Abstract
Targeted nucleases are influential instruments for intervening in genome revision with great accuracy. RNA-guided Cas9 nucleases produced from clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems have noticeably altered the means to modify the genomes of distinct organisms. They can be notably used to facilitate effective genome manipulation in eukaryotic cells by clearly detailing a 20-nt targeting sequence inside its directed RNA. We discuss the most recent advancements in the molecular basis of the type II CRISPR/Cas system and encapsulate applications and elements affecting its use in human cells. We also propose possible applications covering its uses ranging from basic science to implementation in the clinic.
Keywords: CRISPR, crRNA, Cas9, genome editing, human cells, RGEN
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR associated) system has changed to protect microorganisms against unfamiliar invading nucleic acids, mainly DNA from bacteriophages (phages), plasmids, and other mobile segments.1,2 CRISPR/Cas systems have been noted in 47 and 86% of the entire bacterial and archaea genomes.3 Resistance develops when a smaller sequence is taken from the phage or plasmid genome and attached, as a new spacer (guide), to the CRISPR arrays,4 which have short repeats isolated by spacers. In CRISPR systems, a CRISPR RNA (crRNA) with a “spacer” is produced from a longer precursor (pre-crRNA)3,5,6 and blended into a ribonucleoprotein complex of at least one Cas protein.1,5,6,7,8,9,10 These ribonucleoprotein complexes link to, and elicit, the eradication of complementary DNA or RNA from intruding forces.11
CRISPR/Cas systems are categorized into three major types (I-III), and then additionally into subtypes (types III-A and III-B).12 Various types have commonalities, yet can have several factors that make them distinctive, such as in crRNA production or the origin of the target (RNA or DNA). Current studies have started to clarify the method of identifying target protospacers in the major types of CRISPR/Cas systems. Based on prior studies, researchers believed that precise pairing along the length of the spacer RNA was needed,13,14 but recent outcomes point to the fact that some incompatibilities are allowed, at least by some systems.15,16,17 In type I and II systems, protospacer (a site within the invader genome) adjacent motifs (PAM) are needed for identification,14,18,19,20,21,22 and a limited seed sequence inside the match is necessary in certain subtypes.4,10,16 In type III systems, it is uncertain if a seed sequence is present and no PAMs have been determined.21 Instead, the base-pairing probability between the 5' repeat-derived area of the crRNA (known as the 5' handle) and the sequence around the protospacer target is important to encourage interference, and prevent self-targeting for type III-A systems.23
Type II CRISPR/Cas System
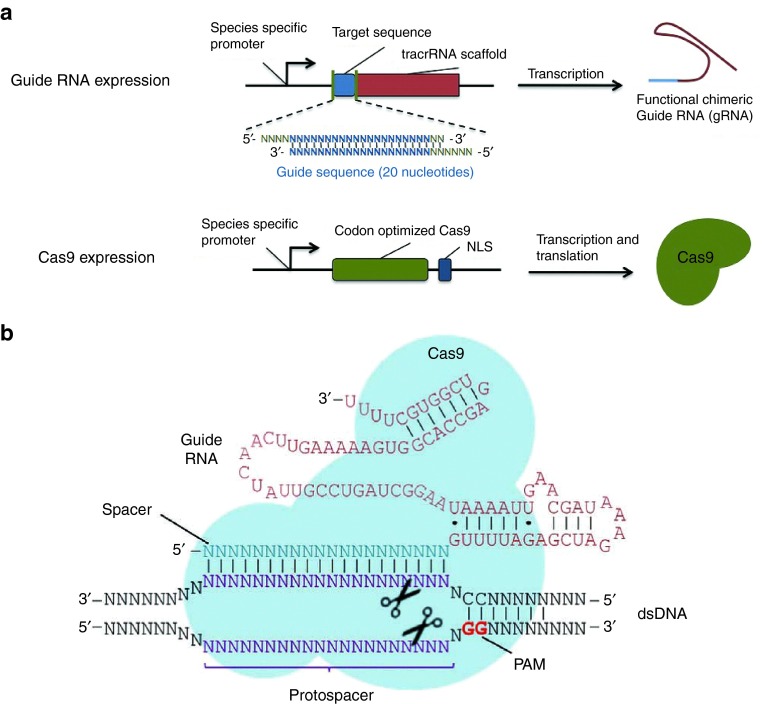
The type II CRISPR/Cas system24 is made up from long pre-crRNA transcribed from the spacer-repeat CRISPR locus, the multifunctional Cas9 protein, and a trans-activating crRNA (tracrRNA) required for processing the pre-crRNA and the creation of the Cas9 complex (Figure 1). The native type II system needs a minimum of three critical parts: RNA-guided Cas9 nuclease, crRNA, and a moderately complementary trans-acting crRNA (tracrRNA).3,6,8 Cas9 (previously referred to Csn1 or Csx12) is the initial crucial part of type II CRISPR/Cas systems and could divide double-stranded DNA (dsDNA) in a sequence-precise fashion.1,2,4 TracrRNA is the second required part of the type IICRISPR/Cas system and is a non-protein-coding RNA for crRNA development and resulting DNA division.5 The processing of crRNAs relies on tracrRNA encoded in the area of CRISPR loci and has a 25-nt-long stretch that is compatible with the crRNA duplicated sequence.3
Figure 1.
Cas9-sgRNA targeting complexes. (a) Expressed tracrRNA-crRNA duplexes and Cas9; (b) The basic S. pyogenes Cas9-sgRNA RNA-guided nuclease complex for eukaryotic genome engineering.58
Type II CRISPR interference involves a multi-step procedure.3 First, tracrRNAs hybridize to replicated areas of the pre-crRNA. Second, endogenous RNase III divides the hybridized crRNA-tracrRNAs, and a subsequent activity eliminates the 5′ bottom of each spacer, producing fully developed crRNAs that stay linked to the tracrRNA and Cas9. The synchronized RNase III and Cas9 activity is required to handle the duplex, and the complementarity of the duplex is essential for RNase III-mediated coprocessing.3 Third, all of the fully developed complex finds an earmarked dsDNA sequence and divides the two strands. The target identification and resulting separation by the crRNA-tracrRNA-Cas9 needs both sequences to be compatible with the spacer and the target “protospacer” sequence as well as the appearance of a suitable protospacer-adjacent motif (PAM) sequence at the 3′bottom of the protospacer sequence (Figure 1).6,10 The PAM is a required targeting part that also acts as a self versus non-self identification system to halt the CRISPR locus from being targeted. Several type II systems have varying PAM prerequisites, which can lower their ease of targeting.7,9 The most popular constructed system thus far, the Streptococcus pyogenes, needs a PAM with the sequence NGG, in which N is a nucleotide,10 and the formation studies of it showed that the PAM motif stays in a base-paired DNA duplex.25
Type II systems may vary in the specifics of pre-crRNA generation and crRNA-tracrRNA processing.8 Current studies revealed various crRNA development procedures in type II systems.1,9 For example, S. pyogenes and Neisseria spp. have unmistakable pre-crRNA transcript characteristics, pre-crRNA processing, and nucleoprotein complexes. In S. pyogenes, a two-step crRNA biogenesis occurs, with an initial division inside the duplicated areas and another division inside the spacers.3 Interestingly, in N. meningitidis, pre-crRNAs are trimmed once at their 3' end to generate 48-nt mature crRNAs via the synchronized activities of RNase III and Cas9, which requires the same as the initial division in S. pyogenes minus the next division inside the spacers.8 These processed RNA parts are put together with Cas9, which creates executive nucleoprotein complexes that aim for and divides the protospacer identified by 20-nt spacer sequences in crRNAs.3,8,26
Application of type II CRISPR/Cas System in Human Cells
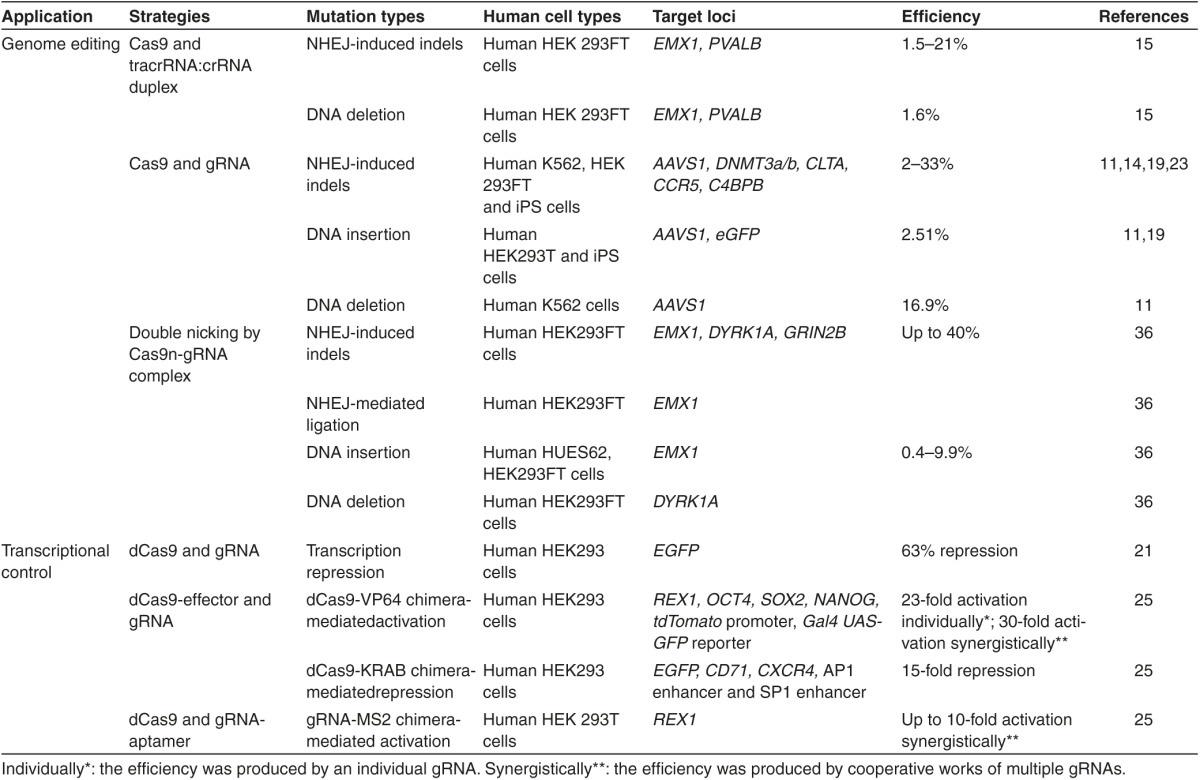
Because of the simplicity and customizability of type II CRISPR systems, host-independent gene-targeting platforms have been created for genome editing and transcriptional control in eukaryotes12,13,14,15,16,17,18,19,27–29 and prokaryotes4,20,21,22 (Table 1). In bacteria, the system can be implemented without the need for adjustments,20 but in human use, this requires the extraction of a human-codon–optimized Cas9 protein with an applicable nuclear localization signal, and the crRNA and tracrRNA to be revealed either by itself or as a single chimera via a RNA polymerase III promoter.15,23,28 In contrast, in vitro-transcribed RNA can be transported specifically to the targeted cell types.14,17 Expressing a chimeric crRNA-tracrRNA, also called a short guide RNA (sgRNA), is the most popular technique because of improved simplicity and hearty targeting, particularly when the sgRNA is not abbreviated.14,15,28 This principal methodology has been implemented in the modification of genomes of many model eukaryotic organisms.12,13,14,15,17,18,23,24,30–32 The recent applications of type II systems can be placed into five groups: native Cas9-mediated genome modifying, Cas9 nickase-mediated genome modification, inactivated or activated Cas9-mediated transcriptional control, high-throughput genome modification and silencing, and separating certain genomic areas of intrigue with a catalytically inactive Cas9 joined with a tag(s) (Table 1).
Table 1. Application of Cas9-based tool in human genome editing and transcriptional control.
Native Cas9-mediated genome editing
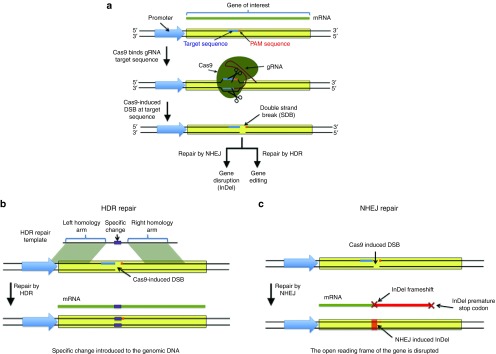
Cas9-mediated genome modification requires two additional steps. First, genomic DNA is divided by Cas9 at a precise area dictated by the 20-nt target identification sequence in crRNA3,10 (Figure 2a). Second, double-strand breaks (DSBs) are ligated by native DNA repair systems,33 template-dependent homology-directed repair (HDR)34 (Figure 2b), or native non-homologous end joining (NHEJ)35 (Figure 2c). To use type II CRISPR/Cas systems, three parts, including the Cas9 protein, tracrRNA, and tailored crRNA, have to be extracted from human cells. Using tailored 20-nt target recognition sequences in a crRNA array, double deletion and/or multiplexed modifying took place in human genomes in one step.15 Simultaneous DSBs with 2 crRNAs in opposition to the EMX1 gene removed 119-bp human genome areas.15 Therefore, three-part CRISPR/Cas9 systems are suitable for achieving earmarked multiplexed modifying in a human genome by easily programming the crRNA array.
Figure 2.
Application of native Cas9-mediated genome editing. Expressed tracrRNA-crRNA duplexes or gRNA is assembled with Cas9, generating executive complexes. These complexes generate breaks in the genome (a) that may trigger homology-directed repair (HDR) when homology-containing dsDNA or ssDNA templates are given (b) to confer precise DNA substitution, or induce error-prone nonhomologous end joining (NHEJ) to rejoin the ends and introduce undefined small deletions and additions (indels) (c).
The tracrRNA:crRNA duplex has been transformed into a molecule, known as a chimeric guide RNA (gRNA), that is 39 to 40 nt in length and has a 20-nt target recognition sequence at the 5' end followed by a gRNA scaffold (or hairpin structure) that keeps the base-pairing encounters inside the tracrRNA:crRNA duplex (Figure 1b).10,28 This advancement is additionally clarified by the employment of type II CRISPR systems in human genome editing. By coexpressing Cas9 nuclease and individualized gRNAs, scientists have gainfully engineered and modified the genomes of humans14,15,26,28,29 (see Table 1). Strikingly, the building of a mammalian expression system with codon-optimized Cas9 and gRNA produced targeted rates of 2 to 25% in different human cells14,15,28 and 36 to 48% in embryonic mouse stem cells.27 By detailing DNA donor templates, expression cassettes of green fluorescent protein have been effectively inserted into the focused on human genome loci14,15,28 and the Cas9-gRNA complex has been used to concurrently interrupt five genes in a single genome.27 Not long ago, with a precise CRISPR-Cas system from Neisseria meningitidis, Thomson et al. showed effective targeting of an endogenous gene in three human pluripotent stem cell (hPSC) lines with homology-directed repair (HDR).36 Thus, the CRISPR/Cas9 system is an effective apparatus for modifying genomes with broad uses in a large variation of hosts.
Cas9 Nickase-mediated genome editing
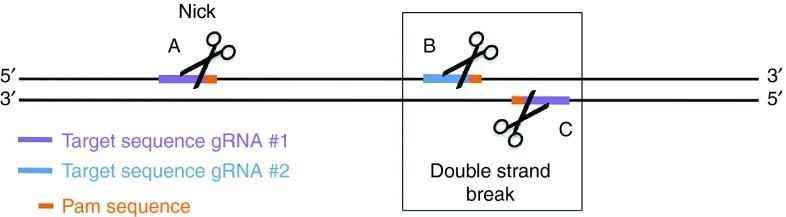
gRNA-guided Cas9n with a RuvC or HNH mutation has the means to produce a nick instead of a DSB in the targeted region.6,15,37,38 Even though single nicks are mainly fixed by the high-fidelity base excision pathway,39 the mixture of nick production and HDR has effectively modified human genomes in the proposed area.15 Initiation of a double nick with a pair of gRNA-directed Cas9ns targeting the opposing strands of the singled-out site has been effectively employed to produce DSBs and NHEJ-induced mutations37,38 (Figure 3). A paired-nicking plan was identified to ease the production of high-efficiency HDR at levels similar to those of original Cas9-mediated HDR and at notably greater rates than individual Cas9n-mediated HDR.37 Interestingly, this paired nicking greatly lowered off-target divisions by 50- to 1,500-fold in human cells without compromising on-target division efficiency.37 Because double nicking has the means to produce anticipated defined overhangs, NHEJ-mediated ligation, by providing double-stranded fixing templates with complementary overhangs, has effectively prompted HDR-independent fragment incorporation in certain areas.37 Additionally, making a pair of double nicks at two sites by four individualized gRNAs effectively eliminated genomic fragments of up to 6 kb in HEK 293FT cells.37 Therefore, multiplex nicking generated by Cas9n can produce high-precision genomic modifications.
Figure 3.
Two different gRNAs (#1 and #2) bind in a particular genomic region. When gRNA #1 and #2 are coexpressed with a Cas9 nickase, single-strand nicks are created in the DNA at (a), (b), and (c). The nick created at (a) is quickly repaired by HDR using the intact compliment strand as a template and no change occurs. The nicks at (b) and (c) are in close proximity (and on opposite strands) and together behave as a DSB.
Inactivated Cas9-based Transcriptional Control
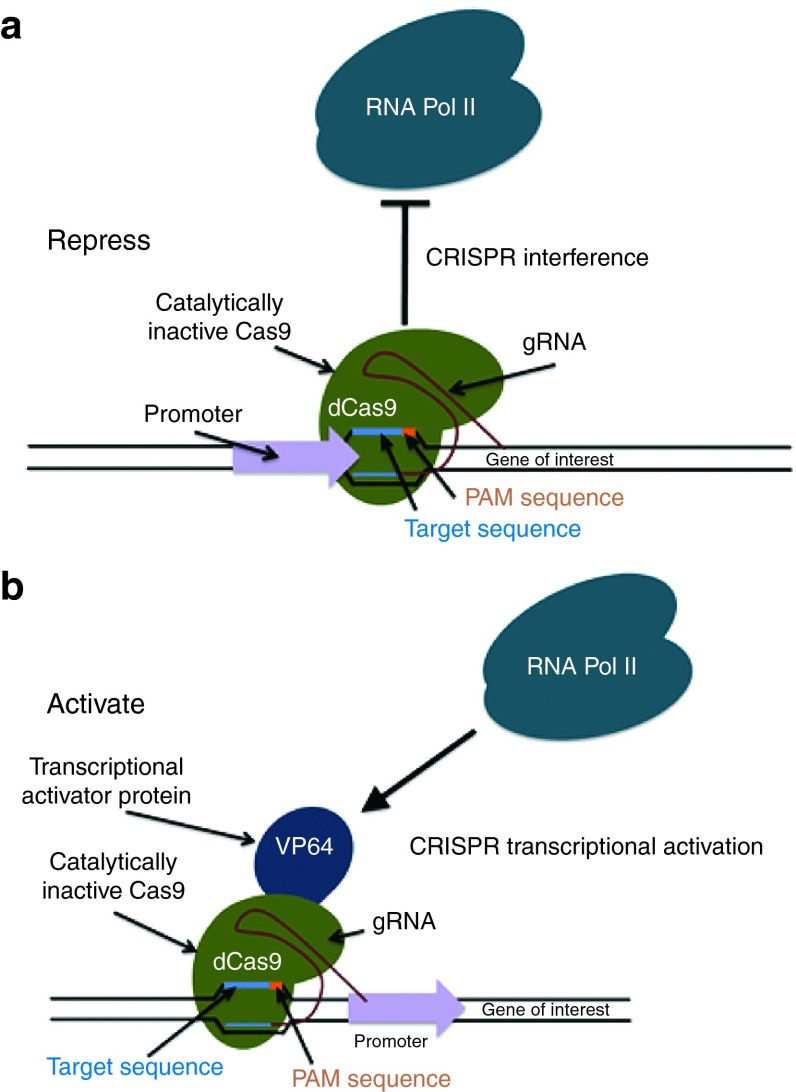
CRISPR/Cas systems have also been created as a novel effortless and multiplexable strategy for transcriptional control without changing the target gene sequence that is named CRISPR interference (CRISPRi) (Figure 4a).21 It has a fully inactive dCas9 and individualized gRNA (or tracrRNA:crRNA duplex). As previously mentioned, dCas9 ceases its endonuclease activity, but its means to include gRNA and link to targets is not interrupted. Similar to RNA interference (RNAi), CRISPRi is controlled by base-pairing compatible with identified target areas; however, they use various mechanisms to control gene expression. RNAi expressly produces transcript degradation and/or translation blocking,40 but CRISPRi obstructs transcription commencement and elongation.21 Qi et al. stated that the mechanism of CRISPRi and its first applications in effectively limiting the extraction of singled-out genes in human cells and silencing effects can be encouraged and undone with an anhydrotetracycline-inducible promoter to encourage dCas9 and gRNA expression.21 Through cocustomization of multiple gRNAs, concurrent regulation of several genes became feasible. Merging two gRNAs that are aiming at the identical gene may generate up to 1,000-fold repression.21 Therefore, the CRISPRi targeting technology has the potential to modulate gene expression at the transcriptional level.
Figure 4.
Application of dCas9-based transcriptional control. The dCas9 can be utilized as a platform for DNA transcriptional regulators to repress (a) or activate (b) gene expression by fusing theinactive enzyme (that retains its gRNA-binding ability) to known regulatory domains.
Aside from silencing gene expression, dCas9 has been effectively used to trigger gene expression. dCas9-activator proteins, including dCas9-VP64 and dCas9-p65AD, showed up to a 25-fold improvement in gene expression9 (Figure 4b). Using the dCas9-VP64 chimera in human cells, gRNAs proximal to the initial transcriptional area substantially triggered gene expression, and several gRNAs teamed up to encourage multifold transcriptional improvement.38 Multiple orthogonal dCas9s merged with VP64 also showed substantial transcriptional activation in human cells at levels comparable to that of the corresponding transcription activator-like (TAL)-VP64 activator.4 Thus, the dCas9-gRNA complex has the potential to create sequence-specific transcriptional regulation in various human tumor cell lines and possibly for further heterogeneous epigenetic examination.
Cas9-based High-throughput Forward Genetic Screening
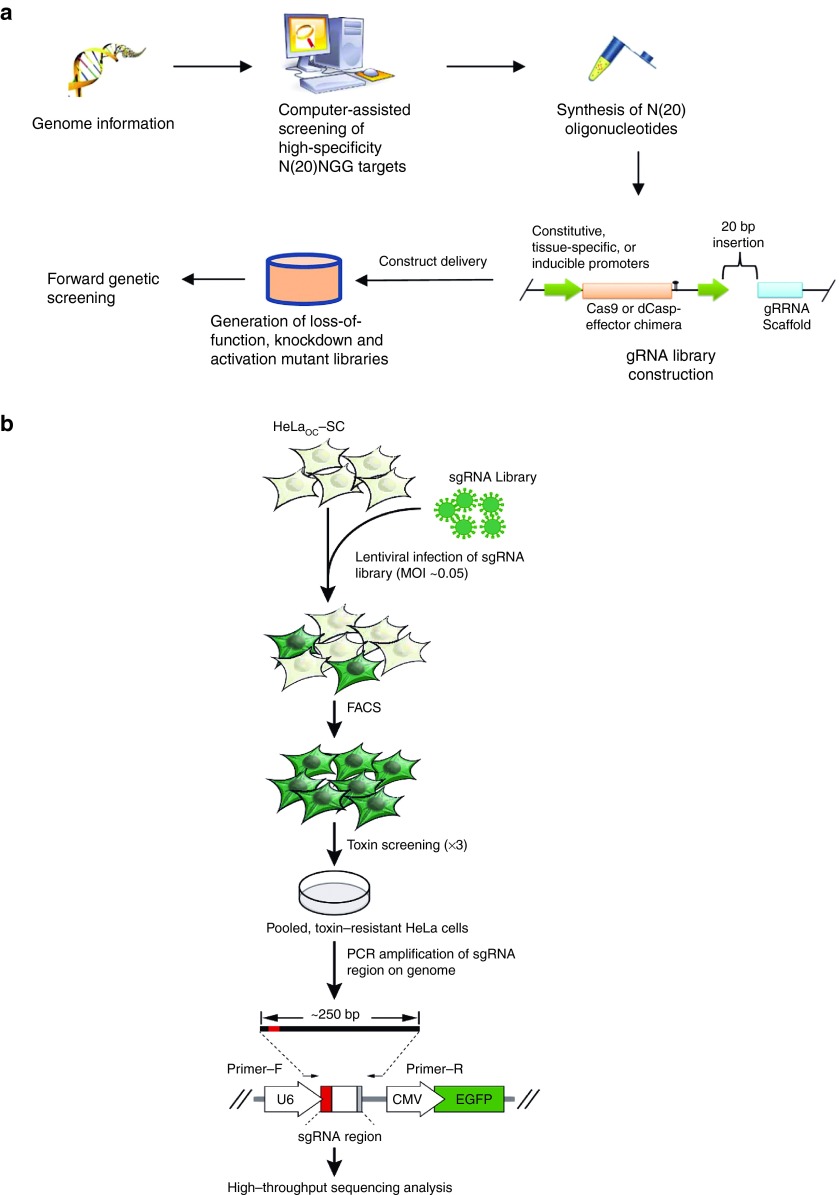
It is an effective tool that uses multifunctional Cas9 variants to produce mutant libraries for screening and identifying genome-scale phenotype-related genetic elements. A customary flowchart to ascertain this goal is shown in Figure 5a.26 For high-throughput targeting, the key is to create high-specificity gRNA libraries. Not long ago, this strategy was effectively implemented in the genetic screening in human cells.41,42,43 Wang et al. identified a pooled, loss-of-function genetic screening method compatible with both positive and negative selection that uses a genome-scale lentiviral single-guide RNA (sgRNA) library.42 sgRNA expression cassettes were stably incorporated into the genome, which allowed a complex mutant pool to be monitored by massive parallel sequencing.42 They used a library of 73,000 sgRNAs to produce knockout collections and carried out screening in two human cell lines. A screen for opposition to the nucleotide analog 6-thioguanine determined all anticipated components of the DNA mismatch repair pathway whereas another for the DNA topoisomerase II (TOP2A) poison etoposide recognized TOP2A, as anticipated, and also cyclin-dependent kinase 6, CDK6. A negative selection screen for necessary genes recognized several gene sets correlating to underlying processes. They also revealed that sgRNA efficiency is linked to certain sequence motifs, allowing for the prediction of more successful sgRNAs.
Figure 5.
Flowchart for Cas9-based high-throughput forward genetic screening. Reasonable and precise N20NGG targetable areas on the other side of the complete genome were chosen using computer-assisted programs. DNA oligonucleotides with N20 sequences were synthesized as well as adjusted for ligation with a construct, which produced a gRNA library that can coexpress Cas9 or a Cas9-effector chimera and varied gRNAs propelled by constitutive, tissue-specific, or inducible promoters (a).26 The gRNA library was then transported to host cells to produce varied mutant libraries for forward genetic screening (b).43 sgRNAs were transported into HeLaOC-SC cells using lentiviral infection with a MOI of 0.05. Library cells steadily expressing sgRNAs were gathered using FACS for green fluorescence 48 hours postinfection. Library screening was performed with three toxin courses, followed by PCR amplification of the sgRNA-coding sequence incorporated into the chromosomes. The purified PCR product then underwent high-throughput sequencing analysis using Illumina's HiSeq 2500.
Shalem et al. demonstrated that lentiviral transportation of a genome-scale CRISPR-Cas9 knockout (GeCKO) library targeting 18,080 genes with 64,751 unique guide sequences permits both negative and positive selection screening in human cells.41 First, they employed the GeCKO library to determine the genes necessary for cell viability in cancer and pluripotent stem cells.41 Next, in a melanoma model, they looked for genes whose loss is connected to the resistance to vemurafenib, a therapeutic RAF inhibitor.41 Not long ago, Zhou et al. also documented the creation of a pinpointed CRISPR/Cas-based lentiviral library in human cells and a technique for gene identification based on functional screening and high-throughput sequencing analysis.43 Using knockout library screens, they effectively determined the host genes required for the intoxication of cells by anthrax and diphtheria toxins, which were affirmed by functional confirmation. All together, these outcomes instituted Cas9/sgRNA screens as an influential tool for systematic genetic analysis in mammalian cells (Figure 5b). In addition, if dCas9 or a dCas9-effector chimera is implemented, knockdown or activation mutant libraries will be produced. Then mutants of interest can be screened with suitable methodologies. Compared to loss-of-function mutant libraries, the knockdown or activation mutant libraries have an unrivaled advantage in the study of deadly genes.
Influential Factors of Crispr/Cas Application in Human Cells
Cas9 is a critical part. Mutation of catalytic sites, inaccurate subcellular localization, or wrong Cas9 dosage can all affect genome modification. In eukaryotic cells, prokaryote-derived Cas9 is typically merged with a nuclear location signal (NLS) at the N or C terminus, or both, to direct protein translocation into the nucleus.13,14,15,18,21,23,27,28,36 Current studies determined that the inclusion of a 32-amino-acid linker between the NLS and Cas9 improved genomic DNA division activity. This might be due to greater subcellular localization because the NLS peptide would be concealed or protected during Cas9 folding without the linker.29 Codon optimization is also required for making functional Cas9 in a heterogeneous expression system.14,15,28 An additional major class of determinants is the RNA part. For tracrRNAs, S. pyogenes produced two types of precursor tracrRNA species. Examination of heterogeneous cells revealed that the smaller tracrRNA was more potent15; however, studies using human and mouse cells revealed that division efficiencies of gRNAs were either less than those of RNA duplexes or imperceptible when the same protospacer was the aim. This indicates that some unidentified cellular factors or RNA characteristics may have an impact on modifying efficiency.
CRISPRi has been systematically examined,21,26 and multiple items, including those mentioned prior, have been determined to impact dCas9-based transcriptional control. First, CRISPRi-mediated prevention of transcriptional elongation offers strand specificity.21,22 gRNAs singling out a nontemplate DNA strand showed greater repression regulation than those aimed at the template strand. Second, the silencing efficiency is inversely correlated with the span of the target from the translation start codon. Third, an augmentative silencing effect could be viewed when two or more gRNAs link to individual earmarked areas on 1 gene21,38; however, if they link to overlapping areas, repression is suppressed. Fourth, CRISPRi-mediated prevention of transcriptional commencement has local effects, but not on strand specificity. Finally, for dCas9 effector-dependent transcriptional regulation, performance also showed location and accumulation effects.38
comparison of targeted genetic-engineering tools
A group of genome-editing platforms have been created recently, including zinc-finger nucleases (ZFNs),44,45,46,47 transcription activator–like effector nucleases (TALENs),47,48,49,50,51 and the RNA-guided CRISPR-Cas EndoNuclease system (RGEN).14,15,16,20,23,28 The initial two platforms implement a method of tethering endonuclease catalytic domains to modular DNA-binding proteins for prompting singled out DNA double-stranded breaks (DSBs) at certain genomic loci. In contrast, Cas9 is a nuclease governed by small RNAs through Watson-Crick base pairing with target DNA,6,10 portraying a system that is markedly simpler to design and is highly specific, effective, and well-suited for high-throughput and multiplexed gene modifying for an assortment of cell types and organisms.15,17,20,27,28 The ZiFiT Targeter program was designed as an tool to easily find possible sgRNA:Cas9 targeting areas in a specific genome.17 RGEN editing occurs at a homologous efficiency as ZFN or TALEN, and as of the present, no off-site targeting was noted.14 Another benefit of RGENs is that the CRISPR interference results in a selection for positive clones since wild-type sequences are frequently singled out until the mutated donor DNA is incorporated by HR and limits the interference process.13 Other characteristics of the RGEN method show their limitations and disadvantages in genome editing. One issue with RGENs is their reliance on a small PAM sequence, which limits the selection of possible target sequences. Furthermore, the span of the crRNA and its seed sequence stunt the range of target sequences and, thus, an optimal design of the sgRNA is needed for effective editing.14,15,17,28 ZFN and TALEN systems need protein-engineered steps, but the two systems are highly tunable by changing the particular domains for sequence-specific identification of the target. Furthermore, libraries of various proteins already exist,52 which lowers the time and expenses associated with the production of novel functional proteins.14,53 The modular formation of the two systems (TALEN and ZNF) produces a highly variable instrument in which the effector domain can be merged to an assortment of various proteins (e.g., transposases, nucleases, and transcriptional regulators).54,55,56 ZFN- and TALEN-based gene therapy methods have been examined for clinical use. Additional advancements, including reviewing recent advances in reorganizing chromosomes with RGENs, will expand the number of potential applications in gene therapy.53,57,58
Toward Cas9 Therapeutics
Taking into account the vast utility of Cas9 for the regulation and modification of complex biological systems, could the Cas9 system end up being just as useful as a basis for novel treatments? Two central routes to Cas9-mediated therapeutic interventions were documented by Mali et al.58 The first requires singling out genome modification to resolve genetic disorders59,60,61 and to potentially interrupt intruding viral genomes. The second implements Cas9nuclease-null fusions for targeted genome regulation in a fashion much like the use of small-molecule therapeutics, except that both repression and activation modalities would be accessible. One can imagine implementing such a method to amend epigenetic misregulation of gene expression, to regulate inflammation and autoimmunity, and also to limit transcription of viral genes or even viral coreceptors in defenseless cell types.
However, several technical hurdles must be addressed before Cas9-based treatments become a reality. First, Cas9-encoding cassettes must be effectively transported to singled out cells in vivo. Unfortunately, Cas9 proteins are substantial; the frequently used Cas9 protein from S. pyogenes is 1,368 amino acids long. Employing smaller Cas9 orthologs, some of which are <1,100 amino acids, or engineering a nominal Cas9 by eliminating domains irrelevant to the planned intent could enable effective packaging into limited-capacity viral vectors (adeno-associated viruses, adenoviral vectors, and lentiviruses)62,63,64,65 for explicit in vivo transport. The creation of strictly regulated expression vectors or modalities that allow both transient and controlled release of targeting reagents will be crucial to limit the resulting Cas9-mediated functions to certain tissues.
Aside from the issue of gene delivery, the greatest hurdle to therapeutic applications is the relatively low specificity of Cas9 binding. Because off-target activity comes with the risk of oncogenesis and even highly specific processes will ultimately lead to catastrophic results when applied to enough cells, addressing this issue is of the greatest importance. Mali et al.58 have discussed some prospective methods that include necessitating cooperativity via offset nicking and biasing repair outcomes toward HR versus NHEJ. Colocalizing target and donor DNA through direct guide RNA tethering or Cas9 recombinases and transposases could also be purposeful for this undertaking, particularly in targeting postmitotic cells where function of endogenous HR machinery is anticipated to be greatly declined.
Avoiding a negative immune response is also crucial.58 Traditional immune suppression for the length of therapy is one choice for mitigating the immune response, but this practice becomes less useful with long-term regulatory modifications. A more encouraging method would involve examining the immunogenicity of Cas9 and “humanizing” the pertinent peptide fragments as has been done with antibody therapeutics.66 Finally, the regulatory control allowed by Cas9 may enable the mimicry of strategies implemented by viruses that include interrupting the main histocompatibility complex trafficking machinery.67
It is necessary to create of other synergistic technologies, in particular, efficient, earmarked, and secure in vivo gene-delivery channels to completely understand the therapeutic possibilities of Cas9 proteins. If ex vivo genome targeting demonstrates its potency (for example, in hematopoietic stem cells), then tools that allow exceptionally quick retrieval and screening of altered cells in a community will be crucial.
Outlook
The possibility of CRISPR/Cas systems working in genetic tools has been considered since the recognition of the system as a prokaryotic immune system. Six years after its identification, the initial steps have been made, as sgRNA:Cas9 complexes are used for effective genome modifications13,14,15,16,17,18,20,21,28,36,60 and the institution of gene silencing.21 Creation of RGEN-based genome modification systems to improve the efficiency and to clarify the incidence of off-site targeting of RGENs to motivate this system to become a possible instrument for disease treatment as has been attained with ZFN practices for the treatment of HIV.53 The institution of RGEN genome-editing instruments is only one part, in which CRISPR/Cas immunity reveals the possibility for applications in genetic and biotechnological systems. In the future, the versatility and ease of use offered by Cas9 combined with its sole means to combine RNA, DNA, and protein in a completely programmable manner will establish the basis of a mighty toolset for the perturbation, regulation, and monitoring of complex biological systems.
Acknowledgments
This work was supported by the Science and Technology Development Planing Program of Shandong Province (2014GGH218029), National Science Foundation of China (81572559, 31500027,41106151), and COMRA program (No. DY125-15-R-01). The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- Bhaya, D, Davison, M and Barrangou, R (2011). CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45: 273–297. [DOI] [PubMed] [Google Scholar]
- Makarova, KS, Haft, DH, Barrangou, R, Brouns, SJ, Charpentier, E, Horvath, P et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva, E, Chylinski, K, Sharma, CM, Gonzales, K, Chao, Y, Pirzada, ZA et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, KM, Mali, P, Braff, JL, Moosburner, M, Yaung, SJ and Church, GM (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis, T, Gasiunas, G, Miksys, A, Barrangou, R, Horvath, P and Siksnys, V (2013). crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol 10: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, P, Romero, DA, Coûté-Monvoisin, AC, Richards, M, Deveau, H, Moineau, S et al. (2008). Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y, Heidrich, N, Ampattu, BJ, Gunderson, CW, Seifert, HS, Schoen, C et al. (2013). Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 50: 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R, Lawrence, CM and Wiedenheft, B (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82: 237–266. [DOI] [PubMed] [Google Scholar]
- Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA and Charpentier, E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau, JE, Dupuis, MÈ, Villion, M, Romero, DA, Barrangou, R, Boyaval, P et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71. [DOI] [PubMed] [Google Scholar]
- Li, JF, Norville, JE, Aach, J, McCormack, M, Zhang, D, Bush, J et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo, JE, Norville, JE, Mali, P, Rios, X, Aach, J and Church, GM (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, SW, Kim, S, Kim, JM and Kim, JS (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K and Yang, Y (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6: 1975–1983. [DOI] [PubMed] [Google Scholar]
- Hwang, WY, Fu, Y, Reyon, D, Maeder, ML, Tsai, SQ, Sander, JD et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, SJ, Cummings, AM, Nguyen, JN, Hamm, DC, Donohue, LK, Harrison, MM et al. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, N, Sun, C, Gao, L, Zhu, D, Xu, X, Zhu, X et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W, Bikard, D, Cox, D, Zhang, F and Marraffini, LA (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, LS, Larson, MH, Gilbert, LA, Doudna, JA, Weissman, JS, Arkin, AP et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard, D, Jiang, W, Samai, P, Hochschild, A, Zhang, F and Marraffini, LA (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 41: 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, East, A, Cheng, A, Lin, S, Ma, E and Doudna, J (2013). RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q, Regan, SN, Xia, Y, Oostrom, LA, Cowan, CA and Musunuru, K (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, C, Niewoehner, O, Duerst, A and Jinek, M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T, Li, Y, Van Nostrand, JD, He, Z and Zhou, J (2014). Cas9-based tools for targeted genome editing and transcriptional control. Appl Environ Microbiol 1544–1552. [DOI] [PMC free article] [PubMed]
- Yang, H, Wang, H, Shivalila, CS, Cheng, AW, Shi, L and Jaenisch, R (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B, Zhang, J, Wu, H, Wang, J, Ma, K, Li, Z et al. (2013). Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland, AE, Tzur, YB, Esvelt, KM, Colaiácovo, MP, Church, GM and Calarco, JA (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D, Qiu, Z, Shao, Y, Chen, Y, Guan, Y, Liu, M et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Li, W, Teng, F, Li, T and Zhou, Q (2013). Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31: 684–686. [DOI] [PubMed] [Google Scholar]
- Wyman, C and Kanaar, R (2006). DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40: 363–383. [DOI] [PubMed] [Google Scholar]
- Smith, GR (2001). Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet 35: 243–274. [DOI] [PubMed] [Google Scholar]
- Shuman, S and Glickman, MS (2007). Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol 5: 852–861. [DOI] [PubMed] [Google Scholar]
- Hou, Z Zhang, Y,Propson, NE,Howden, SE, Chu, LF,Sontheimer, EJet al. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences 110: 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Hsu, PD, Lin, CY, Gootenberg, JS, Konermann, S, Trevino, AE et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Aach, J, Stranges, PB, Esvelt, KM, Moosburner, M, Kosuri, S et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov, GL and Hübscher, U (2013). Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res 41: 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, RC and Doudna, JA (2013). Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem, O,Sanjana, NE, Hartenian, E,Shi, X,Scott, DA,Mikkelsen, TS et al.(2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T, Wei, JJ, Sabatini, DM and Lander, ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y, Zhu, S, Cai, C, Yuan, P, Li, C, Huang, Y et al. (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491. [DOI] [PubMed] [Google Scholar]
- Miller, JC, Holmes, MC, Wang, J, Guschin, DY, Lee, YL, Rupniewski, I et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Porteus, MH and Baltimore, D (2003). Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Sander, JD, Dahlborg, EJ, Goodwin, MJ, Cade, L, Zhang, F, Cifuentes, D et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, AJ, Lo, TW, Zeitler, B, Pickle, CS, Ralston, EJ, Lee, AH et al. (2011). Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, M, Cermak, T, Doyle, EL, Schmidt, C, Zhang, F, Hummel, A et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F, Cong, L, Lodato, S, Kosuri, S, Church, GM and Arlotta, P (2011). Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29: 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer, D, Wang, H, Kiani, S, Lai, CS, Gao, Q, Cassady, JP et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon, D, Tsai, SQ, Khayter, C, Foden, JA, Sander, JD and Joung, JK (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y, Kweon, J,Kim, A,Chon, JK,Yoo, JY,Kim, HJet al. (2013). A library of TAL effector nucleases spanning the human genome. Nature Biotechnol 31: 251–252. [DOI] [PubMed] [Google Scholar]
- Ramalingam, S, Annaluru, N and Chandrasegaran, S (2013). A CRISPR way to engineer the human genome. Genome Biol 14: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, AC, Gaj, T, Fuller, RP and Barbas, CF 3rd (2012). Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res 40: 11163–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, T, Mercer, AC, Gersbach, CA, Gordley, RM and Barbas, CF (2011). Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proceedings of the National Academy of Sciences 108: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant, SR, Huang, Y, Akache, B and Kay, MA (2007). Site-directed transposon integration in human cells. Nucleic Acids Res 35: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, A, Wang, Z, Hu, Y, Wu, Y, Luo, Z, Yang, Z et al. (2013). Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Esvelt, KM and Church, GM (2013). Cas9 as a versatile tool for engineering biology. Nat Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov, FD, Miller, JC, Lee, YL, Beausejour, CM, Rock, JM, Augustus, S et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- Lombardo, A, Genovese, P, Beausejour, CM, Colleoni, S, Lee, YL, Kim, KA et al. (2007). Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol 25: 1298–1306. [DOI] [PubMed] [Google Scholar]
- Li, H, Haurigot, V, Doyon, Y, Li, T, Wong, SY, Bhagwat, AS et al. (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, IM and Somia, N (1997). Gene therapy – promises, problems and prospects. Nature 389: 239–242. [DOI] [PubMed] [Google Scholar]
- Grieger, JC and Samulski, RJ. (2005). Adeno-associated virus as a gene therapy vector: vector development, production and clinical applications. InGene Therapy and Gene Delivery Systems (pp. 119–145). Springer: Berlin, Heidelberg. [PubMed]
- Miyoshi, H, Takahashi, M, Gage, FH and Verma, IM (1997). Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proceed Natl Acad Sci USA 94: 10319–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, P and Perricaudet, M (1997). Advances in adenoviral vectors: from genetic engineering to their biology. FASEB J 11: 615–623. [DOI] [PubMed] [Google Scholar]
- Riechmann, L, Clark, M, Waldmann, H and Winter, G (1988). Reshaping human antibodies for therapy. Nature 332: 323–327. [DOI] [PubMed] [Google Scholar]
- Randow, F, MacMicking, JD and James, LC (2013). Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]