Abstract
Precise genome editing in livestock is of great value for the fundamental investigation of disease modeling. However, genetically modified pigs carrying subtle point mutations were still seldom reported despite the rapid development of programmable endonucleases. Here, we attempt to investigate single-stranded oligonucleotides (ssODN) mediated knockin by introducing two orthologous pathogenic mutations, p.E693G for Alzheimer's disease and p.G2019S for Parkinson's disease, into porcine APP and LRRK2 loci, respectively. Desirable homology-directed repair (HDR) efficiency was achieved in porcine fetal fibroblasts (PFFs) by optimizing the dosage and length of ssODN templates. Interestingly, incomplete HDR alleles harboring partial point mutations were observed in single-cell colonies, which indicate the complex mechanism of ssODN-mediated HDR. The effect of mutation-to-cut distance on incorporation rate was further analyzed by deep sequencing. We demonstrated that a mutation-to-cut distance of 11 bp resulted in a remarkable difference in HDR efficiency between two point mutations. Finally, we successfully obtained one cloned piglet harboring the orthologous p.C313Y mutation at the MSTN locus via somatic cell nuclear transfer (SCNT). Our proof-of-concept study demonstrated efficient ssODN-mediated incorporation of pathogenic point mutations in porcine somatic cells, thus facilitating further development of disease modeling and genetic breeding in pigs.
Keywords: CRISPR/Cas9, genetically modified pigs, orthologous point mutation, single-stranded oligonucleotide, somatic cell nuclear transfer
Introduction
As an important livestock, the domestic pig has been widely studied over the last decade. Many genetically engineered pigs have been generated to improve production yield, growth efficiency, and disease resistance.1,2,3 Moreover, pigs were also modified to produce disease models and used as organ donors for xenotransplantation because they share similar genetic, physiological, and anatomical features with human.4 Swine models are important for providing valuable preclinical data of new therapeutic strategies and deciphering the pathogenesis of human disease. Historically, genetically modified pigs were generated either via random integration5 or conventional homologous recombination.6 However, random insertions often cause variable expression levels of the transgene,7 whereas conventional HR targeting is extremely inefficient,8 and the insertion of antibiotic selection cassettes has raised concern about their clinical prospects.
Recently, the rapid development of custom endonucleases, such as Zinc Finger Nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs) and especially, the recent clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) technology, has opened a new era for site-specific genome editing. Zygote microinjection of Cas9/gRNA has been shown to be an efficient strategy for generating gene knockout pigs.9,10 On the other hand, CRISPR-mediated gene editing in porcine fetal fibroblasts (PFFs), combined with somatic cell nuclear transfer (SCNT), could also produce gene knockout pigs with a high efficiency.11,12
Numerous pathogenic point mutations have been identified in traditional genetic studies. Furthermore, genome-wide association studies, a new tool for identifying genetic variants, have successfully identified thousands of disease-associated single-nucleotide polymorphisms. Swine models can be a powerful tool for elucidating the pathogenic effect of these variants. However, gene-edited pigs harboring subtle modifications were rarely reported despite the advent of programmable endonucleases. Recently, efficient site-specific knockin of human cDNA13 and defined base substitution14 were achieved in pigs through CRISPR/Cas9-induced HDR in zygotes. Although direct zygote injection appears to be a simple and popular strategy for generating knockin pigs, it is limited by technical challenges such as undesired mosaic13,14,15 and unpredictable HDR frequency.16 In contrast, SCNT enables a high rate of obtaining transgenic animals via selecting positive donor cells prior to the cloning procedure and no mosaic occurs. Therefore, CRISPR-mediated HDR in PFFs, in combination with SCNT, can be another important strategy for the generation of genetically engineered pigs harboring defined point mutations.
We had previously reported the CRISPR-mediated gene targeting in pigs,12 and the high knockout level encouraged us to extend its application to precise genetic modification. In this study, efficient ssODN-mediated point mutations were achieved at three different loci in PFFs. Interestingly, both the complete HDR and partial HDR were found in single-cell colonies, and the effect of the mutation-to-cut distance on incorporation rate was investigated. Finally, we generated one cloned piglet carrying the orthologous p.C313Y mutation via SCNT and no off-target mutation was detected among potential sites.
Results
The gene-editing strategy for introducing orthologous human mutations into porcine genome
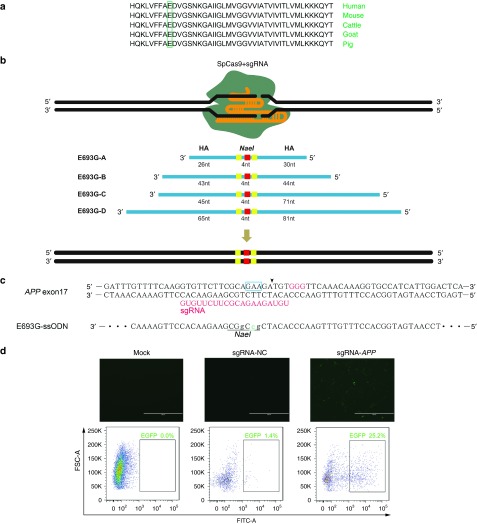
Alzheimer's disease (AD) is characterized by increased amyloid β-protein (Aβ) level in amyloid plaques and amyloid angiopathy. Several pathogenic mutations have been found in the amyloid precursor protein (APP) gene. The Arctic mutation (p.E693G) was firstly found to cause early-onset AD in a Swedish family and was reported to enhance Aβ protofibril formation.17 Moreover, the glutamic acid for mutation is highly conserved among different species (Figure 1a). To introduce the orthologous APPE693G mutation into the porcine genome, we chose a sgRNA at the porcine APP locus whose potential cleavage site is nearest to the intended mutation (Figure 1b,c). A single-strand annealing (SSA) reporter construct containing partially duplicated enhanced green fluorescent protein (EGFP) coding sequences and the sgRNA target site was used to detect the cleavage activity of APP-specific sgRNA (Supplementary Figure S1). Following the cleavage by Cas9/gRNA and subsequent SSA-mediated repair, the functional EGFP cassette and fluorescence would be restored. The result showed that this sgRNA was capable of making double-strand breaks at the target site (Figure 1d). In previous studies, two kinds of templates have been used for custom endonuclease-mediated HDR. Double-strand plasmid donors were used to incorporate large elements18whereas ssODNs were often used for short tag insertions and base substitutions.14 Here, four ssODNs with different length ranging from 60-nt to 150-nt were synthesized (Figure 1b). All ssODN templates harbored one base substitution for the pathogenic mutation and the other two synonymous mutations creating a new NaeI site (Figure 1c). We firstly determined the optimal ssODN dosage for robust HDR by electroporating the ssODN into PFFs with different doses. Three days postelectroporation, genomic DNA was extracted and the purified polymerase chain reaction (PCR) products were digested with NaeI. The restriction fragment length polymorphism (RFLP) results showed that the 3 μmol/l group exhibited a higher HDR frequency than the 0.5 and 1.5 μmol/l group (Figure 2a). A higher concentration of 5 μmol/l presented no obvious enhancement of the HDR efficiency (Supplementary Figure S2). Hence, the dosage of ssODN was fixed on 3 μmol/l in subsequent experiments. To examine the effect of the ssODN length on HDR, different ssODN templates were electroporated into PFFs with the same amount of Cas9/gRNA expression vector. The RFLP analysis revealed that the 120-nt ssODN supported the highest HDR efficiency (more than 10%), whereas further elongation of the ssODN reduced the level of HDR (Figure 2b and Supplementary Figure S2). We analyzed the 150-nt ssODN donor via cloning and Sanger sequencing as described previously. No mutation was detected in any of the 27 clones obtained from the control experiment. However, mutations were detected in 11 out of 29 (~38%) clones harboring the ssODN sequences (Supplementary Figure S2), indicating that the errors during ssODN synthesis may account for the lower HDR efficiency. Recently, the RAD51 agonist RS-1 was reported to dramatically enhance dsDNA-mediated HDR.19,20 In our case, however, no significant improvement of the HDR efficiency was observed with the treatment of RS-1 (Supplementary Figure S3).
Figure 1.
Targeting strategy for introducing orthologous human mutations into porcine genome. (a) The target amino acid for mutation are highly conserved among different species. The green box indicates the target amino acid. (b) Schematic representation of the targeting strategy. Four ssODN donors with different lengths of homology arms were designed. All ssODN templates harbor three point mutations. The red square indicates the mutation for p.E693G, whereas two synonymous mutations creating a NaeI site are shown in yellow. (c) Sequence comparison of porcine APP exon 17 with the ssODN donor. The sgRNA and PAM are shown in red, and the blue box indicates the glutamic acid bases for mutation. The lowercase bases in the ssODN indicate targeted mutations, while the new NaeI restriction site is underlined. The potential cleavage site is indicated by an arrowhead. (d) Functional validation of Cas9/gRNA by the single-strand annealing (SSA) assay. The EGFP-SSA reporter was cotransfected with the APP-specific Cas9/gRNA vector or the control Cas9/gRNA vector. The expression of EGFP was detected by fluorescence microscope and flow cytometry (scale bar = 1,000 μm).
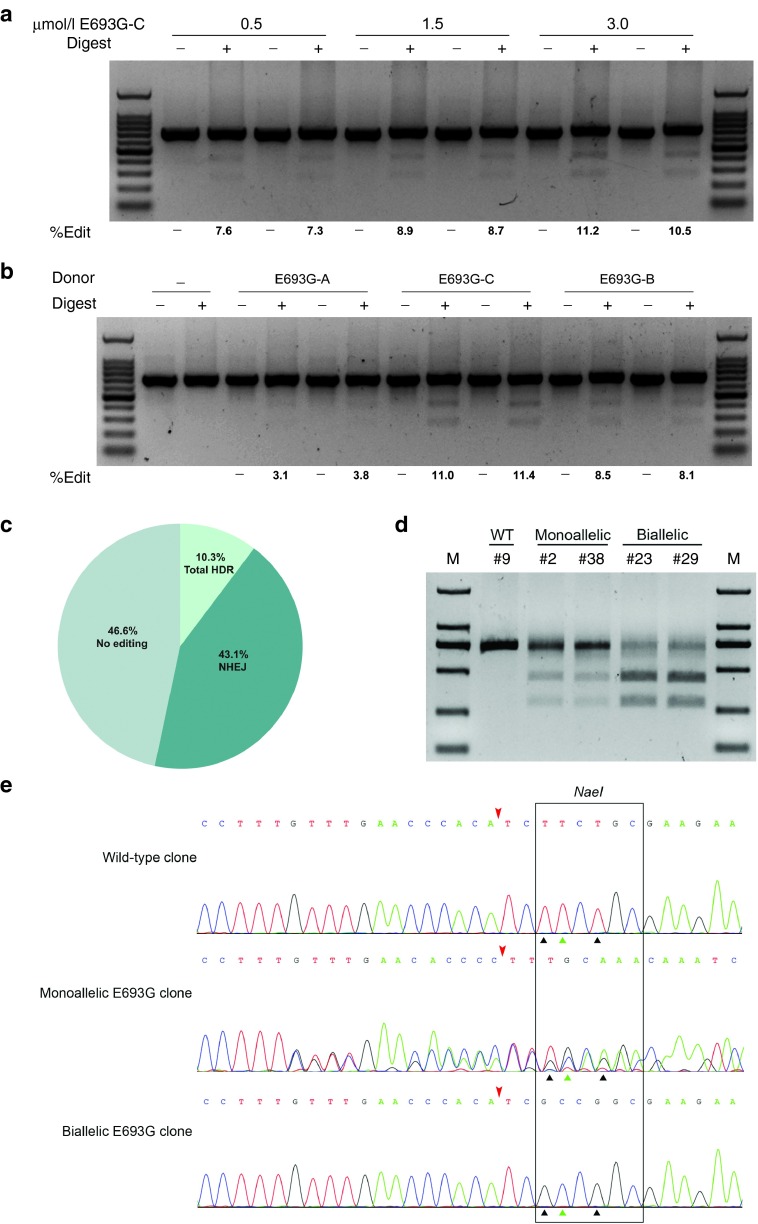
Figure 2.
CRISPR/Cas9-based ssODN-mediated knockin of point mutations at porcine APP locus. (a) The 120-nt ssODN donor was transfected into porcine fetal fibroblasts (PFFs) with different doses to determine the optimal dosage for homology-directed repair (HDR). (b) Four ssODN donors with different lengths of homology arms were transfected into PFFs. The level of HDR was analyzed by the RFLP assay and is shown at the bottom of each gel. The electroporation was performed in biological duplicate. (c) PFF colonies derived from one single cell were isolated by the limiting dilution method. The mutation patterns (NHEJ, HDR, or no editing) for all colonies were shown. (d) RFLP analysis of colonies harboring monoallelic or biallelic HDR alleles. Purified PCR products encompassing the target regions were digested with NaeI. (e) Sequencing chromatogram of representative HDR colonies. The Cas9 cleavage sites were marked with red arrowheads. The green triangles indicate the mutation for p.E693G, while black triangles indicate the synonymous mutations.
We then sought to obtain single-cell colonies carrying the precise Arctic mutation, which would be suitable donor cells of SCNT for generating gene-modified pigs. The PFFs electroporated with the Cas9/gRNA and ssODN were plated via limiting dilution, and single-cell colonies were picked nine days later. A total of 35 colonies were picked among which 29 colonies were recovered. The overall mutation patterns were determined by genotyping each colony (Figure 2c). The whole mutation rate reached 53.4% whereas the HDR accounted for 10.3%, which is consistent with the previous RFLP analysis. Both monoallelic and biallelic HDR colonies were identified by sequencing and RFLP analysis (Figure 2d,e). To sum up, our results demonstrated that the ssODNs could serve as an efficient donor for CRISPR/Cas9-assisted precise point mutations in PFFs.
Oligonucleotide-mediated precise gene editing at the porcine LRRK2 locus
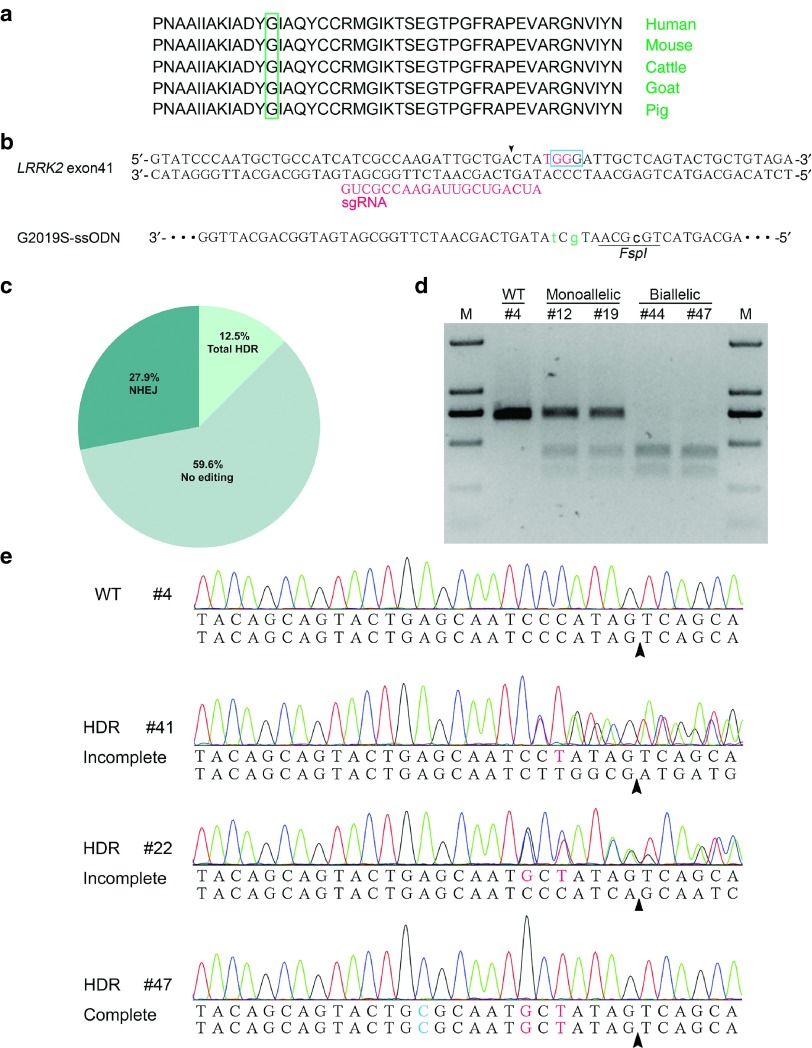
To expand the application of ssODN-mediated seamless gene editing, we decided to introduce a missense pathogenic mutation into the exon41 of porcine LRRK2 locus. The resultant p.G2018S mutation in porcine LRRK2 protein is orthologous to the pathogenic p.G2019S mutation in human (Figure 3a), which has been widely reported in Parkinson's disease patients as one of the most common genetic cause of PD.21 We designed a sgRNA whose protospacer adjacent motif (PAM) overlapped with the glycine codon for mutation (Figure 3b). A 120-nt ssODN was used as the HDR template in which two point mutations account for the p.G2019S mutation and one synonymous mutation creates a new FSPI site (Figure 3b). The Cas9/gRNA plasmid and ssODN were electroporated into PFFs, and the mixed PCR products were sequenced to validate the activity of precise gene editing. The multi-peaks around the potential cutting site revealed the non-homologous end-joining (NHEJ) mutations, whereas the small peaks at the designed site for substitution indicated the HDR events (Supplementary Figure S4). After validating the intended gene editing, we performed limited dilution and identified 52 single-cell colonies via sequencing. The total mutation rate was 40.4% in which the HDR efficiency reached 12.5% (Figure 3c). The RFLP analysis also confirmed the introgression of the FspI site in monoallelic and biallelic HDR colonies (Figure 3d). Interestingly, in addition to the expected HDR colonies harboring all three point mutations in the ssODN template, we also observed the partial HDR in which only one or two point mutations close to the cutting site were incorporated (Figure 3e). This kind of incomplete HDR or imperfect HDR, which has also been reported by other studies,22,23 appears to be dependent on the complex DNA repair processes rather than the errors during ssODN synthesis.
Figure 3.
Efficient incorporation of pathogenic human mutations at porcine LRRK2 locus. (a) The conserved glycine for mutation in LRRK2 protein of different species. (b) Sequence comparison of porcine LRRK2 exon 17 with the corresponding ssODN donor. The glycine bases are labeled with a blue open box, whereas the sgRNA and PAM are shown in red. Three point mutations are indicated by lowercase letters and the FspI restriction site is underlined. (c) Mutation patterns of 52 single-cell colonies were determined by Sanger sequencing. (d) Representative colonies carrying monoallelic or biallelic homology-directed repair (HDR) alleles were confirmed by RFLP assay. (e) Sequencing chromatogram of representative colonies harboring incomplete or complete HDR alleles. Colony #41, #22 and #47 contain one, two and three designed point mutations, respectively.
The mutation-to-cut distance affects the incorporation rate of different point mutations
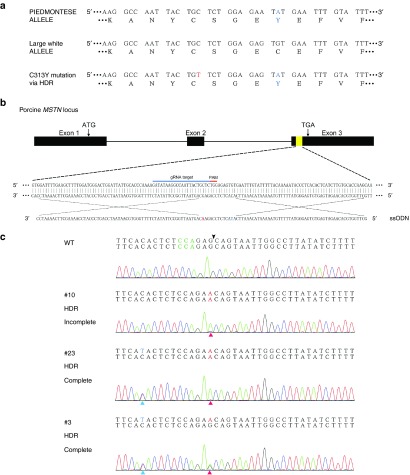
Myostatin, a member of the TGFβ superfamily, mainly acts as a negative regulator of muscle growth. It has been widely studied since the first observation that myostatin-null mice exhibited a dramatic and widespread increase in skeletal muscle mass.24 The muscular hypertrophy phenotype was reported in a child harboring the biallelic point mutation in the noncoding region of the MSTN gene.25 Naturally occurred mutations have also been observed in the MSTN locus of several double-muscled cattle breeds such as Belgian Blue26 and Piedmontese.27,28 Specially, the c.938G>A mutation at the Piedmontese MSTN locus results in the substitution of a highly conserved cysteine to tyrosine (p.C313Y) in the mature region of the protein. This mutation leads to the distortion of the cysteine knot structure, which is required for active conformation and receptor binding.29 To further investigate the effect of the mutation-to-cut distance on incorporation rate, we intended to introduce a missense point mutation into the exon3 of porcine MSTN locus, mimicking the orthologous p.C313Y mutation observed in Piedmontese (Figure 4a). We designed an ssODN template in which the intended nucleotide T is 11 bp away from the potential cleavage site (Figure 4b). In addition, one silent mutation was introduced right at the expected cutting site to block the recleavage activity of CRISPR/Cas9 (Figure 4b). To explore whether the ssODN could achieve the intended mutation, both the MSTN-specific Cas9/gRNA plasmid and the ssODN template were electroporated into PFFs. As expected, the Sanger sequencing of the mixed PCR products revealed intended point mutation characterized by the small T peak beneath the wild-type C peak (Supplementary Figure S5). However, the obvious multi-peaks around the potential cutting site suggested that the majority of the edited PFFs were still repaired via NHEJ.
Figure 4.
Incomplete homology-directed repair (HDR) during the introgression of MSTN p.C313Y mutation. (a) The targeting strategy for introducing the p.C313Y mutation into porcine MSTN locus. The MSTN allele of Piedmontese and Large White are shown. The corresponding amino acid sequences are listed below where the p.C313Y mutation is highlighted in blue. (b) A schematic illustration showing the location of the sgRNA and ssODN. The nucleotide T in blue is responsible for the p.C313Y mutation, while the nucleotide A in red is the silent mutation preventing the recleavage activity on HDR alleles. (c) Genotype and sequencing chromatogram of representative colonies harboring WT allele, incomplete HDR allele or complete HDR allele. The arrowhead indicates the cleavage site of Cas9.
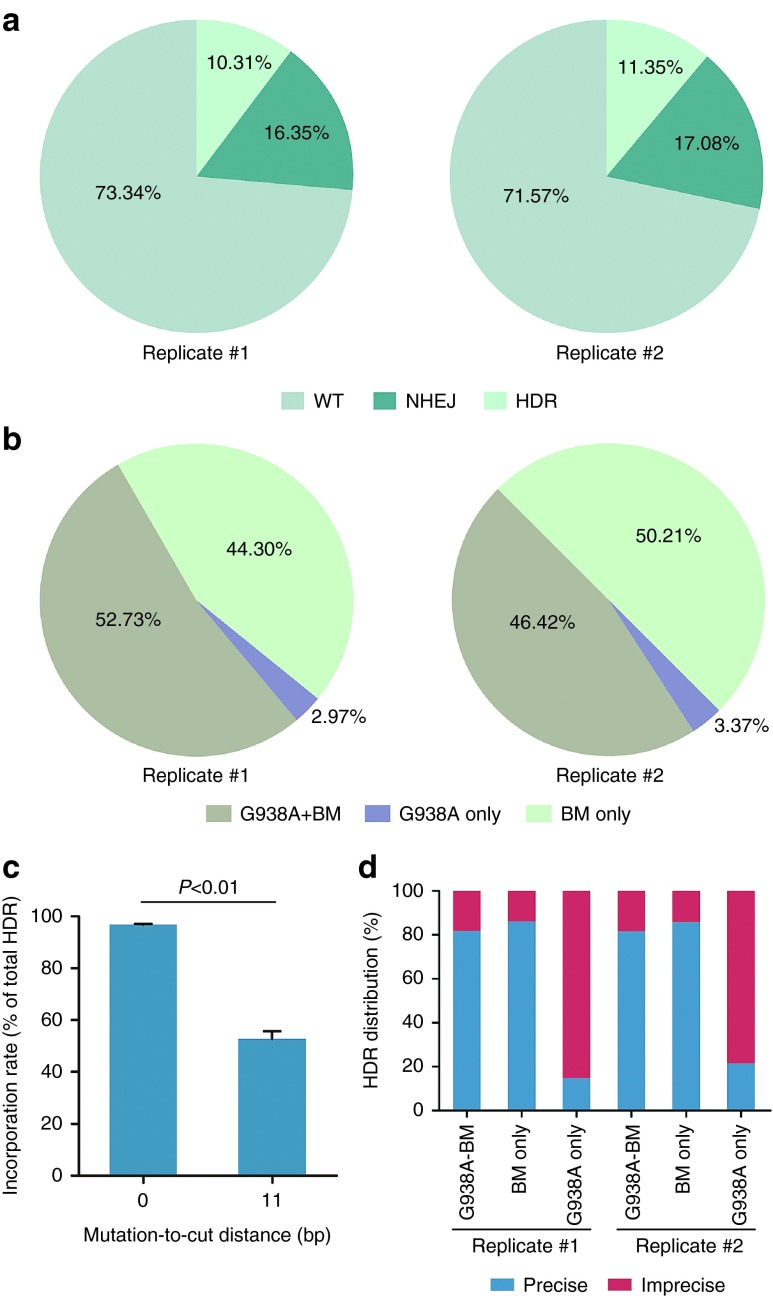
After confirmation of the ssODN-mediated HDR at the porcine MSTN locus, we sought to isolate single-cell colonies carrying defined point mutations. Consistent with the case for p.G2019S mutation, we also observed incomplete HDR in which only the blocking mutation was introduced (Figure 4c and Supplementary Table S4). To further characterize the incorporation rate of the two point mutations, we performed deep sequencing of the mixed PCR amplicons (Figure 5a and Supplementary Figure S6). Indeed, substantial reads carrying only the blocking mutation were detected, indicating that the blocking mutation was incorporated more frequently than the c.938G>A mutation (Figure 5b,c). Our results, combined with previous studies in human cell lines30 and mouse zygotes,22 suggested that the mutation-to-cut distance was a key and universal factor influencing the incorporation rate in mammalian cells. Furthermore, the point mutation near the cutting site exhibited a higher HDR efficiency than the one which is distant from the DSBs. Interestingly, amplicons harboring the c.938G>A mutation but not the blocking mutation, although in a small proportion, were also detected via deep sequencing (Figure 5b). It is possible that the blocking mutation could not entirely prevent the recutting activity on the HDR alleles so that only the c.938G>A mutation was preserved. Indeed, the majority of this kind of amplicons contained additional mutations (Figure 5d), which might indicate the NHEJ repair after precise HDR events.
Figure 5.
The effect of mutation-to-cut distance on incorporation rate of point mutations. (a) MSTN-specific Cas9/gRNA and ssODN donor were transfected into porcine fetal fibroblasts. Deep sequencing was performed to analyze the mutation patterns of mixed PCR amplicons. (b) All homology-directed repair (HDR) amplicons were classified according to different point mutations. Substantial reads carried only the blocking mutation but not the c.938G>A mutation. (c) Incorporation rate of the c.938G>A mutation was significantly lower than that of the blocking mutation (error bars show mean ± standard error of the mean). Statistical analysis was performed using two-tailed Student's t-test. (d) Distribution of precise and imprecise reads for three kinds of HDR amplicons. Different from other kinds of HDR amplicons, the HDR amplicons carrying only the c.938G>A mutation often contained additional mutations.
Generation of gene-modified pigs harboring the orthologous point mutation
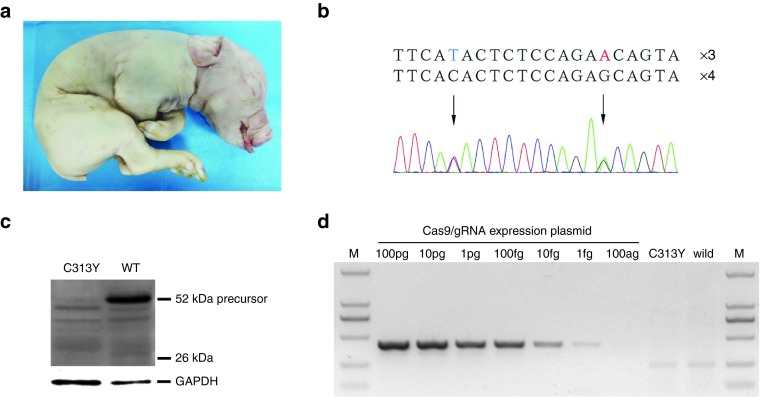
Two colonies carrying the p.C313Y mutation were chosen as the donor cells of SCNT. A total of 930 reconstructed embryos were transferred to four surrogates. One pregnancy was carried to term eventually and gave birth to one stillborn piglet (Figure 6a). As expected, the sequencing result confirmed that the piglet is heterozygous for the intended mutations (Figure 6b), which is consistent with the genotype of the donor cells. To our surprise, the western blot analysis showed significant decrease of myostatin precursor in the cloned pig, indicating the remarkable effect of the heterozygous p.C313Y mutation (Figure 6c). In addition, we screened the MSTN-specific Cas9/gRNA plasmid to examine whether these foreign transgenes existed in the genome of the cloned pig. The highly sensitive primers could detect as low as 1 fg plasmid DNAs. However, the correct transgene band was not detected in the mutant pig (Figure 6d), indicating no integration of the targeting vector into the genome during electroporation and subsequent cloning procedure.
Figure 6.
Generation of gene-modified pigs harboring the orthologous point mutation via SCNT. (a) Photograph of the newborn piglet. (b) Genotype of the piglet was determined by Sanger sequencing. One allele harbors the intended point mutations while the other remains wild-type. (c) The expression level of myostatin in the cloned piglet was analyzed by western blotting. (d) Detection of Cas9/gRNA construct in the cloned piglet by genomic PCR. MSTN-specific Cas9/gRNA expression vector was used as the positive control. The genomic DNA of wild-type Large White was used as the negative control.
The off-target effect has always been a main concern hindering the widespread application of programmable endonucleases. To examine whether off-target mutations occurred in our cloned piglet, 15 potential off-target sites (OTS) were selected from the porcine genome (Supplementary Figure S7). Specific primers were designed to amplify the corresponding regions, and the sequencing result showed that no mutation occurred in any OTS (Supplementary Figure S7).
Discussion
Here, by optimizing the dosage and length of ssODN donors, we improved the incorporation rate of specific point mutations at porcine APP locus. The length of ssODN determines the homology between the template and the target site, which is crucial for HDR. Longer ssODNs can maintain sufficient homology by preventing from exonuclease degradation. However, longer ssODNs are likely to increase the risk of incorrectly synthesized oligonucleotides, and the possible secondary structure can decrease the amount of donor available for HDR.31,32 Indeed, we found that the 120-nt ssODN donor presented the highest HDR efficiency, whereas further elongation failed to increase the frequency. Furthermore, substantial mutations detected in the 150-nt ssODN suggest that, at least in this case, the incorrectly synthesized oligonucleotides may reduce the level of HDR. Hence, rationally designing the length of ssODN donors is important for robust HDR.
A previous research showed that a combined use of TALEN and the ssODN templates introduced a point mutation into porcine MSTN locus at a frequency of 3%.33 In the same study, ssODN-mediated modification was also investigated using the CRISPR system. However, the level of HDR induced by 90-nt donors varied significantly between two loci. During preparation of this manuscript, Yang et al.34 reported the generation of genetically humanized pigs via CRISPR-based ssODN-mediated gene editing. Using a 99-nt ssODN donor, they achieved a HDR efficiency of 5.6% by drug selection. Nevertheless, the use of antibiotics has increasingly raised concern about the safety of animal products. In comparison, the total HDR efficiency exceeds 10% for all three loci in our work, indicating the reliability of ssODN-mediated HDR without the use of selection markers.
The transfection efficacy is a basic and important factor affecting the activity of gene editing. Although we previously improved the transfection efficacy of pEGFP-N1 to approximately 90% in PFFs, a larger plasmid such as the Cas9/gRNA constructs used in this study may exhibit a lower efficacy. It was recently reported that increasing the postpulses recovery time massively improved the transfection efficacy of large plasmids in primary cells.35 Hopefully, this enhancement in transfection efficacy can be an easy way to increase the basic level of CRISPR-mediated genome editing. A few studies reported that the efficiency of Cas9/gRNA-induced HDR was dramatically enhanced by inhibiting key NHEJ pathway proteins.36,37 By using a reporter-based screening method, Yu et al.38 identified small molecules enhancing the efficiency of CRISPR-based HDR. It was also reported that the asymmetry39 and chemical modification40 of ssODNs could affect the level of HDR. All these strategies should be validated in PFFs to further increase the ssODN-mediated HDR.
Several studies have found that the mutation-to-cut distance plays an important role in ssODN-mediated base substitutions.22,41 Here, we conducted the direct efficacy comparison between point mutations with different mutation-to-cut distances. Indeed, we observed a lower incorporation rate for point mutations distal to the cleavage site. Surprisingly, a mutation-to-cut distance of 11 bp caused a significant difference in HDR efficiency between two point mutations, suggesting an even more critical role of the mutation-to-cut distance than expected. Considering the restriction of the PAM motif, the mutation-to-cut distance is likely to be detrimental for CRISPR/Cas9-mediated single base-pair modifications. In this case, different genome editing tools such as other CRISPR systems or Cas9 orthologs with diverse PAM motifs can be used alternatively. Of note, very short homology arm in DNA plasmids could drive efficient site-specific insertions, likely by a MMEJ-based mechanism.42 It is possible that the incomplete oligonucleotide templates containing very short homology arms were involved in the DNA repair processes. Based on the observations, we here hypothesize that ssODN-mediated HDR occurs through two rounds of annealing as described in yeast,43,44 and the central homology arm between point mutations results in the incomplete HDR (Supplementary Figure S8). In this model, after resection of the 5' terminal of the DSB, the 3' terminal of the ssODN anneals to the homologous region on the 3' terminal single strand of the DSB. The nonhomologous sequence is clipped away, and DNA synthesis occurs to copy the remaining sequence of the ssODN donor. The second annealing then takes place between the newly synthesized sequence and the other 3' end of the DSB after removal of the ssODN. Similarly, following clipping the nonhomologous region, gap filling and ligation, the repair is completed. In the case of a short mutation-to-cut distance, the distal homology arm predominates, and the DNA repair processes are prone to the complete HDR. In contrast, when a long mutation-to-cut distance exists, the central homology arm predominates and the incomplete HDR occurs. Of note, the exonuclease degradation may create truncated ssODN donors. In this situation, although the repair event is complete in terms of the donor used, the resultant allele contained only partial point mutations (Supplementary Figure S8). According to our hypothesis, adequate elongation or chemical modification of the ssODN may increase the level of complete HDR by preserving the distal homology arms.39,40
The effect of different mutations on myostatin expression is somewhat complex. A previous study reported that the expression of myostatin varies dramatically in mutant single-cell colonies. The monoallelic mutation in one colony leads to a higher level of expression where the mutant protein was predicted to induce the negative feedback regulation of myostatin expression.45 However, the monoallelic MSTN mutation in Meishan pigs resulted in a decreased level of expression.46 In our case, we speculated that the mutant protein binds to the wild-type myostatin as a dominant negative mutant, and the resultant multimers are prone to the intrinsic protein degradation pathway. The mechanism underlying the unusual expression resulting from different myostatin mutations needs to be studied further.
Early studies47,48 indicated that CRISPR/Cas9 showed a low specificity in human cell lines. However, recent studies49,50 using the whole-genome sequencing suggested that rare off-target mutations occurred in Cas9-modified pluripotent stem cells and mice. These distinct observations may result from the following two factors: (i) different off-target sites chosen for detection since the off-target effect is site-dependent, (ii) differences in chromatin structure and epigenomic landscape between cell types. Hence, a more comprehensive and systematic evaluation of the specificity profile is still required for its further application in disease modeling and gene therapy. In this study, we assessed the off-target activity in the cloned piglet by screening 15 potential off-target sites and no mutation was detected. We cannot rule out potential off-target mutagenesis since the genome-wide analysis was not performed, however, the risk of embryonic lethality resulting from potential off-target mutagenesis can be reduced by mixing several positive colonies during SCNT. Of note, cell injury in SCNT and abnormal reprogramming may also cause embryonic lethality. Considering that only one piglet was delivered, it is hard to conclude whether the off-target mutations should account for the death. Nevertheless, the reliable HDR efficiency achieved in this work allows efficient isolation of positive colonies. We believe that live-born edited piglets can be generated by performing more SCNT and embryo transfer experiments.
In conclusion, we have demonstrated that the ssODNs could serve as an efficient HDR donor at Cas9-induced DSBs in PFFs. Pathogenic point mutations were efficiently incorporated into different loci of the porcine genome at a frequency of more than 10%. Both complete and incomplete HDR were found in single-cell colonies. The mutation-to-cut effect caused a remarkable difference in HDR efficiency between two point mutations, thus providing a valuable reference for ssODN-mediated gene editing. Finally, we generated one cloned piglet harboring the p.C313Y mutation at MSTN locus via SCNT. Our work expands the application of CRISPR in a more precise manner by oligonucleotide-based introgression of intended point mutations, thus holding great promise for custom disease modeling and precision medicine.
Materials and methods
Ethics statement. All animal studies were approved by the Animal Welfare and Research Ethics Committee at Jilin University, and all procedures were conducted strictly in accordance with the Guide for the Care and Use of Laboratory Animals. All surgeries were performed under anesthesia, and every effort was made to minimize animal suffering.
CRISPR/Cas9 construction and HDR template. The pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang51 (Addgene plasmid # 42230). Two complementary oligonucleotides with appropriate adaptors were synthesized and then annealed in standard Taq buffer (NEB, Beijing, China). The resultant double-strand DNA was ligated to the BbsI sites of the vector backbone to form the intact targeting plasmid. The single-strand oligonucleotides used for HDR were synthesized and purified through PAGE (GENEWIZ, Suzhou, China). The detailed sequences were provided in Supplementary Table S2.
Cell culture. PEFs were derived from 33-day-old Large White fetuses. The fetal bodies without heads, tails, limbs, and viscera were cut into small pieces. Then they were disaggregated in the culture medium containing 200 U/ml collagenase IV (type IV, 260 U/mg, Gibco, Grand Island, NY), 0.0125 mg/ml DNase I (2,000 U/mg, Sigma, St. Louis, MO), 20% fetal bovine serum (Gibco, Grand Island, NY) and 1% penicillin/streptomycin (Gibco) for 4–6 hours. Isolated PFFs were then resuspended and cultured in 10-cm cell culture dishes until sub-confluence. Cells at passage 1 were frozen in fetal bovine serum containing 10% dimethylsulfoxide. The isolated PFFs were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum. The RAD51-stimulatory compound RS-1 (Sigma) was added to the culture media from 10 mmol/l stock solutions in dimethyl sulfoxide.
PFF transfection and selection. Approximately 3 × 106 PFFs were electroporated with 35 μg Cas9/gRNA targeting vector and ssODN template in the context of 250 μl Opti-MEM (Gibco) using BTX ECM 2001 (Harvard Bioscience, Holliston, MA). The electroporation parameters for 2 mm gap cuvettes were described previously.12 To explore ssODN-mediated HDR, the electroporated PFFs were cultured at 37.0 °C for 72 hours and then trypsinized for genomic extraction. Target regions were PCR amplified using the following parameters: 94 °C for 3 minutes; 94 °C for 15 seconds, 62 °C for 10 seconds, 68 °C for 15 seconds, for 30 cycles; 68 °C for 5 minutes (Supplementary Table S1).
For no dug selection of sing cell colonies, the electroporated PFFs were plated into 10-cm cell culture dishes at an appropriated density after 3 days of culture. Individual cell colonies were picked and cultured in 24-well plates for recovery. A small part of each colony was lysed to provide templates for genotyping. The target regions surrounding intended point mutations were amplified and then sequenced. Amplicons with multi-peaks were cloned into the pLB vector (Tiangen, Beijing, China) to determine the exact sequence for each allele.
SSA assay to detect the activity of Cas9/gRNA construct. The EGFP-SSA reporter was constructed by our laboratory. Briefly, the multiple cloning site (MCS) was inserted between two EGFP elements containing 191 bp homology sequences. The short PCR product flanking the sgRNA target site was cloned to the MCS of EGFP-SSA reporter by HindIII and BamHI. To perform the SSA assay, 20 μg APP-specific Cas9/gRNA targeting vector and 20 μg SSA reporter were coelectroporated into 3 × 106 PFFs. The Cas9/gRNA vector with a scramble target site was used as the negative control. The expression of restored EGFP was measured by fluorescence microscope and flow cytometry 72 hours later.
Analysis of HDR efficiency via RFLP. Editing efficiency was monitored by restriction enzyme digestion. Briefly, edited cells were harvested 3 days postelectroporation and the genomic DNA was extracted using a TIANamp Genomic DNA Kit (Tiangen, Beijing, China). The target regions were PCR amplified using the following parameters: 94 °C for 5 minutes, 35 cycles of 94 °C for 30 seconds, 62 °C for 30 seconds and 72 °C for 35 seconds, and one cycle of 72 °C for 5 minutes. Approximately 250 ng of purified PCR products were digested with NaeI or FspI in CutSmart Buffer (NEB, Beijing, China). After 2 hours of incubation at 37 °C, the product was resolved on a 1.5% agarose gel stained with ethidium bromide. Gel images were analyzed using Quantity One (Bio-Rad, Hercules, CA). The frequency of HDR was calculated using the following equation (b + c / a+ b + c) × 100, where “a” is the band intensity of DNA substrate while “b” and “c” are the band intensity of cleavage products.
Deep sequencing analysis of the ssODN-mediated HDR. DNA libraries were prepared from the PCR products amplified by a Fast HiFidelity PCR Kit (Tiangen, Beijing, China). The final quality-ensured libraries were sequenced on Illumina Hiseq 2500 for 150 bp pair-end reads (Biomarker Technologies, Beijing, China). The raw sequencing reads were filtered to remove low-quality reads with the following criteria: (i) reads with adaptors; (ii) reads with more than 10% N bases; (iii) reads with more than 50% Q<10 bases (those with a sequencing quality value less than 10). Trimmed reads were aligned with the 185 bp reference sequence and finally analyzed to identify indels, HDR or other types of mutations.
Somatic cell nuclear transfer and piglet genotyping. Positive colonies harboring the intended HDR alleles were selected as donor cells of SCNT, which was mainly carried out in accordance with Lai et al.1,8 Briefly, cumulus oocyte complexes were matured at 39.0 °C for 40 hours in the maturation medium after removing the follicles. Then the first polar body was aspirated from mature oocytes by a fine glass pipette. At last, the donor cells were fused with enucleated oocytes using a BTX electrofusion equipment. The reconstructed embryos were then cultured for approximately 16 hours before transferring into the synchronized recipient pigs. The pregnancy was monitored by ultrasonography 35 days later and the cloned piglet was delivered by eutocia.
The genomic DNA extracted from the tail was analyzed via PCR and subsequent sequencing. The PCR amplicons were also ligated to the pLB vector to confirm the precise sequence of both alleles.
Western blotting analysis. Frozen muscle samples were ground and then resolved in the lysis buffer. The resultant protein samples were quantified by a BCA protein assay kit (Beyotime, Haimen, China). The proteins were then separated by SDS-PAGE before transferring to the nitrocellulose membrane. The blots were incubated with a primary antibody against myostatin (sc-6884, Santa Cruz Biotech, Santa Cruz, CA), washed, and then incubated with a horseradish peroxidase-labelled anti-goat secondary antibody. The bands were detected with an ECL-Plus western blotting reagent (Beyotime, Haimen, China).
Detection of transgene in the cloned pig. Primers for the amplification of the MSTN-specific Cas9/gRNA plasmid were designed, and genomic PCR was performed using the following parameters: 94 °C for 5 minutes, 35 cycles of 94 °C for 30 seconds, 62 °C for 30 seconds and 72 °C for 25 seconds, and one cycle of 72 °C for 5 minutes. To prepare the positive control, MSTN-specific Cas9/gRNA vector was diluted in a series of 10 fold from 100 ag to 100 pg. Genomic DNA from the wild-type Large White was used as a negative control.
Off-target analysis. Potential off-target sites were predicted by scanning the porcine genome according to previous experience.52,53 The genomic DNA of the cloned piglet was assessed for mutagenesis at these off-target sites using PCR and sequencing. The PCR parameters were 94 °C for 3 minutes; 94 °C for 15 seconds, 60 °C for 10 seconds, 68 °C for 15 seconds, for 30 cycles; 68 °C for 5 minutes. Primers for off-target analysis were listed in Supplementary Table S3.
SUPPLEMENTARY MATERIAL Figure S1. Schematic representation of the SSA-EGFP reporter. Figure S2. Further increase in the concentration or length of the ssODN failed to improve the level of HDR at the APP locus. Figure S3. No obvious improvement of the HDR efficiency was observed with the treatment of RS-1. Figure S4. Confirmation of the p.G2019S mutation in mixed PFFs. Figure S5. Confirmation of the p.C313Y mutation in mixed PFFs. Figure S6. Most frequent indel amplicons detected by deep sequencing. Figure S7. Detection of Cas9-induced off-target mutagenesis in the cloned piglet. Figure S8. Hypothesis of the incomplete HDR based on a two SSA model. Table S1. Sequence of primers used in this study. Table S2. Sequence of ssODN donors used in this study. Table S3. Primers for PCR amplification of the off-target sites. Table S4. Summary of HDR alleles with different point mutations.
Acknowledgments
We would like to thank Xue Chen, Tingting Yu, and Chunyi Lu for technical assistance. This work was supported by Special Funds for Cultivation and Breeding of New Transgenic Organisms (No. 2013ZX08006), the National Natural Science Foundation of China (Grant No. 30871841), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, No. IRT1248). The authors declare no competing financial interests.
Supplementary Material
References
- Lai, L, Kang, JX, Li, R, Wang, J, Witt, WT, Yong, HY et al. (2006). Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol 24: 435–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghia-Akli, R, Fiorotto, ML, Hill, LA, Malone, PB, Deaver, DR and Schwartz, RJ (1999). Myogenic expression of an injectable protease-resistant growth hormone-releasing hormone augments long-term growth in pigs. Nat Biotechnol 17: 1179–1183. [DOI] [PubMed] [Google Scholar]
- Hu, S, Qiao, J, Fu, Q, Chen, C, Ni, W, Wujiafu, S et al. (2015). Transgenic shRNA pigs reduce susceptibility to foot and mouth disease virus infection. Elife 4: e06951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, RS, Lorson, M, Ross, JW, Whyte, JJ and Walters, E (2013). Genetically engineered pig models for human diseases. Annu Rev Anim Biosci 1: 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W, Li, Z, Huang, Y, Han, Y, Yao, C, Duan, X et al. (2014). Generation of AQP2-Cre transgenic mini-pigs specifically expressing Cre recombinase in kidney collecting duct cells. Transgenic Res 23: 365–375. [DOI] [PubMed] [Google Scholar]
- Kong, Q, Hai, T, Ma, J, Huang, T, Jiang, D, Xie, B et al. (2014). Rosa26 locus supports tissue-specific promoter driving transgene expression specifically in pig. PLoS One 9: e107945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, W, de Jong, J, Pindyurin, AV, Pagie, L, Meuleman, W, de Ridder, J et al. (2013). Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927. [DOI] [PubMed] [Google Scholar]
- Lai, L, Kolber-Simonds, D, Park, KW, Cheong, HT, Greenstein, JL, Im, GS et al. (2002). Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295: 1089–1092. [DOI] [PubMed] [Google Scholar]
- Hai, T, Teng, F, Guo, R, Li, W and Zhou, Q (2014). One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y, Du, Y, Shen, B, Zhou, X, Li, J, Liu, Y et al. (2015). Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep 5: 8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X, Xin, J, Fan, N, Zou, Q, Huang, J, Ouyang, Z et al. (2015). Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K, OuyangH, Xie, Z, Yao, C, Guo, N, Li, M et al. (2015). Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci Rep 5: 16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J, Wang, Y, Jiang, J, Zhou, X, Song, L, Wang, L et al. (2015). Production of human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the zygotes. Sci Rep 5: 16705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X, Wang, L, Du, Y, Xie, F, Li, L, Liu, Y et al. (2016). Efficient generation of gene-modified pigs harboring precise orthologous human mutation via CRISPR/Cas9-induced homology-directed repair in zygotes. Hum Mutat 37: 110–118. [DOI] [PubMed] [Google Scholar]
- Lillico, SG, Proudfoot, C, Carlson, DF, Stverakova, D, Neil, C, Blain, C et al. (2013). Live pigs produced from genome edited zygotes. Sci Rep 3: 2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, CB, Sheets, TP, Lillico, SG and Telugu, BP (2016). Engineering large animal models of human disease. J Pathol 238: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsberth, C, Westlind-Danielsson, A, Eckman, CB, Condron, MM, Axelman, K, Forsell, C et al. (2001). The ‘Arctic' APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation. Nat Neurosci 4: 887–893. [DOI] [PubMed] [Google Scholar]
- Ruan, J, Li, H, Xu, K, Wu, T, Wei, J, Zhou, R et al. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci Rep 5: 14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder, J, Salsman, J and Dellaire, G (2015). Nuclear domain ‘knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res 43: 9379–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J, Yang, D, Xu, J, Zhu, T, Chen, YE, Zhang, J (2016). RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 7: 10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, HN, Byers, B, Cord, B, Shcheglovitov, A, Byrne, J, Gujar, P et al. (2011). LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 8: 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui, M, Miyado, M, Igarashi, M, Tamano, M, Kubo, A, Yamashita, S et al. (2014). Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep 4: 5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménoret, S, De Cian, A, Tesson, L, Remy, S, Usal, C, Boulé, JB et al. (2015). Homology-directed repair in rodent zygotes using Cas9 and TALEN engineered proteins. Sci Rep 5: 14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron, AC, Lawler, AM and Lee, SJ (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387: 83–90. [DOI] [PubMed] [Google Scholar]
- Schuelke, M, Wagner, KR, Stolz, LE, Hübner, C, Riebel, T, Kömen, W et al. (2004). Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688. [DOI] [PubMed] [Google Scholar]
- Fries, R, Hanset, R and Georges, M. (1997). A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74. [DOI] [PubMed] [Google Scholar]
- Kambadur, R, Sharma, M, Smith, TP and Bass, JJ (1997). Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7: 910–916. [DOI] [PubMed] [Google Scholar]
- McPherron, AC and Lee, S-J (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci 94: 12457–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, C, Thomas, M, Langley, B, Sharma, M and Kambadur, R (2002). Single cysteine to tyrosine transition inactivates the growth inhibitory function of Piedmontese myostatin. Am J Physiol Cell Physiol 283: C135–C141. [DOI] [PubMed] [Google Scholar]
- Paquet, D, Kwart, D, Chen, A, Sproul, A, Jacob, S, Teo, S et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533: 125–129. [DOI] [PubMed] [Google Scholar]
- Howden, SE, Maufort, JP, Duffin, BM, Elefanty, AG, Stanley, EG and Thomson, JA (2015). Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Reports 5: 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L, Guell, M, Byrne, S, Yang, JL, De Los Angeles, A, Mali, P et al. (2013). Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41: 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, W, Carlson, DF, Lancto, CA, Garbe, JR, Webster, DA, Hackett, PB et al. (2013). Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci USA 110: 16526–16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y, Wang, K, Wu, H, Jin, Q, Ruan, D, Ouyang, Z et al. (2016). Genetically humanized pigs exclusively expressing human insulin are generated through custom endonuclease-mediated seamless engineering. J Mol Cell Biol 8: 174–177. [DOI] [PubMed] [Google Scholar]
- Lesueur, LL, Mir, LM and André, FM (2016). Overcoming the specific toxicity of large plasmids electrotransfer in primary cells in vitro. Mol Ther Nucleic Acids 5: e291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, VT, Weber, T, Wefers, B, Wurst, W, Sander, S, Rajewsky, K et al. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Maruyama, T, Dougan, SK, Truttmann, MC, Bilate, AM, Ingram, JR and Ploegh, HL (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C, Liu, Y, Ma, T, Liu, K, Xu, S, Zhang, Y et al. (2015). Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, CD, Ray, GJ, DeWitt, MA, Curie, GL and Corn, JE (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34: 339–344. [DOI] [PubMed] [Google Scholar]
- Renaud, JB, Boix, C, Charpentier, M, De Cian, A, Cochennec, J, Duvernois-Berthet, E et al. (2016). Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep 14: 2263–2272. [DOI] [PubMed] [Google Scholar]
- Bialk, P, Rivera-Torres, N, Strouse, B and Kmiec, EB (2015). Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS One 10: e0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade, S, Tsubota, T, Sakane, Y, Kume, S, Sakamoto, N, Obara, M et al. (2014). Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 5: 5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici, F, Snipe, JR, Chan, GK, Gordenin, DA and Resnick, MA (2006). Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol Cell Biol 26: 7645–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi, K, Kunihiro, Y, Kaneko, T, Nagahora, H, Voigt, B and Mashimo, T (2016). ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7: 10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J, Song, Z, Yu, S, Cui, D, Wang, B, Ding, F et al. (2014). Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PloS One 9: e95225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L, Tang, M, Yang, J, Wang, Q, Cai, C, Jiang, S et al. (2015). Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci Rep 5: 14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y, Foden, JA, Khayter, C, Maeder, ML, Reyon, D, Joung, JK et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick, TJ, Fine, EJ, Antico, CJ and Bao, G (2013). CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41: 9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C, Gore, A, Yan, W, Abalde-Atristain, L, Li, Z, He, C et al. (2014). Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15: 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V, Shen, B, Zhang, W, Hodgkins, A, Keane, T, Huang, X et al. (2015). Off-target mutations are rare in Cas9-modified mice. Nat Methods 12: 479. [DOI] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft, B, van Duijn, E, Bultema JB, Waghmare, SP, Zhou, K, Barendregt, A et al. (2011). RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 108: 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Scott, DA, Weinstein, JA, Ran, FA, Konermann, S, Agarwala, V et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.