Comparative analyses of the maize reference B73 genome assembly and the newly assembled PH207 genome and their transcriptomes provide insights into variation between heterotic groups of elite maize.
Abstract
Intense artificial selection over the last 100 years has produced elite maize (Zea mays) inbred lines that combine to produce high-yielding hybrids. To further our understanding of how genome and transcriptome variation contribute to the production of high-yielding hybrids, we generated a draft genome assembly of the inbred line PH207 to complement and compare with the existing B73 reference sequence. B73 is a founder of the Stiff Stalk germplasm pool, while PH207 is a founder of Iodent germplasm, both of which have contributed substantially to the production of temperate commercial maize and are combined to make heterotic hybrids. Comparison of these two assemblies revealed over 2500 genes present in only one of the two genotypes and 136 gene families that have undergone extensive expansion or contraction. Transcriptome profiling revealed extensive expression variation, with as many as 10,564 differentially expressed transcripts and 7128 transcripts expressed in only one of the two genotypes in a single tissue. Genotype-specific genes were more likely to have tissue/condition-specific expression and lower transcript abundance. The availability of a high-quality genome assembly for the elite maize inbred PH207 expands our knowledge of the breadth of natural genome and transcriptome variation in elite maize inbred lines across heterotic pools.
INTRODUCTION
Access to genome assemblies has ushered in a new era in plant biology beginning with the genome assembly of the model species Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000). Since that time, over 80 Arabidopsis genome assemblies have been completed, revealing genome content and allelic variation, the importance of genotype-specific annotation, and the regulation of gene expression (http://1001genomes.org; Gan et al., 2011). Rice (Oryza sativa) is the only plant species other than Arabidopsis with a “gold standard” genome assembly (International Rice Genome Sequencing Project, 2005). As with Arabidopsis, multiple de novo rice genome assemblies have now been generated, and through these assemblies, several megabases of sequences present in only one individual have been identified, many of which were annotated to contain genes (Schatz et al., 2014). Beyond these studies, extensive structural variation, including differences in gene copy number and presence/absence variation, has been documented in many species (Brunner et al., 2005; Morgante et al., 2007; Ossowski et al., 2008; Gore et al., 2009; Springer et al., 2009; Weigel and Mott, 2009; Lai et al., 2010; Swanson-Wagner et al., 2010; Cao et al., 2011; Gan et al., 2011; Chia et al., 2012; Hansey et al., 2012; Anderson et al., 2014; Hirsch et al., 2014; Schatz et al., 2014; Hirakawa et al., 2015; Hardigan et al., 2016).
In maize (Zea mays), comparative genome hybridization revealed pervasive copy number variation (CNV) and presence/absence variation (PAV) between two heterotic inbred lines (B73 and Mo17), including an ∼2.6-Mb region on chromosome 6 that was present in B73 and absent in Mo17 (Springer et al., 2009; Beló et al., 2010). In an expanded panel of 34 maize and teosinte lines examined using comparative genome hybridization, nearly 4000 instances of CNV/PAV were observed (Swanson-Wagner et al., 2010). Likewise, the first-generation maize HapMap (haplotype map) estimated that the B73 genome represents only ∼70% of the total low-copy sequence in the maize pan-genome (Gore et al., 2009), and the second-generation maize HapMap also identified pervasive structural variation (Chia et al., 2012). Resequencing of six elite maize inbred lines identified several hundred genes that exhibit PAV, and in some cases, show heterotic group specificity (Lai et al., 2010). Transcriptome profiling of maize lines has also revealed extensive differences in transcriptome content. Profiling of 21 diverse inbred lines identified 1321 nonreference loci, of which 145 were heterotic group specific (Hansey et al., 2012). Transcript profiling of seedling tissue from 503 diverse inbred lines was used to characterize the maize pan-genome and pan-transcriptome (Hirsch et al., 2014), and nearly 9000 novel loci absent in the B73 reference genome assembly were identified. Furthermore, a genome-wide association study using transcript presence/absence and transcript abundance as the independent variable showed complementary and unique loci compared with those obtained from a genome-wide association study using single nucleotide polymorphism (SNP) markers (Hirsch et al., 2014).
Maize has been intensively bred in the hybrid seed industry for nearly a century. Breeding efforts in maize focus on improving inbred lines that combine well to produce high-yielding hybrids and have resulted in the development of distinct heterotic germplasm pools. Indeed, elite maize inbred lines have been generated that combine to form hybrid genotypes that yield 70-fold more than the open pollinated varieties from which they originated (Troyer, 2006). Increases in yield performance continue at a consistent rate, indicating persistence of extensive genetic variation in elite maize germplasm. Maize heterotic pools consist primarily of a stiff stalk heterotic pool from which the existing reference maize genome assembly, B73, is derived and non-stiff stalk heterotic pools. In the past 20 years, Iodent germplasm has been a main contributor to the non-stiff stalk heterotic pool. Iodent germplasm used today originated from Pioneer Hi-Bred International germplasm in the 1940s (Troyer, 1999) and now permeates across proprietary breeding programs. A key founder line to Iodent germplasm is the inbred line PH207. The hybrid generated by crossing B73 and PH207 produces high parent heterosis across a number of phenotypic traits (Table 1). Among the 788 U.S. Plant Variety Protection/utility patent commercial maize inbred lines registered between 2009 and 2013, B73 (stiff stalk) was in the pedigree of 327 lines and PH207 (Iodent) was in the pedigree of 441 lines (Mikel, 2011). Among stiff stalks, 92% had B73 as an ancestor, and 91% of the non-stiff stalk lines were descendants of the Iodent PH207.
Table 1. Phenotypic Variation between B73, PH207, and the F1 Reciprocal Hybrids between B73 and PH207.
| Trait | B73 | B73 × PH207 | PH207 × B73 | PH207 |
|---|---|---|---|---|
| Fresh aboveground biomass (g) | 0.93 | 1.43 | 1.25 | 0.97 |
| Fresh belowground Biomass (g) | 0.83 | 1.09 | 1.33 | 0.91 |
| Plant height (cm) | 12.84 | 17.32 | 17.30 | 13.82 |
| Root length (cm) | 24.66 | 29.84 | 30.01 | 26.97 |
Phenotypic measurements were collected on greenhouse-grown plants at the seedling vegetative 1 developmental stage (Abendroth et al., 2011).
A de novo genome assembly of the maize inbred line B73 was released in 2009 (Schnable and Ware et al., 2009), and to date this has been the only publicly available complete maize whole genome assembly. Previous studies of diversity in maize have been conducted in the context of this single reference genome assembly, which has the potential to introduce a reference bias. Here, we present the assembly of elite inbred line PH207, as well as a comparative analysis between the genomes and transcriptomes of these elite maize inbred lines, to further our understanding of natural variation present in elite maize inbred lines that have been selected to combine and produce high-yielding hybrids. This study provides a high-quality assembly to interrogate genome and transcriptome variation in maize, an important crop and model species, and to begin to understand how this genomic variation contributes to the phenotypic diversity and heterosis that is observed between elite inbred lines.
RESULTS
Assembly of the PH207 Genome
Over 550 Gb of short-read sequences from the inbred line PH207 were generated through whole-genome shotgun sequencing using paired-end (PE), mate-pair (MP), and TruSeq synthetic long-read genomic libraries with estimated fragment sizes ranging from 330 bp to 15 kb (Supplemental Table 1). On the basis of 23-mer analysis, the genome size of PH207 was estimated to be ∼2.45 Gb (Supplemental Figure 1), which is comparable to the 2.3 Gb estimated genome size of B73 reported by the maize genome sequencing consortium (Schnable et al., 2009). Using the estimated size of 2.45 Gb, the short-read sequences generated for the PH207 assembly provide ∼230× theoretical coverage of the genome.
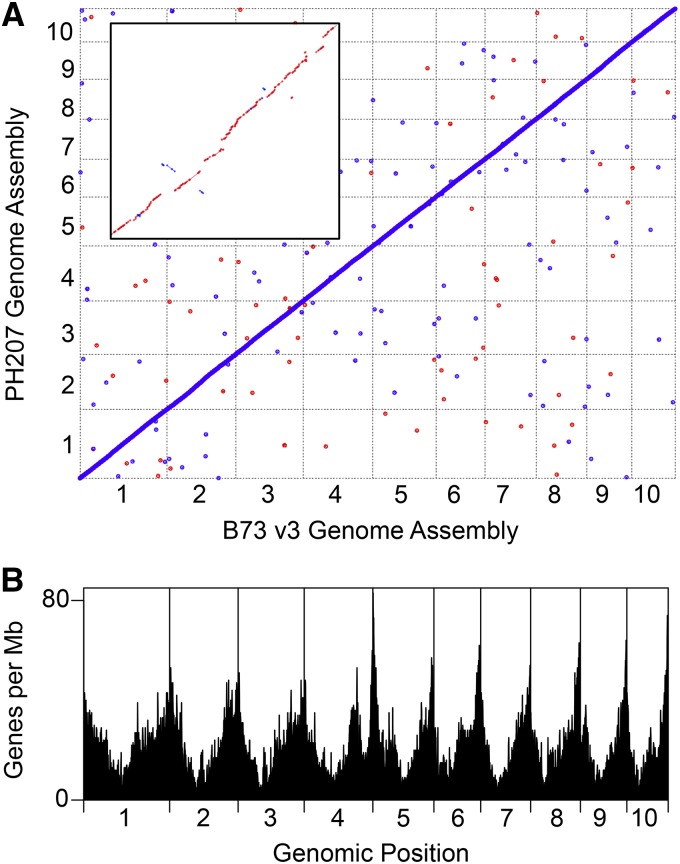
A de novo assembly with an N50 scaffold size of ∼654 kb with ∼16% unfilled gaps was generated (Supplemental Table 2). Most of the gaps spanned repetitive regions that could not be assembled into scaffolds. The total size of the assembly was 2.1 Gb, of which 1.7 Gb was ungapped genomic sequence and 0.4 Gb was unfilled gaps. The PH207 de novo assembled scaffolds were then organized into pseudomolecules based on the B73 reference genome assembly using alignment of the scaffolds to the B73 assembly. In total, 1.99 Gb of the PH207 assembly was placed into the pseudomolecules based on alignment to the B73 reference assembly. Lu et al. (2015) described a genotyping-by-sequencing anchoring pipeline that utilized linkage disequilibrium mapping coupled with machine learning to generate a set of 4.4M tags that can be used as genetic anchors. These 4.4M tags provided a median resolution of ∼10 kb (average tag every 500 bp) and have been shown to be accurate for a given line 95% of the time, with the other 5% most likely reflecting a species consensus position that is different from the line. Overall, ∼54% of the anchors are within 10-kb regions of their true position, 95% within 1 Mb, and 98.6% within 10 Mb (Lu et al., 2015). To assess the quality of the assembly, the PH207 de novo assembled scaffolds were processed using the pan-genome genetic anchor pipeline (Lu et al., 2015) to flag scaffolds that may be the result of chimeras from disparate chromosome locations; ∼21% of the tags could be aligned. Using this method, as well as the alignments of the PH207 scaffolds to the B73 genome, only 120 scaffolds were identified as putative misassemblies and were corrected by splitting them at the junction of the putative misassembly. This marginally decreased the assembly N50 scaffold size to ∼630 kb (Supplemental Table 2). This final assembly has been named Zm-PH207-REFERENCE_NS-UIUC_UMN-1.0 and is available for download at the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.8vj84) and is also available at the Maize Genetics and Genomics Database (http://www.maizegdb.org) and at Phytozome (https://phytozome.jgi.doe.gov). Comparison of the B73 reference genome assembly and the final PH207 genome assembly revealed numerous structural variants between the two genomes. However, few large gaps in the assemblies were observed (Figure 1A).
Figure 1.
Summary of the PH207 Genome Assembly.
(A) Genome-wide comparison of the B73 genome assembly and the PH207 genome assembly. The inset shows a zoomed-in view of the first megabase of chromosome one from each genome. Forward matches, plotted first, are colored red and reverse matches are colored blue.
(B) Density of annotated genes in the PH207 genome assembly.
Several approaches were used to further validate the PH207 assembly completeness and error rate. Genomic sequence reads used to generate the assembly were aligned back to the assembly, and 97.1% of the paired-end short-read sequences could align to at least one position in the genome, demonstrating the completeness of the assembly. For the B73 reference genome assembly, comparable mapping was observed, with 98.0% of paired-end short-read sequences aligning to at least one position in the genome. From these alignments, only 33,812 SNPs or insertion/deletions (InDels) were identified from alignment of the paired-end short-read sequences to the PH207 assembly, which equates to <0.002% of the total genome assembly and indicates a low error rate in the assembly. To evaluate completeness of the genic space, two approaches were used, alignment of RNA-seq reads to the assembly and the Core Eukaryotic Genes Mapping Approach pipeline (Parra et al., 2007). From alignment of RNA-seq reads from six PH207 tissues (leaf blade, root cortical parenchyma, germinating kernel, root tip, whole seedling, and root stele), 96.3% of reads on average could map to at least one position in the genome, and 87.1% mapped to a single unique position (Supplemental Figure 2). Alignment of B73 RNA-seq reads from these same tissues to the B73 genome assembly resulted in 92.2% of reads mapping to at least one position on average. The Core Eukaryotic Genes Mapping Approach pipeline was run for both the PH207 and B73 assemblies and revealed similar representation of the gene space between the assemblies (PH207, 91.5% complete genes and 99.2% partial genes; B73, 91.5% complete genes and 98.4% partial genes). Taken together, these analyses demonstrate the high quality nature of the PH207 genome assembly and support its utility in downstream comparative analyses with the existing B73 reference genome assembly.
Gene Annotation of the PH207 Genome
Structural gene annotation of the PH207 genome assembly was performed on all PH207 scaffolds greater than 500 bp using the MAKER-P pipeline (Campbell et al., 2014b). De novo PH207 transcript assemblies of RNA-seq reads from six different tissues (leaf blade, root cortical parenchyma, root stele, germinating kernel, root tip, and whole seedling) were used as transcript evidence to aid in gene identification. Additionally, the predicted rice proteome and UniProtKB/Swiss-Prot plant proteins (minus maize proteins) were aligned to the PH207 genome assembly and used as evidence for gene prediction (Kawahara et al., 2013; UniProt Consortium, 2014). Evidence from other maize accessions was not used in this annotation to ensure that the annotation reflected genotype-specific annotation to reduce bias in downstream comparisons between the PH207 and B73 gene space. In total, we identified 40,557 high-quality gene models in PH207 supported by aligned transcript evidence, protein evidence, or the presence of a Pfam domain (Finn et al., 2014). These gene models were distributed throughout the genome (Figure 1B) in a distribution similar to that observed in the B73 reference genome assembly (Schnable and Ware et al., 2009; Law et al., 2015). The annotation edit distance analysis of the predicted gene set indicated that the gene annotations were generally well supported (Supplemental Figure 3) (Eilbeck et al., 2009; Yandell and Ence, 2012). The annotated transcripts from the PH207 gene set had an average length of 1294 bp, with an N50 size of 1685 bp, slightly smaller than the average annotated transcript in the B73 v3 filtered gene set (1559 bp with an N50 size of 1910). This difference is likely due to better representation of the untranslated regions in B73 resulting from the extensive transcript evidence that was used for the B73 annotation. Indeed, the average predicted protein length of PH207 (353 amino acids) is nearly identical to that of B73 (352 amino acids), suggesting that the PH207 sequence represents a similar coding sequence compared with B73. Regardless, the number of annotated genes in PH207 was very similar to that of the B73 v3 gene set, which contains 39,301 nuclear genes.
Putative functional annotation of the predicted PH207 gene models was generated using transitive annotations based on the best BLAST hit to the Arabidopsis (33,388), Brachypodium distachyon (37,364), rice (37,201), and Sorghum bicolor (37,998) reference genome annotations and the UniRef100 database (38,710). The functional annotations for many of the genes in these plant species are derived from Arabidopsis annotation directly or indirectly. Thus, a plurality of agreement among data sets can in some cases be misleading for assigning functional annotations. Additionally, the multigene family nature of many genes in plant species can result in misannotation based on transitive annotation from best BLAST hits. Gene Ontology terms were also assigned to 18,430 of the PH207 gene models (Supplemental Data Set 1).
Elite Maize Inbred Lines Exhibit Extensive Genome Content Variation Including Massive Expansion of Gene Families
Access to two high-quality genome assemblies allowed us to comprehensively explore genome content variation between heterotic maize inbred lines, both in terms of presence/absence as well as copy number variation. To identify dispensable genes between these two lines, representative transcript sequences (longest transcript) from each genotype were aligned to both genome assemblies. Within the subset of transcripts that could map to their cognate genome, 5291 B73 transcripts did not align to PH207 and 5029 PH207 transcripts did not align to B73. While both the B73 assembly and our PH207 assembly are high quality, these assemblies are not 100% complete (Schnable and Ware et al., 2009; Lai et al., 2010; Hansey et al., 2012; Hirsch et al., 2014). As such, direct comparison of the genome assemblies overestimates the number of PAVs between the two inbred lines. Indeed, approximately half of the PAVs identified through direct comparison of the assemblies had sequence reads from whole-genome sequencing that did not support the PAV classification.
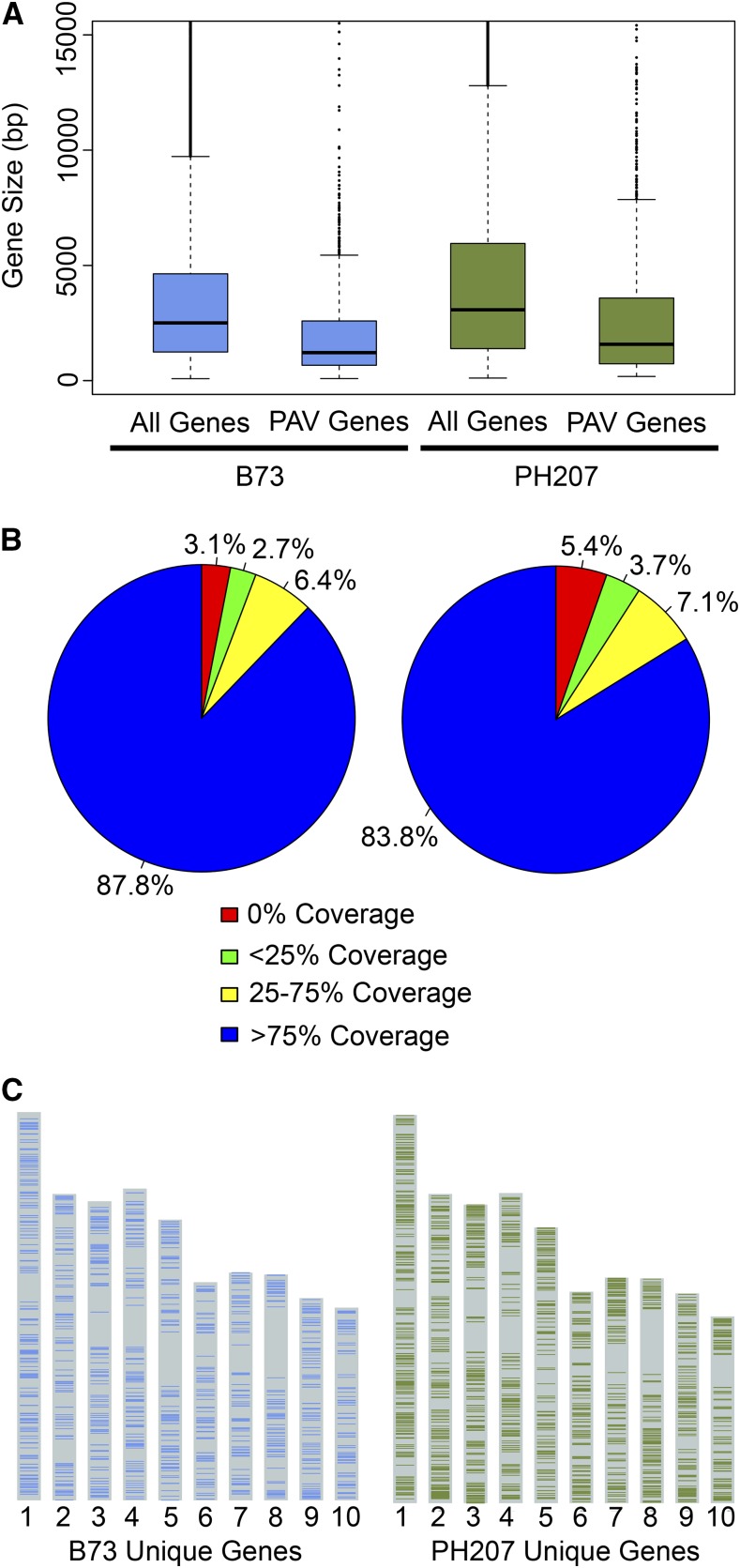
To determine how many genes represent sequences absent in their respective genomes and to provide a conservative estimation of genome PAV between B73 and PH207, all genes from each genotype were categorized as present or absent based on the resequencing data (∼11× average realized coverage from B73 and ∼5× average realized coverage from PH207). If a gene had greater than 75% coverage in its cognate genotype and less than 25% coverage in the reciprocal genotype at a minimum of 1× coverage, it was considered to be a genotype-specific gene. This is a very strict criterion and eliminated a number of genes that are unmappable with short-read sequencing technologies; however, it provides a high confidence method for identifying dispensable genes. Based on this criterion, 1169 genes were B73 specific and 1545 were PH207 specific. An enrichment of cellular response to stress was observed among the enriched Gene Ontology terms within this set of PAV genes (Supplemental Data Set 2). Additionally, dispensable genes tended to be shorter than genes that were present in both genotypes (Figure 2A). While a large number of genes demonstrated clear PAV, there are many genes that appear to be partial deletions in the reciprocal genome (Supplemental Figure 4). Indeed, 12.2% of the B73 genes were partially deleted in PH207 and 16.2% of the PH207 genes were partially deleted in B73 (Figure 2B).
Figure 2.
Genome Content Distribution in Elite Maize Inbred Lines B73 and PH207.
(A) Distribution of gene size for all B73 and PH207 genes and genes showing PAV. The whiskers are plotted at a maximum of 1.5 times the interquartile range away from the end of the box in each direction.
(B) Distribution of partial gene deletions in B73 (right) and PH207 (left) genes relative to the genome in which the gene was annotated based on the ratio of coverage from resequencing data.
(C) Genome-wide distribution of B73 and PH207 unique genes.
It has previously been suggested that dispensable genes provide an extreme case of complementation, supporting the dominance model of heterosis, in which inferior recessive alleles contributed by one parent are complemented by superior dominant alleles contributed by the other parent (Fu and Dooner, 2002; Lai et al., 2010; Swanson-Wagner et al., 2010). Thus, there should be an enrichment of dispensable genes in low recombination regions of the genome (i.e., near centromeres). A significant enrichment of PAV genes in the pericentromeric regions (30.9% of PAV genes) relative to all genes in the pericentromeric regions (25.9%) was observed (χ2 P value > 0.0001). In terms of total gene number, more genes annotated as PAV between B73 and PH207 were located within high recombination regions of the genome (Figure 2C).
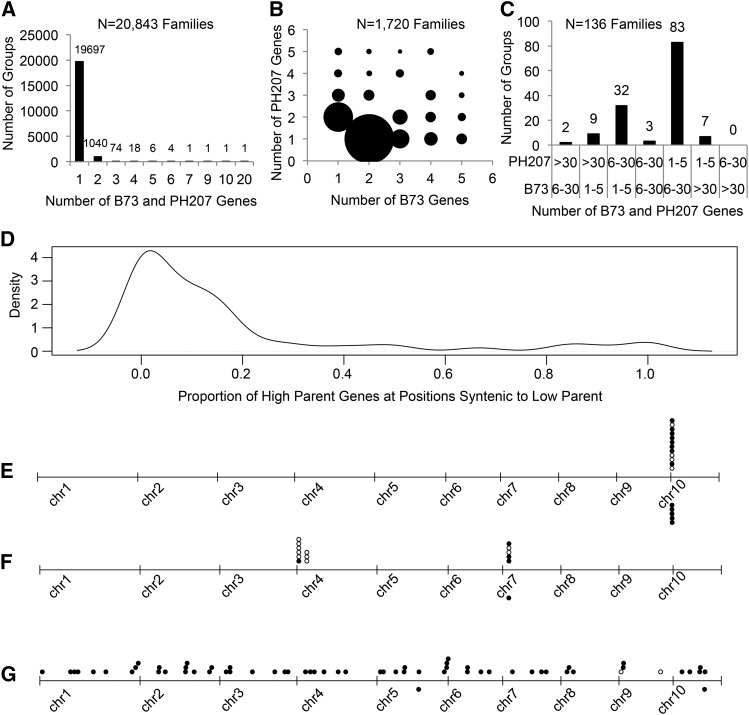
Genome content variation can also arise from differences in CNV leading to variation in gene family size between genotypes. To evaluate the difference in gene family size between B73 and PH207, genes from both genotypes were clustered using the OrthoMCL algorithm (Li et al., 2003; Chen et al., 2007). As was expected, consistent gene family size was observed for the majority of the genes in both genomes, with 19,697 genes showing a direct one-to-one relationship (Figure 3A). Another 1720 families showed only modest changes in gene family size (sizes between 1 and 5 in both individuals; Figure 3B). Interestingly, substantial expansion and contraction of gene families was observed for 136 families, where large expansion was defined as being present in both individuals but having a difference of five or more family members between the genotypes (Figure 3C). In fact, one family in PH207 contained 214 family members, while B73 had only six genes in this family. Within the genotype that had the expanded family, on average 39.7% of the genes within the family were expressed in at least one of six tissues tested (leaf blade, root cortical parenchyma, root stele, germinating kernel, root tip, and whole seedling) when requiring reads to align uniquely. This number is likely an underestimation of the true frequency of expression within these families due to the limitations of unique alignments with short reads in large gene families (Hirsch et al., 2015). Across the families that showed large changes in gene family size, an enrichment of Gene Ontology terms related to stress response was observed, among other functions (Supplemental Data Set 2). A large proportion of the additional gene family members were located in nonsyntenic positions relative to the copies found in the genotype with the smaller family size (Figure 3D). There are a number of helitrons that are currently annotated as genes but are likely nonfunctional and susceptible to deletion. These nonsyntenic expanded families might be a reflection of the action of helitrons. However, examples of expanded families that were completely clustered in a single location (Figure 3E), both clustered and dispersed (Figure 3F), and completely dispersed (Figure 3G) were all observed, indicating that these results cannot be explained simply by the annotation of helitrons or fragments from helitrons as genes. The number of introns was evaluated to determine if the additional copies were primarily the result of retrotransposed transcripts. Within each class (clustered, clustered and dispersed, and dispersed), both genes without introns as well as genes with at least one intron were observed (Figures 3E to 3G), indicating that retrotransposons are not the primary mechanism driving the expansion of these gene families.
Figure 3.
Variation in Gene Family Size in Elite Maize Inbred Lines B73 and PH207.
(A) Distribution of gene families of the same size in B73 and PH207.
(B) Distribution of gene families with moderate changes in gene family size between B73 and PH207.
(C) Distribution of gene families with extreme changes in gene family size between B73 and PH207.
(D) Density plot of the proportion of genes in the inbred line (B73 or PH207) that has expanded gene family members in syntenic positions relative to the reciprocal inbred line, with fewer copies for gene families with extreme changes.
(E) Physical location of genes in a family that had only clustered expansion.
(F) Physical locations of genes in a family that had both clustered and distributed expansion.
(G) Physical locations of genes in a family that had distributed expansion only. In plots (E) to (G), B73 genes are above the axis, and PH207 genes are below the axis. White circles represent genes with no introns, and black circles represent genes with one or more introns.
Genome Content Variation Drives Transcriptional Variation between Elite Maize Inbred Lines
The availability of two maize genome sequence assemblies provided an opportunity to evaluate sources of transcriptional variation between genotypes. We evaluated variation in the transcriptome profiles of six tissues (described above) in the context of both the B73 and PH207 gene models (Supplemental Data Set 3). In total, 84.9% of genes were expressed in at least one of the tissues in at least one of the two genotypes (Table 2). A relatively large number of genes (9.4%) were consistently expressed in a genotype-specific manner, while another 23.3% were genotype specific only in a subset of tissues. Furthermore, 52.5% of genes showed quantitative differential expression between the two genotypes in at least one tissue, and 4.5% of genes showed quantitative differential expression between the two genotypes across all of the tissues. In total, 69.8% of genes showed either quantitative or qualitative variation in transcript abundance in at least one of the six tissues that were evaluated, demonstrating that extensive expression variation is present in the transcriptomes of elite maize inbred lines.
Table 2. Transcriptome Expression Variation across Six Tissues in the Context of B73 and PH207 Annotated Genes.
| B73a | PH207a | |
|---|---|---|
| Expression competence | ||
| Never expressed | 6,180 | 5,885 |
| Expressed in ≥1 tissue | 33,121 | 34,672 |
| Differential expression between genotypes | ||
| Always differentially expressed | 1,655 | 1,968 |
| Sometimes differentially expressed | 18,913 | 19,363 |
| Never differentially expressed | 18,733 | 19,226 |
| Genotype-specific expression pattern | ||
| Always genotype specific if expressed | 3,532 | 3,964 |
| Sometimes genotype specific | 8,719 | 9,910 |
| Never genotype specific | 27,050 | 26,683 |
B73 and PH207 RNA-seq reads were mapped to both their cognate genome and the reciprocal genome, and expression was summarized as read counts in each of the mapping scenarios. Tissues included in this analysis were leaf blade, root cortical parenchyma, root stele, germinating kernel, root tip, and whole seedling.
The total number of genes in each genotype are 39,301 (B73) and 40,557 (PH207).
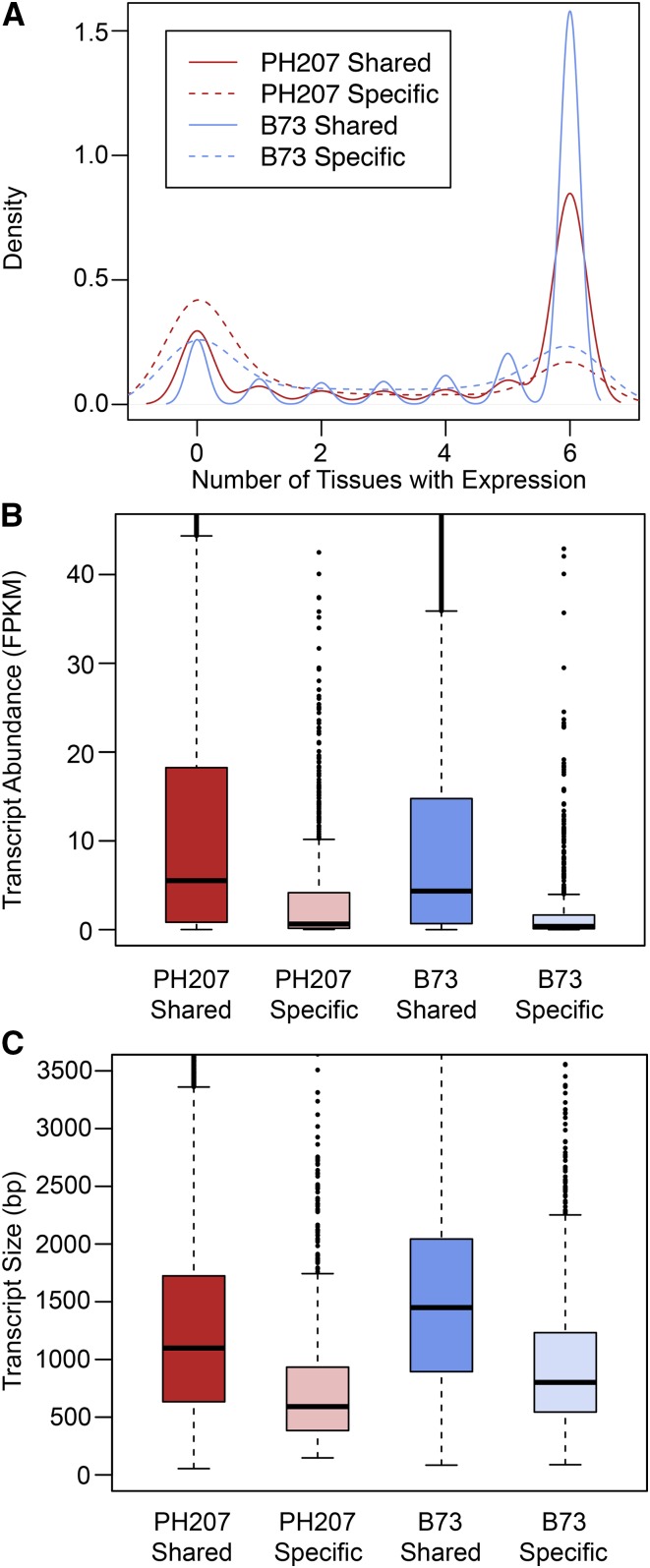
The comparison of the B73 and PH207 genome assemblies provided an opportunity to evaluate the contribution of various types of genomic differences to the extensive transcriptional variation that is present in highly selected complementary maize inbred lines. In total, 9.0% of B73 models and 9.8% of PH207 models showed genotype-specific expression across all of the tissues. For 11.6% of these genes, genomic-level PAV was also observed and was the basis for the observed transcriptional variation. Genes showing genomic-level PAV had more tissue-specific expression (Figure 4A) and had lower average expression when compared with genes that were present in the genomes of both individuals (Figure 4B). This observation is further supported using the B73 gene atlas, which includes transcript abundance estimates for 79 B73 tissues throughout development (Supplemental Figure 5; Stelpflug et al., 2016). Small transcripts (<300 bp) are undersampled and therefore have lower estimated transcript abundance due to technical biases resulting from size selection during library preparation (Hirsch et al., 2015). However, based on the distribution of transcript size for genotype-specific and shared genes, the lower observed expression of genotypic-specific genes was not a product of this bias (Figure 4C). The number of genes that were not expressed in any tissue was slightly higher for single exon genes (29.0%) relative to the total number of nonexpressed genes (18.5%), and there are therefore other contributing factors to this observed distribution.
Figure 4.
Distribution of Expression in B73 and PH207 for Shared and Genotype-Specific Genes.
For each B73 tissue used in this analysis, the average of three biological replicates was used, and the average of two biological replicates was used for PH207 tissues. For both B73 and PH207, the biological replicates consisted of pooled tissue from three plants.
(A) Distribution of number of tissues with expression.
(B) Distribution of average transcript abundance measured in fragments per kilobase of exon models per million fragments mapped (FPKM). The whiskers are plotted at a maximum of 1.5 times the interquartile range away from the end of the box in each direction. For both (A) and (B), expression was measured in leaf blade, root cortical parenchyma, germinating kernel, root tip, whole seedling, and root stele.
(C) Distribution of transcript size in each class of genes.
Extensive quantitative variation in transcript abundance was also observed. Multiple levels of genomic variation that were observed between B73 and PH207 could underlie this variation, such as partial fragmentation of genes (Figure 2B; Supplemental Figure 4), variation in gene family size that may lead to neo- and subfunctionalization at the expression level (Figure 3), and polymorphisms in the promoters between the two genotypes. Of the 5383 B73 and 7991 PH207 genes that showed partial deletions in the reciprocal genome (Figure 2B), 1883 and 2835 were differentially expressed in at least one tissue, respectively. However, transcript abundance estimates and differential expression were calculated assuming common transcript lengths between the genotypes. To test the impact of this on differential gene expression, we calculated coverage-corrected transcript abundance estimates. Based on this correction, 20.5% of the genes that were originally differentially expressed in at least one tissue were no longer differentially expressed in any of the tissues for genes that had 25% or more of the gene model deleted and/or unmappable. These results demonstrate how differences in genome annotation can create biases that can affect downstream analyses such as differential gene expression analysis. Still, a large number of genes that show partial deletion between the two genomes were differentially expressed even after implementing a coverage correction.
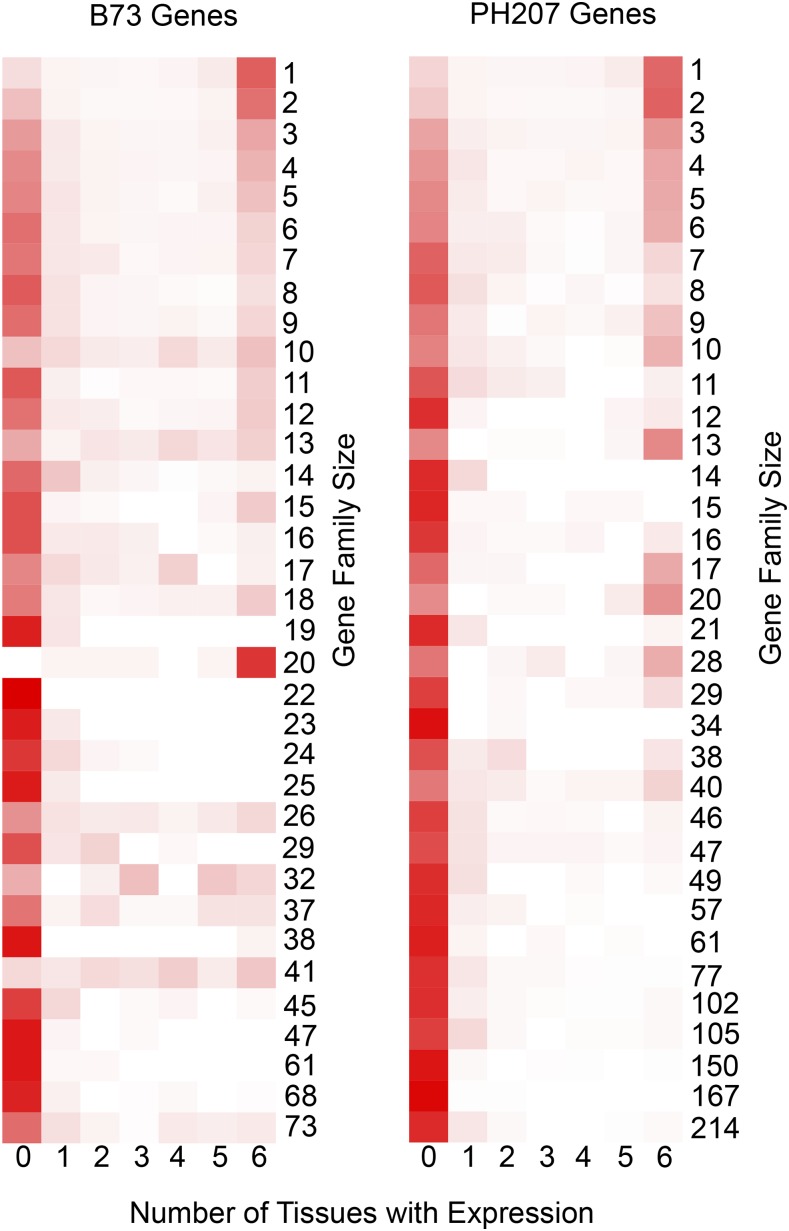
Relationships between copy number variation, transcript abundance, and phenotypic variation have been documented in multiple plant species on a single gene basis (Sutton et al., 2007; Gaines et al., 2010; Cook et al., 2012; Maron et al., 2013; Wang et al., 2015). On a genome-wide scale, within both B73 and PH207, we see a relationship between gene family size and the number of tissues in which expression is observed (Figure 5). As family size increases, the number of tissues in which expression is observed tends to decrease. Additionally, 29.6% of genes that were differentially expressed in at least one tissue were from variably sized gene families.
Figure 5.
Relationship between Gene Family Size and Transcriptional Variation.
Gene families were determined using homologous gene clustering with B73 and PH207 gene models. Heat maps show the proportion of genes in families of size N with expression in zero to six tissues, which include leaf blade, root cortical parenchyma, root stele, germinating kernel, root tip, and whole seedling. White color indicates no genes are expressed and red indicates all genes are expressed.
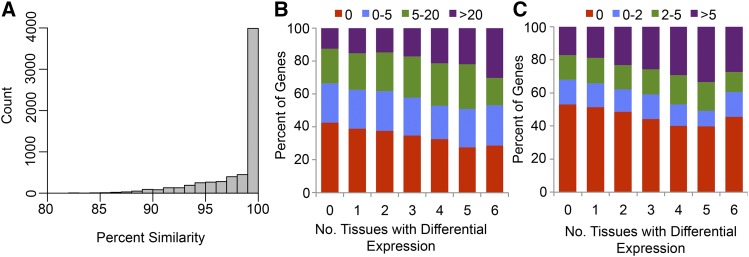
Finally, comparison of the B73 and PH207 assemblies uniquely allowed us to evaluate the impact of context sequence, specifically promoter SNP and InDel variation, on transcriptional variation on a genome-wide scale. For 6379 of the one-to-one genes (Figure 3A), the 1 kb of promoter sequence immediately upstream of the transcription start site could be accurately compared, and the range of sequence similarity between the promoters ranged from 82.1 to 100%, with 4138 having at least one SNP or InDel in the 1 kb of promoter sequence (Figure 6A). Of these 4138, 75.3% of the genes were differentially expressed in at least one tissue. As the number of promoter variants increased, differential expression was observed in a larger number of tissues for both the number of SNPs (Figure 6B) and the number of InDels (Figure 6C), indicating the importance of this local variation in driving transcriptional variation in these elite inbred lines on a genome-wide scale.
Figure 6.
Relationship between Variation in Promoter Sequence and Transcriptional Variation.
(A) Distribution of percentage of similarity of promoter sequences between B73 and PH207 genes in the region 1 kb upstream of the transcription start site for genes that show a one-to-one relationship in the two genome assemblies.
(B) Relationship between number of tissues with differential expression and number of SNPs per kilobase in the promoter sequence.
(C) Relationship between number of tissues with differential expression and number of InDels per kilobase in the promoter sequence.
DISCUSSION
Extensive genome content variation within maize has previously been documented (Gore et al., 2009; Springer et al., 2009; Lai et al., 2010; Swanson-Wagner et al., 2010; Chia et al., 2012; Hansey et al., 2012; Hirsch et al., 2014). However, these studies were limited by ascertainment bias either by investigating only in the context of the reference B73 genome assembly (Springer et al., 2009; Swanson-Wagner et al., 2010) or by the reduced representation approaches that were implemented (Gore et al., 2009; Hansey et al., 2012; Hirsch et al., 2014). Additionally, these studies focused on the broad diversity within maize, while the question of how much variation is present within highly selected elite inbred lines remained unanswered. Access to genome assemblies of the highly selected maize inbred lines B73 and PH207 allowed for a comprehensive analysis of the extensive genome and transcriptome variation present in elite maize germplasm following a century of intense selective pressure to improve grain yield.
The search for large-effect yield genes has been the target of many studies. In some cases, these endeavors have proven fruitful (Park et al., 2014; Weber et al., 2014). However, extensive genome content variation was observed between B73 and PH207 (3.4% of genes show genomic-level PAV). Extensive PAV has previously been observed in other elite maize germplasm (Springer et al., 2009; Lai et al., 2010; Swanson-Wagner et al., 2010; Chia et al., 2012; Hirsch et al., 2014; Lu et al., 2015), and this estimate of PAV between maize inbred lines is quite similar to a previous report where on average 2044 nonreference transcripts were assembled in each of 503 inbred lines (Hirsch et al., 2014). Additionally, we observed extensive transcriptional variation between B73 and PH207, with 32.7% of genes showing transcript PAV in at least one tissue and 52.5% of genes differentially expressed in at least one tissue. The presence of this variation in elite maize inbred lines indicates that there are multiple combinations of genes that can be combined to achieve high-yielding varieties. High yields and heterosis that have been obtained in hybrid varieties to date could be rooted in the extensive genome and transcriptome variation that persists in elite inbred lines from opposite heterotic groups, as has been previously hypothesized (Birchler et al., 2003, 2006, 2010; Song and Messing, 2003; Springer and Stupar, 2007; Lai et al., 2010; Hansey et al., 2012; Yao et al., 2013) and is supported by comparative analysis of B73 and PH207 genome and transcriptome variation.
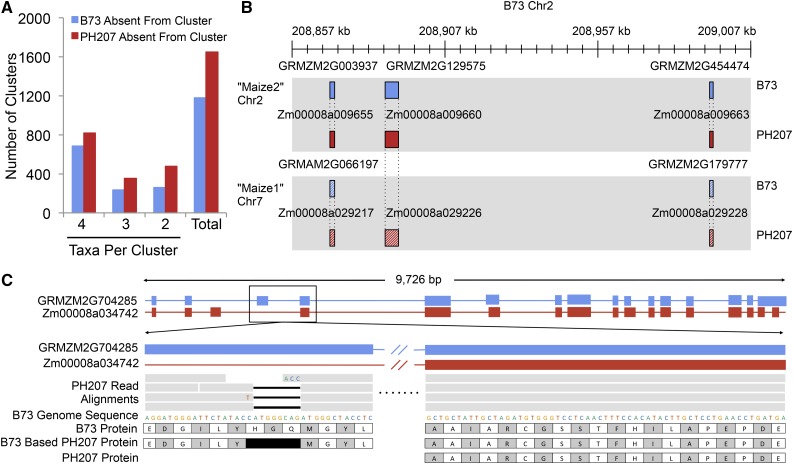
In addition to understanding variation in elite maize inbred lines, comparisons between the B73 and PH207 assemblies also allowed for a number of practical lessons to be learned. Comparative genomics studies between species are often conducted in the context of the reference genome assemblies for each species (Dong et al., 2004; Goodstein et al., 2012; Monaco et al., 2014). However, there is extensive genome content variation between individuals within a species not accounted for in these analyses. Comparison of the B73 and PH207 assemblies revealed the impact of a reference genome centric approach on interspecies comparative genomic studies. Homologous gene clusters were identified between Brachypodium, maize B73, maize PH207, rice, and S. bicolor proteins encoded by the representative transcript (longest) for each gene. Of the subset that included at least one maize protein (24,954 clusters), 2703 clusters contained only a B73 protein and 2025 contained only a PH207 protein sequence (Figure 7A). This finding highlights the importance of expanding comparative genomic studies beyond single reference-to-reference comparisons as the maize pan-genome is substantially larger than the genome sequence represented by B73 and PH207 (Hirsch et al., 2014), and already a large number of new maize homologs have been identified.
Figure 7.
Lessons Learned from Comparisons between Genome Assemblies of Elite Maize Inbred Lines.
(A) Homologous gene clusters between Brachypodium, maize B73, maize PH207, rice, and S. bicolor that contain a maize gene from only B73 or PH207 and at least one other species.
(B) Example of a duplicated gene showing differential fractionation between B73 and PH207. Maize is an ancient tetraploid that has undergone genome fractionation. A previous study identified and classified genes in the B73 genome into two maize subgenomes, Maize1 and Maize2 (Schnable et al., 2011). The middle genes in these syntenic blocks are showing differential fractionation between B73 and PH207, with PH207 retaining both copies and B73 losing the copy in the Maize2 subgenome (gene positions: GRMZM2G003937 chr2:208,869,363..208,870,921; GRMZM2G129575 chr2:208887428..208891800; GRMZM2G454474 chr2:208,993,547..208,994,705; Zm00008a009655 chr2:212,248,102..212,249,027; Zm00008a009660 chr2:212,373,566..212,385,564; Zm00008a009663 chr2: 212,427,728..212,428,519; GRMZM2G066197 chr7:161,658,677..161,660,464; GRMZM2G179777 chr7:161,843,771..161,845,175; Zm00008a029217 chr7:161,948,327..161,949,627; Zm00008a029226 chr7:162,105,478..162,112,256; Zm00008a029228 chr7:162,164,382..162,166,725).
(C) Example of a gene containing a putative frameshift that is compensated by alternative intron/exon boundaries when using annotation specific to the individual. PH207 reads were aligned to the B73 reference genome assembly and putative large effect variants were identified based on the B73 annotation. The putative frameshift variant shown in this figure is corrected for by an alternative gene model in PH207 that was identified using PH207-specific evidence to annotate genes in the PH207 genome assembly.
Within a species, there is a loss of duplicate genes following whole-genome duplication events through a process known as fractionation. Maize is an ancient tetraploid that has undergone genome fractionation. A previous study identified and classified genes in the B73 genome into two maize subgenomes, Maize1 and Maize2, and determined there to be differential fractionation between Maize1 and Maize2 (Schnable et al., 2011). To evaluate the presence of differential fractionation between individuals, the PH207 genome was mined for the presence of missing B73 syntelogs in Maize1 or Maize2. Indeed, there is substantial differential fractionation between B73 and PH207. For example, B73 gene GRMZM2G129575, located in a syntenic block on Chr2 in Maize2, is missing from the Maize1 syntenic block on Chr7 (Schnable et al., 2011). Based on sequence clustering using OrthoMCL, this gene clustered with two PH207 genes, one located on Chr2 and the other on Chr7, both in syntenic blocks with genes neighboring GRMZM2G129575 (Figure 7B). Throughout the genome, over 1265 families were present in two copies in one of the two genomes and one copy in the other genome, likely due to on-going fractionation between these genomes. This supports previous work conducted in the context of the B73 genome assembly (Schnable et al., 2011) showing differential fractionation between individuals following the maize whole genome duplication. This differential fractionation is an important mechanism that generates natural variation within a species and provides the genetic basis for selection to drive genome content variation between maize inbreds from opposite heterotic groups. Additionally, these results highlight the importance of thinking outside of a single reference genome in studies relating to genome fractionation following polyploidization. Interestingly, many of the gene PAVs that were observed between B73 and PH207 were found outside of the Maize1 and Maize2 syntenic blocks, indicating the presence of additional mechanisms driving genome content variation in maize.
Finally, comparison of the two assemblies provided the means to assess two pitfalls that often arise from the use of single reference genomes to make inferences across multiple individuals within a species. There are many biases that exist with RNA-seq for transcript abundance estimates when using a single reference genotype (Hirsch et al., 2015); these biases are further confounded outside of the reference genotype by the alternative gene model structures and partial gene deletions that exist between individuals within a species. For example, transcript abundance estimates for 15.9% of genes were highly influenced by deletion of more than 25% of the gene model in the opposite genome, and 20.5% were falsely identified as differentially expressed in one or more tissues based on this bias. Variable gene models between individuals have also been shown to compensate for large-effect mutations in Arabidopsis (Gan et al., 2011). Based on alignment of PH207 resequencing reads to the B73 genome assembly, 53,377 moderate-to-large effect mutations were identified, of which 12,855 (24.1%) were located in intronic sequences based on the PH207 specific annotation of the PH207 genome assembly (Figure 7C; Supplemental Figure 6).
Access to multiple genome assemblies of elite maize inbred lines has expanded our knowledge on the breadth of natural genome and transcriptome variation that persists in elite maize inbred lines following over a century of intense breeding. This genome-wide characterization of genomic variation and the relationship with transcriptomic variation provides important information in our understanding of the molecular basis of heterosis and the breeding community’s ability to continue to make gains from selection in highly selected maize germplasm. With access to only two whole-genome assemblies, many lessons were learned regarding limitations in interspecies and intraspecies comparisons, transcriptome profiling, and characterization of allelic variation in the context of a single reference genome assembly and annotation that transcend beyond maize.
METHODS
Plant Material and DNA and RNA Extraction
DNA was extracted from maize (Zea mays) inbred lines PH207 (PI 601005; Lot 03ncai01) and B73 (PI 550473; Lot 08ncai02) from seedlings germinated from seed provided by the National Plant Germplasm System Regional Plant Introduction Station in Ames Iowa using a modified CTAB procedure (Murray and Thompson, 1980). Plants were grown under greenhouse conditions (27°C/24°C day/night and 16 h light/8 h dark) in Metro-Mix 300 (Sun Gro Horticulture) with no additional fertilizer and under fluorescent lights. DNA was quantified using picogreen, absorbance at 260 nm/280 nm using a NanoDrop, and gel bands using 1.2% E-Gel.
RNA-seq reads for B73 leaf blade, cortical parenchyma, germinating kernel, root tip, whole seedling, and stele were downloaded from the National Center for Biotechnology Information (accession numbers PRJNA171684 and SRP010680). For each tissue, sequencing reads for three biological replicates were obtained. Each biological replicate consisted of pooled tissue from three individual plants. Tissue sampling and RNA extraction for PH207 leaf blade, cortical parenchyma, germinating kernel, root tip, whole seedling, and stele were conducted as previously described for the comparable B73 tissues (Stelpflug et al., 2016). For each tissue, two biological replicates were collected and processed, again with three individual plants pooled per biological replicate.
DNA and RNA Sequencing
PH207 DNA Sequencing
Shotgun genomic libraries of 300- and 800-bp DNA fragment sizes were prepared using the TruSeq DNA Sample Preparation Kit version 2 according to the manufacturer’s protocol (Illumina). A third shotgun library was made using the same kit from DNA template fragments size selected from ∼350 to ∼450 bp with no PCR amplification (PCR-free). This fragment size was designed to produce a sequencing overlap of the fragments to be sequenced on the MiSeq as PE sequencing 250 nucleotides per end, thus creating an opportunity to produce “stitched” reads of ∼350 to 400 nucleotides in length. Multiple MP libraries per jump were made with the objective to increase sequence diversity and genome coverage. Four separate MP libraries were constructed for each of the 3- and 8-kb jumps, and two MP libraries for the 15-kb jump using the Illumina Nextera Mate-Pair Sample Preparation Kit (Illumina).
The 300- and 800-bp shotgun libraries and the MP libraries were sequenced on an Illumina HiSeq 2000 as 100-nucleotide PE reads. The pool of MP libraries was further sequenced on an Illumina HiSeq 2500 as 150-nucleotide PE reads. The PCR-free shotgun library was sequenced on an Illumina MiSeq as 250-nucleotide reads. All sequencing was conducted at the Roy J. Carver Biotechnology Center (Urbana, IL) at the University of Illinois, except for the 300-nucleotide PE genomic library, which was sequenced at Dow AgroSciences (Indianapolis, IN).
The synthetic long-read libraries were constructed, sequenced, and assembled by Illumina Sequencing Services. The PH207 genomic libraries were made using the TruSeq Synthetic Long-Read DNA library preparation kit materials and workflow (http://www.illumina.com/index-d.html). Each library consists of a bar-coded pool of 384 indexes (one per each well of 10-kb genomic DNA template fragments), and each of the 10 libraries was sequenced individually on one HiSeq 2000 lane as 100-nucleotide PE reads.
B73 DNA Sequencing
A 300-bp DNA fragment size shotgun genomic library was prepared using a TruSeq DNA Sample Preparation Kit version 2 according to the manufacturer’s protocol (Illumina). The B73 genomic library was sequenced by Dow AgroSciences on an Illumina HiSeq 2000 as 100-nucleotide PE reads.
PH207 RNA Sequencing
Approximately 5 µg of total RNA was processed for mRNA isolation, fragmented, converted to cDNA, and PCR amplified according to the Illumina TruSeq RNA Sample Prep Kit protocol and sequenced on an Illumina HiSeq 2500 at the Roy J. Carver Biotechnology Center at the University of Illinois as 150-nucleotide PE reads.
PH207 Genome Assembly
Read Preprocessing and Error Correction
For all PH207 genomic libraries, with the exception of TruSeq synthetic long-reads, PCR duplicates were removed using FastUniq software (Xu et al., 2012). The Illumina HiSeq 2000 adaptor AGATCGGAAGAGC was removed, and reads were error corrected using the Corrector_HA module of SOAPdenovo (using kmer size 23 and cutoff of 6) (Luo et al., 2012). For the PCR-free library (MiSeq stitched reads), following adaptor truncation, overlapping reads were merged using FLASH (Magoč and Salzberg, 2011) with a minimal required overlap of 10 bp to create the stitched reads. Processing of MP reads consisted of filtering out putative false mate-pairs by searching for the Nextera linker (10 nucleotides of CTGTCTCTTATACACATCTAGATGTGTATAAGAGACAG) sequence on either end of the MP. Mate pairs for which the linker was not found were sorted into a separate file for restricted scaffolding application. Mate-pairs that did not hit the linker were used only in support of links found with the filtered MPs but were not used to create links independently. The TruSeq synthetic long-read assembly pipeline application was processed by Illumina (Illumina IGN FastTrack Long Reads version 41) to create synthetic long-read builds that represent long contiguous template fragments.
PH207 de Novo Scaffold Assembly
In the first step of assembly, SOAPdenovo v1.05 (Luo et al., 2012) was used to construct a De Bruijn graph of contigs from the single-end reads of the PE library using very conservative settings (no bubble merge and no repeat masking, but removing low coverage kmers and edges). A kmer of size 63 bp was optimal for De Bruijn graph construction. To scaffold the contigs of the De Bruijn graph, nonrepetitive contigs within the graph were identified and assembled into scaffolds based on mapping information of the single end reads, followed by application of synthetic long reads to resolve long repeats in a similar way. The mapping of the single-read ends (mapping without gaps) and long-read builds facilitated scaffolding by linking contigs mapping to the same read.
Scaffolding was completed using a directed graph containing scaffolds longer than 200 bp as nodes, and edges were based on the PE and MP links as vertices. Erroneous connections were filtered out to generate unconnected subgraphs that were ordered into scaffolds. PE reads were used to find reliable paths in the graph for additional repeat resolving. This was accomplished through searching the De Bruijn graph for a unique path connecting pairs of reads mapping to two different scaffolds. At this phase of scaffolding, scaffolds had an average size of thousands of bases and almost no gaps. The scaffolds were then furthered ordered and linked using the MP libraries, estimating gaps between the contigs according to the distance of MP links. Linking scaffolds with MP reads required confirmation of at least three filtered MPs or at least one filtered MP with supporting confirmation from two or more filter failed MPs where the Nextera adaptor was not found. Scaffolds shorter than 200 bp were masked and links between nonrepetitive contigs mapping to the same scaffolds were united, generating a directed scaffold graph. In agreement with previous reports, a significant amount of erroneous MPs was observed. Since these pairs link between long nonbranched components in the scaffold graph, the scaffolding procedure identified the nonbranched components and filtered out the rare connections between them.
Reference Guided Construction of Pseudomolecules
Further ordering of scaffolds was achieved through scaffold alignment to the B73 reference genome using BWA-SW version 0.6.1 (Li and Durbin, 2010) requiring alignment quality of at least one. The most probable genomic location was assigned to the ordered scaffold based on the alignments of the unordered scaffolds that generated it. The ordered scaffolds were then placed into pseudomolecules to maximize synteny between the PH207 and B73 genomes. Finally, small scaffolds that mapped inside the unfilled gap of ordered scaffolds replaced these gaps if MP links existed between them.
TruSeq synthetic long reads were aligned to scaffolds generated without usage of the TruSeq synthetic long reads. Scaffolds were identified that had a significant alignment of greater than 30 kb to two different locations greater than 10 Mb apart on the B73 reference genome. For those scaffolds where the block aligning to the alternative locations was between two blocks that aligned to the same location, it was assumed to be a translocation between the two genomes, and it was considered unlikely there were two misassemblies at the same region. If a block aligned to one location followed by a block aligning to another distant genomic location, it was considered a misassembly. Suspicious scaffolds were further resolved using a previously defined genotyping-by-sequencing anchor tag pipeline (Lu et al., 2015), and those flagged by both criteria were manually reviewed for misassembly.
To assess the completeness of the genome assembly, PH207 genomic paired-end reads were cleaned using Cutadapt version 1.8.1 (Martin, 2011) requiring a minimum length of 70 and a minimum quality of 10, and aligned to the PH207 genome assembly using Bowtie version 2.2.4 (Langmead and Salzberg, 2012) with default parameters as single-end reads to determine the percentage of reads that could map to the assembly. Variants were called using Samtools mpileup (Li et al., 2009) with a minimum depth of three and a maximum depth of 200. Only variants exceeding a quality score of 20 were retained. To assess the completeness of the gene space in the assemblies, PH207 RNA-seq reads were aligned to the PH207 genome assembly and B73 RNA-seq reads were aligned to the B73 version 3.21 genome assembly (ftp://ftp.ensemblgenomes.org/pub/plants/release-21). Reads were cleaned using Cutadapt version 1.8.1 (Martin, 2011) requiring a minimum length of 75 nucleotides and a minimum quality of 20. All reads were trimmed to 75 nucleotides using fastx_trimmer within the FASTX toolkit version 0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html) for consistency between the B73 and PH207 reads. Reads were aligned using Bowtie2 version 2.2.4 (Langmead and Salzberg, 2012) and TopHat2 version 2.0.12 (Kim et al., 2013) with the maximum number of multihits set to 20, minimum intron size of 10 bp, and maximum intron size of 60,000 bp. Additionally, the Core Eukaryotic Genes Mapping Approach pipeline version 2.4 (Parra et al., 2007) was run using default parameters. Comparison of the B73 and PH207 genome assemblies was completed using the NUCmer program within MUMmer version 3.23 (Kurtz et al., 2004). The B73 version 3.21 assembly was used for the comparison, and the minimum length of a cluster match was set to 5000 for the genome-wide plot and 250 for the regional view of chromosome 1.
PH207 Genome Annotation
Gene Model Structural Annotation
The MAKER genome annotation pipeline was used to generate gene annotations (Campbell et al., 2014a, 2014b; Law et al., 2015). The PH207 genome assembly was first masked using RepeatMasker (http://www.repeatmasker.org) and a maize custom repeat library (Law et al., 2015). Six PH207 transcript assemblies (leaf blade, root cortical parenchyma, root stele, germinating kernel, root tip, and whole seedling) generated using Trinity (Grabherr et al., 2011), the predicted rice (Oryza sativa) proteome and UniProtKB/Swiss-Prot plant proteins (minus maize proteins) were used as evidence during each stage of the MAKER pipeline (Kawahara et al., 2013; UniProt Consortium, 2014). Initially, a Hidden Markov Model (HMM) was trained for the SNAP ab initio gene prediction program using high-quality transcript assembly alignments as gene proxies (Korf, 2004). MAKER was run using the initial SNAP HMM, and these gene predictions were used to train SNAP a second time. MAKER was run again using the second SNAP HMM to make gene predictions, and these gene models were used to train an HMM for the Augustus gene prediction program (Stanke and Waack, 2003). MAKER was run a final time using both the second SNAP HMM and the Augustus HMM. The genes identified by MAKER included models with and without transcript or protein alignment evidence. The predicted proteins from the MAKER gene set were analyzed with hmmscan to identify Pfam domains (Eddy, 2011; Finn et al., 2014). A high-quality gene set was created using all gene predictions that were supported by transcript or protein evidence or that coded for a protein with a Pfam domain. Using this analysis pipeline, a single transcript was annotated for each locus.
Gene Model Functional Annotation
All PH207 transcripts were functionally annotated using transitive annotation based on best BLAST his from Arabidopsis thaliana TAIR10 (Lamesch et al., 2012), Brachypodium distachyon v2.1 (International Brachypodium Initiative, 2010), O. sativa release 7 (Ouyang et al., 2007), and Sorghum bicolor v1.4 (Paterson et al., 2009) annotations. For each species, TBLASTN was used within WUBLAST (v2.0) (Altschul et al., 1990b) to search PH207 protein sequences against a nucleotide database for each species. The results were filtered to retain only the top hit requiring an E-value < 1e-5 to retain the hit. The same criteria were used to assign a UniRef100 release 2015_05, April 29, 2015 (Suzek et al., 2007), identifier to each PH207 protein, except that BLASTP was used. The -goterms option of InterProScan version 5.0 (Zdobnov and Apweiler, 2001) was used to assign Gene Ontology terms to each PH207 transcript.
Genome Content Variation
To identify genotype-specific genes from each assembly, B73 version 3.21 and PH207 transcript sequences were aligned to both the B73 version 3.21 and PH207 genome assemblies using GMAP version 2012-06-02 (Wu and Watanabe, 2005). A transcript with an alignment with >85% coverage and >85% identity to its cognate genome and the reciprocal genome were considered present in both genomes, those with >85% coverage and >85% identity to its cognate genome and <85% coverage and <85% identity to the reciprocal genome were considered absent in the reciprocal genome, and those with <85% coverage and <85% identity to its cognate genome were not classified. Due to gaps in the genome assemblies, PAVs were also determined using mapping of B73 and PH207 resequencing reads to both the B73 version 3.21 and PH207 genome assemblies. B73 and PH207 resequencing reads were mapped to the B73 version 3.21 assembly and the PH207 assembly using Bowtie2 version 2.2.3 (Langmead and Salzberg, 2012) requiring a mapping quality score greater than 20. The portion of the bases with coverage were determined using the intersect program within BEDTools version 2.19.0 (Quinlan and Hall, 2010). A bimodal distribution of coverage was observed, with the majority of genes having <25% coverage or greater than 75% coverage. Thus, any gene that had >75% of positions covered by its own reads and <25% of positions covered by the opposite genotype reads was considered PAV. This method was also used to classify partially deleted genes and the percentage of the gene that was deleted. For partial gene deletions, the percentage of the gene that was deleted was calculated as the percentage of the gene with coverage in the reciprocal genome divided by the percent of the gene with coverage in the cognate genome. The comparison of gene density in high and low recombination regions was determined using a χ2 test with previously determined boundaries based on previously described B73/Mo17 near isogenic lines (Swanson-Wagner et al., 2010; Eichten et al., 2011). Boundary coordinates were converted to B73 version 3.21 coordinates using NCBI BLASTN with 400 bp of context sequence.
To identify gene families that were expanded or contracted in PH207 relative to B73, an all-versus-all blast was performed using WU BLASTN (Altschul et al., 1990a) with the B73 version 3.21 transcripts and PH207 transcripts, requiring a minimum E-value of 1e-10 and allowing up to 5000 hits per sequence. The transcripts were clustered using OrthoMCL version 1.4 (Li et al., 2003; Chen et al., 2007) in mode 4 with default parameters to identify putative paralogous/homologous gene families between the two genotypes. This analysis was also used to identify high confidence one-to-one genes between the two individuals for subsequent analyses. The enrichment of Gene Ontology terms was analyzed using AgriGO (Du et al., 2010) using Fisher’s exact test to calculate an enrichment P value, with a value of <0.05 being considered significant after using the Yekutieli method for multiple test correction.
Promoter SNP and InDel variation was explored for one-to-one genes that were located on the same chromosome within 20 Mb of each other between the two genome assemblies and had at least 1 kb of promoter sequence upstream of the transcription start site in both assemblies (n = 18,798). Sequences with one or more Ns in either assembly were removed, resulting in 12,537 promoter comparisons. For each pairwise comparison, the 1 kb of promoter sequences from each assembly was aligned using NCBI BLASTN version 2.2.28 (Altschul et al., 1990b). Promoter comparisons with less than 700 bp alignments (n = 6114) were removed from downstream analysis. Alignments were parsed to identify SNPs and InDels using the blastn2snp.jar scripts within Jvarkit (http://dx.doi.org/10.6084/m9.figshare.1425030).
Transcriptional Analysis
All RNA-seq reads were trimmed to 75 nucleotides to remove low-quality bases and aligned to both the B73 version 3.21 reference genome assembly and the PH207 genome assembly using Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012) and TopHat2 version 2.0.10 (Kim et al., 2013). The minimum intron size was set to 5 bp, and the maximum intron size was set to 60,000 bp. All other mapping parameters were set to the default values. Transcript abundance estimates (measured as fragments per kilobase of exon model per million fragments mapped) were determined using Cufflinks2 version 2.1.1 (Trapnell et al., 2010) with a minimum and maximum intron size of 5 and 60,000 bp, respectively, and providing genome assemblies for bias correction. Transcript abundance counts for the representative transcript (longest transcript) were generated with HTSeq version 0.6.1p1 (Anders et al., 2015) at the gene level using the union mode and a minimum mapping quality of 20 with non-strand-specific counting (–stranded no). Corrected counts by genome coverage were determined by taking the raw counts divided by the percent exon coverage from the resequencing data rounded to the nearest whole number. A gene was defined as expressed in a sample if it had a count of one or more. Differential expression analysis was conducted using DESeq2 version 1.8.2 (Love et al., 2014) within R version 3.2.1 (R Development Core Team, 2011) using Bioconductor version 3.1 (Gentleman et al., 2004) with default parameters. Genes with an adjusted P value < 0.05 and a fold change >2 in either direction were considered differentially expressed. Differential expression analysis was conducted for B73 RNA-seq reads mapped to the B73 genome assembly versus PH207 RNA-seq reads mapped to B73 genome assembly as well as for B73 RNA-seq reads mapped to the PH207 genome assembly versus PH207 RNA-seq reads mapped to the PH207 genome assembly within each tissues. Differential expression analysis was conducted in this way because of our documented difference in untranslated region representation between the annotation for B73 and PH207, which would bias expression downwardly in PH207 one-to-one genes relative to the corresponding B73 gene model.
Comparative Genomics Analysis
Homologous grass genes that were present in only one assembly (either B73 or PH207) were identified using OrthoMCL version 1.4 (Li et al., 2003; Chen et al., 2007). An all-versus-all BLAST analysis was performed using WU BLASTP (Altschul et al., 1990a) requiring a minimum E-value of 1e-10 and allowing up to 5000 hits per sequence with the Brachypodium 2.1 (International Brachypodium Initiative, 2010), maize B73 version 3.21, maize PH207, O. sativa release 7 (Ouyang et al., 2007), and S. bicolor v1.4 (Paterson et al., 2009) representative protein sequences defined as the protein encoded by the longest transcript. Homologous gene families were identified by OrthoMCL version 1.4 in mode 4 with default parameters.
To evaluate differential fractionation in B73 and PH207 following the maize whole-genome duplication, the previously defined B73 Maize1 and Maize2 classifications (Schnable et al., 2011) from B73 v2 were converted to B73 v3 coordinates using CrossMap (Zhao et al., 2014), and overlapping gene models were identified. The converted Maize1 and Maize2 classifications were compared with the homologous gene clustering generated using OrthoMCL (Li et al., 2003; Chen et al., 2007) with the B73 version 3.21 and PH207 nucleotide sequences described above.
Analysis of Deleterious Mutations
PH207 genomic reads were cleaned using Cutadapt version 1.8.1 (Martin, 2011) requiring a minimum length of 70 nucleotides and a minimum quality of 10. Cleaned reads were aligned to the B73 version 3.21 reference assembly using Bowtie version 2.2.4 (Langmead and Salzberg, 2012) with default parameters as single end reads. Variants were called using Samtools mpileup (Li et al., 2009) with a minimum depth of three and a maximum depth of 200. Only variants exceeding a quality score of 20 were retained. Variants were then annotated via SnpEff version 4.1 H (Cingolani et al., 2012) using default parameters. The annotated VCF file was filtered to retain only the predicted high and moderate impact variants that were located within the representative transcript of one-to-one genes based on the B73 model. As defined by the VCF annotation standard, high impact effects included frameshift variants, splice acceptor/donor variants, start/stop lost, and stop gained. Moderate impact predicated effects included in-frame InDels and missense variants. BLASTN 2.2.25+ (Altschul et al., 1990b) was used to align B73 genic sequence from one-to-one genes to the PH207 assembly. Variants were parsed from the alignment to determine the position of annotated variants from the Bowtie alignment in the context of the PH207 assembly by scanning for each variant in the parsed BLAST output. Only variants that had a consistent alignment position between the Bowtie and BLAST alignment algorithms were retained. The “intersect” sub-command within the BEDTools suite version 2.17.0 (Quinlan and Hall, 2010) was used to determine whether the retained variants were located within PH207 exon sequences.
Accession Numbers
Sequence data from this article can be found in the Sequence Read Archive at the National Center for Biotechnology Information under accession number PRJNA258455. The PH207 genome multifasta file, GFF annotation file, transcript multifasta file, and protein multifasta file are available for download from the Dryad repository (http://dx.doi.org/10.5061/dryad.8vj84) and are also available at the Maize Genetics and Genomics Database (http://www.maizegdb.org) and at Phytozome (https://phytozome.jgi.doe.gov). Additionally, the PH207 genome assembly results as Integrative Genomics Viewer (Broad Institute) tracks are available for public access at http://nrgene.com.
Supplemental Data
Supplemental Figure 1. Distribution of 23-mers in the PH207 sequence reads.
Supplemental Figure 2. PH207 and B73 RNA-seq read mapping statistics.
Supplemental Figure 3. Annotation edit distance curve for the PH207 MakerP annotation.
Supplemental Figure 4. Resequencing-based approach to identify partial gene deletions.
Supplemental Figure 5. Density distribution of expression in B73 and PH207 for shared and genotype-specific genes.
Supplemental Figure 6. Distribution of predicted impact of PH207 variants.
Supplemental Table 1. Reads generated from the PH207 genome assembly.
Supplemental Table 2. Assembly statistics at each progressive assembly step.
Supplemental Data Set 1. Functional annotation of the PH207 gene models.
Supplemental Data Set 2. Enriched Gene Ontology terms for B73 and PH207 genes that are members of gene families that have expanded or contracted between the two genotypes and genes that show PAV between the two genotypes.
Supplemental Data Set 3. Transcript abundance estimates for six distinct tissues in the inbred lines B73 and PH207.
Acknowledgments
This work was funded in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494), by Dow AgroSciences, and by the National Science Foundation (Grant IOS-1126998 to K.L.C.). The Minnesota Supercomputing Institute at the University of Minnesota provided computational resources that contributed to the research results reported in this article. C.D.H. was supported by a National Science Foundation National Plant Genome Initiative Postdoctoral Fellowship in Biology Fellowship (Grant 1202724). A.B.B. was supported by the DuPont Pioneer Bill Kuhn Honorary Fellowship.
AUTHOR CONTRIBUTIONS
C.N.H., K.K., N.M.S., E.B., C.R.B., N.d.L., S.M.K., and M.A.M. designed the study. A.G.H., C.J.F., and C.L.W. acquired data. C.N.H., C.D.H., A.B.B., M.J.B., I.S., O.B., D.S.-T., K.B., F.L., C.J.F., and K.L.C. analyzed and interpreted data. C.N.H. and M.A.M. wrote the manuscript. All authors read and approved the final manuscript.
Glossary
- PAV
presence/absence variation
- CNV
copy number variation
- SNP
single nucleotide polymorphism
- PE
paired-end
- MP
mate-pair
- InDel
insertion/deletion
Footnotes
Articles can be viewed without a subscription.
References
- Abendroth L.J., Elmore R.W., Boyer M.J., Marlay S.K. (2011). Corn Growth and Development. (Ames, IA: Iowa State University Extension). [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990a). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990b). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.E., Kantar M.B., Kono T.Y., Fu F., Stec A.O., Song Q., Cregan P.B., Specht J.E., Diers B.W., Cannon S.B., McHale L.K., Stupar R.M. (2014). A roadmap for functional structural variants in the soybean genome. G3 (Bethesda) 4: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Beló A., Beatty M.K., Hondred D., Fengler K.A., Li B., Rafalski A. (2010). Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor. Appl. Genet. 120: 355–367. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Auger D.L., Riddle N.C. (2003). In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Yao H., Chudalayandi S. (2006). Unraveling the genetic basis of hybrid vigor. Proc. Natl. Acad. Sci. USA 103: 12957–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Yao H., Chudalayandi S., Vaiman D., Veitia R.A. (2010). Heterosis. Plant Cell 22: 2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S., Fengler K., Morgante M., Tingey S., Rafalski A. (2005). Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.S., Holt C., Moore B., Yandell M. (2014a). Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48: 4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.S., et al. (2014b). MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chen F., Mackey A.J., Vermunt J.K., Roos D.S. (2007). Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J.M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D.E., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209. [DOI] [PubMed] [Google Scholar]
- Dong Q., Schlueter S.D., Brendel V. (2004). PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 32: D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (2011). Accelerated Profile HMM Searches. PLOS Comput. Biol. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., Foerster J.M., de Leon N., Kai Y., Yeh C.T., Liu S., Jeddeloh J.A., Schnable P.S., Kaeppler S.M., Springer N.M. (2011). B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiol. 156: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K., Moore B., Holt C., Yandell M. (2009). Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Dooner H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines T.A., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 107: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S. (2009). A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansey C.N., Vaillancourt B., Sekhon R.S., de Leon N., Kaeppler S.M., Buell C.R. (2012). Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS One 7: e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan M.A., et al. (2016). Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 28: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., et al. (2015). Survey of genome sequences in a wild sweet potato, Ipomoea trifida (H. B. K.) G. Don. DNA Res. 22: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.D., Springer N.M., Hirsch C.N. (2015). Genomic limitations to RNA sequencing expression profiling. Plant J. 84: 491–503. [DOI] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. (2004). Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., et al. (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42: 1027–1030. [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., et al. (2015). Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 167: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., et al. (2015). High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 6: 6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron L.G., et al. (2013). Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110: 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Mikel M.A. (2011). Genetic composition of contemporary U.S. commercial dent corn germplasm. Crop Sci. 51: 592–599. [Google Scholar]
- Monaco M.K., et al. (2014). Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 42: D1193–D1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., De Paoli E., Radovic S. (2007). Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10: 149–155. [DOI] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., et al. (2007). The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 35: D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Schatz M.C., et al. (2014). Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 15: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Song R., Messing J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., Stupar R.M. (2007). Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res. 17: 264–275. [DOI] [PubMed] [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Waack S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (suppl. 2): ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Stelpflug S.C., Sekhon R.S., Vaillancourt B., Hirsch C.N., Buell C.R., de Leon N., Kaeppler S.M. (2016). An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16. [DOI] [PubMed] [Google Scholar]
- Sutton T., Baumann U., Hayes J., Collins N.C., Shi B.J., Schnurbusch T., Hay A., Mayo G., Pallotta M., Tester M., Langridge P. (2007). Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449. [DOI] [PubMed] [Google Scholar]
- Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. (2007). UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23: 1282–1288. [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner R.A., Eichten S.R., Kumari S., Tiffin P., Stein J.C., Ware D., Springer N.M. (2010). Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer A.F. (1999). Background of U.S. hybrid corn. Crop Sci. 39: 601–626. [Google Scholar]
- Troyer A.F. (2006). Adaptedness and heterosis in corn and mule hybrids. Crop Sci. 46: 528–543. [Google Scholar]
- UniProt Consortium (2014). Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42: D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47: 944–948. [DOI] [PubMed] [Google Scholar]
- Weber R.L., et al. (2014). Expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean. BMC Plant Biol. 14: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Mott R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Xu H., Luo X., Qian J., Pang X., Song J., Qian G., Chen J., Chen S. (2012). FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7: e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M., Ence D. (2012). A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13: 329–342. [DOI] [PubMed] [Google Scholar]
- Yao H., Dogra Gray A., Auger D.L., Birchler J.A. (2013). Genomic dosage effects on heterosis in triploid maize. Proc. Natl. Acad. Sci. USA 110: 2665–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. (2001). InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. [DOI] [PubMed] [Google Scholar]
- Zhao H., Sun Z., Wang J., Huang H., Kocher J.P., Wang L. (2014). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30: 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]