Abstract
Breast cancer (BC) results in ~40,000 deaths each year in the United States and even among survivors treatment of the disease may have devastating consequences, including increased risk for heart disease and cognitive impairment resulting from the toxic effects of chemotherapy. Aptamer-mediated drug delivery can contribute to improved treatment outcomes through the selective delivery of chemotherapy to BC cells, provided suitable cancer-specific antigens can be identified. We report here the use of capillary electrophoresis in conjunction with next generation sequencing to develop the first vitronectin (VN) binding aptamer (VBA-01; Kd 405 nmol/l, the first aptamer to vitronectin (VN; Kd = 405 nmol/l) , a protein that plays an important role in wound healing and that is present at elevated levels in BC tissue and in the blood of BC patients relative to the corresponding nonmalignant tissues. We used VBA-01 to develop DVBA-01, a dimeric aptamer complex, and conjugated doxorubicin (Dox) to DVBA-01 (7:1 ratio) using pH-sensitive, covalent linkages. Dox conjugation enhanced the thermal stability of the complex (60.2 versus 46.5°C) and did not decrease affinity for the VN target. The resulting DVBA-01-Dox complex displayed increased cytotoxicity to MDA-MB-231 BC cells that were cultured on plasticware coated with VN (1.8 × 10−6mol/l) relative to uncoated plates (2.4 × 10−6 mol/l), or plates coated with the related protein fibronectin (2.1 × 10−6 mol/l). The VBA-01 aptamer was evaluated for binding to human BC tissue using immunohistochemistry and displayed tissue specific binding and apparent association with BC cells. In contrast, a monoclonal antibody that preferentially binds to multimeric VN primarily stained extracellular matrix and vessel walls of BC tissue. Our results indicate a strong potential for using VN-targeting aptamers to improve drug delivery to treat BC.
Keywords: aptamer, breast cancer, SELEX, vitronectin
Introduction
Nearly a quarter of a million women will be diagnosed with breast cancer (BC) every year in the United States, and despite decades of intense research and advances in treatment over 40,000 women will die of the disease each year.1 The discovery of amplification of the Her2/Neu gene in a large portion of breast cancer patients and the advent of specific therapy using the antibody Herceptin revolutionized the treatment of breast cancer, extending survival significantly.2 However, only ~25% of breast cancers overexpress this marker limiting the utility of this treatment and underscoring the necessity for further research to improve prognosis for the majority of women.3
One of the most successful ways to improve the prognosis for BC patients is early detection of the disease. Patients that begin treatment at stage I approach 100% 5-year survival while those diagnosed at stage IV have < 25% 5-year survival.1 The only current method used for breast cancer screening is mammography, which can reduce the mortality of breast cancer by 32% in women in their 60s.4 However, mammography is time consuming, intrusive to the patient, can result in over diagnosis, and can contribute to increased risk of developing cancer.5,6,7 Unlike prostate cancer in men where Prostate Specific Antigen blood concentration can be monitored and used for screening and diagnostic purposes, there are no equivalent blood monitoring tests for BC. Blood screening tests are favorable over radiographic imaging for multiple reasons, including: no radiation exposure to the patient, lack of subjective diagnosis based on image analysis, faster and less intrusive to the patient, as well as the ability to check multiple markers simultaneously. Currently, a variety of targets are being investigated as alternatives to mammography. Tumor-derived DNA, circulating stem cells, and MUC1, a protein associated with increased BC progression, are all being examined for use in BC screenings; however, these markers are generally only useful at later stages of BC development.8,9,10 Mammaprint, a screening of 70 genes related to BC, can be used for diagnostic purposes and can be used to help predict if a cancer will progress or become metastatic.11 Recently, miRNA's have been shown to have early diagnostic potential for identifying BC before it is in an advanced stage, but a definitive blood screen is still unavailable, with multiple new candidates being pursued.12
Vitronectin (VN) is being investigated as a possible biomarker that can be used for diagnostic purposes in BC. VN is a highly glycosylated protein produced primarily in the liver and secreted into the blood.13 VN can exist as a monomer, which predominates in the serum, or as a multimer.14,15 One of VN's major roles is in wound healing and clotting. High levels of VN are stored in platelets and upon activation VN is released and prevents clot degradation via binding of plasminogen activator inhibitor-116. However, VN also binds to many other receptors including αVβ3, an integrin that plays a role in macrophage activation, angiogenesis, and tumor progression.17,18 VN is also known to become incorporated into the extracellular matrix (ECM) and serves as a support for cell motility and adhesion.19 Of particular interest in this regard is the ability of VN to bind to the urokinase receptor (uPAR, CD87), which is also involved in plasminogen activation.20 Once uPAR binds to VN it appears to induce cell adhesion and cancer migration through Rac activation.21,22 VN binding appears to be key in inducing these effects via uPAR.23,24 Because of this, uPAR has been investigated as a potential biomarker for BC. It has been demonstrated that uPAR expression is increased as the disease progresses and may have prognostic value.25 VN itself has also been investigated as a possible cancer-promoting protein as well. Circulating serum VN has been shown to increase with BC progression.26 Additionally, increased VN staining can be observed around the vessels of ductal cancer containing breast tissue.27 Analysis of mRNA from BC displaying high VN staining revealed that neither malignant nor normal breast tissue expressed VN, suggesting it was being supplied by infiltrating cells in breast tissue.27 Further, VN was shown to be a driving factor in BC cancer stem cell differentiation, and when VN binding to αVβ3 was blocked BC cells failed to form tumors in vivo.28 These results make VN a promising candidate as not only a possible prognostic marker for BC, but also as a therapeutic target used to treat the disease by binding to circulating VN and localizing to areas where malignant tissues are incorporating it into the surrounding ECM.
In order to identify a molecule that could differentiate between VN and fibronectin (FN), a structurally similar protein, we developed an aptamer using a variant of Systematic Evolution of Ligands by Exponential Enrichment (SELEX)29 that included capillary electrophoresis (CE) selection and use of next generation sequencing (NGS). Aptamers are single-stranded DNA or RNA molecules that fold into distinct 3D conformations that can recognize proteins, small molecules, and even metal ions.30,31 Aptamers have advantages over antibodies in that they are chemically stable, can undergo multiple freeze-thaw cycles, can be denatured and refolded, are nonimmunogenic, and production can be scaled with reasonable cost.32 They can also be modified with drugs, dyes, or other molecules to alter their function and stability.33,34 We utilized CE-NGS SELEX to identify an aptamer against VN.35,36 Herein we describe the selection, characterization, and drug delivery capability of a novel aptamer against VN, VBA-01, and its dimer DVBA-01, using CE-NGS SELEX.
Results
Aptamer selection using CE-NGS SELEX
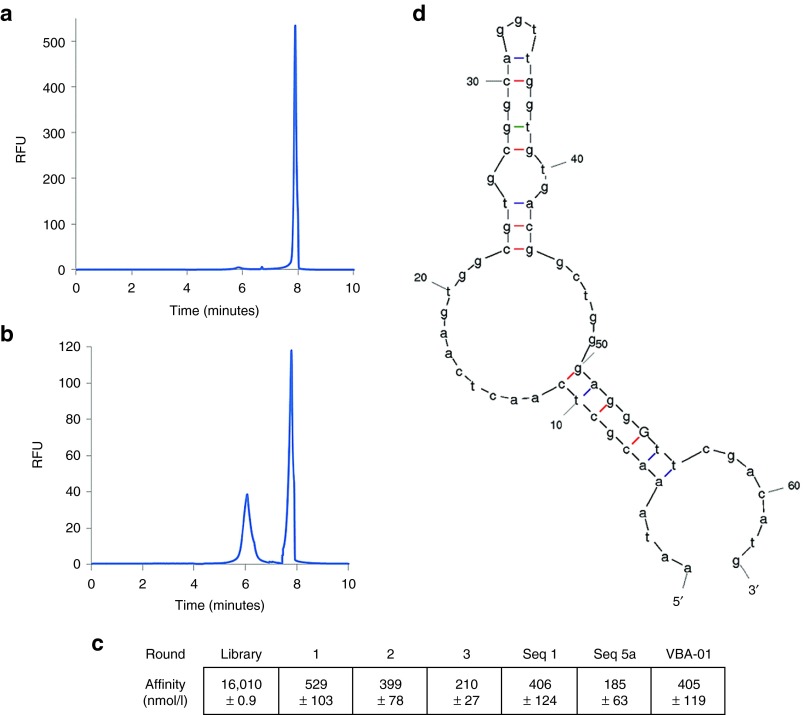
The use of capillary electrophoresis enabled us to rapidly enrich a pool of 1.5 × 1013 unique sequences for only those that bound VN, with no detectable binding to FN. The use of CE, specifically the well-established nonequilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) technique, allows for researchers to monitor the Kd of each selection step based on the areas beneath peaks corresponding to bound and unbound aptamer in the resulting electropherogram.38 The initial random library had a Kd of ~16 µmol/l for VN before any selection took place (Figure 1a). After a single negative selection against FN and positive selection for VN, we increased the affinity for VN by 30-fold (Kd ≈ 500 nmol/l; Figure 1b). Each successive round of selection increased the affinity of the pool by approximately twofold, resulting in a 200 nmol/l Kd for the final library (Figure 1c). After the selection process was completed, the pool was converted to dsDNA and underwent NGS, which identified 143,845 unique sequences. We selected the top seven unique sequences (S1–S7) to test for VN binding using CE (see Supplementary Figure S1). S1 and a closely related sequence (which differed only by the addition of another C to a CCC sequence, and was likely an error in PCR or sequencing), accounted for 2.7% of the total reads in the data set. Sequence S2 accounted for 0.78%, with S3–S7 accounting for 0.14% to 0.08% of total reads. After Kd evaluation, we identified sequence 1 (S1), the most populous sequence identified in NGS, as the aptamer with the lowest Kd of the selected sequences (Kd ≈ 400 nmol/l).
Figure 1.
Aptamer selection monitored by NECEEM. (a) Electropherogram of the starting library against VN. (b) Electropherogram of the aptamer library after a single round of positive SELEX. (c) Kd of various aptamer libraries and individual aptamer candidates. (d) Structure of VBA-01 as predicted by mFold. SELEX, Systematic Evolution of Ligands by Exponential Enrichment; VN, vitronectin.
In order to identify sequences similar to S1 we analyzed other aptamer sequences identified using NGS according to 8-base reading frames and identified all of the aptamers that contained any of these reading frames. These sequences were then compiled in CLUSTAL OMEGA v1.2 software and clustered into families according to sequence similarity. Using a phylogenic tree to differentiate the families (see Supplementary Figure S2), we then selected 10 sequences from these groups. We avoided multiple selections from the same group in order to avoid overly similar sequences (see Supplementary Figure S1), although two sequences were selected from the largest family, as well as from a highly variable family. The Kd for each of these 10 aptamer candidates was then determined by CE. We identified three aptamers from the candidates that displayed low nanomolar affinity for VN: S3a, S5a, and S8a. Kd analysis by CE showed that Sequence 5a (S5a) had the lowest binding affinity of these modified aptamers (Figure 1c). Trimming S5a aptamer sequence in from the 3′ and 5′ sides revealed that 10 bases could be removed from both ends before any significant reduction in affinity was observed. The final Kd of this aptamer, VBA-01, was found to be 405 nmol/l and its secondary structure was modeled using mFold (Figure 1d).
Biophysical characterization of aptamer complexes
We covalently linked Dox with DNA using a pH-sensitive bond. However, this chemistry requires dsDNA. Aptamers with double-stranded character were formed by appending either CGGC-dA12-GCCG or CGGC-dT12-GCCG to the 3′ terminus of VBA-01. This allows for the formation of the dimeric DVBA-01 (Figure 2) and the monomeric VBA-01-dT, which consists of VBA-01 with a CGGC-dA16-GCCG tail base paired to a complement 20mer oligonucleotide. Both DVBA-01 and VBA-01 displayed a similar Tm of ~45°C. When DVBA-01 was covalently modified with Dox, the Tm increased to ~60°C. Similar levels of stabilization resulting from Dox conjugation have been observed previously,34 and may result from Dox intercalation.39 DVBA-01 bound seven equivalents of Dox (Figure 3a). An additional observation was that when DVBA-01 reacted with Dox, all the drug was bound to the complex, indicating that DVBA-01 may be capable of binding additional Dox molecules. Dox-loaded DVBA-01 and DVBA-01-Dox were assessed for binding affinity against VN using CE as described above. DVBA-01 had an apparent Kd of 485 nmol/l, and DVBA-01-Dox had a Kd of 28 nmol/l (Figure 3b).
Figure 2.
Structure of DVBA-01, with possible Dox binding sites highlighted in red.
Figure 3.
Effects of Dox conjugation on aptamer properties. (a) Physical characteristics of VBA-01 and its derivatives. Dox:Ap is the ratio of Dox to aptamer in the DVBA-01-Dox complex. (b) UV-VIS spectrum of DVBA-01-Dox. The inset expands the 400–600 nm range of the spectrum to show the absorbance from Dox by the DVBA-01-Dox complex. Dox, doxorubicin.
VN-specific cytotoxicity of DVBA-01-Dox
The 96-well plates coated with VN, FN, or uncoated were used to assay the ability of DVBA-01-Dox to specifically kill BC cells in a VN-rich environment. Plates coated with 10 µg/ml of VN facilitated increased killing of MDA-MB-231 cells treated with DVBA-01-Dox when compared with FN or untreated plates (Figure 4). The IC50 was reduced by ~25% compared with cells plated on uncoated or FN treated plates (Figure 4b). When the cytotoxicity data over a range of concentrations is evaluated, there is a modest difference between cells cultured on VN and FN/untreated plates (Figure 4a). However, a distinct difference can be seen between the treatments when cells are treated near the IC50 value (Figure 4c). Cells on VN-coated plates are much more sensitive to DVBA-01-Dox than cells cultured on plates lacking the target. In order to confirm that VN is responsible for the increased activity, we pretreated coated plates for 1 hour at 4°C with 4 µmol/l DVBA-01-Dox before washing and plating MDA-MB-231 cells. When the assay was performed in this manner, there is much greater cytotoxicity for cells cultured on VN-treated plates compared with controls (Figure 4d), confirming that DVBA-01-Dox is binding to VN and is delivering Dox to the cells.
Figure 4.
Aptamer: vitronectin interaction promotes targeted drug delivery. (a) Dose effect curve of DVBA-01-Dox effect on MDA-MB-231 cells seeded on coated plates and then treated with drug. (b) IC50 values of the drugs based on treatment after seeding determined by Calcusyn. (c) Cytotoxicity data of cells treated with 2 μmol/l DVBA-01-Dox showing the difference resulting from plate coating. (d) Cytotoxicity data from plates that were treated with DVBA-01-Dox, washed, and then seeded with cells. Dox, doxorubicin.
Tissue binding
In order to ascertain if VBA-01 could bind to VN in a tumor environment, we stained breast cancer tissue samples. Tumors were sectioned and probed with either an anti-VN antibody or the VBA-01 aptamer with a biotinylated 3′ terminus. Anti-VN was detected by using an antimouse secondary antibody linked to horseradish peroxidase, while VBA-01 was detected with an antibiotin linked horseradish peroxidase antibody. Immunohistochemistry showed specific binding of both anti-VN and VBA-01; however, they stain different regions of the tumor (Figure 5). Anti-VN appears to bind the ECM, where VN can become multimerized. Anti-VN also appears to heavily stain the vessel wall. VBA-01 does not bind the ECM, but appears to bind directly to the tumor cells, suggesting that VBA-01 may be specific for monomeric VN over the multimeric VN present in ECM. This binding was confirmed with fluorescence microscopy where VBA-01 binding was visualized using a streptavidin-Texas Red conjugate (Figure 5d). Fluorescence microscopy shows that the aptamer binds directly to tumor cells, with much lower binding to ECM. This high specificity for tumor cell-bound VN over ECM-incorporated VN may play an important role in differentiating between normal VN incorporation, and tumors that bind high levels of the blood protein.
Figure 5.
Comparative staining of breast cancer tissue by the VBA-01 aptamer and the NBP2-20866 VN antibody. (a) No antibody control. (b) VBA-01 staining of BC cells with nuclear exclusion. (c) Antibody stains ECM and collagen-rich regions of tissue. (a-c) are consecutive sections from a single specimen. (d) Immunofluorescence confirming VBA-01 staining (shown in red) of BC cells with nuclear exclusion relative to DAPI (blue). BC, breast cancer; DAPI, 4,6-diamidino-2-2-phenylindole; ECM, extracellular matrix.
Discussion
Breast cancer kills over 40,000 women each year necessitating the search for new treatments. VN is a blood protein secreted by the liver and has been shown to have BC-promoting activity, as well as having increased expression during BC progression and being present at high levels in BC tissue relative to healthy tissue. These properties make VN an attractive candidate for drug development. VN can be used for two purposes with respect to BC treatment. First, because it is primarily a blood protein with concentrations in the low mg/l range, it can be used as both a measure of disease progression and potentially as a carrier protein for delivery. Albumin, the most abundant blood protein, has been used as a drug carrier molecule to favorably alter pharmacokinetic properties of a variety of drugs.40 We hypothesize that VN may also act as a blood carrier protein similar to albumin; however, this was not explored in this work. The second use of VN in BC treatment is as a drug targeting agent. Because VN is upregulated in BC and appears to accumulate in malignant breast tissue, it can be used as a molecular target for the disease.26 With this in mind we have developed an aptamer specific to VN, VBA-01, using a variant of SELEX which utilizes CE and NGS to rapidly select the aptamer in fewer rounds than was possible using non-CE based approaches to aptamer identification.
In our aptamer selection protocol we aimed to select an aptamer that had high specificity for VN over FN. In order to accomplish this, our first selection was a negative selection against FN. In order to further expedite SELEX, a phosphorylated primer was used, which could be removed enzymatically after each round of polymerase chain reaction (PCR), thus eliminating the need for column binding and denaturation used to collect ssDNA in other selection protocols. An advantage of using CE for selection is that the affinity of the library can be monitored during selection. This method of SELEX resulted in reducing the Kd of the library by 30× after a single round of SELEX (Figure 1a,b). Each successive round increased the affinity by a factor of two, which suggests that the majority of poor binding sequences are removed early in the SELEX process (Figure 1c). However, the DNA:VN ratio remained unchanged throughout the selection, and reducing this ratio may have favored stronger binders, increasing the affinity of future rounds further.
Once we completed our rounds of selection, dsDNA was produced from the aptamer library and was sequenced using NGS. The data obtained by NGS showed that there were only a few sequences that were expressed as a significant percentage of all sequences. We hypothesize that additional rounds of selection may be useful in amplifying some of the less common sequences that are tight binders, but each round of PCR introduces bias. Some sequences may be expressed at high levels despite relatively low affinity to the target. This can occur when many rounds of PCR are performed, effectively selecting for sequences that are more amenable to PCR than other sequences, which may bind more tightly but may respond to PCR at a lower rate.35,41,42 Regardless, we selected the seven most populous sequences for further analysis. S1, the most populous sequence, had the highest binding affinity of the selected sequences, with a Kd ≈ 400 nmol/l. While this binding is weaker than the final library Kd of ~200 nmol/l, it is still appropriate for specific binding. However, this result suggests that many of the less populous sequences had higher affinity than the more populous sequences, indicating that additional rounds of SELEX may have resulted in an aptamer with a lower Kd. In order to identify if there were sequences similar to S1, we broke the random region of the aptamer into eight base pair reads and identified any aptamer sequence that possessed those reads anywhere in their sequence. This resulted in about 1,500 sequences. CLUSTAL OMEGA was then used to group the 1,500 aptamer candidates into eight families and one sequence was selected from each family along with an additional sequence from each of the largest and most variable families. Binding analysis of these sequences revealed that sequence 5a had the highest affinity (200 nmol/l), which was similar to the library affinity immediately before NGS sequencing. This further supports the finding that some of the less populous sequences were strong binders. For future studies we suggest at least three rounds of SELEX at the initial selection conditions, with additional rounds of a more stringent selection, such as reducing target protein concentration, in order to amplify stronger binders more successfully. To form our final aptamer, VBA-01, we trimmed the aptamer from both termini, removing 20 bases of the aptamer, which increased the Kd to 400 nmol/l. This reduction in affinity is not entirely unexpected, and suggests that the ends of the aptamer may have been playing some role in stabilizing the binding conformation.
After finalizing VBA-01, we assembled the dimeric DVBA-01 and doxorubicin modified dimer, DVBA-01-Dox, by synthesizing the aptamers with either poly-dT or poly-dA tails, which, when mixed, result in an AT base pair bridge linking each aptamer, as we have described previously.34 Dimers were made in part because we hypothesized that a multivalent aptamer would have a higher affinity for VN than a monomer, since it could bind two VN molecules simultaneously, and if one was released the second would remain bound, thus increasing the apparent affinity. However, upon examination of the Kd using CE we found that the affinity was relatively unchanged versus the monomer and was actually slightly reduced, from ~400 nmol/l for the monomer to ~500 nmol/l for the dimer. We believe that this demonstrates that our dimer is unable to bind multiple VN molecules simultaneously despite possessing two binding aptamers. A possible reason for this is that once the dimer binds a single VN molecule, the second aptamer is blocked from binding. This could occur if our dA-dT base pair bridge is not sufficiently long enough to allow room for multiple VN molecules.
We assessed DVBA-01-Dox for its drug binding capacity and found that DVBA-01-Dox was capable of binding at least seven equivalents of drug. Because this was the ratio used in our reactions we believe that the aptamer incorporated all of the Dox that was available. The characteristic absorbance of Dox is apparent in the 500–600 nm range, can be used to quantify the amount of Dox loaded into the aptamer, as we have previously described.34 While more Dox could theoretically be loaded into the aptamer, the drug can have an effect on the shape of the aptamer and possibly reduce the affinity for VN. We found that Dox incorporation stabilized DVBA-01 against temperature-dependent melting, indicating that Dox was having a structural effect on the aptamer. When DVBA-01 and DVBA-01-Dox were assessed for binding affinity to VN, we discovered that the affinity of DVBA-01-Dox was greatly increased over unmodified DVBA-01. It is possible that Dox stabilizes the aptamer in a binding conformation, increasing the affinity of the complex for VN.
After establishing by way of CE studies that VBA-01 and its variants could bind to VN in solution, we sought to establish that we could use the aptamer complex, DVBA-01-Dox, to deliver drugs to cancer cells in a VN-dependent manner. Because VN is produced and released by the liver and may be bound by BC cells, we used protein-coated plates to simulate what may happen in vivo where BC cells bind to and accumulate VN26. We observed increased cytotoxicity against MDA-MB-231 cells plated on VN plates compared with FN or uncoated plates. We hypothesize that BC cells plated with VN receive a higher dose of Dox than other cells because the aptamer binds to the VN on the plate and stays with the cells even after they are washed, increasing the effective dose those cells are exposed to. This effect is especially apparent when cells are treated near the IC50 value of 2 μmol/l. However, we believe that during the 3 hours of incubation, cells were taking up the drug complex due to nonspecific mechanisms such as endocytosis. In order to test this, we coated plates with either VN, FN, collagen, or left them uncoated. These plates were then treated with DVBA-01-Dox, washed, and then plated with cells. When the assay is performed in this manner cells will only be exposed to drug if the complex is binding to the protein. This assay shows that VN-coated plates have much more cytotoxicity than FN, collagen, or uncoated plates, consistent with DVBA-01-Dox binding to VN preferentially over the other substrates. We believe that these data show the feasibility of using the DVBA-01-Dox aptamer to deliver drug to BC cells in a VN-rich environment, and lay the foundation for future in vivo experiments, where VN blood binding may also increase the specificity of the complex.
Once we established the effects of VBA-01 and its derivatives in vitro we sought to confirm that VBA-01 could detect VN in BC tissue samples from ductal carcinoma in situ (DCIS) patients. Sections of fresh tumor samples were stained with either VBA-01 or a VN specific antibody, and assessed for binding. VBA-01 used for these experiments was synthesized with a biotin label, which was detected by an antibiotin secondary antibody. By comparing the antibody and aptamer-stained sections, major differences immediately become apparent. Antibody-stained sections display heavy staining of the ECM and almost no staining of cellular bodies (Figure 5). It is known that VN can become multimerized and incorporated into the ECM of surrounding BC cells. However, aptamer-stained sections show almost exclusive cellular staining (Figure 5). We also synthesized VBA-01 with a fluorescent tag, which does not require a secondary antibody for detection and this confirmed the binding pattern described above. We believe that this results from VBA-01 binding to monomeric VN molecules, which are bound to a VN receptor molecule (possibly uPAR or αVβ3). When VN multimerizes the conformation of the protein is altered, resulting in altered antibody affinity and cellular function. Because our selection protocol utilized monomeric VN, which is the form of the protein most prominent in circulation but not in the ECM, we likely identified an aptamer that recognizes an epitope that becomes obscured once the protein is mulitmerized. We believe that this will be an advantage in an in vivo scenario, where multimerized VN is present in ECM throughout the body, especially in the vasculature where it plays a role in heparin binding.43 An aptamer that recognizes multimeric VN could find itself bound to vasculature well before it could locate to the tumor site, whereas an aptamer against the monomer would be able to use the VN to locate to BC cells, which are binding the protein in large quantities.
We have described the selection of a novel aptamer using a combined CE and NGS SELEX process. This allowed us to select an aptamer with mid- to low-nanomolar affinity in only three rounds of selection, which traditionally has taken a dozen or more rounds. NGS allowed us to refine our aptamer rationally, and eliminated the need for tedious cloning and sequencing as well. We believe that CE-NGS SELEX has the potential to decrease the time required to identify aptamers against protein targets and should become a preferred method by which to identify new aptamers. Once we identified our VN specific aptamer, VBA-01, we formed a dimer and covalently modified it with Dox for drug delivery. This also significantly increased the affinity of the aptamer for its target, and resulted in VN-dependent cell killing. VN is a developing molecular target for BC, both as a biomarker, which can be used for diagnosis, and as a target for therapy. The VN specific aptamer VBA-01 further advances this field of study.
Materials and methods
Aptamer selection. Aptamer selection was achieved by CE-NGS SELEX to select for and identify aptamer sequences. A DNA library containing ≈ 2 × 1019 unique sequences was used as the basis for all SELEX studies with 1.5 × 1013 sequences used for the initial selection. Each candidate sequence included set primer sequences on the 5′ and 3′ ends and 40 random nucleotides in the center of the oligonucleotide (5′-AGCTCAGAATAAACGCTCAA(N)40TTCGACATGAGGCCGGATC-3′). Primers used for PCR were 5′-Phos-GATCCGGCCTCATGTCGAA-3′ and 5′- GGGAGCTCAGAATAAACGCTCAA-3′. FN, VN, and aptamers were all prepared to 50 µmol/l in pH 8.2, 50 mmol/l Tris-HCl buffer. Candidate aptamers were folded by heating to 95°C for 10 minutes and snap-cooled by placing them directly in an ice bath for 15 minutes and the pool of candidate aptamers was mixed 1:1 with protein at a final concentration of 500 nmol/l. The mixtures were allowed to incubate at 4°C for 30 minutes prior to CE experiments. Selections were performed using a Beckman Coulter P/ACE MDQ CE system (Beckman Coulter, Brea, CA) with laser-induced fluorescence (LIF) detection, with excitation/emission set to 488/520 nm. 32 KARAT software was used for CE control and analysis. An uncoated, fused-silica capillary (with a 60.2 cm total length (50.0 cm from inlet to detector) and 75 μm internal diameter) was used for the CE studies. The capillary and inlet/outlet vials were filled with separation buffer, which was composed of 31 mmol/l Tris plus 500 mmol/l Gly (pH 8.2), along with SYBR gold dye (Thermo Fisher, Gainesville, FL at a final dilution of 1:100,000 in the buffer (from the as-received 10,000 × concentrate) for DNA labeling. Samples were injected at 4 psi for 5 seconds (~ 180 nl sample injected) and separations were carried out at 18 kV. Fractions of either bound DNA (during positive selection steps) or unbound DNA (during negative selection steps) were collected into 20 µl of sample buffer (Tris-HCl) using a collection voltage of 10 kV.
The aptamer candidates were first evaluated for binding to FN as a negative selection step. The bound fraction of aptamers was discarded, and the remaining sequences were mixed with VN and rerun through CE SELEX. The bound aptamers were then collected, and unbound DNA was discarded. CE fraction collection proceeded according to a method previously developed in our laboratory.37 Aptamers were PCR-amplified with phosphorylated forward primer using Taq DNA polymerase, and degraded to single stranded DNA using lambda exonuclease according to manufacturer protocols. Single-stranded aptamers were collected using a QIAquick PCR purification kit. Positive SELEX was performed for two additional rounds, with a final negative selection against FN.
After the initial selection rounds were complete, the aptamer pool was converted to blunt end dsDNA by amplification with T4 DNA polymerase. The selected and amplified dsDNA was sequenced by Nanomedica, LLC using the Ion Torrent PGM.
The seven most populous sequences identified by NGS were then synthesized (see Supplementary Figure S1). Aptamer candidates were folded and evaluated for binding under identical conditions that were used for aptamer selection. Sequence 1 (S1), the most populous sequence identified by NGS, had the lowest Kd of these initial aptamers selected and was used for further aptamer refinement. Sequences other than S1 also displayed affinity in the mid-to-high nanomolar range (S5 and S6), indicating that there are other aptamers that bind VN despite having significantly different sequences. To identify aptamers that were similar to S1, nonprimer bases of S1 were divided into eight base reading frames, which were then compared with the entire library of sequences identified by NGS. Any sequence that shared an eight-base sequence with S1 was added to a new library of candidate aptamers, for a total of 1,582 unique candidate aptamers. The list of candidate aptamers was analyzed using CLUSTAL OMEGA v1.2; (EMBL), which grouped the candidate aptamers by sequence similarity. The aptamer groups were visualized with a phylogeny tree with average distance using percent identity to construct the tree. This was divided into eight “families” based on sequence similarity. After clustering, we selected 10 aptamers from separate families, with two aptamers selected from the largest family and two selected from a highly variable family. These aptamers were synthesized and evaluated by CE for binding affinity. We identified three aptamers with low nanomolar affinity: S3a, S5a, and S8a. We then chose to further develop S5a, as it had a similar structure to S1, which was the most populous sequence originally identified by NGS. S5a was trimmed from the 3′ and 5′ ends until binding affinity began to decrease significantly, resulting in a final length of 63 bases. This trimmed aptamer, VBA-01 was used for binding and toxicity studies.
VN-specific aptamer synthesis. All DNA aptamer sequences were synthesized by Integrated DNA Technologies. (IDT; Coralville, IA). Aptamers were reconstituted in sterile 50 mmol/l Tris buffer (pH 8.2) to a concentration of 100 µmol/l. All aptamers were synthesized with either a dA16 or T16 single-stranded tail at the 3′-terminus, in addition to a 3′ phosphorothioate bond to increase nuclease resistance. DVBA-01 was formed by prefolding the aptamers and mixing the two monomers in a 1:1 ratio followed by 15 minutes incubation at 4°C. The DVBA-01 used for these studies (unless otherwise indicated) included the sequence dCGGCA16GCCG at the 3′ end. The secondary structure for the DVBA-01 was calculated using mFold (www.unafold.rna.albany.edu/?q=mfold).
Synthesis of doxorubicin conjugated aptamers. The covalent complex of DVBA-01 with Dox was prepared by mixing 250 µl of a 50 µmol/l solution with a Dox-formaldehyde solution prepared upon incubation of a 0.37% formaldehyde solution in Dulbecco's phosphate buffered saline (DPBS) without Calcium or Magnesium, pH 7.4. The resulting covalent bond is pH-sensitive with Dox released under acidic conditions.34 The reaction was proceeded in the dark at 4°C for 48 hours. The solution was extracted once with 300 µl of phenol:chloroform followed by two additional extractions with 300 µl chloroform. The aqueous phase was then ethanol-precipitated and the pellet rinsed 2× with 70% ethanol and once with absolute ethanol and dried under reduced pressure. The red pellet was resuspended in 100 µl diH2O. Yields were typically >90% based on DNA recovery.
Determination of Dox:DNA ratios. DNA samples were prepared in DPBS and the absorbance was measured from 200–800 nm using a Beckman Coulter DU-800 spectrophotometer (Beckman-Coulter). A standard curve of Dox was established between 1 µmol/l and 10 µmol/l by using the absorbance at 494 nm at 85°C. To assess the amount of Dox covalently bound to DNA, the samples were heated to 85°C before measuring the absorbance at 494 and 260 nm. The 260 nm wavelength was used to determine the DNA content in the sample and the 494/260 ratio was used to determine the Dox:DNA ratio.
Temperature-dependent UV studies. Temperature-dependent UV absorption spectra were obtained using a Beckman Coulter DU-800 UV-Vis spectrophotometer. Samples of DVBA-01, DVBA-01-Dox, VBA-01, and VBA-01-Dox were prepared in DPBS. The temperature was increased at a rate of 0.7°C/min over the range 20– 85°C and absorbance at 260 nm was measured for each sample (400 µl and 1 µmol/l).
Cell maintenance. MDA-MB-231 cells were obtained from the Cell and Viral vector Core Laboratory at Wake Forest School of Medicine. All cells were maintained with Dulbecco's modified essential medium (DMEM) (Gibco; Thermo Fisher) with 10% fetal bovine serum (FBS) (Geminii Bio-Products, Sacramento, CA). All cells were kept at 5% CO2 at 37°C.
Cell viability assays. Cells were seeded at a density of 5,000 cells/well in 96-well plates and incubated at 37°C under 5% CO2. For cells cultured on protein-coated plates, 96-well plates were prepared by adding 100 µl of 10 µg/ml of the appropriate protein in PBS to each well. Plates were sealed with Parafilm and stored at 4°C overnight. Plates were warmed to room temperature before removing excess protein solution and plating cells. The following day, cells were washed with FBS-free media and treated for 3 hours with drugs. After treatment, cells were washed with media containing 10% FBS and incubated for 72 hours. Cell viability was assessed indirectly by measuring the adenosine triphosphate (ATP) amounts using CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer's protocol. For experiments where plates were treated before cells were cultured, protein-coated or uncoated plates were treated with drugs for 1 hour at 4oC before washing with PBS. After plates were washed, cells were plated as stated above. Cells were incubated 72 hours prior to being assayed for viability.
Histology. Frozen samples of patient-derived breast tissue from patients with breast cancer were obtained from the Wake Forest Tissue Bank, sectioned, and fixed using 10% acetone for 10 minutes at −20°C. Sections were then blocked for 15 minutes using 5% bovine serum albumin (BSA). After blocking, sections were washed 3× with PBS. Sections were treated with either 1 μmol/l of biotin labeled VBA-01, or a 1:750 dilution of the VTN antibody NBP2-20866 (Novus Biological, Littleton, CO), both in 1.5% BSA in PBS, overnight at 4°C. Sections were washed in PBS for 1 hour before an antibiotin horseradish peroxidase conjugated antibody was added to aptamer-stained sections, or antimouse horseradish peroxidase conjugated antibody was added to antibody stained sections. Sections incubated for 1 hour at room temperature before being washed 3× with PBS and developed. Sections were stained with Nova red and counterstained with hematoxylin.
SUPPLEMENTARY MATERIAL Figure S1. Sequences of the various aptamers identified via NGS and tested for affinity with CE. Figure S2. Phylogeny of the ≈ 1500 sequences related to S1, compiled by CLUSTAL OMEGA.
Acknowledgments
This work was supported by NIH-NCI P30 012197 (B Pasche), the Tumor Tissue and Pathology shared resource, and the WFSM BTCOE. FCM was supported in part by 1R01 NS069964 (NIH) and P20 DK097806 (NIDDK). Boyacioglu is grateful for a fellowship from the Scientific and Technological Research Council of Turkey (TUBITAK).
Supplementary Material
References
- Howlader, N, Noone, AM, Krapcho, M, Garshell, J, Miller, D, Altekruse, SF et al. (eds.). (2015). SEER Cancer Statistics Review, 1975–2012. National Cancer Institute: Bethesda, MD, http://seer.cancer.gov/csr/1975_2012/. Based on November 2014 SEER data submission, posted to the SEER web site, April 2015. [Google Scholar]
- Baselga, J (2001). Clinical trials of Herceptin (trastuzumab). Eur J Cancer 37 (Suppl 1): 18–24. [PubMed] [Google Scholar]
- Emi, Y, Kitamura, K, Shikada, Y, Kakeji, Y, Takahashi, I and Tsutsui, S (2002). Metastatic breast cancer with HER2/neu-positive cells tends to have a morbid prognosis. Surgery 131(1 Suppl): S217–S221. [DOI] [PubMed] [Google Scholar]
- Pace, LE and Keating, NL (2014). A systematic assessment of benefits and risks to guide breast cancer screening decisions. JAMA 311: 1327–1335. [DOI] [PubMed] [Google Scholar]
- Tupper, R and Holm, K (2014). Screening mammography and breast cancer reduction: examining the evidence. J Nurse Practitioners 10: 721–728. [Google Scholar]
- Berrington de González, A and Reeves, G (2005). Mammographic screening before age 50 years in the UK: comparison of the radiation risks with the mortality benefits. Br J Cancer 93: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, MJ and Mainprize, JG (2011). Risk of radiation-induced breast cancer from mammographic screening. Radiol 258: 98–105. [DOI] [PubMed] [Google Scholar]
- Cristofanilli, M, Hayes, DF, Budd, GT, Ellis, MJ, Stopeck, A, Reuben, JM et al. (2005). Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol 23: 1420–1430. [DOI] [PubMed] [Google Scholar]
- Dawson, SJ, Tsui, DW, Murtaza, M, Biggs, H, Rueda, OM, Chin, SF et al. (2013). Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Keshaviah, A, Dellapasqua, S, Rotmensz, N, Lindtner, J, Crivellari, D, Collins, J et al. (2007). CA15-3 and alkaline phosphatase as predictors for breast cancer recurrence: a combined analysis of seven International Breast Cancer Study Group trials. Ann Oncol 18: 701–708. [DOI] [PubMed] [Google Scholar]
- Mook, S, Knauer, M, Bueno-de-Mesquita, JM, Retel, VP, Wesseling, J, Linn, SC et al. (2010). Metastatic potential of T1 breast cancer can be predicted by the 70-gene MammaPrint signature. Ann Surg Oncol 17: 1406–1413. [DOI] [PubMed] [Google Scholar]
- Cuk, K, Zucknick, M, Heil, J, Madhavan, D, Schott, S, Turchinovich, A et al. (2013). Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer 132: 1602–1612. [DOI] [PubMed] [Google Scholar]
- Seiffert, D, Crain, K, Wagner, NV and Loskutoff, DJ (1994). Vitronectin gene expression in vivo. Evidence for extrahepatic synthesis and acute phase regulation. J Biol Chem 269: 19836–19842. [PubMed] [Google Scholar]
- Zhuang, P, Chen, AI and Peterson, CB (1997). Native and multimeric vitronectin exhibit similar affinity for heparin. Differences in heparin binding properties induced upon denaturation are due to self-association into a multivalent form. J Biol Chem 272: 6858–6867. [DOI] [PubMed] [Google Scholar]
- Wilkins-Port, CE and McKeown-Longo, PJ (1998). Degradation of distinct forms of multimeric vitronectin by human fibroblasts. Biochim Biophys Acta 1404: 353–366. [DOI] [PubMed] [Google Scholar]
- Podor, TJ, Campbell, S, Chindemi, P, Foulon, DM, Farrell, DH, Walton, PD et al. (2002). Incorporation of vitronectin into fibrin clots. Evidence for a binding interaction between vitronectin and gamma A/gamma' fibrinogen. J Biol Chem 277: 7520–7528. [DOI] [PubMed] [Google Scholar]
- Wilder, RL (2002). Integrin alpha V beta 3 as a target for treatment of rheumatoid arthritis and related rheumatic diseases. Ann Rheum Dis 61 Suppl 2: ii96–ii99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, CC (2003). Integrin alpha v beta 3 as a therapeutic target for blocking tumor-induced angiogenesis. Curr Drug Targets 4: 123–131. [DOI] [PubMed] [Google Scholar]
- Casaroli Marano, RP and Vilaró, S (1994). The role of fibronectin, laminin, vitronectin and their receptors on cellular adhesion in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci 35: 2791–2803. [PubMed] [Google Scholar]
- Wei, Y, Waltz, DA, Rao, N, Drummond, RJ, Rosenberg, S and Chapman, HA (1994). Identification of the urokinase receptor as an adhesion receptor for vitronectin. J Biol Chem 269: 32380–32388. [PubMed] [Google Scholar]
- Noh, H, Hong, S and Huang, S (2013). Role of urokinase receptor in tumor progression and development. Theranostics 3: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, M, Takimoto, S, Montel, V and Gonias, SL (2009). The urokinase receptor promotes cancer metastasis independently of urokinase-type plasminogen activator in mice. Am J Pathol 175: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, CD, Ferraris, GM, Andolfo, A, Cunningham, O and Sidenius, N (2007). uPAR-induced cell adhesion and migration: vitronectin provides the key. J Cell Biol 177: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzoli, V, Ferraris, GM and Sidenius, N (2013). Direct evidence of the importance of vitronectin and its interaction with the urokinase receptor in tumor growth. Blood 121: 2316–2323. [DOI] [PubMed] [Google Scholar]
- Kotzsch, M, Luther, T, Harbeck, N, Ockert, D, Lutz, V, Noack, F et al. (2000). New ELISA for quantitation of human urokinase receptor (CD87) in cancer. Int J Oncol 17: 827–834. [DOI] [PubMed] [Google Scholar]
- Kadowaki, M, Sangai, T, Nagashima, T, Sakakibara, M, Yoshitomi, H, Takano, S et al. (2011). Identification of vitronectin as a novel serum marker for early breast cancer detection using a new proteomic approach. J Cancer Res Clin Oncol 137: 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaboe, M, Offersen, BV, Christensen, A and Andreasen, PA (2003). Vitronectin in human breast carcinomas. Biochim Biophys Acta 1638: 72–82. [DOI] [PubMed] [Google Scholar]
- Hurt, EM, Chan, K, Serrat, MA, Thomas, SB, Veenstra, TD and Farrar, WL (2010). Identification of vitronectin as an extrinsic inducer of cancer stem cell differentiation and tumor formation. Stem Cells 28: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- McKeague, M, DeRosa, MC, McKeague, M and DeRosa, MC (2012). Challenges and opportunities for small molecule aptamer development, challenges and opportunities for small molecule aptamer development. J Nucleic Acids 2012: e748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzesinski, J and Ciesiolka, J (2005). Characterization of structure and metal ions specificity of Co2+-binding RNA aptamers. Biochem 44: 6257–6268. [DOI] [PubMed] [Google Scholar]
- Foy, JW, Rittenhouse, K, Modi, M and Patel, M (2007). Local tolerance and systemic safety of pegaptanib sodium in the dog and rabbit. J Ocul Pharmacol Ther 23: 452–466. [DOI] [PubMed] [Google Scholar]
- Ng, EW, Shima, DT, Calias, P, Cunningham, ET Jr, Guyer, DR and Adamis, AP (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5: 123–132. [DOI] [PubMed] [Google Scholar]
- Boyacioglu, O, Stuart, CH, Kulik, G and Gmeiner, WH (2013). Dimeric DNA aptamer complexes for high-capacity-targeted drug delivery using pH-sensitive covalent linkages. Mol Ther Nucleic Acids 2: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze, T, Wilhelm, B, Greiner, N, Braun, H, Peter, F, Mörl, M et al. (2011). Probing the SELEX process with next-generation sequencing. PLoS One 6: e29604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, KR, Gagliano, J, Xiao, J, Libby, K, Saito, S, Yu, G et al. (2015). Combining capillary electrophoresis and next-generation sequencing for aptamer selection. Anal Bioanal Chem 407: 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, KR, Saito, S, Gagliano, J and Colyer, CL (2014). Facilitating aptamer selection and collection by capillary transient isotachophoresis with laser-induced fluorescence detection. J Chromatogr A 1368: 183–189. [DOI] [PubMed] [Google Scholar]
- Berezovski, M, Nutiu, R, Li, Y and Krylov, SN (2003). Affinity analysis of a protein-aptamer complex using nonequilibrium capillary electrophoresis of equilibrium mixtures. Anal Chem 75: 1382–1386. [DOI] [PubMed] [Google Scholar]
- Stuart, CH, Horita, DA, Thomas, MJ, Salsbury, FR Jr, Lively, MO and Gmeiner, WH (2014). Site-specific DNA-doxorubicin conjugates display enhanced cytotoxicity to breast cancer cells. Bioconjug Chem 25: 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz, F (2008). Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release 132: 171–183. [DOI] [PubMed] [Google Scholar]
- Tolle, F, Wilke, J, Wengel, J and Mayer, G (2014). By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS One 9: e114693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, WH, Bair, T, Wyatt Thiel, K, Dassie, JP, Rockey, WM, Howell, CA et al. (2011). Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid Ther 21: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker, W, Hess, S, Vischer, P and Preissner, KT (1993). Binding and processing of multimeric vitronectin by vascular endothelial cells. J Histochem Cytochem 41: 1823–1832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.