Abstract
Disease-causing splicing mutations can be rescued by variants of the U1 small nuclear RNA (U1snRNAs). However, the evaluation of the efficacy and safety of modified U1snRNAs as therapeutic tools is limited by the availability of cellular and animal models specific for a given mutation. Hence, we exploited the hyperactive Sleeping Beauty transposon system (SB100X) to integrate human factor IX (hFIX) minigenes into genomic DNA in vitro and in vivo. We generated stable HEK293 cell lines and C57BL/6 mice harboring splicing-competent hFIX minigenes either wild type (SChFIX-wt) or mutated (SChFIXex5-2C). In both models the SChFIXex5-2C variant, found in patients affected by Hemophilia B, displayed an aberrant splicing pattern characterized by exon 5 skipping. This allowed us to test, for the first time in a genomic DNA context, the efficacy of the snRNA U1-fix9, delivered with an adeno-associated virus (AAV) vector. With this approach, we showed rescue of the correct splicing pattern of hFIX mRNA, leading to hFIX protein expression. These data validate the SB100X as a versatile tool to quickly generate models of human genetic mutations, to study their effect in a stable DNA context and to assess mutation-targeted therapeutic strategies.
Keywords: coagulation factor IX, Sleeping Beauty, SB100X, splicing
Introduction
Intervention at the pre-mRNA splicing level is emerging as a promising therapeutic strategy to treat genetic disorders.1,2 Increasing attention has been given to the U1 small nuclear RNA (U1snRNA) that, in the initial splicing steps, mediates the recognition of the donor splice site (5′ss) by the small nuclear ribonucleoprotein U1 (snRNP U1).3,4 Studies in various cellular models of human splicing mutations indicated the potential therapeutic effect of engineered U1snRNAs to rescue aberrant splicing caused by mutations at 5′ss, a relatively frequent cause of human diseases.5,6 In the recent years, modified U1snRNAs have been exploited to correct splicing mutations causing severe coagulation factor VII deficiency and Hemophilia B (HB) using cellular or animal models expressing episomal minigenes.7,8,9,10,11 In fact, the thorough evaluation of U1snRNA-mediated correction strategies in vivo requires the use of mouse models, which are not always available. Here, we exploited the hyperactive Sleeping Beauty transposon system (SB100X)12,13 as a tool to quickly and efficiently develop cellular and mouse models for the c.519A>C splicing mutation (here indicated as hFIXex5-2C) in the F9 gene, previously reported in patients affected by HB14. This single nucleotide mutation falls in exon 5 5′ss and causes exon 5 skipping, leading to the production of an aberrant mRNA.15 Previous data obtained by exploiting episomal minigene assays in cellular models revealed that this aberrant mRNA variant, as well as other HB-causing mutations inducing exon 5 skipping, can be efficiently rescued with a second-generation of modified U1snRNA (Exon-Specific U1, ExSpeU1), designed to target intronic sequences downstream of the exon 5 5′ss,15 which are able to recruit the spliceosome machinery and improve exon definition.16 To evaluate the efficacy of the selected ExSpeU1 fix9 (U1-fix9) at correcting the hFIXex5-2C mutation in a stable chromatin context we exploited the SB100X17 to induce the integration of splicing-competent human FIX transgene variants (SChFIX) into the genome of both human embryonic kidney cells (HEK293) and C57BL/6 mice. The integration of the desired splicing variants in the genome is crucial to allow stable gene expression as compared with episomal plasmids, which lead to transient gene expression.18 Moreover, it has the advantage of mimicking a more physiological condition in terms of transcription and splicing regulation since these mechanisms can be both influenced by the chromatin organization.19,20 Notably, successful integration of either the wild type (SChFIX-wt) or the mutated (SChFIXex5-2C) expression cassettes in vitro and in vivo was rapidly and efficiently achieved. This allowed us to subsequently assess the efficacy of adeno-associated virus (AAV) vectors expressing the snRNA U1-fix9 at correcting the hFIX splicing pattern.
Results
Generation of stable HEK293 cell clones expressing hFIX splicing variants with the SB transposon technology
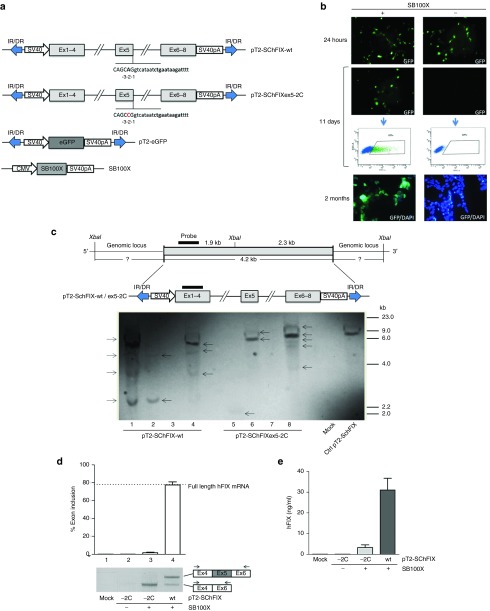
To rapidly create in vitro models of splicing mutations causative of HB, we used the SB100X system.17,21 We obtained stable cell clones expressing splicing-competent hFIX cassettes either wild type (SChFIX-wt) or carrying the splicing variant hFIXex5-2C (SChFIXex5-2C).15 Each splicing-competent construct was based on the hFIX cDNA region spanning from exon 1 to 4, exon 5 flanked by a splicing-functional region from introns 4 and 5, and the remaining cDNA from exon 6 to 8 (Figure 1a). The pT2-SChFIX transposon plasmids were obtained by cloning the expression-cassettes between the terminal inverted repeats (IR/DR) recognized by the hyperactive transposase SB100X,22 provided in trans (Figure 1a). An additional transposon plasmid encoding for enhanced green fluorescence protein (pT2-eGFP) was used as a marker to sort transfected cells. The pT2-SChFIX variants (wt and ex5-2C) were transfected into HEK293 cells together with the pT2-eGFP at a specific molar ratio (5:1). All the transposons were transfected with or without the SB100X-expressing plasmid, and the transfection efficiency was confirmed 24 hours post-transfection (Figure 1b). Eleven days post-transfection eGFP-positive (eGFP+) cells were observed only upon transposase transfection (Figure 1b). eGFP+ cells were then sorted and plated in limiting dilution to obtain single stable clones. The eGFP expression in the HEK293 stable clones was confirmed 2 months after transfection (Figure 1b). To evaluate the efficiency of the transposon integration in the HEK293 genome, we performed a Southern blot analysis on genomic DNA samples extracted from SChFIX-wt and SChFIXex5-2C clones (Figure 1c). For each SChFIX variant, eGFP+ HEK293 clones nonexpressing hFIX were used as negative controls (Figure 1c, lanes 3 and 7). In the clones harboring SChFIX-wt (Figure 1c, lanes 1, 2, and 4) some hybridization bands were detected, from a minimum of 2 (Figure 1c, lane 2) to a maximum of 4 (Figure 1c, lane 1), indicating integration of multiple copies of the transposon plasmid. In the clones harboring SChFIXex5-2C (Figure 1c, lanes 5, 6, and 8) up to 5 bands were detected. Genomic DNAs extracted from untransfected HEK293 cells (Figure 1c, Mock) and linearized pT2-SChFIX plasmid (Figure 1c, Ctrl pT2-SChFIX) were used as negative and positive controls, respectively. We also estimated by real time qPCR the transposon copy number of some HEK293 clones (two for each variant) and we found between 2 and 10 copies of transposon per cellular genome (see Supplementary Figure S5). Next, we extracted total RNA from HEK293 clones and performed reverse transcription followed by PCR (RT-PCR) analyses using specific primers in exon 4 and 6 to amplify hFIX transcripts (Figure 1d). As expected from the poor natural definition of hFIX exon 5 that has a weak 5′ donor splice site,15 we observed two mRNA variants (the full-length one and the aberrant one resulting from exon 5 skipping) even in HEK293 clones harboring the SChFIX-wt cassette (Figure 1d, lane 4). Conversely, cell clones harboring the mutated SChFIXex5-2C cassette showed almost exclusively the presence of the aberrant form of the hFIX mRNA (Figure 1d, lane 3). The quantification of the correct splicing of exon 5 was performed by densitometry analysis and was established at ~78% of the total transcript in the SChFIX-wt versus ~3% for the mutated SChFIXex5-2C (Figure 1d). It is important to highlight that the aberrant transcript lacking exon 5 retains the reading frame and encodes a deleted hFIX form that, albeit with reduced efficiency, is secreted by the cells. The results achieved by anti-hFIX ELISA assay confirmed this and showed that hFIX protein levels reflected full-length mRNA levels (35 and 3 ng/ml, for the SChFIX-wt and the SChFIXex5-2C, respectively; Figure 1e). No transcript was detected in the un-transfected cells (Mock) and in the cells transfected with the transposon plasmids in the absence of the transposase enzyme (Figure 1d). These results indicate that the SB100X can be employed to rapidly develop stable in vitro models of splicing mutations.
Figure 1.
Generation of HEK293 cellular models of the hFIXex5-2C splicing mutation. (a) Scheme representing the transposon and transposase-expressing plasmids used. (b) eGFP fluorescence in HEK293 cells transfected with the pT2-eGFP and cotransfected (+) or not (−) with the SB100X. eGFP+ HEK293 cells were sorted as represented in the fluorescence-activated cell sorting (FACS) plots, and analyzed 2 months post-transfection to evaluate eGFP expression by fluorescence microscopy. DAPI staining was used to label cell nuclei. (c) Scheme representing the transposon cassettes integrated in the HEK293 genomic DNA. The position of the probe (black rectangle) and XbaI restriction enzyme used for the Southern blot analysis are depicted. The blotting of the genomic DNAs from HEK293 stable clones expressing the wild-type (lanes 1–4) and the mutated (lanes 5–8) cassettes is shown. Positive control is represented by the linearized pT2-SChFIX transposon plasmid (Ctrl pT2-SChFIX). Negative control is represented by genomic DNA extracted from both eGFP+ HEK293 clones non-expressing hFIX (lanes 3 and 7) and untransfected HEK293 cells (Mock). The position of the molecular weight marker is indicated. Arrows point at the detected bands. (d) RT-PCR and densitometry analyses on the total RNA extracts from SChFX-wt and ex5-2C HEK293 clones (lanes 3 and 4). The RT-PCR products were resolved by electrophoresis on 2% agarose gel. The position of the primers used for the RT-PCR is depicted. The quantification of exon 5 inclusion in the mRNA is expressed as percentage relative to the amount of full-length transcript (that includes exon 5) found in SChFIX-wt HEK293 clones by RT-PCR (dotted line). (e) Quantification of hFIX protein levels in the media of the different HEK293 stable clones performed by anti-hFIX ELISA. Results are expressed as mean ±SD derived from three independent experiments. Negative controls in the RT-PCR and ELISA assays are represented by HEK293 transfected with the transposon plasmids in the absence of the transposase enzyme (lane 2) and by HEK293 cells transfected with the lipofectamine vehicle alone (Mock, lane 1). Student's t-test, *P < 0.05, **P < 0.01. DAPI, 4,6-diamidino-2-2-phenylindole; ELISA, enzyme-linked immunosorbent assay; eGFP, enhanced green fluorescence protein; HEK293, human embryonic kidney cells; SChFIX, splicing-competent human FIX transgene variants; RT-PCR, reverse-transcription followed by polymerase chain reaction.
Plasmid- and AAV vector-mediated delivery of the snRNA U1-fix9 restores correct hFIX splicing in vitro
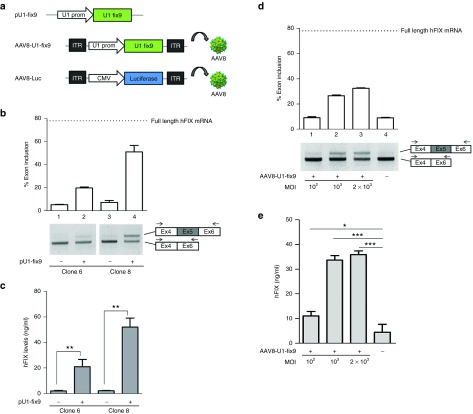
Next, we evaluated the ability of the U1-fix915 to rescue the splicing defect of the mutant hFIXex5-2C expressed from the integrated hFIX minigene. HEK293 cells stably expressing the SChFIXex5-2C cassette were transiently transfected with a plasmid encoding the modified U1-fix9 (pU1-fix9, Figure 2a). This resulted in detection of correctly spliced hFIX transcripts, up to 45% of the wild type, as revealed by RT-PCR and densitometry analysis (Figure 2b). This was paralleled by an increase in hFIX protein levels (3–45 ng/ml) in conditioned media from HEK293 SChFIXex5-2C clones treated with pU1-fix9, with some variability depending on the clone analyzed (Figure 2c). To test the U1-fix9 rescue efficacy in a gene transfer setting, cell clones expressing the SChFIXex5-2C variant were transduced with an AAV8 vector expressing U1-fix9 (AAV8-U1-fix9) under the control of the endogenous promoter of the U1snRNA gene (Figure 2a). The transduction of stable SChFIXex5-2C HEK293 clones was carried out at increasing multiplicity of infection (MOI, from 102 to 2 × 103). The transduction efficiency of HEK293 at the highest MOI (2 × 103) was established by infecting the cells with an AAV vector encoding for eGFP as reporter gene, and found to be ~70% (data not shown). At 24 hours post vector treatment we observed a dose-dependent rescue of the correct mRNA splicing up to ~32% (Figure 2d), reflected also in an increase of protein levels (Figure 2e). No effect on the splicing pattern was observed when SChFIXex5-2C HEK293 clones were transduced with an AAV8 control vector expressing the luciferase transgene (AAV8-Luc, Figure 2a and Supplementary Figure S1). These results validate our in vitro model for the study of rescue of splicing mutants. They also prove that AAV vector-mediated gene delivery of U1-fix9 can rescue FIX splicing mutants in the context of chromosomal DNA.
Figure 2.
Assessment of the U1-fix9 mediated rescue of hFIX splicing mutations in HEK293 stable clones. (a) Schematic representation of the snRNA U1-fix9 expression cassettes used. An AAV8 vector expressing the firefly luciferase under the control of the CMV promoter (AAV8-Luc) was used as control. In panels (b) and (d) RT-PCR and densitometry analyses on the total RNA of SChFIXex5-2C HEK293 clones transfected with the pU1-fix9 plasmid or transduced with the AAV8-U1-fix9. RT-PCR products were resolved by electrophoresis on 2% agarose gel. Negative controls are represented by untransfected or untransduced clones (−). The position of the primers used for the RT-PCR analyses is depicted. The quantification of exon 5 inclusion is performed as described above. Panels (c) and (e) show the quantification of hFIX protein levels in the media of SChFIXex5-2C HEK293 clones. Results are expressed as mean ±SD and derive from three independent experiments. Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.001. AAV, adeno-associated virus; HEK293, human embryonic kidney cells; SChFIX, splicing-competent human FIX transgene variants; SD, standard deviation; snRNA, small nuclear RNA; RT-PCR, reverse-transcription polymerase chain reaction; U1-fix9, ExSpeU1 fix9.
Generation of mouse models of hFIX splicing variants by transposon technology
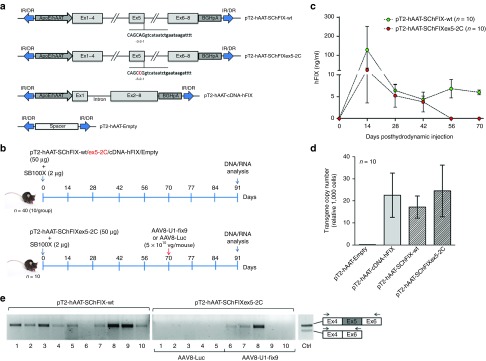
To create mouse models of hFIX splicing variants, we optimized the SChFIX expression cassettes to achieve hepatocyte-specific expression of the wild-type and mutant hFIX transgenes. To this aim, the pT2-SChFIX plasmids described above were modified to include the hepatocyte-specific hAAT promoter and the bovine growth hormone poly-A signal (Figure 3a). A transposon plasmid carrying the hFIX-cDNA without introns and an empty plasmid served as positive and negative controls, respectively (Figure 3a). Each transposon plasmid was hydrodynamically injected into the tail vein of 8-week old C57BL/6 wt mice (50 µg of plasmid DNA/mouse; n = 10 per group) together with the SB100X transposase-expressing plasmid (2 µg of plasmid DNA/mouse) (Figure 3b). To assess the stability of expression of the transgenes from mouse hepatocytes, plasma levels of circulating hFIX in treated mice were measured by enzyme-linked immunosorbent assay (ELISA) assay every 14 days for 3 months (Figure 3c and Supplementary Figure S2a). Positive control mice injected with the plasmid carrying the hFIX-cDNA showed stable protein levels (about 1.5 μg/ml) starting from 1 month posthydrodynamic injection, while no hFIX was detected in plasma mice that received the negative control plasmid (Supplementary Figure S2a). Stable levels of hFIX expression were observed in mice treated with the SChFIX-wt variant (10 ng/ml at 2 months posthydrodynamic injection; Figure 3c), though these levels were lower than those achieved in mice injected with the full-length hFIX-cDNA (Supplementary Figure S2a). The difference in expression levels between the SChFIX-wt and the hFIX-cDNA control likely reflects the low efficiency of splicing of the SChFIX-wt expression cassette.15,23 In this experiment no hFIX expression was detectable 2 months posthydrodynamic injection in mice receiving the mutant SChFIXex5-2C cassette (Figure 3c). Next, we measured the transgene copy number in mouse liver samples, which resulted to range from 17 to 24 copies per 1,000 cells across all treatment groups except for the negative control, lacking the target sequence of the qPCR used in the assay (Figure 3d). Noticeably, we observed a direct correlation between the hFIX expression levels and the transgene copy number in liver, with mice expressing higher levels displaying more than 30 copies/1,000 cells (Supplementary Figure S4). To assess the efficiency of U1-fix9 snRNA at rescuing the mutation in vivo, mice harboring the SChFIXex5-2C construct were split into two subgroups (n = 5/group) 2 months after hydrodynamic injection and treated with either an AAV8-U1-fix9 or a control AAV8-Luc (Figure 2a). Each AAV vector was delivered at a dose of 5 × 1010 vg/mouse via the tail vein. Mice were then monitored for circulating hFIX levels over a total period of 20 days and then killed to evaluate the effect of the U1-fix9 expression on hFIX mRNA splicing pattern in hepatocytes by RT-PCR analysis. As expected, the full-length hFIX mRNA variant was detected in mice injected with the hFIX-cDNA positive control (Supplementary Figure S2b) and in animals that received the SChFIX-wt (Figure 3e). No correctly-spliced hFIX mRNA was detectable in mice injected with the mutated transposon and then treated with the control AAV8-Luc vector (Figure 3e), while three out of five mice treated with the AAV8-U1-fix9 vector showed expression of the correctly spliced form of the hFIX mRNA (Figure 3e). The variability in the efficiency of rescue of splicing observed likely reflects the different levels of liver transduction achieved with hydrodynamic DNA delivery. This in turn drives the quantity of template pre-mRNA available for AAV vector mediated splicing rescue. Despite the correction of the splicing defect, no hFIX was detectable in plasma of treated mice. No humoral response against hFIX was observed in AAV8-U1-fix9 treated animals (data not shown). These results demonstrate that it is feasible to create mouse models of specific human splicing mutations and use them to test rescue strategies. Based on these preliminary results, we then proceeded toward the optimization of our protocol to increase the robustness of our model and to detect the rescue of splicing mutations at both the mRNA and protein levels.
Figure 3.
Generation of mouse models for the human FIXex5-2C splicing mutation and analyses of rescue by systemic delivery of AAV8-U1-fix9. (a) Scheme representing the transposon plasmids used in vivo. Transposon plasmids used as controls harbor the hFIX cDNA (pT2-hAAT-cDNA-hFIX, positive control) or a spacer DNA (pT2-hAAT-Empty). (b) In vivo hepatic gene delivery using the SB100X transposase. Transposons (50 µg/each) were coinjected with SB100X-encoding plasmid (2 µg) by hydrodynamic injection in C57BL/6 mice (n = 10 per group). Blood samples were collected every 14 days posthydrodynamic injection and at day 91 mice were sacrificed for the subsequent DNA and mRNA analyses (upper panel). Mice group hydrodynamically injected with the pT2-hAAT-SChFIXex5-2C was split in two different subgroups (n = 5) and injected via tail vein with AAV8-U1-fix9 or AAV8-Luc (5 × 1010 vg/mouse) at day 70 (lower panel). (c) Circulating hFIX levels measured by anti-hFIX ELISA on mouse plasma samples and analyzed in triplicate. Mean values of hFIX concentration ± SD are shown at each time point. (d) Copy number of the transposon cassettes in C57BL/6J mouse liver DNA determined by qPCR. Values reported are mean ± SEM derived from samples analyzed in triplicate. (e) RT-PCR analyses on total RNA extracted from the liver of mice expressing the SChFIX-wt cassette (pT2-hAAT-SChIX-wt) and the SChFIXex5-2C cassette (pT2-hAAT-SChFIXex5-2C) treated with the AAV8-Luc (lanes 1–5) or with the AAV8-U1-fix9 (lanes 6–10). A positive control is represented by PCR amplification of the pT2-hAAT-SChFIX-wt plasmid (lane Ctrl). The RT-PCR products were resolved by electrophoresis on 2% agarose gel. The position of the primers used for the RT-PCR analyses is depicted. AAV, adeno-associated virus; ELISA, enzyme-linked immunosorbent assay; HEK293, human embryonic kidney cells; qPCR, quantitative PCR; RT-PCR, reverse-transcription polymerase chain reaction; SChFIX, splicing-competent human FIX transgene variants; SB100X, Sleeping Beauty transposon system; SD, standard deviation; SEM, standard error of the mean; snRNA, small nuclear RNA; U1-fix9, ExSpeU1 fix9.
An optimized protocol for the generation of mouse models by transposon technology allows the evaluation of the rescue of hFIX splicing mutations at mRNA and protein levels
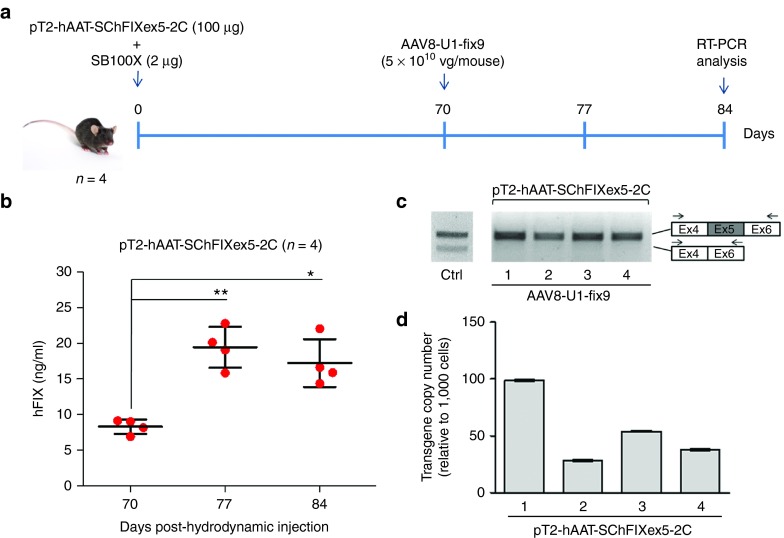
To allow the detection of circulating hFIX protein after rescue of the splicing mutant by AAV8-U1-fix9 gene transfer, we modified the protocol to optimize both transgene delivery and integration. In particular, we doubled the transposon/transposase ratio by increasing the dose of pT2-hAAT-SChFIXex5-2C transposon plasmid (100 µg of plasmid DNA/mouse), while maintaining constant the quantity of the SB100X plasmid (2 µg/mouse) (Figure 4a). We hydrodynamically injected 8-week old C57BL/6 wt mice (n = 4) and monitored the levels of hFIX protein in the plasma every 2 weeks up to day 70 (Supplementary Figure S3 and Figure 4b). Notably, hFIX levels remained stable from day 42 to day 70 allowing us to establish a pretreatment baseline (Figure 4b and Supplementary Figure S3). The residual hFIX protein levels can be explained by the fact that the transcript lacking exon 5 maintains the reading frame and encodes low levels of a deleted form of hFIX protein, as previously confirmed in vitro (Figure 1e and Figure 2c,e). To rescue the splicing mutation in the injected mice, we treated them intravenously with the AAV8-U1-fix9 (5 × 1010 vg/mouse). Plasma samples from treated animals were collected at days 77 and 84 (Figure 4a) to measure circulating hFIX transgene levels. Notably, we detected a significant increase (up to 20 ng/ml, t-test P < 0.05) in hFIX plasma levels in mice treated with AAV8-U1-fix9 vector at both time points (Figure 4b), thus reflecting the rescue of full-length hFIX mRNA expression, as confirmed by RT-PCR (Figure 4c). Moreover, in this optimized experimental setting we measured an increased number of transposon copies in mouse liver (up to 99 copies/1,000 cells, Figure 4d) that may lead to a higher availability of target mRNA template. Thus, by using an improved SB100X-based protocol we established an in vivo model of hFIX splicing mutations sensitive enough to measure rescue mediated by AAV vector-mediated U1-fix9 gene transfer both at mRNA and protein level.
Figure 4.
Optimization of the protocol used generate the mouse model of the human FIXex5-2C splicing mutation and analyses of the rescue by systemic delivery of AAV8-U1-fix9. (a) Schematic representation of the optimized protocol used in C57BL/6 mice: 100 µg of the pT2-hAAT-SChFIXex5-2C transposon plasmid and 2 µg of the SB100X-expressing plasmid were delivered by hydrodynamic injection. Mice were followed for 70 days and then injected via tail vein with the AAV8-U1-fix9 (5 × 1010 vg/mouse). Blood samples were collected every 2 weeks posthydrodynamic injection up to 84 days. (b) Circulating hFIX levels measured by anti-hFIX ELISA on plasma samples and analyzed in triplicate. Mean values of hFIX concentration ± SD are shown at each time point. t-test, *P-value < 0.05, **P-value < 0.01. (c) RT-PCR analyses of hFIX mRNA on mouse liver samples. A positive control is represented by PCR amplification of the pT2-hAAT-SChFIX-wt plasmid (lane Ctrl). RT-PCR products were separated by electrophoresis on 2% agarose gel. The position of the primers used for the RT-PCR analyses is depicted. (d) Copy number of the transposon cassettes in C57BL/6J mouse liver DNA determined by qPCR. Values reported are mean ± SD derived from samples analyzed in triplicate. AAV, adeno-associated virus; ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative PCR; RT-PCR, reverse transcription polymerase chain reaction; SChFIX, splicing-competent human FIX transgene variants; SB100X, Sleeping Beauty transposon system; SD, standard deviation; U1-fix9, ExSpeU1 fix9.
Discussion
In this study, we successfully exploited the SB100X system to generate models harboring the single point mutation hFIXex5-2C affecting the definition and inclusion of hFIX exon 5 in the mature mRNA. Notably, this allowed us to subsequently test a therapeutic strategy based on the modified U1-fix9. We initially developed an in vitro model of the hFIXex5-2C splicing mutation using HEK293, taking advantage of the fact that these cells lack endogenous hFIX gene expression, as revealed by both RT-PCR and ELISA analyses. The use of the SB100X system has allowed to rapidly establish HEK293 clones by finely tuning the ratio transposon/transposase and to avoid unwanted effects like: (i) overproduction inhibition, i.e., elevated concentration of transposase enzyme inhibits the transposition reaction12,24 and (ii) potential cytotoxicity due to residual integration events of transposase expression vectors that may lead to permanent transposase expression and uncontrolled transposition.25 Other optimization of the experimental settings in vitro consisted in the use of two separate transposon plasmids (one expressing eGFP and the other the desired hFIX variant) instead of a single transposon carrying the two cassettes in cis, thus allowing to maximize the rate of transposition. Indeed, previous studies indicated that transposition efficiency decreases at approximately a logarithmic rate as a function of length and that transposon around 6 kb retain only 50% of the maximal efficiency.24 Hence, we employed separate plasmids in order to work with transposons < 5 kb in length. Using this experimental set-up the transposition occurred efficiently in HEK293 cells and was stably maintained thereafter, meaning that, after a transient episomal phase, the transgene is integrated in the genome, as clearly demonstrated by Southern blot analysis. Importantly, analysis at mRNA level demonstrated that HEK293 stable clones expressing the hFIX mutant cassette (SChFIXex5-2C) reproduce the aberrant splicing pattern characterized by exon 5 skipping, as previously reported using episomal minigene assays.15 This is crucial to quantify the inclusion of hFIX exon 5 in the mature mRNA and the rescue of hFIX protein expression induced by the snRNA U1-fix9 delivered to HEK293 clones by either transfection or AAV-mediated gene transfer. In summary, we demonstrated the feasibility of using the SB100X-technology to generate cellular models of disease-causing splicing mutation that would be useful to both study the biological effect of these mutations in a defined system and evaluate the efficacy of therapeutic agents. Here, we also report the optimization of the SB100X technology to generate mouse models expressing hFIX splicing variants specifically in hepatocytes. Interestingly, upon transposon and transposase hydrodynamic injection in mice we observed a spike in hFIX transgene expression at day 14 postinjection, followed by a decrease and a stabilization of the levels measured from day 28 (Figure 3c, Supplementary Figures S2a and S3). It is plausible to speculate that this is due to the presence of episomal plasmid maintained for a few weeks after the hydrodynamic injection, which is probably completely lost 1 month postinjection, as previously reported in other studies using mutant or inactive transposases.17 Mice expressing the hFIX-cDNA showed very high expression in the supraphysiologic range at day 14 (~18 µg/ml), which remained high and stable at ~1.5 µg/ml starting from day 28 and until day 90, supporting productive transposition and consequent long-lasting expression of the transgene. In our study it was possible to observe a good correlation between the gene copy number in mouse hepatocytes and hFIX expression. Indeed, mice showing the highest circulating hFIX levels displayed also the highest transposon copy number (> 30 copies/1,000 cells), while mice showing low hFIX levels displayed lower transposon integration rate (1–10 copies/1,000 cells). Overall, the mean copy number among the groups was consistent and quantified around 20 copies/1,000 cells, meaning that on average the integration rate in this experimental conditions is ~2% of the entire liver. An optimized protocol conducted by exploiting a 50:1 transposon/transposase ratio permitted us to increase the integration rate at on an average 5–6% of the liver and to better evaluate the AAV8-U1-fix9 mediated rescue, since it allowed us to compare hFIX protein levels in mouse plasma before and after the treatment. As reported from previous studies SB100X integration can be considered fairly random into chromosomes,17,26 with roughly 35% of the transposon insertions occurring in transcribed regions,27,28 thus supporting in our setting a reduced probability of harmful effects due to insertional mutagenesis. Interestingly, at a similar integration rate among the injected groups no correlation to hFIX levels was observed, possibly depending on the nature of the hFIX transposon variant integrated. Mice injected with the pT2-hAAT-SChFIX-wt exhibited far lower hFIX levels than mice injected with the pT2-hAAT-cDNA-hFIX optimized cassette.29,30,31 The difference in hFIX transgene expression is due to the fact that while both cassettes produce the wild-type hFIX transcript, the splicing-competent cassette harbors exon 5 that is partially skipped. This is consistent with the analysis of the splicing pattern of SChFIX-wt in vitro, showing two forms of spliced hFIX mRNA. In vivo, in liver samples from SChFIX-wt injected animals, the aberrant form of hFIX mRNA was not detectable by RT-PCR analysis, which might be due to the intrinsic instability of the aberrant form. Consistently, animals expressing SChFIXex5-2C did not show the presence of any hFIX transcript, while mice injected with the AAV8-U1-fix9 showed a rescued full-length hFIX mRNA and protein expression. The remarkably low variability in the hFIX transgene expression levels in vivo after U1-fix9 splicing rescue likely reflects the random pattern of transposon integration in the host genome.13,27 Additionally, the optimization of the hydrodynamic delivery protocol also helped reducing the variability of expression levels within treatment groups. Therefore, even if in this experimental setting the rescue obtained is not informative in therapeutic terms, the proposed approach has permitted to establish the efficacy of the U1-fix9 delivered by an AAV vector from a qualitative point of view. Importantly, here we report for the first time for newly created in vitro and in vivo models of splicing mutations the efficacy of the snRNA U1-based therapeutic approach in a genomic context, a condition that is interlaced with the splicing process.19,28,29 As compared with viral systems (both integrating and nonintegrating),32 the SB100X permits to by-pass several drawbacks such as costs and time-consuming manufacturing procedures and at the same time to easily assess the integration efficiency in different experimental settings by evaluating the transgene copy number. On the other hand, as compared with plasmid-based episomal systems, the SB100X provides stable gene expression allowing to perform long-term studies and repeated tests (e.g., those required for drug development).33 An alternative to our approach is the use of genome editing tools for DNA manipulation.34 These have the advantage of introducing a desired mutation in a very specific position of the genome but they also rely on homologous recombination mechanisms to insert a donor DNA template,34 which is not easy to achieve especially in nondividing cells.35,36 Therefore, the transposon represents a good alternative to stably express a specific transgene when no precise genomic modification is required. Our data on modelling of a F9 splicing mutation and its rescue via U1snRNA gene therapy support the SB100X system as a novel, simple and time-saving approach to generate cellular and animal models of human diseases. This system allows for the investigation of the molecular mechanisms (e.g., dominant mutations37) or for the assessment of tailored therapeutic strategies. This technology is also helpful to generate models of numerous disorders characterized by highly heterogeneous mutational patterns, such as HB, for which the development of personalized therapies (i.e., splicing-switching molecules37; readthrough-inducing drugs for nonsense mutations38; chemical/pharmacological chaperones for missense mutations39; etc.) is actively investigated.
Materials and methods
Plasmids, vector backbone constructs, and AAV vectors. All constructs used for the creation of the cellular models were cloned into the pT2-SVNeo plasmid (Addgene #26553, Cambridge, MA). The SB100X expressing plasmid was obtained from Addgene (Addgene #34879, Cambridge, MA). The pT2-SChFIX-wt and pT2-SChFIXex5-2C were obtained by PCR amplification of the splicing-competent hFIX cassette from the plasmid pBSK-FIX IVS4-5 and pBSK-FIX IVS5-2C 15, respectively. The pT2-eGFP plasmid was obtained by PCR amplification of the eGFP coding-sequence which was subsequently cloned into the pT2-SVNeo. The pT2-hAAT-SChFIX-wt/ex5-2C were obtained by blunt-end cloning of the SC-hFIX cassettes from the previously described pT2-variants between the SB100X IR/DR and under the control of the human apolipoprotein E (ApoE) control region followed by the liver specific α1-antitrypsin promoter (hAAT) and the bovine growth hormone polyadenylation signal. The pT2-hAAT-Empty vector was created by removing the SChFIX cassette from the pT2-hAAT-SChFIX-wt. The pT2-hAAT-cDNA-hFIX control was generated by cloning of the hFIX-cDNA cassette from the pAAV-hFIX plasmid23 into the IR/DR of the pT2-SVNeo. The AAV8-U1-fix9 was created by cloning the U1 expression-cassette from a previous pU1-fix915 into a pSMD2-ApoE-hAAT40 backbone containing the AAV2 ITRs sequences for the subsequent vector preparations, made a previously described.41 The AAV vector titration was performed by qPCR performed in ABI PRISM 7900 HT Sequence Detector using Absolute ROX mix (Taqman, Thermo Fisher Scientific, Waltham, MA).
Cell culture and generation of HEK293 stable clones. The human embryonic kidney HEK293 cell lines were maintained under 37°C, 5% CO2 condition in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mmol/l GlutaMAX (Thermo Fisher Scientific, Waltham, MA). HEK293 cells were seeded into T75 flasks 3 days before transfection (2 × 106 cells/flask). Cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) with pT2-trasposon plasmids and the SB100X-expressing plasmid at a molar ratio of 5:1. Eleven days post-transfection, eGFP+ cells were sorted on a BD Facs Aria II sorter (BD Biosciences, San Jose, CA) and expanded in a T25 flask. Cells were then plated at a density of 10 cells/ml on 148 cm2 petri dishes for the isolation of stable clones. Several eGFP+ clones were picked for each construct by using cloning cylinders (Sigma-Aldrich, Saint-Quentin Fallavier, France) in which 0.2 ml of 5% trypsin-phosphate buffered saline (PBS) solution was added. Single colonies were transferred to a 24-well and expanded for subsequent analysis.
Evaluation of the hFIX expression. The hFIX expression in HEK293 eGFP+ clones has been evaluated at the RNA and secreted protein level. The total RNA was extracted from cells at confluency from a six-well plate (around 1.2 × 106 cells) with TRIzol Reagent (Thermo Fisher Scientific) and treated with DNaseI (Ambion, Thermo Fisher Scientific). The cDNA was generated by RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer's recommended protocol. hFIX transcript was detected by PCR with primer hFIX Ex4F (5′-ATTCCTATGAATGTTGGTGTCCCT-3′) and hFIX Ex6R (5′-GGGTGCTTTGAGTGATGTTATCCAA-3′). The conditions used for the PCR were 94°C for 5 minutes for the initial denaturation, 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 30 seconds for 30 cycles and 72°C for 10 minutes for the final extension. The evaluation of the protein levels was performed by a Factor IX Antigen Kit (Affinity Biologicals, Ancaster, ON, Canada) on the media samples according to the manufacturer's recommended protocol.
Evaluation of the gene copy number in HEK293 stable clones by Southern blot and qPCR analysis. The integration of the hFIX transgene in HEK293 stable clones was examined by Southern blot analysis. The genomic DNA was extracted from the cells by Gentra Puregene Cell Kit (Qiagen, Valencia, CA), and digested overnight with XbaI by restriction enzyme (Thermo Fisher Scientific). After electrophoresis on agarose gel 0.7% at 35V 4–5 hours the digested DNA was transferred to a Nylon membrane through iBlot Dry blotting System (Thermo Fisher Scientific). The membrane was subsequently denatured in a 1.5 mol/l NaCl/0.5 mol/l NaOH solution. The probe was made by amplification of a 390 bp sequence of the hFIX transgene with AccuPrime Taq polymerase (Thermo Fisher Scientific) and with primer Probe F (5′-TGCAGCGCGTGAACATGATC-3′) and R (5′-CTAATTCACAGTTCTTTCCTTCAA-3′). The denatured DNA product was labeled with alkaline phosphatase trough Amersham AlkPhos Direct Labeling and Detection Systems (GE Healthcare, Velizy-Villacoublay, France) according to the manufacturer's instructions. The hybridization with the membrane was carried out overnight at 37°C. The day after membrane was washed and hybridized blots were detected by adding Enhanced Chemifluorescence ECF substrate and by scanning at Storm Imager 840 (Molecular Dynamics, GMI, Ramsey, MN) in chemifluorescence mode, 100 microns, 650 V. The copy number of the hFIX transgenes was established by qPCR with SYBR Green Dye (Bio-Rad, Marnes-la-Coquette, France), with primers hFIX Ex1F (5′- CCTCATCACCATCTGCCTTT-3′) and hFIX Ex2R (5′-ATACCTCTTTGGCCGA TTCA-3′); ALB F (5′- GCTGTCATCTCTTGTGGGCTGT -3′) and ALB R (5′- ACTCATGGGAGCTGCTGGTTC -3′) to amplify the albumin (ALB) gene. The transposon copy number per cellular genome was established by plotting the CT values obtained on a standard curve (from 1.0 to 3 × 105 copies of the transposon plasmid) and normalized to the number of genomic ALB copies.
In vitro rescue of splicing mutants by transfection and transduction. HEK293 cell clones were seeded into 6-well plates the day before transfection (0.3 × 106 cells/well) and then transfected with 2 µg of the pU1-fix9. The evaluation of the hFIX mRNA and protein levels was performed 48 hours after transfection, as described above. For the transduction HEK293 cell clones seeded into 12-well plates the day before (0.2 × 106 cells/well) were infected with AAV8-U1-fix9 or AAV8-Luc at different MOI (102, 103, or 2 × 103). Cells in each well were incubated in water jacketed incubators at 37°C, 5% CO2 overnight with the addition of 500 µl of DMEM with 2% FBS. The day after the total RNA was extracted from the cells and hFIX mRNA and protein levels assessed as described above.
Animal procedures. Animal protocols were approved by Genethon's Ethical Committee and conducted by the certified operators according to the guidelines and laws regulating animal experimentation under agreement number CE12-037. Eight-weeks-old mice C57BL/6 wt mice were used for in vivo delivery of plasmids by hydrodynamic injection of DNA.42 Blood samples were collected from the retro-orbital plexus in heparin coated capillary tubes (Sarstedt, Nümbrecht, Germany).
Vector genome copy number, hFIX antigen, and transgene mRNA levels. Human FIX antigen and protein levels in mouse plasma were evaluated by Factor IX Antigen Kit (Affinity Biologicals, Ancaster, ON, Canada) as previously described.43 To extract nucleic acids from animal organs, entire livers were homogenized by using T10 Ultra Turrax Homogenizer (Hielscher-Ultrasound Technologies, Ringwood, NJ) in PBS-solution, while the muscles were homogenized in PBS-solution by using MP FastPrep-24 Tissue and Cell Homogenizer (MP Biomedicals, Santa Ana, CA), for 60 seconds at 6.5 m/s. The genomic DNA was extracted from homogenate samples by the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Life Science, Meylan Cedex, France). The copy number of the hFIX transgenes was established by qPCR with SYBR Green Dye (Bio-Rad, Marnes-la-Coquette, France), with primers hAAT F (5′-GGCGGGCGACTCAGATC-3′) and hAAT R (5′-GGGAGGCTGCTGGTGAATATT-3′); and Ttn F (5′-AAAACGAGCGGTGACATGAGC-3′) and Ttn R (5′-TTCAGTCATGCTAGCGCTCC-3′) to amplify the mouse titin (TTN) gene. The transgene copy number, relative to 1,000 cells, was established by plotting the CT values obtained on a standard curve (from 5.0 to 5 × 106 copies) and normalized to the TTN control gene.
SUPPLEMENTARY MATERIAL Figure S1. Treatment of HEK293 SChFIXex5-2C clones with the AAV8-Luc negative control at increasing MOI. Figure S2. Quantification of hFIX levels in mice hydrodynamically injected with pT2-hAAT-cDNA-hFIX or the pT2-hAAT-Empty cassettes and analyses of hFIX mRNA by RT-PCR on total RNA extracts from the liver homogenates of mice expressing hFIX-cDNA. Figure S3. Quantification of the hFIX levels in mice hydrodynamically injected with the pT2-hAAT-SChFIXex5-2C. Figure S4. Correlation between transposon copy number and transgene expression in the mouse liver. Figure S5. Copy number of the transposon cassettes in HEK293 cell clones determined by qPCR.
Author contributions
E.B., P.C., F.M., and M.P. designed the experiments, analyzed results, and wrote the manuscript. E.B. performed the experiments. F.C. produced the AAV vectors. M.F., G.R., P.S., P.C. and L.v.W. contributed to experimental activities.
Acknowledgments
This work was supported by Genethon and by the European Union Marie Sodowska Curie C.I.G. grant no. 33628 to F.M, by Telethon Italy (GGP14190) to M.P., and by the Young Research Fellowship 2014, University of Ferrara, to E.B. We thank Perry Hackett (University of Minnesota, MN) and Zsuzsanna Izsvak (MDC, Berlin, Germany) for providing us with the pT2/SVNeo and the pCMV(CAT)T7-SB100 original plasmids. We thank also Franco Pagani (ICEGB, Trieste, Italy) and Dario Balestra (University of Ferrara, Italy) for providing us with the pBSK-FIX IVS 4-5 and the U1-fix9 expression cassette. M.P. is founder of the start-up company Raresplice. F.M. is inventor in patents and patent applications describing the AAV technology. All the other authors declare no conflict of interest associated with this work.
Supplementary Material
References
- Garcia-Blanco, MA, Baraniak, AP and Lasda, EL (2004). Alternative splicing in disease and therapy. Nat Biotechnol 22: 535–546. [DOI] [PubMed] [Google Scholar]
- Havens, MA, Duelli, DM and Hastings, ML (2013). Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA 4: 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, F, Forné, T, Antoine, E, Tazi, J, Brunel, C and Cathala, G (1996). Involvement of U1 small nuclear ribonucleoproteins (snRNP) in 5′ splice site-U1 snRNP interaction. J Biol Chem 271: 23985–23991. [DOI] [PubMed] [Google Scholar]
- Will, CL, Rümpler, S, Klein Gunnewiek, J, van Venrooij, WJ and Lührmann, R (1996). In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res 24: 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino, NA and Cooper, TA (2003). Pre-mRNA splicing and human disease. Genes Dev 17: 419–437. [DOI] [PubMed] [Google Scholar]
- Buratti E, Chivers M, Královičová J, et al. (2007). Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res 35: 4250–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinotti, M, Balestra, D, Rizzotto, L, Maestri, I, Pagani, F and Bernardi, F (2009). Rescue of coagulation factor VII function by the U1 + 5A snRNA. Blood 113: 6461–6464. [DOI] [PubMed] [Google Scholar]
- Pinotti, M, Bernardi, F, Dal Mas, A and Pagani, F (2011). RNA-based therapeutic approaches for coagulation factor deficiencies. J Thromb Haemost 9: 2143–2152. [DOI] [PubMed] [Google Scholar]
- Balestra, D, Faella, A, Margaritis, P, Cavallari, N, Pagani, F, Bernardi, F et al. (2014). An engineered U1 small nuclear RNA rescues splicing defective coagulation F7 gene expression in mice. J Thromb Haemost 12: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajnik, M, Rogalska, ME, Bussani, E, Barbon, E, Balestra, D, Pinotti, M et al. (2016). Molecular basis and therapeutic strategies to rescue Factor IX variants that affect splicing and protein function. PLoS Genet 12: e1006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra, D, Barbon, E, Scalet, D, Cavallari, N, Perrone, D, Zanibellato, S et al. (2015). Regulation of a strong F9 cryptic 5′ss by intrinsic elements and by combination of tailored U1snRNAs with antisense oligonucleotides. Hum Mol Genet 24: 4809–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed, H, Izsvák, Z, Walisko, O and Ivics, Z (2004). Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther 9: 292–304. [DOI] [PubMed] [Google Scholar]
- Hackett, PB, Ekker, SC, Largaespada, DA and McIvor, RS (2005). Sleeping beauty transposon-mediated gene therapy for prolonged expression. Adv Genet 54: 189–232. [DOI] [PubMed] [Google Scholar]
- Factor IX Variant Database (http://www.factorix.org).
- Fernandez Alanis, E, Pinotti, M, Dal Mas, A, Balestra, D, Cavallari, N, Rogalska, ME et al. (2012). An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Hum Mol Genet 21: 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalska, ME, Tajnik, M, Licastro, D, Bussani, E, Camparini, L, Mattioli, C et al. (2016). Therapeutic activity of modified U1 core spliceosomal particles. Nat Commun 7: 11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés, L, Chuah, MK, Belay, E, Jerchow, B, Manoj, N, Acosta-Sanchez, A et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41: 753–761. [DOI] [PubMed] [Google Scholar]
- Herweijer, H, Zhang, G, Subbotin, VM, Budker, V, Williams, P and Wolff, JA (2001). Time course of gene expression after plasmid DNA gene transfer to the liver. J Gene Med 3: 280–291. [DOI] [PubMed] [Google Scholar]
- Schwartz, S and Ast, G (2010). Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J 29: 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S, Meshorer, E and Ast, G (2009). Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995. [DOI] [PubMed] [Google Scholar]
- Ivics, Z, Hackett, PB, Plasterk, RH and Izsvák, Z (1997). Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510. [DOI] [PubMed] [Google Scholar]
- Cui, Z, Geurts, AM, Liu, G, Kaufman, CD and Hackett, PB (2002). Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol 318: 1221–1235. [DOI] [PubMed] [Google Scholar]
- Manno, CS, Pierce, GF, Arruda, VR, Glader, B, Ragni, M, Rasko, JJ et al. (2006). Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 12: 342–347. [DOI] [PubMed] [Google Scholar]
- Geurts, AM, Yang, Y, Clark, KJ, Liu, G, Cui, Z, Dupuy, AJ et al. (2003). Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 8: 108–117. [DOI] [PubMed] [Google Scholar]
- Galla, M, Schambach, A, Falk, CS, Maetzig, T, Kuehle, J, Lange, K et al. (2011). Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res 39: 7147–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák, Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z et al. (2002).. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem 277: 34581–34588. [DOI] [PubMed] [Google Scholar]
- Izsvák, Z and Ivics, Z (2004). Sleeping beauty transposition: biology and applications for molecular therapy. Mol Ther 9: 147–156. [DOI] [PubMed] [Google Scholar]
- Liu, G, Geurts, AM, Yae, K, Srinivasan, AR, Fahrenkrug, SC, Largaespada, DA et al. (2005). Target-site preferences of Sleeping Beauty transposons. J Mol Biol 346: 161–173. [DOI] [PubMed] [Google Scholar]
- Miao, CH, Ohashi, K, Patijn, GA, Meuse, L, Ye, X, Thompson, AR et al. (2000). Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol Ther 1: 522–532. [DOI] [PubMed] [Google Scholar]
- Brinster, RL, Allen, JM, Behringer, RR, Gelinas, RE and Palmiter, RD (1988). Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA 85: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, RD, Sandgren, EP, Avarbock, MR, Allen, DD and Brinster, RL (1991). Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA 88: 478–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, MA, Glorioso, JC and Naldini, L (2001). Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med 7: 33–40. [DOI] [PubMed] [Google Scholar]
- Bajaj, S, Sakhuja, N, Singla, D, Bajaj Principal S (2012). Stability testing of pharmaceutical products. J Appl Pharm Sci 2: 129–138. [Google Scholar]
- Maeder, ML and Gersbach, CA (2016). Genome-editing technologies for gene and cell therapy. Mol Ther 24: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, VT, Weber, T, Wefers, B, Wurst, W, Sander, S, Rajewsky, K et al. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Jasin, M and Rothstein, R (2013). Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman, J, Jearawiriyapaisarn, N and Kole, R (2009). Therapeutic potential of splice-switching oligonucleotides. Oligonucleotides 19: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel-Wolfrum K, Möller F, Penner I, Baasov T, Wolfrum U (2016). Targeting nonsense mutations in diseases with translational read-through-inducing drugs (TRIDs). BioDrugs 30: 49–74. [DOI] [PubMed] [Google Scholar]
- Cortez L and Sim V (2014). The therapeutic potential of chemical chaperones in protein folding diseases. Prion 8: 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, RO, Spratt, SK, Lagarde, C, Bohl, D, Kaspar, B, Sloan, B et al. (1997). Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther 8: 1891–1900. [DOI] [PubMed] [Google Scholar]
- Ayuso, E, Mingozzi, F, Montane, J, Leon, X, Anguela, XM, Haurigot, V et al. (2010). High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther 17: 503–510. [DOI] [PubMed] [Google Scholar]
- Suda, T and Liu, D (2007). Hydrodynamic gene delivery: its principles and applications. Mol Ther 15: 2063–2069. [DOI] [PubMed] [Google Scholar]
- Mingozzi, F, Anguela, XM, Pavani, G, Chen, Y, Davidson, RJ, Hui, DJ et al. (2013). Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med 5: 194ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.