60:69–73. doi: 10.1159/000030214. [DOI] [PubMed] [Google Scholar]
- 9.Khan S, Thamilselvan S. Nephrolithiasis: a consequence of renal epithelial cell exposure to oxalate and calcium oxalate crystals. Mol Urol. 2000;4:305–312. [PubMed] [Google Scholar]
- 10.Bihl G, Meyers A. Recurrent renal stone disease—advances in pathogenesis and clinical management. Lancet. 2001;358:651–656. doi: 10.1016/S0140-6736(01)05782-8. [DOI] [PubMed] [Google Scholar]
- 11.Asselman M, Verkoelen C. Crystal-cell interaction in the pathogenesis of kidney stone disease. Curr Opin Nephrol. 2002;12:271–276. doi: 10.1097/00042307-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Barbas C, Garcías A, Saavedra L, Muros M. Urinary analysis of nephrolithiasis markers. J Chromatogr B. 2002;781:433–455. doi: 10.1016/s1570-0232(02)00557-3. [DOI] [PubMed] [Google Scholar]
- 13.Moe O. Kidney stones: pathophysiology and medical management. Lancet. 2006;367:333–344. doi: 10.1016/S0140-6736(06)68071-9. [DOI] [PubMed] [Google Scholar]
- 14.Ramello A, Vitale C, Marangella M. Epidemiology of nephrolithiasis. J Nephrol. 2000;13:45–50. [PubMed] [Google Scholar]
- 15.Alexander R, Hemmelgarn B, Wiebe N, Bello A, Morgan C, Samuel S, Klarenbach S, Curhan G, Tonelli M. Kidney stones and kidney function loss: a cohort study. BMJ. 2012;345:e5287. doi: 10.1136/bmj.e5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgkinson A. Oxalic acid in biology and medicine. Academic Press; New York: 1977. [Google Scholar]
- 17.Allison MJ, Cook HM, Milne DB, Gallagher S, Clayman RV. Oxalate degradation by gastrointestinal bacteria from humans. J Nutr. 1986;116:455–460. doi: 10.1093/jn/116.3.455. [DOI] [PubMed] [Google Scholar]
- 18.Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 19.Hokama S, Honma Y, Toma C, Ogawa Y. Oxalate-degrading Enterococcus faecalis. Microbiol Immunol. 2000;44:235–240. doi: 10.1111/j.1348-0421.2000.tb02489.x. [DOI] [PubMed] [Google Scholar]
- 20.Ren Z, Pan C, Jiang L, Wu C, Liu Y, Zhong Z, Ran L, Ren F, Chen X, Wang Y. Oxalate-degrading capacities of lactic acid bacteria in canine feces. Vet Microbiol. 2011;152:368–373. doi: 10.1016/j.vetmic.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Daniel SL, Hartman PA, Allison MJ. Intestinal colonization of laboratory rats with Oxalobacter formigenes. Appl Environ Microbiol. 1987;53:2767–2770. doi: 10.1128/aem.53.12.2767-2770.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turroni S, Vitali B, Bendazzoli C, Candela M, Gotti R, Federici F, Pirovano F, Brigidi P. Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J Appl Microbiol. 2007;103:1600–1609. doi: 10.1111/j.1365-2672.2007.03388.x. [DOI] [PubMed] [Google Scholar]
- 23.Miller AW, Kohl KD, Dearing MD. The gastrointestinal tract of the white-throated woodrat (Neotoma albigula) harbors distinct consortia of oxalate-degrading bacteria. Appl Environ Microbiol. 2014;80:1595–1601. doi: 10.1128/AEM.03742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidhu H, Holmes R, Allison M, Peck A. Direct quantification of the enteric bacterium Oxalobacter formigenes in human fecal samples by quantitative competitive-template PCR. J Clin Microbiol. 1999;37:1503–1509. doi: 10.1128/jcm.37.5.1503-1509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann D, Kelly J, Curhan G, Anderson T, Dretler S, Preminger G, Cave D. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197–1203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campieri C, Campieri M, Bertuzzi V, Swennen E, Matteuzzi D, Stefoni S, Pirovano F, Centi C, Ulisse S, Famularo G. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 2001;60:1097–1105. doi: 10.1046/j.1523-1755.2001.0600031097.x. [DOI] [PubMed] [Google Scholar]
- 27.Turroni S, Bendazzoli C, Dipalo SC, Candela M, Vitali B, Gotti R, Brigidi P. Oxalate-degrading activity in Bifidobacterium animalis subsp. lactis: impact of acidic conditions on the transcriptional levels of the oxalyl coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes. Appl Environ Microbiol. 2010;76:5609–5620. doi: 10.1128/AEM.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieske JC, Tremaine WJ, De Simone C, O’Connor HM, Li X, Bergstralh EJ, Goldfarb DS. Diet, but not oral probiotics, effectively reduces urinary oxalate excretion and calcium oxalate supersaturation. Kidney Int. 2010;78:1178–1185. doi: 10.1038/ki.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidhu H, Allison M, Peck A. Identification and classification of Oxalobacter formigenes strains by using oligonucleotide probes and primers. J Clin Microbiol. 1997;35:350–353. doi: 10.1128/jcm.35.2.350-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange J, Wood K, Wong H, Otto R, Mufarrij P, Knight J, Akpinar H, Holmes R, Assimos D. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology. 2012;79:1286–1289. doi: 10.1016/j.urology.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharlamb V, Schelker J, Francois F, Jiang J, Holmes RP, Goldfarb DS. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. J Endourol. 2011;25:1781–1785. doi: 10.1089/end.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan S, Richardson A, Kaul P, Holmes R, Allison M, Stewart C. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol. 2002;68:3841–3847. doi: 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoppe B, Von Unruh G, Blank G, Rietschel E, Sidhu H, Laube N, Hesse A. Absorptive hyperoxaluria leads to an increased risk for urolithiasis or nephrocalcinosis in cystic fibrosis. Am J Kidney Dis. 2005;46:440–445. doi: 10.1053/j.ajkd.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Sidhu H, Allison M, May Chow J, Clark A, Peck A. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J Urol. 2001;166:1487–1491. [PubMed] [Google Scholar]
- 35.Hoppe B, Beck B, Gatter N, Von Unruh G, Tischer A, Hesse A, Laube N, Kaul P, Sidhu H. Oxalobacter formigenes: a potential tool for the treatment of primary hyperoxaluria type 1. Kidney Int. 2006;70:1305–1311. doi: 10.1038/sj.ki.5001707. [DOI] [PubMed] [Google Scholar]
- 36.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol. 2011;300:G461–G469. doi: 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palgi N, Ronen Z, Pinshow B. Oxalate balance in fat sand rats feeding on high and low calcium diets. J Comp Physiol B. 2008;178:617–622. doi: 10.1007/s00360-008-0252-1. [DOI] [PubMed] [Google Scholar]
- 38.Belenguer A, Ben Bati M, Hervás G, Toral PG, Yáñez-Ruiz DR, Frutos P. Impact of oxalic acid on rumen function and bacterial community in sheep. Animal. 2013;7:940–947. doi: 10.1017/S1751731112002455. [DOI] [PubMed] [Google Scholar]
- 39.Knight J, Deora R, Assimos DG, Holmes RP. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis. 2013;41:187–196. doi: 10.1007/s00240-013-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung J, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirley EK, Schmidt-Nielsen K. Oxalate metabolism in the pack rat, sand rat, hamster, and white rat. J Nutr. 1967;91:496–502. doi: 10.1093/jn/91.4.496. [DOI] [PubMed] [Google Scholar]
- 42.Miller A, Oakeson K, Dale C, Dearing M. The effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.00216-16. doi: 10.1128/AEM.00216-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol Lett. 2014;17:1238–1246. doi: 10.1111/ele.12329. [DOI] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso J, Lauber C, Walters W, Berg-Lyons D, Lozupone C, Turnbaugh P, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bokulich N, Subramanian S, Faith J, Gevers D, Gordon J, Knight R, Mills D, Caporaso J. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2012;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 48.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohl KD, Skopec MM, Dearing MD. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conserv Physiol. 2014;2:cou009. doi: 10.1093/conphys/cou009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grehan M, Brody T, Leis S, Campbell J, Hazel M, Wettstein A. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J Clin Gastroenterol. 2010;44:551–561. doi: 10.1097/MCG.0b013e3181e5d06b. [DOI] [PubMed] [Google Scholar]
- 51.Khoruts A, Dicksved J, Jansson J, Sadowsky M. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 52.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genet Res. 2010;20:1411–1419. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge E, Dutta A, Dutta S, Fricke W. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One. 2013;8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langille M, Meehan C, Koenig J, Dhanani A, Rose R, Howlett S, Beiko R. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. doi: 10.1186/s40168-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Germino G, Kirkali Z. Urinary stone disease: research challenges and opportunities. NIDDK; [Accessed 10 January 2016]. 2015. http://www.niddk.nih.gov/news/events-calendar/Pages/Urinary-Stone-Disease-Research-Challenges-Opportunities_04-2015.aspx#tab-minutes. [Google Scholar]
- 56.Trinchieri A. Epidemiology of urolithiasis: an update. Clin Cas Min Bone Metab. 2008;5:101–106. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.