MPK4 interacts with and phosphorylates MYB75 to increase MYB75 stability and anthocyanin biosynthesis.
Abstract
Light is a major environmental cue affecting various physiological and metabolic processes in plants. Although plant photoreceptors are well characterized, the mechanisms by which light regulates downstream responses are less clear. In Arabidopsis thaliana, the accumulation of photoprotective anthocyanin pigments is light dependent, and the R2R3 MYB transcription factor MYB75/PAP1 regulates anthocyanin accumulation. Here, we report that MYB75 interacts with and is phosphorylated by MAP KINASE4 (MPK4). Their interaction is dependent on MPK4 kinase activity and is required for full function of MYB75. MPK4 can be activated in response to light and is involved in the light-induced accumulation of anthocyanins. We show that MPK4 phosphorylation of MYB75 increases its stability and is essential for light-induced anthocyanin accumulation. Our findings reveal an important role for a MAPK pathway in light signal transduction.
INTRODUCTION
Higher plants absorb light and convert it through photosynthesis into chemical energy required for virtually all physiological processes (Smith, 1982; Yi and Deng, 2005). However, excessive light absorption leads to photoinhibition (repression of photosynthesis) and accumulation of reactive oxygen species, which perturb cellular metabolism and reduce yield (Long et al., 1994; Foyer et al., 1994; Steyn et al., 2002). Plants have multiple mechanisms to sense light and adjust energy capture to avoid radiation damage (Yi and Deng, 2005). One such mechanism produces nonphotosynthetic pigments in vegetative tissues that function as light screens. Anthocyanins are among the best studied such pigments (Grotewold, 2006). Their accumulation confers deep red to purple colors to plant tissues, and they greatly reduce the irradiation of chloroplasts by light (Neill and Gould, 2000; Albert et al., 2009). In addition, anthocyanins are likely to scavenge reactive oxygen species produced under light stress (Gould, 2004).
The biosynthesis of anthocyanins, a subgroup of plant flavonoids, can be divided into three stages: (1) the initial, general phenylpropanoid reactions involving phenylalanine ammonia-lyase, cinnamate-4-hydroxylase, and 4-coumaroyl CoA:ligase; (2) the early flavonoid reactions catalyzed by chalcone synthase (CHS), chalcone isomerase, flavanone 3-hydroxylase (F3H), and flavonoid 3′-hydroxylase; and (3) the late anthocyanin-specific reactions involving dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucose:flavonoid 3-O-glycosyl-transferase (UF3GT) (Holton and Cornish, 1995; Shi and Xie, 2014). Expression of the structural genes of the anthocyanin biosynthetic pathway is controlled by WD40, MYB, and basic helix-loop-helix (bHLH) transcription factors (TFs) (Gonzalez et al., 2008; Shi and Xie, 2014). In Arabidopsis thaliana, the ternary WD40-bHLH-MYB complex, composed of R2R3 MYB TFs (MYB75, MYB90, MYB113, and MYB114), bHLH TFs (TT8, GL3, and EGL3), and the WD40 protein TTG1, regulates anthocyanin biosynthesis by enhancing expression of the late anthocyanin biosynthetic genes DFR, LDOX, and UF3GT (Dooner et al., 1991; Gonzalez et al., 2008; Shi and Xie, 2014). Overexpressing the MYB TFs increases anthocyanin levels in Arabidopsis (Borevitz et al., 2000; Gonzalez et al., 2008). Therefore, it seems that these MYB TFs are limiting factors regulating anthocyanin biosynthesis (Maier et al., 2013). In Arabidopsis seedlings, the expression of MYB75 predominates over the other three MYBs, indicating that MYB75, also known as PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), is a key TF in activating anthocyanin biosynthetic genes (Gonzalez et al., 2008). The identity of MYB75/PAP1 was confirmed by comparison of natural allelic variation and by insertion mutant analyses (Teng et al., 2005).
In Arabidopsis, anthocyanins only accumulate in the light (Maier et al., 2013). In agreement with its function in regulating anthocyanin biosynthesis, MYB75/PAP1 is regulated by light at both the transcriptional and posttranslational levels. Consistent with its key role in light-induced anthocyanin biosynthesis, MYB75/PAP1 gene expression is induced by light and precedes the expression of MYB90/PAP2 and the structural genes (CHS, DFR, F3H, and LDOX) (Cominelli et al., 2008). However, overexpression of MYB75/PAP1 is insufficient on its own to stimulate anthocyanin accumulation in the dark (Maier et al., 2013; Li et al., 2014). In the dark, MYB75/PAP1 is targeted for degradation by the CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYTOCHROME A (COP1/SPA) E3 ubiquitin ligase. When plants are exposed to light, COP1/SPA is inactivated and MYB75/PAP1 is stabilized and able to enhance anthocyanin biosynthesis (Maier et al., 2013). Accordingly, anthocyanins accumulate even in the dark in cop1 and spa mutants (Deng et al., 1991; Hoecker et al., 1998; Laubinger et al., 2004). However, light can still enhance anthocyanin accumulation in cop1 mutants because illuminated cop1 and spa mutants accumulate much higher levels of anthocyanins than the wild type. These E3 ligase mutants are therefore hyperresponsive to light (Maier and Hoecker, 2015). This indicates that there is an additional regulatory process involved in light-induced anthocyanin accumulation.
Plants use light not only as their energy source, but also as a cue to adapt to changing environments (Sheerin et al., 2015). Such cues are perceived by receptors/sensors and transduced to physiological responses via intracellular signaling pathways (Kholodenko, 2006). Mitogen-activated protein kinase (MAPK) cascades represent a major set of such pathways. A canonical MAPK cascade consists of three sequentially phosphorylated protein kinases, a MAPK kinase kinase (MAP3K), a MAPK kinase (MAP2K), and a MAPK (Widmann et al., 1999). Activated MAPKs then phosphorylate effector/substrate proteins to induce appropriate responses. Their substrates are diverse and include TFs (Mao et al., 2011). Plant MAPKs regulate numerous processes, including abiotic and biotic stress responses, hormonal signaling, and developmental programs (Rodriguez et al., 2010; Meng and Zhang, 2013). The Arabidopsis genome encodes 20 MAPKs including MPK3, MPK6, and MPK4 with functions in responses to various extracellular stimuli (Colcombet and Hirt, 2008). Interestingly, these three MAPKs are activated by light (Lee, 2015). However, the roles of MAPKs in light responses and the mechanisms by which MAPKs may regulate light signaling remain largely unknown.
Here, we report that MPK4 regulates the light-induced accumulation of anthocyanins. Light can activate MPK4, and activated MPK4 interacts with the transcription factor MYB75. Phosphorylation of MYB75 by MPK4 increases MYB75 stability, which is essential for anthocyanin accumulation in response to light. Our work thus reveals an important role of a MAPK in light signaling.
RESULTS
MPK4 Interacts with the MYB Transcription Factor MYB75
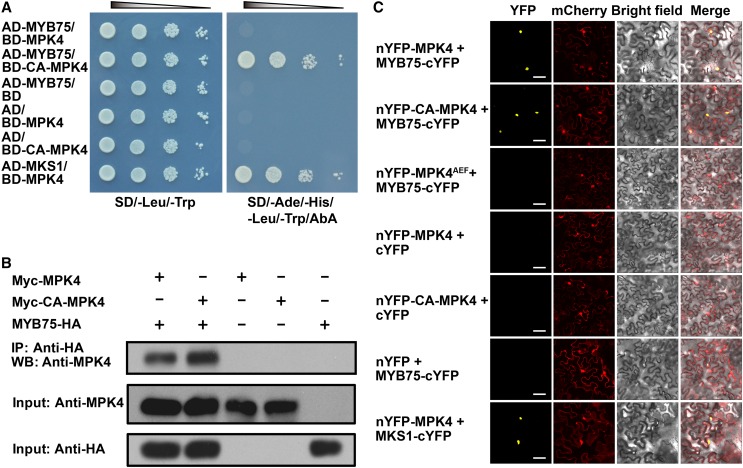
Arabidopsis MPK4 regulates cellular responses including innate immunity, hormone signaling, and cytokinesis (Petersen et al., 2000; Kosetsu et al., 2010). We aimed to identify MPK4 substrate proteins to link MPK4 to downstream functions. For example, we previously identified MAP KINASE SUBSTRATE1 (MKS1) as an MPK4 substrate by yeast two-hybrid (Y2H) screens using native MPK4 as bait (Andreasson et al., 2005). However, phosphorylation-mediated activation of MAPKs is generally transient, as is their interaction with downstream substrate proteins. This makes it difficult to identify MAPK substrates by genome wide proteomic or yeast two-hybrid approaches (Feilner et al., 2005). To circumvent this difficulty, we exploited a recently described, constitutively active MPK4 form (CA-MPK4), which results from two mutations (D198G/E202A) and retains its substrate specificity and physiological functions (Berriri et al., 2012). In control Y2H assays, when a BD-CA-MPK4 bait construct in plasmid pGBKT7 was cotransformed with the empty pGADT7 prey vector, expression of selectable markers was not activated (Figure 1A). We then screened an Arabidopsis cDNA library to isolate potential MPK4 substrates. A total of 1.5 × 107 clones were screened representing 4.6-fold redundancy of the Clontech library used. Among 157 positive clones isolated, two encoded full-length MYB75 (At1g56650), a MYB family transcription factor. Directed yeast two-hybrid assays confirmed the interaction between CA-MPK4 and MYB75. Interestingly, the native form of MPK4 did not interact detectably with MYB75 (Figure 1A).
Figure 1.
Interaction of MYB75 with MPK4 Is Dependent on MPK4 Kinase Activity.
(A) Constitutively active MPK4 (CA-MPK4), but not native MPK4, interacts with MYB75 in yeast. Serial dilutions of transformed yeast cells were spotted on the indicated amino acid dropout agar plates. Cotransformed MPK4 and MKS1 were used as a positive control. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. SD, synthetically defied medium; AbA, Aureobasidin A.
(B) Coimmunoprecipitation of MPK4 and CA-MPK4 with MYB75. MYB75-HA was coexpressed with Myc-MPK4 and Myc-CA-MPK4 in N. benthamiana leaves by agroinfiltration. Leaf lysates were immunoprecipitated with anti-HA beads and stained with anti-MPK4. One-twentieth of the input proteins was stained with the same antibodies as loading control. “+” and “−” denote presence and absence of the protein in each sample, respectively.
(C) A BiFC assay in N. benthamiana leaves showing that MYB75 interacts with MPK4 and CA-MPK4, but not with kinase-dead MPK4AEF. mCherry was coexpressed as a transformation control. Yellow indicates a positive interaction signal, and red indicates signals from mCherry. Bars = 50 μm.
To test whether MYB75 interacts with MPK4 in vivo, we performed coimmunoprecipitation experiments in Nicotiana benthamiana. Immunoprecipitates of transiently expressed MYB75-HA in leaves transformed with MYB75-HA and Myc-CA-MPK4 or Myc-MPK4 were found to contain MPK4 (Figure 1B). Native MPK4 was found to be associated with MYB75, most probably due to the basal activity of MPK4 in planta. The MYB75 and MPK4 interaction was further confirmed by bimolecular fluorescence complementation (BiFC) assays on N. benthamiana leaves (Figure 1C). A direct interaction between CA-MPK4 or MPK4 and MYB75 was observed in nuclei. However, no interaction was found between MYB75 and MPK4AEF, a kinase-dead form with two mutations (T201A/Y203F) in its activation loop (Petersen et al., 2000). Taken together, these data indicate that MYB75 interacts with MPK4 in a kinase activity-dependent manner.
MPK4 Is Involved in High Light-Induced Anthocyanin Accumulation
The activation tag mutant pap1-D (production of anthocyanin pigment 1-Dominant), harboring insertion of 35S enhancer adjacent to the MYB75/PAP1 gene, constitutively overexpresses MYB75/PAP1 (Borevitz et al., 2000). Since MYB75 can interact with MPK4, we tested whether MPK4 regulates anthocyanin accumulation. As a control, MYB75 knockout mutants were generated in the Col-0 ecotype using the CRISPR-Cas9 system and are referred to here as myb75-c (Supplemental Figure 1).
In Arabidopsis, anthocyanin accumulation only occurs in the light (Maier et al., 2013) and the levels of anthocyanin accumulation are light intensity dependent (Rowan et al., 2009; Shi and Xie, 2010). In this study, we maintained Arabidopsis seedlings under weak light of 40 µmol m−2 s−1 (hereafter called low light), a control light intensity at which wild-type plants accumulate very low levels of anthocyanins (Shi and Xie, 2010; Maier and Hoecker, 2015) (Figures 2A and 2B). To induce anthocyanin biosynthesis, seedlings were then shifted to moderate high light (175 µmol m−2 s−1, hereafter called high light). To test whether the moderate high light induces a stress response, we analyzed transcript levels of general stress-responsive genes RESPONSIVE TO DESICCATION 29A (RD29A), EARLY RESPONSIVE TO DEHYDRATION10 (ERD10), LATE EMBRYOGENESIS ABUNDANT14 (LEA14), and KIN1 (At5g15960) (Kimura et al., 2003) under low and moderate high light conditions. Expression of these stress genes was only slightly stimulated by moderate high light (Supplemental Figure 2), supporting that our moderate high light regimen did not cause much stress to Arabidopsis plants.
Figure 2.
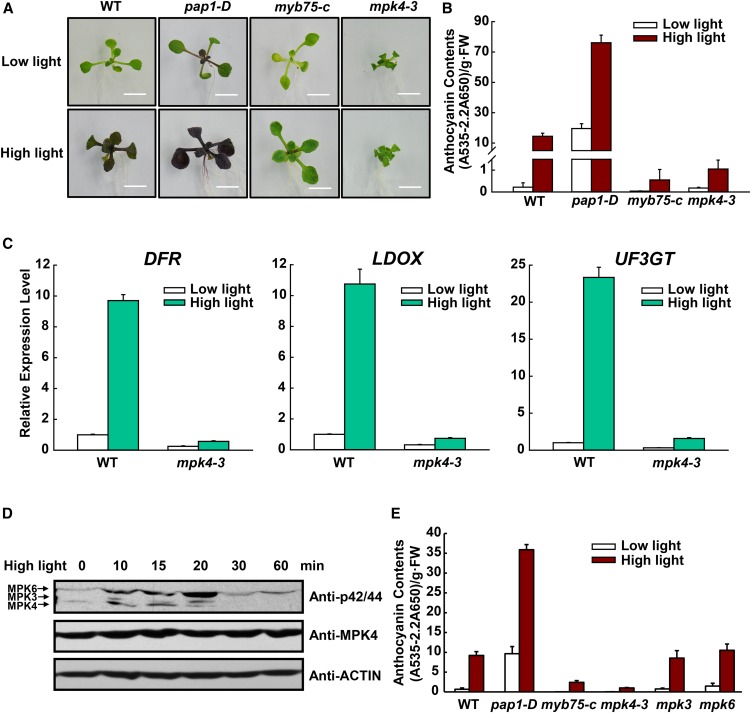
MPK4 Is Involved in High Light-Induced Anthocyanin Accumulation.
(A) Twelve-day-old Arabidopsis seedlings of the wild type, pap1-D, myb75-c, and mpk4-3 grown on plates under low light or moderate high light (high light). Bars = 0.5 cm.
(B) Anthocyanin contents of the seedlings in (A). FW, fresh weight. Error bars represent sd of three replicates (representing three separate extractions from separate samples of pooled aboveground tissue of seedlings grown on one plate for each genotype and treatment).
(C) Quantitative real-time PCR analysis of DFR, LDOX, and UF3GT transcript levels in 12-d-old wild-type and mpk4-3 seedlings grown on plates exposed to low light and moderate high light (high light) for 9 h, respectively. Results were normalized to ACTIN8, and expression levels of the genes in the wild type under low light were set at one unit. Error bars indicate sd of three replicates (as described above).
(D) High light-induced MAPK activation. Twelve-day-old seedlings were exposed to high light and collected at the indicated time points. MAPK activity was analyzed by immunoblot with Phospho-p44/42 MAPK (Erk1/2) antibody (top panel), the level of MPK4 was determined by immunoblotting (middle panel), and ACTIN was used as a protein loading control (bottom panel).
(E) High light-induced anthocyanin accumulation is not affected in the mpk3 and mpk6 mutants. Twelve-day-old Arabidopsis seedlings were grown on plates under low light and moderate high light (high light). FW, fresh weight. Error bars represent sd of three replicates (as described above).
High light exposure induced anthocyanin accumulation in the wild type and even more so in pap1-D seedlings. In contrast, anthocyanin accumulation was minimal in the mpk4-3 and myb75-c mutants (Figures 2A and 2B). The expression of anthocyanin biosynthetic genes was also greatly reduced in mpk4-3 under high light (Figure 2C). Plants homozygous for three different loss-of-function MPK4 alleles, mpk4-1, mpk4-2, and mpk4-3, all exhibited similarly compromised accumulation (Supplemental Figure 3). This indicates that their compromised anthocyanin accumulation is not limited to a specific MPK4 mutation and is independent of Arabidopsis ecotypes. Moreover, the wild-type MPK4 gene rescued the mpk4 phenotype (Supplemental Figure 3).
The mpk4 mutants exhibit autoimmunity because the MPK4 pathway is guarded by the immune receptor SUMM2, which is regulated by SUMM1 (Kong et al., 2012; Zhang et al., 2012). To test whether mpk4 autoimmunity affects anthocyanin accumulation, we analyzed their accumulation in mpk4-3 summ1 and mpk4-3 summ2 double mutants. Their anthocyanin contents were only slightly higher than that of the mpk4 mutant, but greatly lower than that of summ1 and summ2 (Supplemental Figure 3). This indicates that the anthocyanin-deficient phenotype of the mpk4 mutant is mostly not due to mpk4 autoimmunity and confirms that MPK4 functions in light-induced anthocyanin accumulation.
We next checked whether MPK4 is activated in response to light. Shifts from low light to high light led to rapid activation of MPK4, reaching its highest level at 15 min. Interestingly, MPK3 and MPK6 were also activated in response to high light (Figure 2D), confirming a recent report (Lee, 2015). We then investigated whether MPK3 and MPK6 are also involved in anthocyanin accumulation. As shown in Figure 2E, high light-induced anthocyanin accumulation was unaffected in mpk3 or mpk6 mutants. Since the mpk3 mpk6 double mutant is embryo lethal, we could not analyze anthocyanin accumulation in the double mutant. Thus, the possible roles of MPK3 and MPK6 in light signaling may be addressed in future. In addition, MPK11 is the closest homolog of MPK4. High light-induced anthocyanin accumulation was also unaffected by MPK11 mutation (Supplemental Figure 4). This indicates that MPK11 is not required for anthocyanin accumulation.
Interaction with MPK4 Is Required for MYB75 Function in Anthocyanin Accumulation
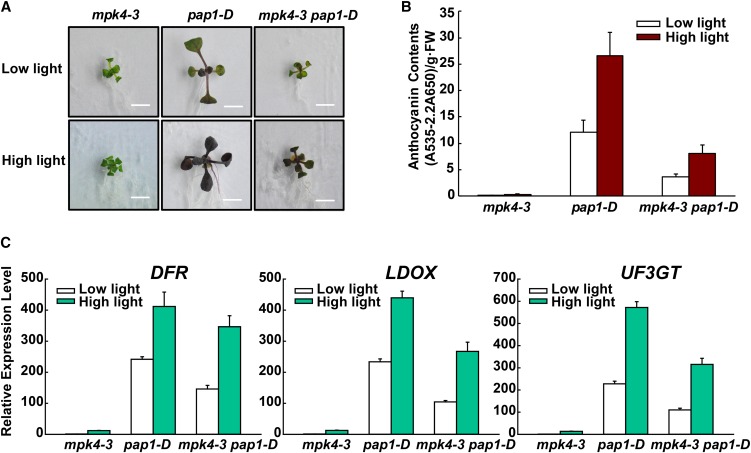
To more directly confirm that MPK4 is required for MYB75-meditated anthocyanin accumulation, the pap1-D mutant was crossed with mpk4-3 to generate the mpk4-3 pap1-D double mutant. The anthocyanin content of the mpk4-3 pap1-D double mutant was significantly lower than that of pap1-D (Figures 3A and 3B). Consistent with this, transcript levels of the anthocyanin biosynthetic genes DFR, LDOX, and UF3GT were also lower in the mpk4-3 pap1-D double mutant than in the pap1-D mutant (Figure 3C). These results support a functional interaction between MPK4 and MYB75.
Figure 3.
MPK4 Is Required for MYB75-Mediated Anthocyanin Accumulation.
(A) Twelve-day-old Arabidopsis seedlings of mpk4-3, pap1-D, and mpk4-3 pap1-D were grown on plates under low light and moderate high light (high light). Bars = 0.5 cm.
(B) Anthocyanin contents of the seedlings in (A). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(C) Quantitative real-time PCR analysis of DFR, LDOX, and UF3GT transcript levels in 12-d-old mpk4-3, pap1-D, and mpk4-3 pap1-D seedlings grown on plates exposed to low light and moderate high light (high light) for 9 h, respectively. Results were normalized to ACTIN8, and expression levels of the genes in mpk4-3 under low light were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
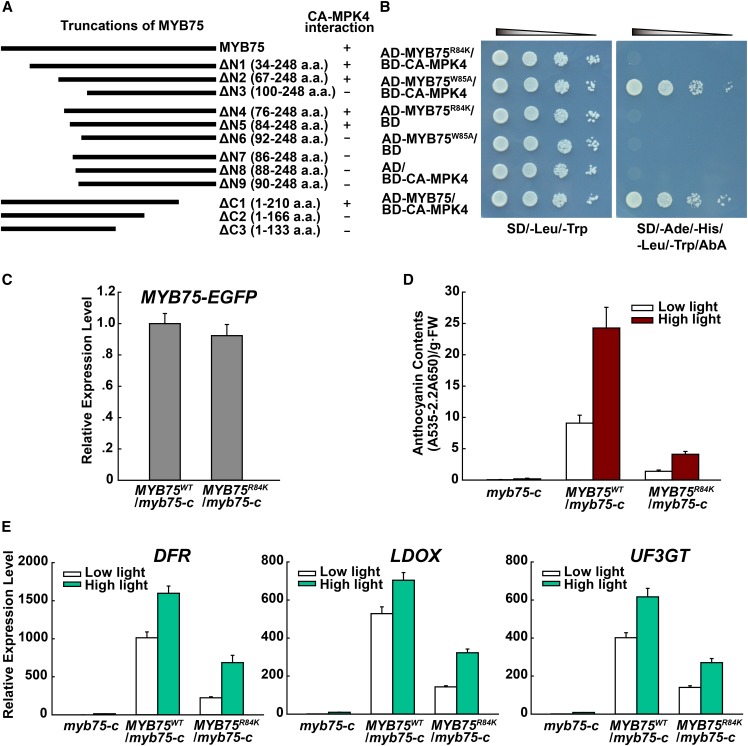
To identify the domains of MYB75 interacting with MPK4, a directed Y2H analysis was performed with CA-MPK4 and a set of N- and C-terminal truncations of MYB75. This indicated that both the N- and C-terminal regions of MYB75 are necessary for interaction with MPK4. We focused on the N-terminal region and narrowed its interaction to two residues, Arg-84 and Trp-85 (Figure 4A). Mutation of these two residues (Arg-84 to Lys and Trp-85 to Ala) showed that the Arg-84-to-Lys mutation abolished the interaction between MYB75 and CA-MPK4 (Figure 4B).
Figure 4.
Interaction with MPK4 Is Required for MYB75 Function in Anthocyanin Accumulation.
(A) Schematic representation of the MYB75 truncations and their interaction with CA-MPK4 in yeast two-hybrid assays. The numbers indicate the positions of the amino acids in the constructs. “+” and “−” denote positive and negative results for interaction, respectively.
(B) Yeast two-hybrid assay showing that mutation R84K of MYB75, but not W85A, abolishes its interaction with CA-MPK4.
(C) Quantitative real-time PCR analysis showing comparable expression levels of the transgenes MYB75WT and MYB75R84K in 35S:MYB75WT/myb75-c and 35S:MYB75R84K/myb75-c transgenic seedlings. The forward and reverse primers for MYB75 and EGFP, respectively, were designed to detect transgene transcripts of MYB75WT and MYB75R84K fused with EGFP, but not endogenous MYB75 transcripts. Results were normalized to ACTIN8, and expression levels of the genes in 35S:MYB75WT/myb75-c transgenic seedlings were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
(D) Anthocyanin contents of 12-d-old Arabidopsis seedlings of myb75-c, 35S:MYB75WT/myb75-c, and 35S:MYB75R84K/myb75-c grown on plates under low light and moderate high light (high light). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(E) Quantitative real-time PCR analysis of DFR, LDOX, and UF3GT transcript levels in 12-d-old seedlings grown on plates exposed to low light and moderate high light (high light) for 9 h, respectively. Results were normalized to ACTIN8, and expression levels of the genes in myb75-c under low light were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
To further examine the importance of the interaction between MYB75 and MPK4 for anthocyanin accumulation, we generated transgenic plants expressing MYB75R84K and MYB75WT driven by the constitutive 35S promoter in the myb75-c mutant background. The expression levels of these two transgenes were similar, but the anthocyanin level of MYB75R84K transgenic plants was much lower than that of MYB75WT plants (Figures 4C and 4D). Consistent with this, the levels of DFR, LDOX, and UF3GT transcripts were also lower in MYB75R84K transgenic plants compared with MYB75WT plants (Figure 4E).
R84 is located in the R3 repeat of the MYB domain of MYB75, which is also responsible for EGL3 binding (Zimmermann et al., 2004). The loss-of-function phenotype of plants expressing MYB75R84K could therefore be due to loss of DNA binding and/or to loss of EGL3 interaction. To examine this, we first analyzed the interaction between MYB75R84K and EGL3 in the Y2H system and found that the R84K mutation did not alter the interaction (Supplemental Figure 5). Furthermore, electrophoretic mobility shift assays revealed that the R84K mutation did not affect the binding of MYB75 to its target gene promoter (Supplemental Figure 6). Taken together, these data support the idea that interaction with MPK4 is important for MYB75 to regulate anthocyanin biosynthesis.
MPK4 Phosphorylates MYB75
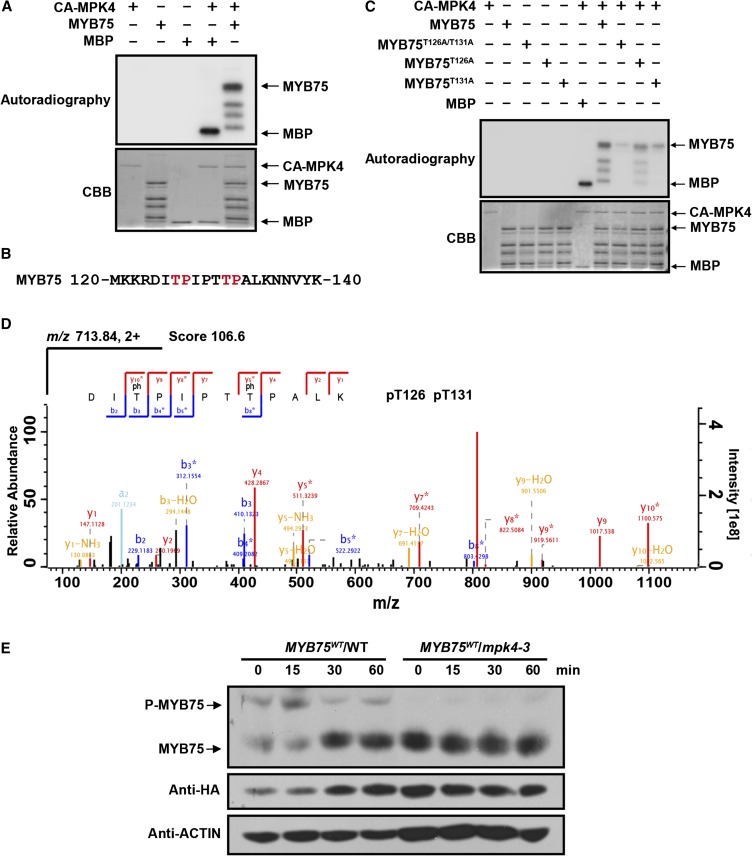
To investigate whether MYB75 can be phosphorylated by MPK4, His-tagged CA-MPK4 and MYB75 recombinant proteins were purified and used in in vitro kinase assays. CA-MPK4 was able to phosphorylate the standard MAP kinase substrate myelin basic protein, which confirms the previous report that CA-MPK4 is a constitutively active form of MPK4 (Berriri et al., 2012). MYB75 protein was also strongly phosphorylated by CA-MPK4 (Figure 5A). There are two potential MAPK phosphorylation sites (with a Ser or Thr residue followed by a Pro residue) in MYB75, Thr-126, and Thr-131 (Figure 5B). To initially identify the phosphorylation site(s) of MYB75, these two Thr sites were mutated to Ala individually and together. MYB75T126A, MYB75T131A, and MYB75T126A/T131A recombinant proteins were prepared and their phosphorylation by CA-MPK4 was examined. The T126A or T131A single substitutions had little effect on the phosphorylation of MYB75. However, when both sites were mutated (T126A/T131A), phosphorylation of MYB75 was greatly reduced (Figure 5C).
Figure 5.
MPK4 Phosphorylates MYB75.
(A) Phosphorylation of recombinant MYB75 by CA-MPK4. CA-MPK4 was used to phosphorylate purified His-tagged MYB75. Side-by-side control reactions using myelin basic protein (MBP) as a substrate confirmed the kinase activity of CA-MPK4. Phosphorylated MYB75 was visualized by autoradiography after gel electrophoresis.
(B) Putative MAPK phosphorylation sites (marked in red) in the MYB75 protein sequence.
(C) Mutation of the putative MAPK phosphorylation sites greatly reduces the phosphorylation of MYB75 by CA-MPK4. Recombinant MYB75, MYB75T126A, MYB75T131A, and MYB75T126A/T131A were incubated with CA-MPK4. Phosphorylated MYB75 was visualized by autoradiography after gel electrophoresis.
(D) LC-MS/MS analysis showing that MYB75 Thr-126 and Thr-131 are phosphorylated. The sequence of the doubly charged peptide ion at m/z 713.84, score 106.6, matches DIpTPIPTpTPALK of MYB75. “b” and “y” denote peptide fragment ions retaining charges at the N and C terminus, respectively. The subscript numbers indicate positions in the identified peptide. pT indicates phosphorylated Thr.
(E) In vivo phosphorylation of MYB75. Twelve-day-old seedlings of 35S:MYB75WT/WT and 35S:MYB75WT/mpk4-3 were exposed to high light for the indicated times. Protein extracts were separated on SDS-PAGE gels with Phos-tag reagent, and MYB75 was detected with anti-HA antibody. Due to the low MYB75 levels in mpk4-3, double the amount of total protein extract was loaded for the mpk4-3 samples than from the wild type for comparative purposes.
To further characterize the MPK4 phosphorylation sites on MYB75, we enriched the phosphorylated peptides of recombinant MYB75 by TiO2 chromatography and analyzed them by liquid chromatography-tandem mass spectrometry (LC-MS/MS). This identified several phosphopeptides (Supplemental Table 1). The most abundant phosphopeptide contained Thr-126 and Thr-131 as phosphorylation sites (Figure 5D). Thr-130 was also found to be phosphorylated (Supplemental Figure 7). However, Thr-130 is not followed by a Pro residue and is unlikely to be a MAPK phosphorylation site. It is therefore possible that the Thr-130 phosphorylation detected was produced by the activity of an Escherichia coli kinase. These results imply that MYB75 Thr-126 and Thr-131 are the major residues phosphorylated by MPK4.
To test whether MPK4 is able to phosphorylate MYB75 in vivo, the 35S:MYB75-HA transgene was transformed into mpk4-3 heterozygotes. First, transgenic plants expressing MYB75-HA were identified by RT-PCR and immunoblotting with anti-HA antibody. Plants homozygous for the MYB75-HA transgene and heterozygous for mpk4-3 were then used to produce segregating, doubly homozygous progeny for further analysis. Total protein was extracted from these plants at different time points exposed to high light. MYB75 phosphorylation was detected using SDS-PAGE gels containing Phos-tag, a ligand that retards the electrophoretic mobility of phosphorylated proteins. The mobility shift of MYB75 bands was revealed by immunoblotting with anti-HA antibody. Retardation of MYB75 from the wild-type transgenic plants was detected and peaked at 15 min after exposure to high light. However, retardation due to phosphorylation of MYB75 was largely absent in the mpk4-3 background (Figure 5E). These data indicate that high light induces in vivo phosphorylation of MYB75 and that this phosphorylation is mediated by MPK4. Phosphorylated MYB75 was also detected under low light, suggesting that MPK4 maintains partial activity under this condition.
Phosphorylation of MYB75 Enhances Anthocyanin Accumulation
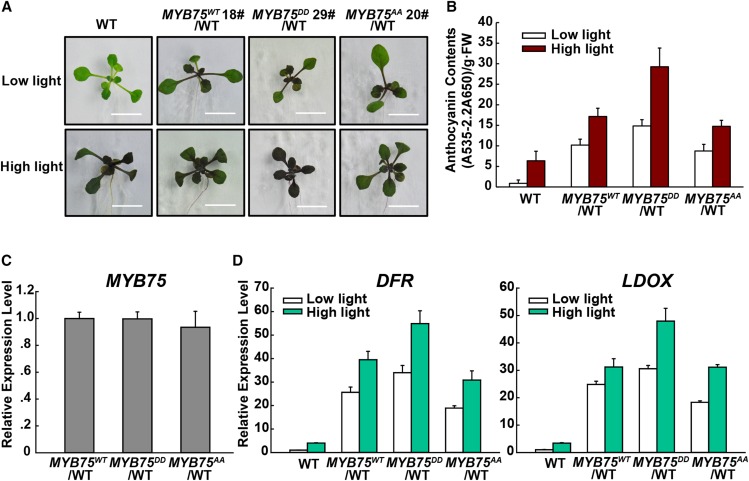
To further confirm that MPK4-mediated phosphorylation is required for MYB75 function, we generated expression vectors with the native form (MYB75WT), phosphomimic MYB75DD (Thr-126/Thr-131 mutated to Asp), and phosphodeficient MYB75AA (Thr-126/Thr-131 mutated to Ala) under control of the constitutive 35S promoter. We used the 35S promoter rather than the native MYB75 promoter as suggested in a previous report (Maier et al., 2013). Wild-type plants expressing MYB75DD accumulated significantly more anthocyanin under both low light and high light conditions (Figures 6A and 6B), despite the comparable expression levels of the MYB75DD, MYB75WT, and MYB75AA transgenes (Figure 6C). In addition, the anthocyanin levels of MYB75AA and MYB75WT transgenic plants were not significantly different (Figure 6B). As shown in Figure 6D, the expression levels of the anthocyanin biosynthetic genes DFR and LDOX paralleled their anthocyanin contents. These data indicate that MYB75 functions downstream of MPK4 in regulating anthocyanin accumulation.
Figure 6.
Phosphorylation of MYB75 Enhances Its Function in Anthocyanin Accumulation.
(A) Twelve-day-old Arabidopsis seedlings of wild-type, 35S:MYB75WT/WT, 35S:MYB75DD/WT, and 35S:MYB75AA/WT transgenic seedlings grown on plates under low light and moderate high light (high light). Bars = 1 cm.
(B) Anthocyanin contents of the seedlings in (A). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(C) Quantitative real-time PCR analysis showing comparable MYB75 expression levels in 35S:MYB75WT/WT, 35S:MYB75DD/WT, and 35S:MYB75AA/WT transgenic seedlings. Primers were designed to detect both transgenic and endogenous MYB75 transcripts. Results were normalized to ACTIN8, and expression levels of the genes in the 35S:MYB75WT/WT transgenic seedlings were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
(D) Quantitative real-time PCR analysis of DFR and LDOX transcript levels in 12-d-old wild type, 35S:MYB75WT/WT, 35S:MYB75DD/WT, and 35S:MYB75AA/WT transgenic seedlings grown on plates exposed to low light and moderate high light (high light) for 9 h, respectively. Results were normalized to ACTIN8, and expression levels of the genes in the wild type under low light were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
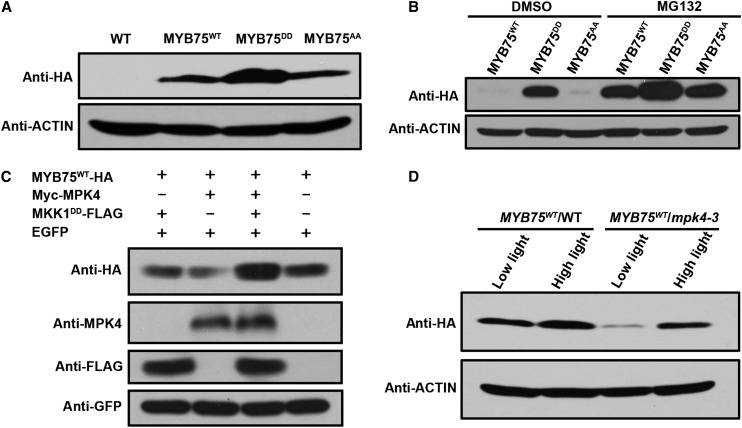
Phosphorylation by MPK4 Increases the Stability of MYB75
Since the phosphomimic and phosphodeficient alterations in MYB75 did not affect its binding to the target gene promoter (Supplemental Figure 6), we compared MYB75 protein levels in wild-type plants expressing MYB75WT, MYB75DD, and MYB75AA. As shown in Figure 7A, MYB75DD accumulated to higher levels in the transgenic plants although the mRNA levels of the three MYB75 variants were comparable (Figure 6C). Since the stability of MYB75 has been shown to be regulated by light (Maier et al., 2013), we examined the levels of MYB75 proteins in dark-adapted seedlings to further demonstrate whether MYB75 phosphorylation affects its stability. This showed that MYB75WT and MYB75AA were almost undetectable in dark-adapted seedlings, whereas MYB75DD accumulated to high levels (Figure 7B). This indicates that the phosphomimic alteration increased MYB75 stability. In addition, when we treated dark-adapted seedlings with 100 µM MG132, an inhibitor of the 26S proteasome, the levels of MYB75WT and MYB75AA increased greatly, whereas that of MYB75DD was only slightly increased above its already high level (Figure 7B). These findings indicate that MYB75 is degraded via the 26S proteasome and that phosphorylation of MYB75 opposes degradation and thus increases MYB75 stability. MYB75AA and MYB75WT exhibited similar protein stability and effects on anthocyanin accumulation (Figures 6B and 7A) in the transgenic plants. One explanation for these results is that overexpression of these proteins saturates the activity of MPK4.
Figure 7.
Phosphorylation of MYB75 by MPK4 Increases Its Stability.
(A) Immunoblot analysis showing accumulation of the MYB75 variants in 35S:MYB75WT, 35S:MYB75DD, and 35S:MYB75AA transgenic seedlings in the wild-type background under low light. The levels of these MYB75 variants were determined by immunoblotting with anti-HA antibody. ACTIN was used as loading control.
(B) Phosphomimic alteration increases MYB75 stability. Dark-adapted, transgenic seedlings expressing 35S:MYB75WT, 35S:MYB75DD, and 35S:MYB75AA were treated with MG132 or DMSO for 12 h in darkness. The levels of these MYB75 variants were determined by immunoblotting with anti-HA antibody. ACTIN was used as loading control.
(C) Coexpression of MPK4 and MKK1DD increases the stability of MYB75. MYB75WT-HA was coexpressed with Myc-MPK4 and MKK1DD-FLAG in the leaves of N. benthamiana by agroinfiltration. EGFP was coexpressed as an internal control. “+” and “−” denote presence and absence of the protein in each sample.
(D) MPK4 regulates the stability of MYB75 in planta. 35S:MYB75WT/WT and 35S:MYB75WT/mpk4-3 were segregated from the same transformed mpk4-3 heterozygous plants. Twelve-day-old seedlings grown on plates were exposed to low light or moderate high light (high light) for 9 h. Immunoblot analysis showing the differential accumulation of MYB75WT in the wild-type and mpk4-3 backgrounds under both low light and moderate high light (high light).
MKK1 is the MAPK kinase upstream of MPK4 (Gao et al., 2008; Qiu et al., 2008). MKK1DD is a constitutively active, phosphomimic form of the MKK1 generated by mutating the conserved serine and threonine residues in the kinase activation loop to aspartate (Asai et al., 2002). We transiently expressed MYB75WT, MPK4, and MKK1DD in leaves of N. benthamiana. As seen in Figure 7C, MYB75 accumulated to a higher level only in the presence of MKK1DD and MPK4. This again indicates that MPK4-mediated phosphorylation stabilizes MYB75.
We then examined MYB75 protein levels in the wild type and mpk4-3 mutants expressing the MYB75WT transgene. The levels of MYB75 were greatly decreased in mpk4-3 under both low light and high light conditions compared with the wild type (Figure 7D). This confirms that MPK4 regulates MYB75 stability. Interestingly, MYB75 could still be induced by light in the mpk4-3 mutant, suggesting that additional factors also regulate its stability.
MPK4 and COP1 Have Opposing Effects on Anthocyanin Accumulation
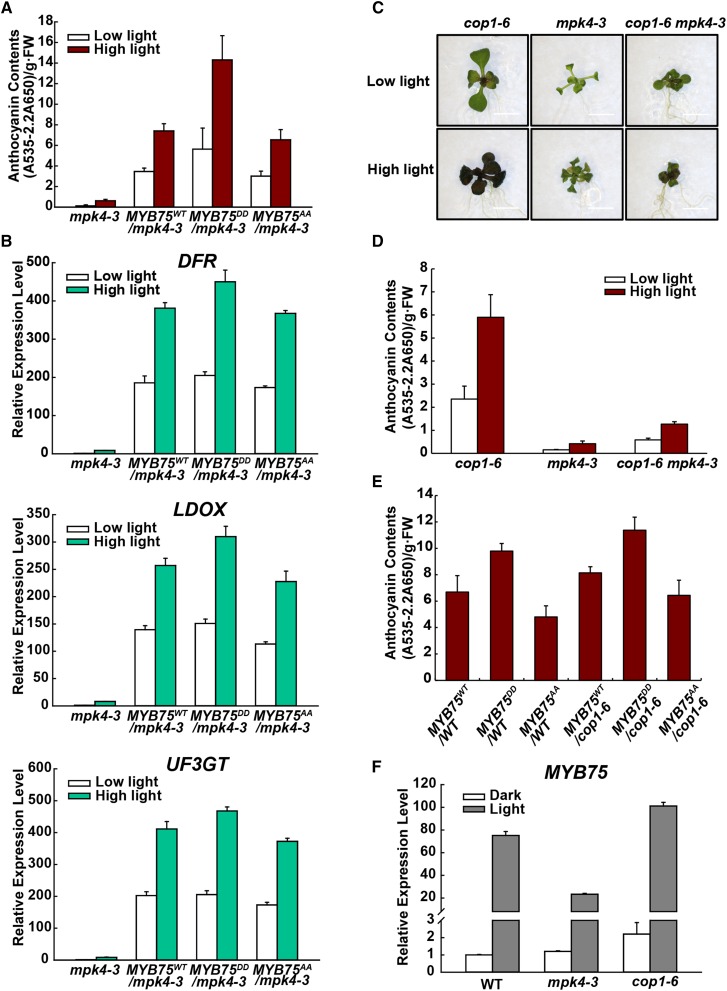
Transgenic plants expressing MYB75WT, MYB75DD, and MYB75AA in the mpk4-3 background were also tested for high light-induced anthocyanin accumulation. mpk4-3 plants expressing MYB75DD accumulated more anthocyanins than MYB75WT and MYB75AA transgenic plants (Supplemental Figure 8; Figure 8A). The expression pattern of DFR, LDOX, and UF3GT transcripts was consistent with these anthocyanin levels (Figure 8B). However, these plants still retained high light responsiveness, implying the existence of an MPK4-independent pathway also regulated by light.
Figure 8.
MPK4 and COP1 Have Opposing Effects on Anthocyanin Accumulation.
(A) Anthocyanin contents of 12-d-old Arabidopsis seedlings of mpk4-3, 35S:MYB75WT/mpk4-3, 35S:MYB75DD/mpk4-3, and 35S:MYB75AA/mpk4-3 transgenic seedlings grown on plates under low light and moderate high light (high light). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(B) Quantitative real-time PCR analysis of DFR, LDOX, and UF3GT transcript levels in 12-d-old mpk4-3, 35S:MYB75WT/mpk4-3, 35S:MYB75DD/mpk4-3, and 35S:MYB75AA/mpk4-3 transgenic seedlings grown on plates exposed to low light and moderate high light (high light) for 9 h, respectively. Results were normalized to ACTIN8, and expression levels of the genes in mpk4-3 under low light were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
(C) Twelve-day-old Arabidopsis seedlings of cop1-6, mpk4-3, and cop1-6 mpk4-3 grown on plates under low light and moderate high light (high light). Bars = 0.5 cm.
(D) Anthocyanin contents of the seedlings in (C). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(E) Anthocyanin contents of 12-d-old Arabidopsis seedlings of the 35S:MYB75WT, 35S:MYB75DD, and 35S:MYB75AA transgenic lines in the wild-type and cop1-6 backgrounds grown on plates under low light. FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(F) Quantitative real-time PCR analysis of MYB75 expression in wild-type, mpk4-3, and cop1-6 seedlings. Seedlings were placed in the dark or continuous low light for 4 d. Results were normalized to ACTIN8, and expression levels of the genes in wild type in the dark were set at one unit. Error bars indicate sd of three replicates (as described in Figure 2B).
The E3 ubiquitin ligase COP1 mediates ubiquitination and degradation of MYB75 in the dark (Maier et al., 2013). Light inactivates COP1 and thereby stabilizes MYB75 (Osterlund et al., 2000; Maier et al., 2013). Increased anthocyanin accumulation is a phenotype of cop1 mutants (Deng et al., 1991). The increased anthocyanin level in cop1 mutants requires MYB75, indicating that COP1 functions upstream of MYB75 (Maier et al., 2013). To investigate whether COP1 is required for the effect of MPK4 on anthocyanin accumulation, we generated a cop1-6 mpk4-3 double mutant. Remarkably, anthocyanin accumulation was greatly decreased in the double mutant (Figures 8C and 8D). This indicates that MPK4 does not simply function through COP1 to regulate anthocyanin accumulation.
Plants expressing MYB75WT, MYB75DD, and MYB75AA were also generated in the cop1-6 background by crossing with wild-type transgenic plants. Anthocyanin levels were higher in the transgenic plants in cop1-6 mutant background than in wild-type background, and the MYB75DD expressing plants appeared to accumulate still more anthocyanin (Figure 8E). This indicates that MPK4 and COP1 function at least partially independently. Taken together, the data suggest that MPK4 and COP1 have opposing effects on MYB75 stability and thereby together regulate light-induced anthocyanin accumulation.
MYB75 transcripts are upregulated by light (Cominelli et al., 2008) and are affected by the COP1/SPA complex (Maier et al., 2013). To investigate whether MPK4 regulates MYB75 at the transcriptional level, transcripts of MYB75 were measured in wild-type, mpk4-3, and cop1-6 seedlings. The levels of MYB75 transcripts were only slightly higher in dark-grown mpk4-3 than in the wild type but were greatly increased in dark-grown cop1-6 (Figure 8F). This indicates that MPK4 may not directly regulate MYB75 at the transcriptional level. However, MYB75 transcript levels were strongly downregulated in mpk4-3 in low light (Figure 8F). This again indicates that MPK4 is involved in light signaling and is consistent with a dual regulation of anthocyanin accumulation by MPK4 and by the COP1/SPA complex. Therefore, MPK4 regulates the function of MYB75 both by directly affecting MYB75 protein stability and by indirectly affecting its transcript levels.
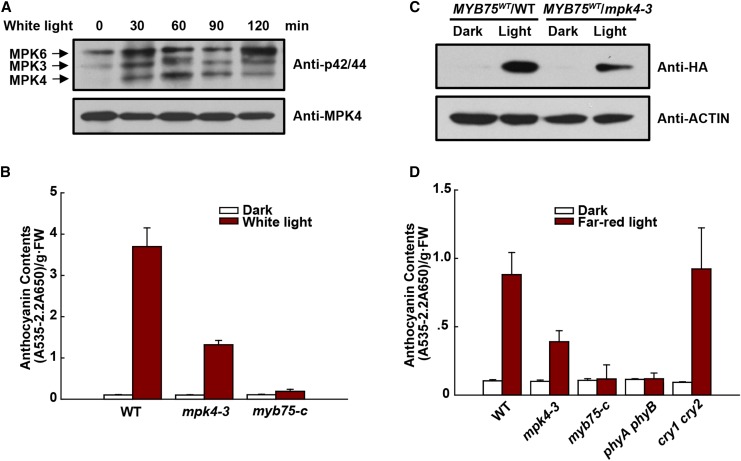
MPK4 Regulates Light-Induced Anthocyanin Accumulation
To further confirm that MPK4 is involved in light responses, we investigated its activation and role in anthocyanin accumulation under far-red and weak white light. Wild-type and mpk4-3 seedlings were grown in the dark for 4 d and then exposed to weak white light (40 µmol m−2 s−1) for various times. Exposure to light led to rapid activation of MPK4, MPK3, and MPK6 (Figure 9A). This exposure induced accumulation of low levels of anthocyanin in wild-type seedlings, but this accumulation was again greatly decreased in mpk4-3 seedlings (Figure 9B). Moreover, we compared MYB75 protein levels in wild-type and mpk4-3 seedlings expressing MYB75WT grown in the dark and under weak light (Figure 9C). In the dark, MYB75 protein in the wild type was almost undetectable, but it accumulated under weak light. In contrast, MYB75 protein levels in mpk4-3 were much lower than in the wild type. Interestingly, MYB75 protein levels could still be induced by light in mpk4-3, suggesting that factors in addition to MPK4 are involved in regulating the stability of MYB75.
Figure 9.
MPK4 Is Involved in Light-Induced Anthocyanin Accumulation.
(A) Weak white light-induced MAPK activation. Four-day-old dark-grown wild-type seedlings were exposed to weak white light (40 μmol m−2 s−1) for the indicated times. MAPK activity was analyzed by immunoblotting with Phospho-p44/42 MAPK (Erk1/2) antibody (top panel), and MPK4 protein was determined by immunoblotting (bottom panel).
(B) Anthocyanin contents of 8-d-old seedlings of the wild type, mpk4-3, and myb75-c grown on plates under continuous dark or weak white light (40 μmol m−2 s−1). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
(C) Protein levels of MYB75 in the wild type and mpk4-3 grown under weak white light. 35S:MYB75WT/WT and 35S:MYB75WT/mpk4-3 were segregated from mpk4-3 heterozygous plants. Eight-day-old seedlings grown on plates under weak white light were dark-adapted for 4 d and then placed in the dark or continuous weak white light for another 9 h. The levels of MYB75 were determined by immunoblotting with anti-HA antibody.
(D) Anthocyanin contents of 8-d-old seedlings of the wild type, mpk4-3, myb75-c, phyA phyB, and cry1 cry2 grown on plates under continuous dark and far-red light (10 μmol m−2 s−1). FW, fresh weight. Error bars represent sd of three replicates (as described in Figure 2B).
Far-red light with a fluence rate of around 10 µmol m−2 s−1 rapidly activated MPK4, MPK3, and MPK6 in wild-type seedlings but not in a phyA phyB double mutant seedlings (Supplemental Figure 9). This suggests that MPK4 functions downstream of these photoreceptors. Similarly, far-red-light-induced anthocyanin accumulation was greatly reduced in mpk4-3 seedlings and almost abolished in myb75-c and in the phyA phyB seedlings (Figure 9D). In contrast, the blue light receptor double mutant cry1 cry2 was still fully responsive to far-red light (Figure 9D). Together these findings show that MPK4 is required for light-induced anthocyanin accumulation.
DISCUSSION
Light is a predominant environmental stimulus affecting plant development and growth (Jiao et al., 2007). Being sessile, plants have evolved complicated mechanisms to sense, respond, and adapt to varying light conditions (Xu et al., 2015). Although plant photoreceptors are well characterized (Galvão and Fankhauser, 2015), downstream signaling processes are less clear. In Arabidopsis, anthocyanin accumulation is light-regulated (Chalker-Scott, 1999; Maier et al., 2013) and the transcription factor MYB75 activates the expression of anthocyanin biosynthetic genes (Borevitz et al., 2000; Gonzalez et al., 2008). In this study, we report that MYB75 is a substrate of the MAP kinase MPK4 and that phosphorylation of MYB75 by MPK4 is required for light-induced anthocyanin accumulation. Our work demonstrates an important role for a MAP kinase in the light response.
MAPK signaling specificity is to a large extent conferred by their diverse substrates (Mao et al., 2011). The identification of MAPK substrates thus provides mechanistic insight into their regulatory functions (Meng and Zhang, 2013). However, only a few plant MAPK substrates have been identified because both MAPK activation and their interactions with substrates are transient and rapid. We reasoned that a constitutively active MAPK form (CA-MAPK) could largely overcome these transient features. Our identification of MYB75 as an MPK4 substrate successfully demonstrates this new approach to identify MAPK substrates through Y2H screening using CA-MAPK as bait. Our approach should be applicable to different MAPKs in many species.
We used several strategies to show that MYB75 phosphorylation by MPK4 increases MYB75 stability (Figure 7). MYB75 has been reported to be targeted by the COP1/SPA E3 ubiquitin ligase for degradation in the dark. Light represses COP1/SPA activity (Maier et al., 2013), thereby stabilizing MYB75 protein (Maier et al., 2013; Li et al., 2014). It was therefore possible that MYB75 phosphorylation by MPK4 prevents MYB75 recognition by COP1, thereby enhancing MYB75 stabilization. However, our genetic analysis of a cop1-6 mpk4-3 double mutant revealed that MPK4 primarily functions independently of COP1 (Figure 8). Previous studies with cop1 and spa mutants also demonstrated that they accumulate low levels of anthocyanins in darkness, but are hyperresponsive to light and accumulate anthocyanins to higher levels in light compared with the wild type (Maier and Hoecker, 2015). Similarly, the mpk4-3 mutant also retained light-induced responses (Figures 3 and 8), and MYB75 protein levels could still be induced by light in the mpk4-3 mutant (Figures 7 and 9). However, the light-induced accumulation of anthocyanins in cop1-6 single mutants was greatly reduced in cop1-6 mpk4-3 double mutants (Figure 8). It is therefore most likely that MPK4 and COP1/SPA antagonistically regulate MYB75 protein levels. This dual regulation may provide more flexible and precise control of MYB75 protein levels such that anthocyanin accumulation in vegetative organs is optimized for protective light screening versus photosynthetic light harvesting (Albert et al., 2009).
Light-induced phosphorylation of TFs occurs mostly at Ser/Thr sites (Hardtke et al., 2000; Ni et al., 2013). Arabidopsis casein kinase II (CKII) is activated in darkness and phosphorylates HY5, which becomes less susceptible to degradation (Hardtke et al., 2000). However, this CKII activity is reduced in light (Hardtke et al., 2000). In contrast, the activation of Arabidopsis MAPK cascade components in response to light has recently been described. For example, high light-induced expression of CSD1 and CSD2 was found to be mediated by the MAPK kinase MKK5 (Xing et al., 2013). In addition, the MKK3-MPK6 module was shown to be activated by blue light in a MYC2-dependent manner and to be involved in blue light-mediated seedling development (Sethi et al., 2014). Most recently, the mkk3-1 mutant was found to be impaired in responses to red light (Lee, 2015). Our results demonstrate that MPK4 plays a role in light-induced anthocyanin accumulation, probably through phosphorylation of MYB75. Even though MPK4 was not much activated in the dark (Figure 9A), we cannot exclude the possibility that MPK4 constitutively phosphorylates MYB75 but exerts its affect mainly in light-grown seedlings due to the low MYB75 protein accumulation in darkness. Since the MYB75 knockout mutant retains light responsive anthocyanin accumulation (Figure 2B), it is most likely that other proteins in addition to MYB75, such as MYB90/PAP2, are involved in light-induced anthocyanin accumulation. However, MPK4 does not interact with PAP2 in yeast (Supplemental Figure 10). Thus, the mpk4-3 mutant and the cop1-6 mpk4-3 double mutant are still able to respond to light. Moreover, there may be additional MPK4 substrates that affect anthocyanin biosynthesis. In agreement with this, MYB75DD could not fully rescue the anthocyanin-deficient phenotype of the mpk4-3 mutant (Figures 6 and 8).
We conclude that the involvement of MAPKs in light-induced responses appears to be a general mechanism deserving further study. Such a study might link MAPKs to signaling pathways controlled by specific light receptors and/or to more or less specific stress response pathways triggered by changes in light intensity. Moreover, the production of anthocyanin is promoted by various extracellular stimuli including nutrient depletion, sucrose, jasmonic acid, cold, pathogen attack, and wounding (Chalker-Scott, 1999; Teng et al., 2005; Shi and Xie, 2014; Gan et al., 2014). Since MPK4 is known to play an important role in plant immunity and jasmonic acid signaling (Petersen et al., 2000), and to be activated by cold, osmotic stress, and wounding (Teige et al., 2004; Droillard et al., 2004; Schweighofer et al., 2007), MPK4 may substantially contribute to anthocyanin accumulation induced by these signals. Intriguingly, the production of anthocyanins in the dark-grown cop1-6 mutant is enhanced by jasmonic acid treatment (Li et al., 2014). This implies the existence of a COP1-independent pathway in light-independent induction of anthocyanins. Further work is required to establish the general role of MPK4 in anthocyanin biosynthesis.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants and transgenic lines were all in Col-0, except the mpk4-1 mutant (Petersen et al., 2000) in the Ler background. The mpk4-3 (Gao et al., 2008), pap1-D (Borevitz et al., 2000), summ1 (Kong et al., 2012), summ2 (Zhang et al., 2012), cop1-6 (McNellis et al., 1994), phyA phyB (Liu et al., 2001), and cry1 cry2 (Mockler et al., 1999) mutants have been described previously. The myb75-c mutant was generated using the CRISPR-Cas9 system following standard procedures (Shan et al., 2013), and Cas9-free T3 plants were used.
Arabidopsis seeds were surface sterilized and plated on half-strength Murashige and Skoog medium supplemented with 2% sucrose. Plates were stratified at 4°C for 3 d and transferred to a growth chamber at 22°C under different light conditions. White light was supplied by white fluorescent bulbs (TSZJD2-T5-28W; TOPSTAR), and far-red light was supplied by LED light sources (model E-30LEDL3; Percival Scientific). For high light-induced analyses, plates were exposed to low light (40 μmol m−2 s−1 white light) or high light (175 µmol m−2 s−1 white light) with a 16-h-light/8-h-dark photoperiod. For far-red light and weak white light-induced analyses, plates were exposed to continuous 40 μmol m−2 s−1 white light or 10 μmol m−2 s−1 far-red light for 8 d. MG132 treatments were performed on 12-d-old seedlings grown under low light with a 16-h-light/8-h-dark photoperiod. Seedlings were dark-adapted for 1 d and then treated with 100 μM MG132 or DMSO (control) for 12 h in the dark (Liu et al., 2013).
Plasmid Construction and Plant Transformation
For recombinant protein expression, the full-length MYB75 and CA-MPK4 coding regions were amplified and cloned into the pET-28a vector in frame with an N-terminal 3×His epitope tag. MYB75T126A, MYB75T131A, and MYB75T126A/T131A were generated using a site-directed mutagenesis kit (Takara) and confirmed by sequencing. A 1.5-kb genomic fragment of MYB75 including the coding region without stop codon was fused 3′ with a 3×HA tag, then cloned into the pCambia1300 binary vector to generate the 35S:MYB75-HA (MYB75WT) construct. The MYB75DD and MYB75AA constructs were produced by site-directed mutagenesis. Primers for plasmid construction are listed in Supplemental Table 2. These constructs were transformed into heterozygous mpk4-3 plants by Agrobacterium tumefaciens (strain GV3101) via floral dip (Clough and Bent, 1998). Heterozygous mpk4-3 plants homozygous for the transgenes with similar expression levels were obtained in the T3 generation. Wild-type and homozygous mpk4-3 T4 segregants homozygous for the MYB75WT, MYB75DD, and MYB75AA transgenes were then selected for experiments.
Yeast Two-Hybrid Assays
Assays were performed using the Matchmaker Gold System (Clontech) according to the manufacturer’s instructions. The CA-MPK4 coding region was cloned into pGBKT7 vector as bait to screen an Arabidopsis cDNA library from Clontech. For directed yeast two-hybrid assays, MYB75 coding sequence was cloned in pGADT7 as an activation domain fusion.
BiFC Assays
Genomic sequences of MYB75 and the coding sequences of MPK4, CA-MPK4, and MPK4AEF were individually cloned into binary YFP BiFC vectors (Song et al., 2011) to generate MYB75-cYFP and nYFP-MPK4/CA-MPK4/MPK4AEF constructs. Agrobacterium carrying the constructs were coinfiltrated into leaves of Nicotiana benthamiana. The plants were kept in darkness for 48 h before detecting YFP fluorescence with a Leica confocal laser scanning microscope (Leica Microsystems).
Coimmunoprecipitation Assay
MYB75-HA and Myc-MPK4 or Myc-CA-MPK4 were transiently expressed in N. benthamiana by Agrobacterium infiltration. Protein was extracted and resuspended in IP buffer (100 mM Tris-Cl, pH 7.4, 75 mM NaCl, 1 mM EDTA, 0.05% SDS, 0.1% Triton X-100, 10% glycerol, protease inhibitor cocktail [Roche], and PhosSTOP [Roche]). Coimmunoprecipitation was performed as previously described (Fiil et al., 2008).
Transient Expression Assay
Agrobacterium carrying the indicated constructs was coinfiltrated into leaves of N. benthamiana. The plants were kept in darkness for 48 h, and total proteins were extracted for immunoblotting.
Recombinant Protein Expression and Purification
Recombinant proteins were expressed in Escherichia coli strain BL21 (DE3) and induced with 0.5 mM IPTG (isopropylthio-β-galactoside) overnight at 16°C. The proteins were purified with HisTrap column on an AKTA purifier automatic chromatograph (GE Healthcare) and concentrated with a 10 K filter (Millipore).
In Vitro Phosphorylation Assay
Assays were performed as previously described (Andreasson et al., 2005). Briefly, recombinant MYB75 or its mutant forms were mixed with recombinant CA-MPK4 protein at a ratio of 2:1 in kinase reaction buffer (20 mM Tris-HCl, pH 7.4, 2 mM EGTA, 30 mM MgCl2, 1 mM DTT, and PhosSTOP phosphatase inhibitor) and 50 μM cold ATP and [γ-32P]ATP (2 μCi per reaction). Reactions were performed at 30°C for 60 min and stopped by the addition of SDS sample buffer. Proteins were separated on 10% SDS-PAGE gels and phosphorylated proteins visualized by autoradiography.
LC-MS/MS Analysis
To prepare samples for mass spectrometry analysis, ∼2 mg of recombinant MYB75 protein was mixed with 1 mg recombinant CA-MPK4 protein in kinase reaction buffer with 1×PhosSTOP phosphatase inhibitor (Roche) and 50 μM cold ATP. Reactions were incubated at 30°C for 60 min and then digested with trypsin. Phosphopeptides were enriched with TiO2 and analyzed with a Q Exactive LC-MS/MS system (Thermo Scientific). MS/MS spectra were analyzed with MaxQuant (version 1.5.1.2), and the identified phosphorylated sites were confirmed manually.
Protein Extraction and Immunoblot Analysis
Arabidopsis tissues were ground in liquid nitrogen and total protein extracted with protein extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 0.5% SDS, 5% glycerol, 2.5 mM DTT, protease inhibitor cocktail [Roche], and PhosSTOP [Roche]) (Su et al., 2013). Protein concentration was determined by the Bradford assay. Immunoblotting was performed following standard procedures. Antibodies used were as follows: anti-HA (Cwbiotech, catalog no. CW0260M; 1:1000 dilution), anti-FLAG (Sigma-Aldrich, catalog no. F1804; 1:1000 dilution), anti-MPK4 (Sigma-Aldrich, catalog no. A6979; 1:1000 dilution), anti-ACTIN (Abmart, catalog no. M20009L; 1:2500 dilution), peroxidase-conjugated goat anti-mouse IgG antibody (Sigma-Aldrich, catalog no. A4416; 1:10,000 dilution), and goat anti-rabbit IgG antibody (Sigma-Aldrich, catalog no. A6154; 1:10,000 dilution).
Phos-Tag Gel Electrophoresis
Total proteins were extracted with buffer containing 50 mM Tris-HCl, pH 7.5,100 mM NaCl, 1 mM DTT, 0.5% Triton X-100, 0.1% SDS, and 10% glycerol supplemented with protease inhibitor cocktail (ETDA-free; Roche) and PhosSTOP (Roche). Proteins were separated on 10% SDS-PAGE gels containing 50 mM Phos-tag (Wako Chemicals) and 100 mM MnCl2 as previously described (Xie et al., 2014).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay was performed as previously described (Mao et al., 2011). A synthetic oligonucleotide (5′-GACGAGGTAACCACCACGTGTTATTTCTTA-3′) containing the MYB75/PAP1 cis-regulatory element from the promoter of the DFR gene (Dare et al., 2008) was used as probe. The probe was radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase. One micrograms of recombinant protein corresponding to each MYB75 variant was incubated with 1 pmol labeled DNA probe for 30 min at room temperature in binding buffer [10 mM Tris, pH7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 10 mM EDTA, 50 ng/mL poly(dI·dC), and 0.05% Nonidet P-40] in the presence or absence of unlabeled competitor DNA (10-, 50-, and 100-fold of labeled probes). The protein-DNA complexes were separated on 5% native PAGE gels in half-strength TBE buffer. Following electrophoresis, gels were dried on filter paper and visualized by autoradiography.
MAPK Assay
For high light-induced MAPK activity, seedlings were grown on 0.5× MS plates for 12 d under 40 μmol m−2 s−1 white light and then exposed to moderate high light (175 µmol m−2 s−1) for the indicated times. For far-red light or weak white light-induced MAPK activity, seedlings were grown in darkness for 4 d and then exposed to far-red light (10 μmol m−2 s−1) or white light (40 μmol m−2 s−1) for the indicated times. Proteins were extracted in lysis buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 15 mM EGTA, 100 mM NaCl, 2 mM DTT, 1 mM sodium molybdate, 30 mM β-glycero-phosphate, 0.1% NP-40, protease inhibitor cocktail [Roche], and PhosSTOP [Roche]). MAPK activities were detected by immunoblotting with Phospho-p44/42 MAPK (Erk1/2) antibody (Cell Signaling) as previously described (Boutrot et al., 2010).
Real-Time PCR Analysis
Total RNA was extracted from aboveground tissue of seedlings with TRNzol reagent (Tiangen) and treated with RNase-free DNase, then reverse transcribed to synthesize first-strand cDNA using PrimeScript RT reagent lit (TaKaRa). Real-time PCR analysis was performed using SYBR Premix Ex Taq II (TaKaRa) and run in a Bio-Rad CFX96 PCR system (Bio-Rad Laboratories). Normalized expression levels were calculated with CFX Manager Software (Bio-Rad) using the 2−ΔΔCT method. ACTIN8 was used as an internal reference gene. The primers used are listed in Supplemental Table 2.
Anthocyanin Measurement
Anthocyanin was measured as previously described with some modifications (Lange et al., 1971; Wu et al., 2014). Arabidopsis seedlings were quickly weighed, boiled for 3 min in extraction solution (propanol:HCl:H2O = 18:1:81), and incubated at room temperature in the dark for at least 2 h. Samples were centrifuged, and the absorbance of the supernatants was measured at 535 and 650 nm. Anthocyanin content was calculated as (A535 − 2.2A650)/g·FW.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: MYB75 (AT1G56650), MPK3 (AT3G45640), MPK4 (AT4G01370), MPK6 (AT2G43790), MPK11 (AT1G01560), RD29A (AT5G52310), ERD10 (AT1G20450), LEA14 (AT1G01470), KIN1 (AT5G15960), DFR (AT5G42800), LDOX (AT4G22880), UF3GT (AT5G54060), EGL3 (AT1G63650), ACTIN8 (AT1G49240), COP1 (AT2G32950), MKS1 (AT3G18690), PHYA (AT1G09570), PHYB (AT2G18790), CRY1 (AT4G08920), and CRY2 (AT1G04400).
Supplemental Data
Supplemental Figure 1. Characterization of the myb75-c Mutants Generated Using the CRISPR-Cas9 System.
Supplemental Figure 2. Expression Levels of General Stress Genes under Low Light and Moderate High Light Conditions.
Supplemental Figure 3. Mutations of MPK4 Lead to Compromised Anthocyanin Accumulation.
Supplemental Figure 4. MPK11 Is Not Involved in High Light-Induced Anthocyanin Accumulation.
Supplemental Figure 5. R84K Mutation of MYB75 Does Not Change Its Interaction with EGL3 in Yeast.
Supplemental Figure 6. Specific Mutations of MYB75 Do Not Affect Its DNA Binding Activity.
Supplemental Figure 7. LC-MS/MS Analysis of in Vitro Phosphorylation of MYB75.
Supplemental Figure 8. Expression Levels of the MYB75 Variant Transgenes in the mpk4-3 Mutant.
Supplemental Figure 9. Far-Red Light Does Not Activate MAPKs in the phyA phyB Mutant.
Supplemental Figure 10. MPK4 Does Not Interact with MYB90/PAP2 in Yeast.
Supplemental Table 1. Phosphopeptides Identified in MYB75 by Mass Spectrometric Analysis.
Supplemental Table 2. Sequences of the Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Dongqing Xu for the cop1 mutant seeds, Justin Lee for the mpk11 seeds, Yule Liu for the BiFC vectors, and Peng Xue for mass spectrometry data analysis. This work was supported by grants to J.-L.Q. from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11030500) and the National Natural Science Foundation of China (31371929).
AUTHOR CONTRIBUTIONS
J.-L.Q. conceived the research. S.L. and W.W. performed most of the experiments with help from C.W., R.W., J.G., and K.Y. All authors discussed the results and analyzed data. J.-L.Q., J.M., S.L., and W.W. wrote the manuscript with contributions from all authors.
Glossary
- TF
transcription factors
- MAPK
mitogen-activated protein kinase
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
Footnotes
Articles can be viewed without a subscription.
References
- Albert N.W., Lewis D.H., Zhang H., Irving L.J., Jameson P.E., Davies K.M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60: 2191–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Berriri S., Garcia A.V., Frei dit Frey N., Rozhon W., Pateyron S., Leonhardt N., Montillet J.L., Leung J., Hirt H., Colcombet J. (2012). Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L. (1999). Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 70: 1–9. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226. [DOI] [PubMed] [Google Scholar]
- Cominelli E., Gusmaroli G., Allegra D., Galbiati M., Wade H.K., Jenkins G.I., Tonelli C. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165: 886–894. [DOI] [PubMed] [Google Scholar]
- Dare A.P., Schaffer R.J., Lin-Wang K., Allan A.C., Hellens R.P. (2008). Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Droillard M.J., Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 574: 42–48. [DOI] [PubMed] [Google Scholar]
- Feilner T., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4: 1558–1568. [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Qiu J.L., Petersen K., Petersen M., Mundy J. (2008). Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc. 2008: pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Lelandais M., Kunert K.J. (1994). Photooxidative stress in plants. Physiol. Plant. 92: 696–717. [Google Scholar]
- Galvão V.C., Fankhauser C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34: 46–53. [DOI] [PubMed] [Google Scholar]
- Gan Y., Li H., Xie Y., Wu W., Li M., Wang X., Huang J. (2014). THF1 mutations lead to increased basal and wound-induced levels of oxylipins that stimulate anthocyanin biosynthesis via COI1 signaling in Arabidopsis. J. Integr. Plant Biol. 56: 916–927. [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Gould K.S. (2004). Nature’s swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57: 761–780. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Xu Y., Quail P.H. (1998). SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton T.A., Cornish E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Kholodenko B.N. (2006). Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 7: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Yamamoto Y.Y., Seki M., Sakurai T., Sato M., Abe T., Yoshida S., Manabe K., Shinozaki K., Matsui M. (2003). Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 77: 226–233. [DOI] [PubMed] [Google Scholar]
- Kong Q., Qu N., Gao M., Zhang Z., Ding X., Yang F., Li Y., Dong O.X., Chen S., Li X., Zhang Y. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., Takahashi Y., Hirt H., Machida Y. (2010). The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22: 3778–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Shropshire W., Mohr H. (1971). An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol. 47: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. (2015). Mitogen-activated protein kinase kinase 3 is required for regulation during dark-light transition. Mol. Cells 38: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Jia K.P., Lian H.L., Yang X., Li L., Yang H.Q. (2014). Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 454: 78–83. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang Q., Liu Y., Zhao X., Imaizumi T., Somers D.E., Tobin E.M., Lin C. (2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. USA 110: 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.P., Humphries S., Falkowski P.G. (1994). Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. 45: 633–662. [Google Scholar]
- Maier A., Hoecker U. (2015). COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 10: e970440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., Rubio V., Uhrig J.F., Hülskamp M., Hoecker U. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74: 638–651. [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Mockler T.C., Guo H., Yang H., Duong H., Lin C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082. [DOI] [PubMed] [Google Scholar]
- Neill S., Gould K.S. (2000). Optical properties of leaves in relation to anthocyanin concentration and distribution. Can. J. Bot. 77: 1777–1782. [Google Scholar]
- Ni W., Xu S.L., Chalkley R.J., Pham T.N., Guan S., Maltby D.A., Burlingame A.L., Wang Z.Y., Quail P.H. (2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Rowan D.D., Cao M., Lin-Wang K., Cooney J.M., Jensen D.J., Austin P.T., Hunt M.B., Norling C., Hellens R.P., Schaffer R.J., Allan A.C. (2009). Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 182: 102–115. [DOI] [PubMed] [Google Scholar]
- Schweighofer A., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V., Raghuram B., Sinha A.K., Chattopadhyay S. (2014). A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26: 3343–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., Zhang K., Liu J., Xi J.J., Qiu J.L., Gao C. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688. [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M.Z., Xie D.Y. (2010). Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta 231: 1385–1400. [DOI] [PubMed] [Google Scholar]
- Shi M.Z., Xie D.Y. (2014). Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 8: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1982). Light quality, photoperception, and plant strategy. Annu. Rev. Plant Physiol. 33: 481–518. [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn W.J., Wand S.J.E., Holcroft D.M., Jacobs G. (2002). Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 155: 349–361. [DOI] [PubMed] [Google Scholar]
- Su L., Li A., Li H., Chu C., Qiu J.L. (2013). Direct modulation of protein level in Arabidopsis. Mol. Plant 6: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152. [DOI] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139: 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Jarpe M.B., Johnson G.L. (1999). Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79: 143–180. [DOI] [PubMed] [Google Scholar]
- Wu H.Y., Liu K.H., Wang Y.C., Wu J.F., Chiu W.L., Chen C.Y., Wu S.H., Sheen J., Lai E.M. (2014). AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Chen J., Wang Q., Yang Y. (2014). Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 26: 3077–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Cao Q., Zhang Q., Qin L., Jia W., Zhang J. (2013). MKK5 regulates high light-induced gene expression of Cu/Zn superoxide dismutase 1 and 2 in Arabidopsis. Plant Cell Physiol. 54: 1217–1227. [DOI] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Huq E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20: 641–650. [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.