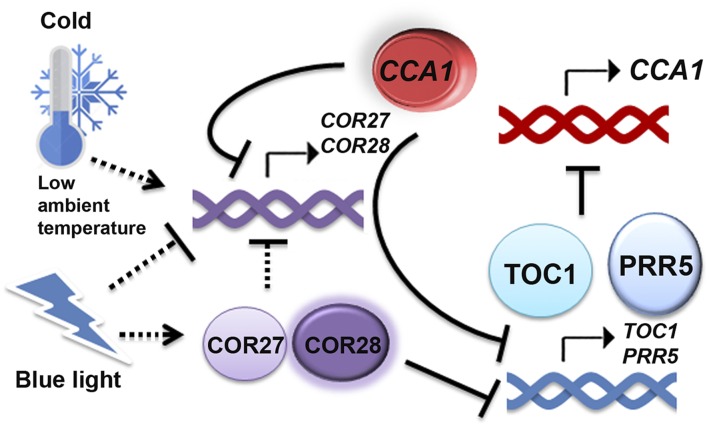
COR27 and COR28 are regulated by light and low temperature and, in turn, regulate the circadian clock, freezing tolerance, and flowering time.
Abstract
Light and temperature are two key environmental signals that profoundly affect plant growth and development, but underlying molecular mechanisms of how light and temperature signals affect the circadian clock are largely unknown. Here, we report that COR27 and COR28 are regulated not only by low temperatures but also by light signals. COR27 and COR28 are negative regulators of freezing tolerance but positive regulators of flowering, possibly representing a trade-off between freezing tolerance and flowering. Furthermore, loss-of-function mutations in COR27 and COR28 result in period lengthening of various circadian output rhythms and affect central clock gene expression. Also, the cor27 cor28 double mutation affects the pace of the circadian clock. Additionally, COR27 and COR28 are direct targets of CCA1, which represses their transcription via chromatin binding. Finally, we report that COR27 and COR28 bind to the chromatin of TOC1 and PRR5 to repress their transcription, suggesting that their effects on rhythms are in part due to their regulation of TOC1 and PRR5. These data demonstrate that blue light and low temperature-regulated COR27 and COR28 regulate the circadian clock as well as freezing tolerance and flowering time.
INTRODUCTION
A major developmental transition in plants is the switch from the vegetative to the reproductive, flowering phase. CO (CONSTANS) and FT (FLOWERING LOCUS T) are among the most important genes that regulate floral initiation in response to photoperiod (Putterill et al., 1995; Kobayashi et al., 1999). CO is a zinc finger transcription factor that promotes flowering by activating FT expression (Onouchi et al., 2000; Samach et al., 2000). FT is an RAF (rapidly accelerated fibrosarcoma) kinase inhibitor-related protein, which acts as a long-distance signal that migrates through the vascular system from leaves to the apical meristem (Lifschitz et al., 2006; Corbesier et al., 2007). The blue light photoreceptor cryptochrome 2 (CRY2) has been shown to activate FT expression in response to blue light by suppressing degradation of the CO protein (Yanovsky and Kay, 2002; Valverde et al., 2004; L.J. Liu et al., 2008), by direct activation of the CIB1 (CRY2-interacting bHLH1) transcription factor (H. Liu et al., 2008, 2013; Y. Liu et al., 2013), and by regulating light entrainment of the circadian clock (Jang et al., 2008).
In addition to CRY2, other photoreceptors such as phytochrome A (phyA), phyB, and the LOV-domain F-box proteins FLAVIN BINDING; KELCH REPEAT1 (FKF1), ZEITLUPE (ZTL), and LOV KELCH PROTEIN2 also regulate the expression of CO and FT and affect flowering time in response to photoperiod (Hayama and Coupland, 2004; Thomas, 2006). phyA and phyB interact with PHYTOCHROME-INTERACTING FACTOR3 (PIF3) to regulate CO and FT expression by both clock-dependent and clock-independent mechanisms (Martínez-García et al., 2000; Leivar and Quail, 2011). FKF1 mediates blue light-dependent degradation of CYCLING DOF FACTOR1 (CDF1) and stabilization of CO, facilitating transcription of FT (Imaizumi et al., 2005; Song et al., 2012). ZTL is the substrate binding subunit of the SCFZTL E3 ubiquitin ligase, which regulates abundance of key proteins that act as circadian oscillator components, TIMING of CAB EXPRESSION1 (TOC1) and PSEUDO-RESPONSE REGULATOR5 (PRR5), to affect expression of a number of flowering-time genes, including FT (Más et al., 2003; Kiba et al., 2007).
One way that light signals could regulate photoperiodic flowering is by regulating the circadian clock. The circadian clock system can be divided conceptually into three parts: inputs that receive environmental cues (light and temperature) to entrain the central oscillator, a central oscillator that generates self-sustained rhythmicity, and outputs that consist of various rhythmic processes. There are multiple transcription feedback loops in the Arabidopsis thaliana circadian clock (Sanchez and Yanovsky, 2013; Hsu and Harmer, 2014; McClung, 2014; Shim and Imaizumi, 2015). In the initially described feedback loop, transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) repress the transcription of TOC1. TOC1 (also known as PRR1) represses transcription of CCA1 and LHY (Huang et al., 2012). CCA1 and LHY also repress transcription of EARLY FLOWERING3 (ELF3), ELF4, LUX ARRHYTHMO (LUX; also known as PHYTOCLOCK1), PRR5, and GIGANTEA (GI), and they all positively regulate expression of CCA1 and LHY except PRR5 (Hazen et al., 2005; Kikis et al., 2005; Onai and Ishiura, 2005; Kamioka et al., 2016). In addition, CCA1 and LHY promote transcription of two TOC1 homologs, PRR7 and PRR9, as they in turn, repress expression of CCA1 and LHY (Farré et al., 2005; Nakamichi et al., 2010).
The circadian clock is involved in not only photoperiodic flowering but also in the cold response and in freezing tolerance. C-REPEAT BINDING FACTORs (CBFs; also known as DEHYDRATION-RESPONSIVE ELEMENT BINDING) are cold-induced transcription factors, and they are sufficient for inducing freezing tolerance (Thomashow, 1999). Cold induction of CBF1-3 is gated by the circadian clock (Fowler et al., 2005), and CBF1-3 are direct targets of CCA1 and LHY (Dong et al., 2011). In addition, the evening element (EE; AATATC) is a conserved motif in the promoter of cold-inducible genes (Mikkelsen and Thomashow, 2009; Maruyama et al., 2012). PRR5, PRR7, and PRR9 were also reported to be involved in modulating diurnal expression of cold-responsive genes and freezing tolerance. The prr5-10 prr7-10 prr9-11 triple mutant showed increased CBF expression and increased freezing tolerance (Nakamichi et al., 2009). Similarly, the toc1-101 mutant also showed increased expression of CBF3 and increased freezing tolerance (Keily et al., 2013). Cold may also affect the clock function, as it is reported that CBF1 binds directly to the LUX promoter to regulate the transcription of LUX (Chow et al., 2014).
There is limited information about how light and low temperature coordinate to regulate the circadian clock and photoperiodic flowering. COLD-REGULATED GENE27 (COR27) and COR28 were identified as cold-responsive genes from Arabidopsis transcriptome profiling, and the expression of COR27 was shown to be clock regulated (Fowler and Thomashow, 2002; Mikkelsen and Thomashow, 2009). The biological functions of COR27 and COR28 are unknown. Here, we show that COR27 and COR28 are regulated by both low temperature and light, representing a trade-off between flowering and freezing tolerance. COR27 and COR28 are clock regulated, and they are direct targets of CCA1. Furthermore, COR27 and COR28 are involved in regulating period length in the circadian clock, and they associate with chromatin regions of PRR5 and TOC1 to regulate their transcription. Light- and low temperature-regulated COR27 and COR28 are involved in the regulation of the circadian clock as well as freezing tolerance and flowering time.
RESULTS
COR27 and COR28 Are Regulated by Both Blue Light and Temperature
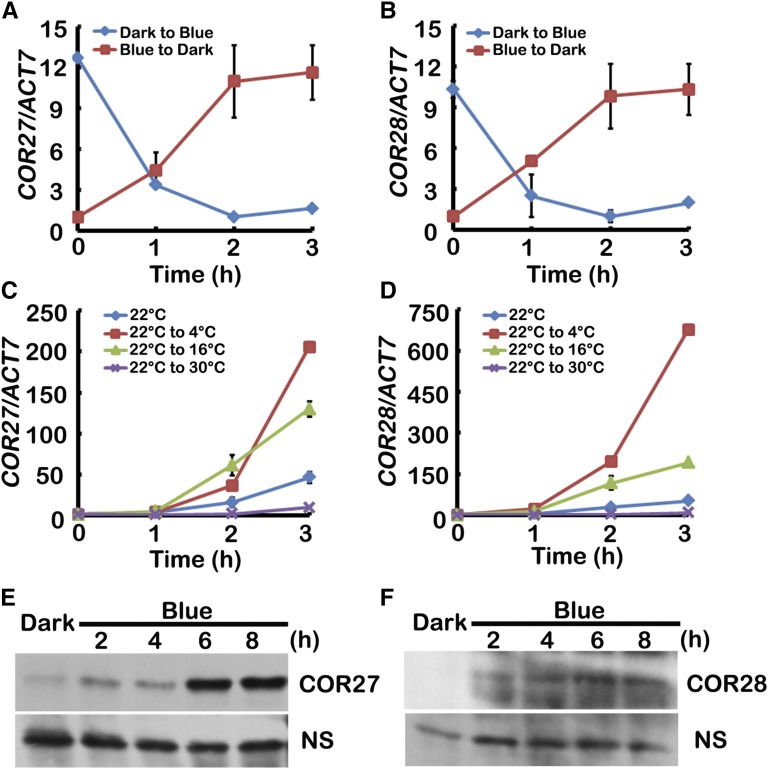
To identify new genes regulated by both light and temperature, we used Genevestigator (https://genevestigator.com/gv), an online platform of genome wide expression profiles, to analyze large groups of genes. We discovered that both COR27 (At5g42900) and COR28 (At4g33980) expression was induced by cold treatment, but repressed by light exposure. To confirm these expression patterns, we performed qPCR to monitor mRNA levels under varying environmental conditions. Transcription of COR27 and COR28 decreased when seedlings were moved from darkness to blue light (40 μmol m−2 s−1) or red light (40 μmol m−2 s−1) but were upregulated when seedlings were moved from blue light to darkness (Figures 1A and 1B; Supplemental Figures 1A and 1B). In addition, transcription of COR27 and COR28 was increased following a cold treatment as seedlings were moved from 22°C to 4°C or 16°C, but was slightly downregulated when seedlings were moved from 22°C to 30°C (Figures 1C and 1D).
Figure 1.
COR27 and COR28 Are Regulated by Both Blue Light and Low Temperature.
(A) and (B) qPCR results showing transcription of COR27 and COR28 are repressed by blue light. Five-day-old seedlings grown at 22°C in CL were transferred to darkness or blue light for 1 d before being transferred to blue light or darkness for the indicated time before sample collection.
(C) and (D) qPCR results showing transcription of COR27 and COR28 are induced by low temperature. Five-day-old seedlings grown in 22°C LD conditions were transferred to 4°C, 16°C, or 30°C at ZT6 for the indicated time before sample collection. Error bars in (A) to (D) represent se of three biological replicates. Expression levels are normalized to the ACT7 mRNA level.
(E) and (F) Immunoblots showing the expression of YFP-COR27 or GFP-COR28 protein in transgenic plants expressing the Pro35S:YFP-COR27 or Pro35S:GFP-COR28 transgene. Samples were fractionated by 10% SDS-PAGE, blotted, and probed with an anti-GFP antibody. NS represents nonspecific band. Plants were grown in 22°C under LD conditions for 5 d and then transferred to darkness for 2 d before transferred to blue light for 2, 4, 6, or 8 h before sample collection.
We then investigated whether blue light or low temperature affected COR27 and COR28 protein expression. Because none of the antibodies generated against COR27 and COR28 recognized endogenous COR27 and COR28 proteins in plants, we used transgenic plants constitutively expressing fluorescently tagged COR27 (Pro35S:YFP-COR27) and COR28 (Pro35S:GFP-COR28) to analyze protein expression. We grew transgenic plants in long-day conditions (LD; 16 h light/8 h dark) for 5 d, transferred those plants into the dark for 2 d, and then exposed them to blue light for various times and measured the levels of COR27 and COR28. Little COR27 or COR28 protein was detected in plants pretreated with darkness, but the level of both COR27 and COR28 increased significantly within 2 h of blue light treatment (Figures 1E and 1F) whereas transcription of COR27 and COR28 decreased (Supplemental Figures 1C and 1D). Temperature did not affect the level of COR27 and COR28 (Supplemental Figures 1E and 1F). Transcription of COR27 and COR28 was repressed by blue light, while expression of their encoded proteins was promoted by blue light, indicating that they may be involved in the transcriptional autoregulation. We tested this hypothesis by measuring transcription of endogenous COR27 and COR28 both in transgenic lines expressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 and in cor27 and cor28 mutants. Transcription of endogenous COR27 and COR28 was dramatically lower in overexpression lines than in the wild type, but the transcription of COR27 is higher in cor28 mutant, and the transcription of COR28 is higher in the cor27 mutant (Supplemental Figures 2A and 2B). These results show that COR27 and COR28 repressed their own as well as each other’s transcription.
COR27 and COR28 Are Involved in Flowering
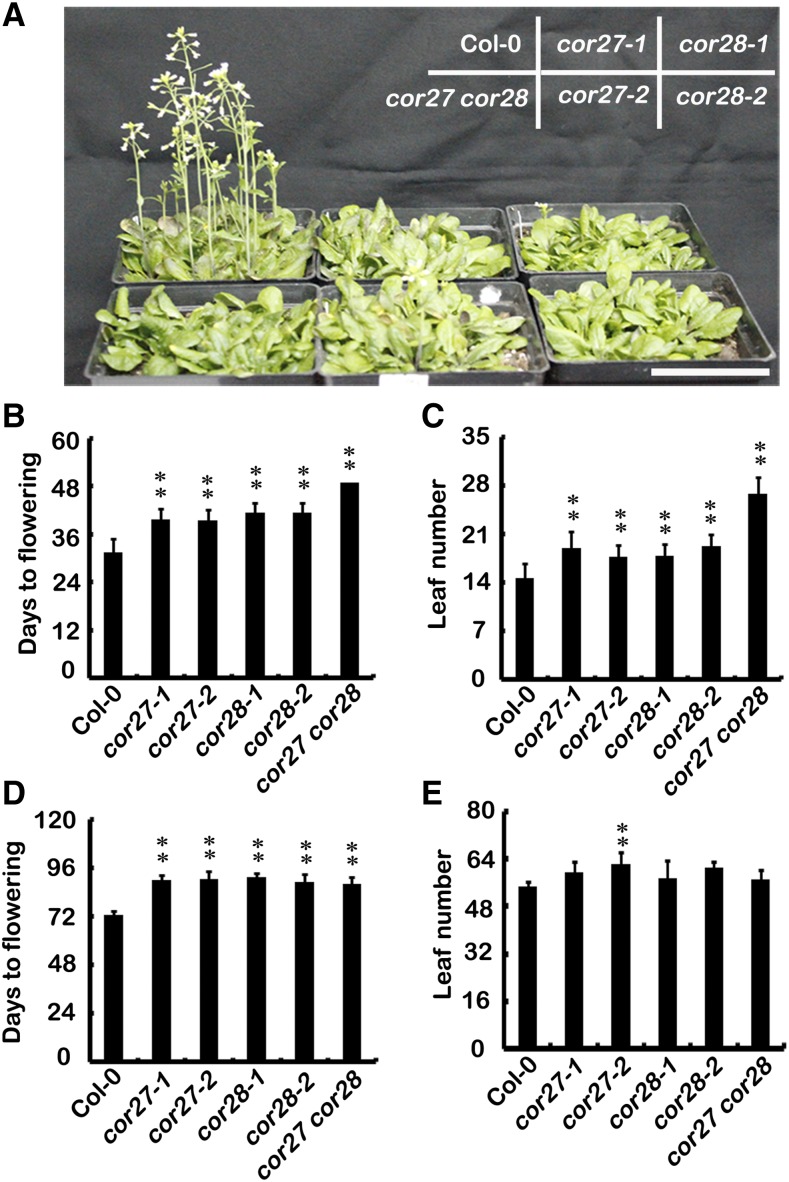
To determine the biological roles of COR27 and COR28, we obtained T-DNA insertion mutants from the Arabidopsis Biological Resource Center, naming them cor27-1, cor27-2, cor28-1, and cor28-2. cor27-1, cor27-2, and cor28-1 are knockdown mutants, and cor28-2 is a null mutant (Supplemental Figures 3A to 3C). The cor27-1, cor27-2, cor28-1, and cor28-2 mutants all showed a statistically significant delay of flowering under LD conditions, as measured either by days to flowering or by number of leaves at flowering (Figures 2A to 2C). We next prepared and examined the cor27 cor28 double mutant, which exhibited a more significant late flowering phenotype than the respective single mutants (Figures 2A to 2C), indicating that COR27 and COR28 function at least partially redundantly in regulating flowering time. We also examined the flowering phenotype under short-day conditions (SD; 8 h light/16 h dark, 22°C). Our results revealed that the cor27, cor28, and cor27 cor28 mutants displayed a subtle phenotype, flowering slightly later than wild-type plants under SD conditions as measured by days to flowering (Figures 2D and 2E). Transgenic plants overexpressing COR27 or COR28 driven by the cauliflower mosaic virus 35S promoter also flowered slightly later than the wild type in LD conditions as measured by days to flowering (Supplemental Figures 4A to 4E). The late flowering phenotype of the cor27-1 mutant was complemented when Pro35S:MYC-COR27 was crossed into cor27-1 (Supplemental Figure 5A). Interestingly, the late flowering phenotype could not be complemented when Pro35S:YFP-COR27 was crossed into cor27-1. We then examined the transcription of COR27 in these two lines and found that the transcription of COR27 in MYC-COR27/cor27-1 was ∼2.5 times greater than in the wild type, whereas the transcription of COR27 in YFP-COR27/cor27-1 was ∼12 times greater than in the wild type (Supplemental Figure 5B). It is possible that the expression of COR27 and COR28 must be well balanced for their proper function, that they may function in a protein complex, and that they cannot function properly when there is too less or too much of them.
Figure 2.
COR27 and COR28 Are Involved in Photoperiodic Flowering.
(A) Representative photos of 38-d-old plants of the genotypes indicated grown in 22°C LD conditions. Bar = 5 cm.
(B) and (C) The quantitative flowering times measured as days to flower (B) and the number of rosette leaves (C) at the day floral buds became visible of genotypes shown in (A). Error bars represent sd (n ≥ 20).
(D) and (E) Flowering phenotype of indicated genotypes grown in 22°C in SD conditions. The quantitative flowering times measured as days to flower (D) and the number of rosette leaves (E) at the day floral buds became visible. Error bars represent sd (n ≥ 20); the asterisks indicate significant differences compared with the wild type under the same treatment conditions (**P < 0.01, *P < 0.05, Student’s t test).
COR27 and COR28 Affect Multiple Flowering Pathways
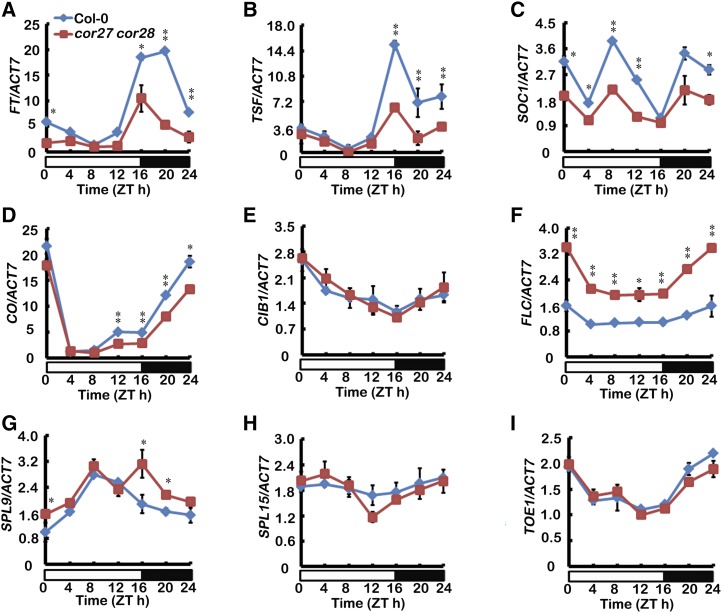
There are at least five distinct pathways controlling flowering in the model plant Arabidopsis, including the photoperiod pathway, the vernalization/thermosensory pathway, the autonomous floral initiation, the gibberellins pathway, and the age pathway (Mouradov et al., 2002; Amasino and Michaels, 2010; Bergonzi et al., 2013; Zhou et al., 2013; Wang, 2014). miR156-SPL (SQUAMOSA PROMOTER BINDING LIKE) controls the age pathway (Wang, 2014). To determine molecular mechanisms of the delayed flowering phenotype of the cor27 cor28 double mutants, we examined the expression of FT, TSF (TWEEN SISTER OF FT), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1). Compared with the wild type, the rhythmic expression of FT, TSF, and SOC1 was substantially reduced in cor27 cor28 double mutant plants (Figures 3A to 3C). To examine whether the cor27 cor28 double mutation affects flowering through the photoperiodic pathway, we examined the expression of CO and CIB1. We found that the expression of CIB1 was not affected much, but the rhythmic expression of CO was reduced in cor27 cor28 double mutant plants (Figures 3D and 3E), indicating that the photoperiodic flowering pathway is affected. FLOWERING LOCUS C (FLC), a convergence point of the autonomous and the vernalization pathways, represses flowering through direct binding to FT and SOC1 chromatin to repress their expression (Helliwell et al., 2006). The transcript level of FLC was elevated in the cor27 cor28 double mutant relative to the wild type (Figure 3F), suggesting that either the autonomous or the vernalization pathway is affected by the cor27 cor28 double mutations. The transcription of SPL9 was slightly higher in the cor27 cor28 double mutants, but the transcription of SPL15 and TOE1 was not changed (Figures 3G to 3I). These results indicated that COR27/COR28 might regulate multiple flowering pathways, so we focused on the photoperiodic pathway because this pathway is regulated by blue light
Figure 3.
COR27 and COR28 Affect the Transcription of FT, SOC1, and Other Genes.
qPCR results showing mRNA expression of FT, TSF, SOC1, CO, CIB1, FLC, SPL9, SPL15, and TOE1 in cor27 cor28 and the wild type grown in LD conditions. Samples were collected from 5-d-old seedlings of the genotypes indicated every 4 h over one day in LD. Expression levels are normalized to the ACT7 mRNA level. Error bars represent se of three biological replicates. The asterisks indicate significant differences compared with the wild type (**P < 0.01, *P < 0.05, Student’s t test).
Mutation of COR27 and COR28 Enhances Freezing Tolerance
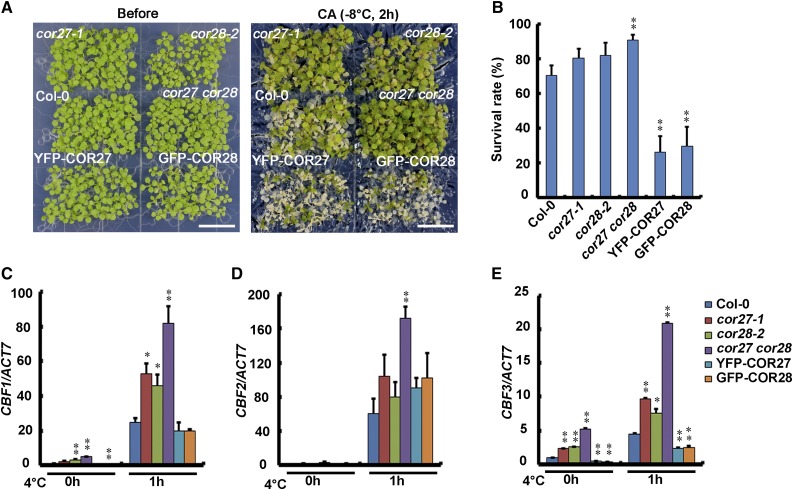
Because COR27 and COR28 are induced by low temperature, we performed freezing stress tests to study their function in freezing tolerance. For this assay, cor27-1 and cor28-2 single mutant plants grown in LD conditions at 22°C were pretreated at 4°C for 1 d, and then the temperature was gradually dropped to −8°C and held at −8°C for 2 h. The mutants showed slightly better freezing tolerance than did the wild type, and the cor27 cor28 double mutant showed significantly more freezing tolerance than the wild type. Therefore, COR27 and COR28 appear to function at least partially redundantly in regulating freezing tolerance. Phenotypes and survival rates after freezing treatment are shown in Figures 4A and 4B. We also found that survival rates of transgenic lines overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 were lower than the wild type (Figures 4A and 4B), whereas the survival rates of MYC-COR27/cor27 were much higher than MYC-COR27 (Supplemental Figure 6). These results indicate that COR27 and COR28 play significant roles in the response to cold temperatures.
Figure 4.
COR27 and COR28 Affect Freezing Tolerance.
(A) and (B) Freezing tolerance assay using cor27-1, cor28-2, cor27 cor28, YFP-COR27, GFP-COR28, and wild-type control seedlings. Eight-day-old seedlings grown at 22°C in LD conditions were cold acclimated (CA) at 4°C for 1 d then frozen at −8°C for 2 h and then transferred to 22°C for 3 d before the measurement of survival rates.
(A) Representative photos of plants of the genotypes indicated after the freezing treatment. Bars = 1.5 cm.
(B) Survival rates of the genotypes indicated after the freezing treatment. Error bars represent se of three biological replicates, and the asterisks indicate significant differences compared with the wild type under the same treatment conditions (**P < 0.01, Student’s t test).
(C) to (E) qPCR results showing expression patterns of CBF1, CBF2, and CBF3 in cor27 cor28 and the wild type following cold treatment. Eight-day-old seedlings grown at 22°C in LD conditions were transferred to 4°C at ZT10 for the indicated time. Expression levels are normalized to the ACT7 mRNA level. Error bars represent se of three biological replicates. The asterisks indicate significant differences compared with the wild type (**P < 0.01, *P < 0.05, Student’s t test).
To confirm that COR27 and COR28 are involved in the cold response, we analyzed expression of CBFs in cor27 and cor28 single mutants, cor27 cor28 double mutants, transgenic lines overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28, and the wild type. When seedlings grown at 22°C in LD conditions for 8 d were transferred to 4°C, transcript abundances of CBF1, CBF2, and CBF3 were elevated in all genotypes investigated. Transcription of CBF1 and CBF3 was significantly higher in cor27 and cor28 single mutants than in the wild type, and even more dramatically higher in cor27 cor28 double mutants than in the wild type (Figures 4C to 4E). The transcription of CBF2 was dramatically higher in cor27 cor28 double mutants than in the wild type, and the transcription of CBF3 was significantly lower in YFP-COR27 and GFP-COR28 overexpression lines than in the wild type (Figures 4D and 4E). These results indicate that COR27 and COR28 are involved in expression regulation of CBFs and also in cold response.
The Circadian Clock Regulates Expression of COR27 and COR28, Which Are Direct Targets of CCA1
COR27 and COR28 are positive regulators of photoperiodic flowering and negative regulators of freezing tolerance; therefore, they may present a trade-off between flowering and freezing tolerance. Because the circadian clock is involved in both photoperiodic flowering and cold response, we hypothesized that COR27 and COR28 might balance flowering and the cold response via the circadian clock.
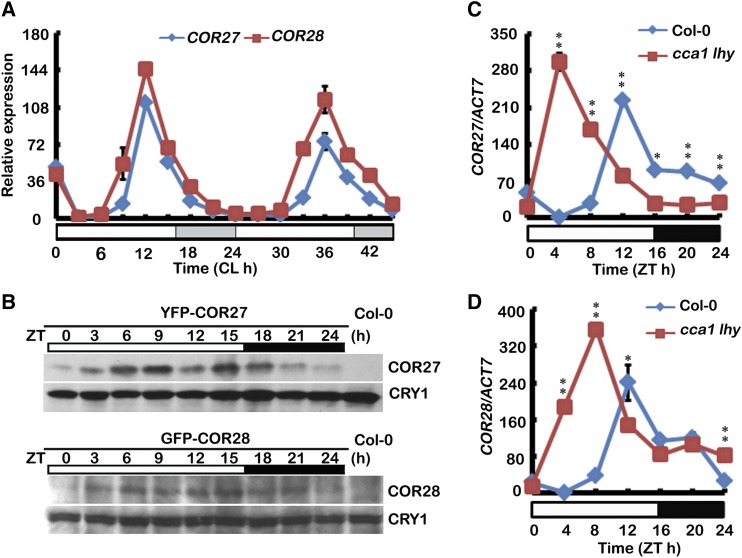
It was reported previously that the transcription of COR27 is regulated by the circadian clock (Mikkelsen and Thomashow, 2009). Our qPCR results demonstrate that both COR27 and COR28 are clock-regulated and expression of both peaks at noon (ZT12) (Figure 5A). In addition, levels of both COR27 and COR28 were also regulated by the clock, both peaking in abundance in the afternoon (ZT15) in LD conditions (Figure 5B). Given that there are EE (AAAATATCT) and EE-like elements (AATATCT) in the COR27 promoter region (Mikkelsen and Thomashow, 2009) and that it was reported that the promoter of COR27 was occupied by CCA1 in a ChIP-seq analysis of plants expressing CCA1-GFP (Nagel et al., 2015), we measured transcription of COR27 and COR28 in the cca1 lhy double mutant. ACTIN7 (ACT7), ASPARTIC PROTEINASE A1 (APA1), and ISOPENTENYL PYROPHOSPHATE:DIMETHYLALLYL PYROPHOSPHATEISOMETASE2 (IPP2) (Endo et al., 2014) were used as internal controls for normalization in our qPCR analysis. Transcription of both COR27 and COR28 was upregulated in the cca1 lhy double mutant in the morning (ZT4 and ZT8) (Figures 5C and 5D; Supplemental Figures 7A to 7D) in LD conditions. Thus, considering that expression of CCA1 and LHY peaks in the morning, CCA1 might directly repress transcription of COR27 and COR28. Our ChIP-PCR (chromatin immunoprecipitation-PCR) assays confirmed that, in vivo, CCA1 was associated with the chromatin of both COR27 and COR28 promoters (Supplemental Figures 7E and 7F). These data indicate that the circadian clock regulates transcription of COR27 and COR28 via CCA1 directly binding to their chromatin and regulating their transcription.
Figure 5.
COR27 and COR28 Are Regulated by CCA1.
(A) and (B) qPCR and immunoblots results showing mRNA and protein expression of COR27 and COR28. Five-day-old seedlings were entrained in a LD cycle, transferred to CL conditions (22°C) (A), or kept in LD (B) and harvested for 2 d (A) or 1 d (B) at 3-h intervals. Expression levels are normalized to the ACT7 mRNA level. Error bars represent sd of three technical replicates. Each experiment was performed at least three times with similar results.
(C) and (D) qPCR results showing expression of COR27 and COR28 in cca1 lhy and the wild type grown in LD conditions. Samples were collected from 5-d-old seedlings of genotypes indicated every 4 h over one day in LD. Expression levels are normalized to the ACT7 mRNA level. Error bars represent se of three biological replicates. (**P < 0.01, *P < 0.05, Student’s t test).
COR27 and COR28 Affect the Period Lengths of Circadian Outputs
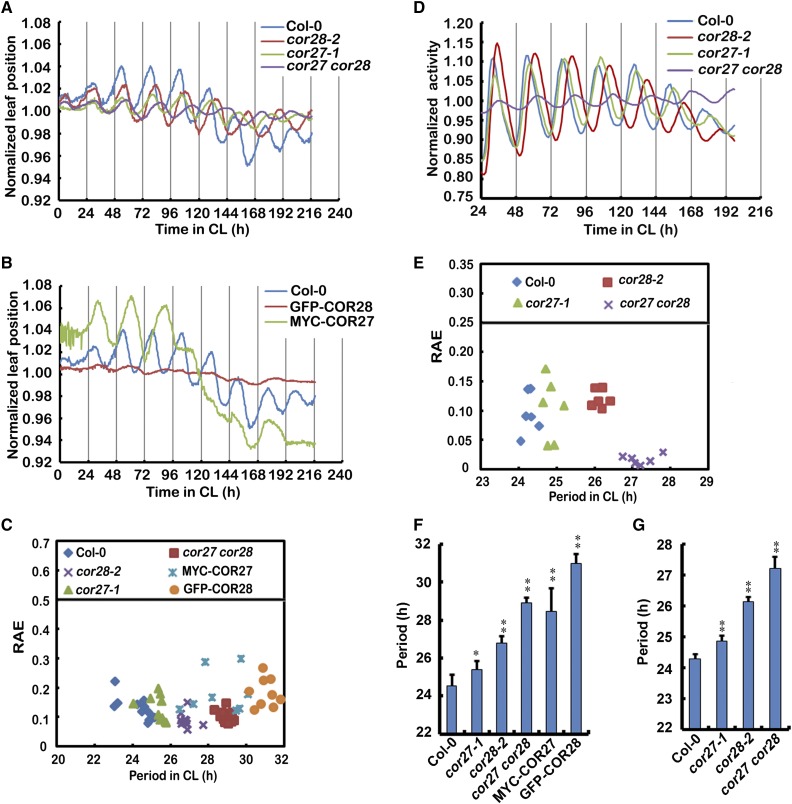
Because COR27 and COR28 are regulated by the clock as direct targets of CCA1, they may be involved in clock regulation. To determine whether COR27 and COR28 affect circadian clock function, we first examined leaf movement rhythms, a well-established circadian response in Arabidopsis (Hicks et al., 1996; Lu et al., 2011). Seedlings were entrained for 10 d in medium-day conditions (MD; 12 h light/12 h dark) and subsequently transferred to constant light (CL). Wild-type plants exhibited a robust rhythmic movement of primary leaves with a free-running period length of 24.3 ± 0.4 h (Figures 6A to 6C). In cor27 or cor28 mutant plants, a robust circadian rhythm of leaf movement was observed, but with a free-running period length of 25.3 ± 0.45 h and 26.8 ± 0.3 h (Figures 6A and 6C). This is ∼1 and 2.5 h longer than that in the wild type. Mutants deficient in both COR27 and COR28 displayed even longer periods than either of the single mutants, having a period 4.9 h longer than seen in wild-type plants. This indicates that COR27 and COR28 function redundantly to regulate period length. Transgenic plants overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 showed significantly longer period phenotypes, which is ∼4.2 h or 6.6 h longer than the wild type, respectively (Figures 6B, 6C, and 6F). To determine the pervasiveness of COR27 and COR28 function in the circadian clock, the circadian reporter ProCCR2:LUC (Strayer et al., 2000) was transformed into cor27, cor28, and cor27 cor28 mutant plants. Luminescence was examined in wild-type and mutant plants entrained for 6 d in LD conditions and then transferred to CL. ProCCR2:LUC expression oscillated with a period length of 24.3 ± 0.2 h in the wild type, 24.9 ± 0.2 h in cor27, 26.1 ± 0.2 h in cor28, and 27.2 ± 0.4 h in the cor27 cor28 mutant (Figures 6D, 6E, and 6G).
Figure 6.
COR27 and COR28 Affect the Period Lengths of Various Circadian Outputs.
(A) and (B) Assay of circadian leaf movement under CL (70 μmol m−2 s−1) conditions. Seedlings were entrained in MD for 10 d and then transferred to CL. Normalized positions (as described in Methods) of primary leaves for the wild type (n = 17), cor27-1 (n = 15), cor28-2 (n = 15), cor27 cor28 (n = 17), MYC-COR27 (n = 16), and GFP-COR28 (n = 15) are shown.
(C) Period and relative amplitude error estimates (RAEs) of the leaf movement rhythms shown in (A) and (B).
(D) ProCCR2:LUC bioluminescence rhythms in indicated genetic backgrounds under CL conditions. Seedlings were entrained in LD conditions for 6 d and then transferred to CL. ProCCR2:LUC activity rhythms were then monitored, and each point is the average of 15 to 20 seedlings normalized as described in Methods.
(E) Period and relative amplitude error estimates of the ProCCR2:LUC bioluminescence rhythms shown in (D).
(F) Period and statistical analysis of the leaf movement rhythms show in (A) and (B). Error bars represent se of data from nine plants.
(G) Period and statistical analysis of the ProCCR2:LUC bioluminescence rhythms shown in (D). Error bars represent se of data from 60 to 90 plants. The asterisks indicate significant differences compared with the wild type (**P < 0.01, *P < 0.05, Student’s t test).
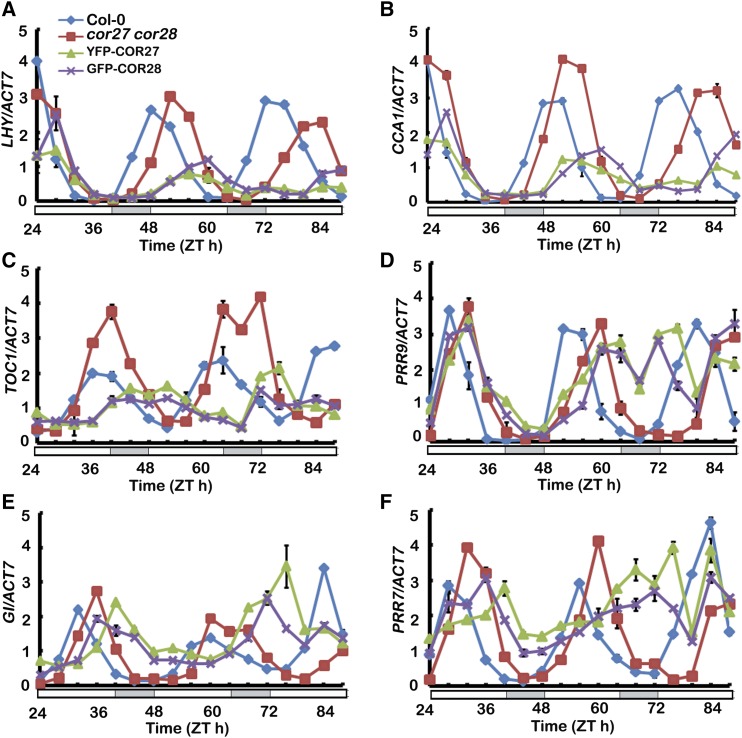
We further confirmed that COR27 and COR28 affect the circadian clock function by analyzing expression of clock genes using qPCR. Seedlings were entrained for 6 d in LD conditions and subsequently transferred to CL. Samples were collected every 4 h for 3 d in the CL condition. All clock genes checked showed longer periods in the cor27 cor28 double mutant and transgenic lines overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 (Figure 7; Supplemental Figure 8). Together, these results show that COR27 and COR28 affect period lengths of circadian output rhythms (leaf movement, ProCCR2:LUC activity, and clock gene transcription), indicating that COR27 and COR28 are involved in regulating period length in the circadian clock. To investigate whether the cold induction of COR27 and COR28 is related to their function in regulation of the clock, we checked the period of cor27 cor28 double mutant and transgenic lines overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 in a temperature entrainment experiment. Seedlings were entrained for 6 d in CL with a diurnal temperature variation of 12 h 22°C/12 h 12°C and subsequently transferred to 22°C in CL. We found that cor27 cor28 double mutants still displayed a significantly longer period phenotype, ∼3 h longer than the wild type. By contrast, transgenic plants overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 showed unrhythmic phenotype (Supplemental Figure 9), indicating that COR27 and COR28 might be involved in the temperature regulation of the circadian clock.
Figure 7.
The cor27 cor28 Double Mutation Lengthens the Free-Running Period of Central Oscillator Gene Expression.
qPCR results showing the expression of the indicated gene in plants of the genotypes indicated. Seedlings were entrained in LD conditions for 6 d before being transferred to CL and collected every 4 h at the indicated times. Expression levels are normalized to the ACT7 mRNA level. Error bars represent sd of three technical replicates. Each experiment was performed at least three times with similar results.
COR27 and COR28 Bind to the Chromatin of PRR5 and TOC1 and Repress Their Expression
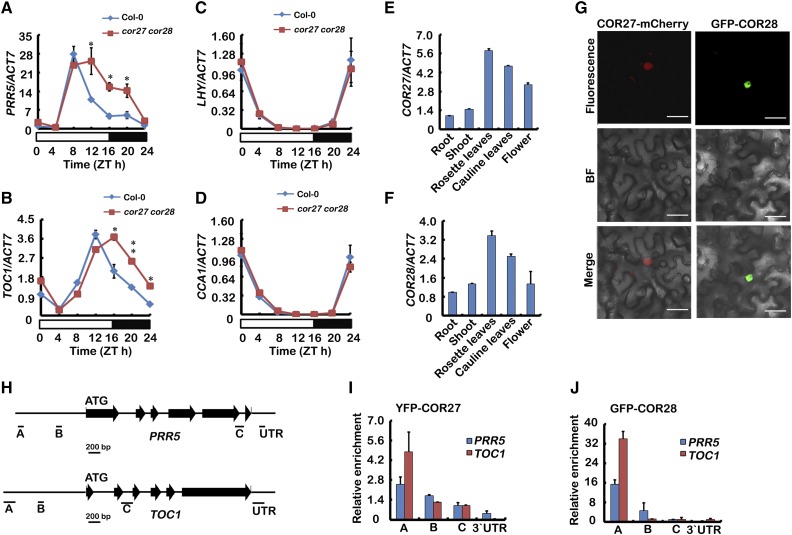
To determine the mechanism of COR27- and COR28-regulated period length in the circadian clock, we measured expression of clock genes in both cor27 cor28 and the wild type in LD conditions. ACT7, APA1, and IPP2 were used as internal controls for normalization in our qPCR analysis. Compared with the wild type, rhythmic expression of TOC1 and PRR5 was substantially increased in cor27 cor28 double plants in the afternoon or night time (Figures 8A and 8B; Supplemental Figures 10A to 10D). Expression of LHY, CCA1, PRR7, and PRR9 was not significantly changed in the cor27 cor28 mutant (Figures 8C and 8D; Supplemental Figures 10E and 10F). To investigate whether the cold-regulated expression of COR27 and COR28 affects their function in the clock regulation, we measured the transcription of PRR5 in cor27 cor28 double mutants following cold treatment. Our results indicate that the transcription of PRR5 is even higher in cor27 cor28 mutants with cold treatment than without cold treatment (Supplemental Figures 10G to 10I). These results suggest that COR27 and COR28 may be negative regulators reducing the cold regulation of PRR5.
Figure 8.
COR27 and COR28 Bind to the Chromatin of PRR5 and TOC1 to Affect Their Expression.
(A) to (D) qPCR results showing expression of PRR5, TOC1, LHY, and CCA1 in cor27 cor28 and the wild type grown in 22°C LD conditions. Samples were collected from 5-d-old seedlings of the genotypes indicated every 4 h over one day in LD. Error bars represent se of three biological replicates. The asterisks indicate significant differences compared with the wild type (**P < 0.01, *P < 0.05, Student’s t test).
(E) and (F) qPCR results showing expression of COR27 and COR28 in different tissues. Expression levels in (A) to (F) are normalized to the ACT7 mRNA level. Error bars represent se of three biological replicates. Each experiment was performed at least three times with similar results.
(G) Subcellular localization of COR27 and COR28 proteins. Epidermal cells of Nicotiana benthamiana leaf were transformed with Pro35S:COR27-mCherry or Pro35S:GFP-COR28. Bars = 10 μm.
(H) Diagram of the gene structures for oscillator genes. Horizontal black lines depict the DNA regions that were amplified by ChIP-qPCR using the indicated primer set.
(I) and (J) Representative result of the ChIP-qPCR assays. ChIP-qPCR assays were performed with an anti-GFP antibody. Plants were grown under LD conditions and harvested at ZT15. The GFP-IP or YFP-IP signal was normalized with the corresponding input signal to get the relative enrichment. Error bars represents sd of three technical repeats. Three independent experiments were performed with similar results.
How could COR27 and COR28 regulate transcription of TOC1 and PRR5? To determine the molecular function of COR27 and COR28, we first analyzed the expression patterns of COR27 and COR28. qPCR analysis indicated that COR27 and COR28 were both highly expressed in rosette leaves, but poorly expressed in root (Figures 8E and 8F). We then examined the cellular localization of COR27 and COR28 and both were detected in the nucleus (Figure 8G). Although COR27 and COR28 could repress transcription of TOC1 and PRR5, we wondered whether they could be physically associating with genomic regions of TOC1 and PRR5 to directly regulate their transcription and therefore performed ChIP-qPCR. ChIP-qPCR assays showed that COR27 and COR28 were associated with chromatin regions of TOC1 and PRR5 promoters and also PRR5’s intragenic regions in vivo (Figures 8H to 8J). COR27 and COR28 are both small proteins with unknown biochemical functions, and they do not have any known DNA binding domains. We employed the random binding site selection assay to examine whether COR27 and COR28 might be sequence-specific DNA binding proteins (He et al., 2005; H. Liu et al., 2008). In this experiment, Escherichia coli-expressed and purified full-length COR27 and COR28 were used to screen for possible interacting DNA sequences. We did not find specific sequences selected by COR27 or COR28 from the pools of random DNAs, so we also performed a yeast one-hybrid analysis and did not detect interaction of COR27 or COR28 with either the PRR5 or the TOC1 promoter (Supplemental Figure 11). Perhaps COR27 and COR28 do not bind to DNA in a sequence-specific manner themselves, but instead physically associate with genomic regions of TOC1 and PRR5 to directly regulate their transcription. These results indicate that COR27 and COR28 may form a protein complex with other transcription factors to associate with chromatin and regulate transcription of clock genes.
DISCUSSION
COR27 and COR28 Are Regulated by Light, Temperature, and the Circadian Clock
Light and temperature are two key environmental signals that profoundly affect plant growth and development, but how these two abiotic factors integrate remains largely unknown. It was previously reported that the red light response was temperature dependent and that it promoted hypocotyl elongation at 27°C but repressed hypocotyl extension at 17°C and 22°C (Johansson et al., 2014). Blue light and CRYs were reported to be required for temperature compensation of the circadian clock (Gould et al., 2013), and phyB and CRY1 were shown to be critical for controlling growth in high temperature (Foreman et al., 2011). We showed recently that the blue light photoreceptor CRY1 interacted directly with PIF4 in a blue light-dependent manner to repress transcriptional activation by PIF4 and that multiple plant photoreceptors (CRY1 and phyB) and ambient temperature can mediate morphological responses through the same signaling component, PIF4 (Ma et al., 2016). Here, we showed that the cold-responsive genes COR27 and COR28 are induced not only by cold but also by reduced ambient temperature (Figure 1), indicating that they are involved in both cold response and ambient temperature response. Blue light and red light repress transcription of COR27 and COR28, whereas blue light stabilizes the proteins (Figure 1), indicating that they are intricately regulated by light. COR27 and COR28 are also direct targets of CCA1, which binds to their chromatin to repress their transcription in the morning (Figure 5; Supplemental Figure 7). COR27 and COR28 are regulated by two key environmental signals and the circadian clock, suggesting that they are key components integrating external light, temperature signals, and the internal circadian clock.
COR27 and COR28 Regulate Freezing Tolerance and Flowering
It was reported that COR27 was rapidly induced in response to low temperature through a CBF-independent pathway (Mikkelsen and Thomashow, 2009). COR28 was also reported to be a cold-responsive gene (Fowler and Thomashow, 2002). COR27 and COR28 share 40.7% nucleic acid sequence identity and 22.7% amino acid similarity with each other, and their biological functions are unknown. Here, we show that they work redundantly in regulating both photoperiodic flowering and freezing tolerance (Figures 2 and 4), and they may present a trade-off between flowering (development) and freezing tolerance because they are positive regulators of flowering but negative regulators of freezing tolerance. How could COR27 and COR28 balance development and freezing tolerance? The circadian clock allows plants to anticipate and prepare for regular environmental changes, thus providing them with an adaptive advantage (Dodd et al., 2005; Greenham and McClung, 2015). The circadian clock is involved in both photoperiodic flowering and cold response. Clock-regulated CDFs repress CO transcription in the morning (Andrés and Coupland, 2012; Romera-Branchat et al., 2014; Song et al., 2015), and, during long days, FKF1 and GI mediate blue light-dependent degradation of CDF1 and stabilization of the CO protein to facilitate transcription of FT (Imaizumi et al., 2005; Song et al., 2012). ZTL regulates the abundance of TOC1 and PRR5, affecting expression of a number of flowering time genes, including FT (Más et al., 2003; Kiba et al., 2007). The circadian clock is also involved in the cold response because the EE (AATATC) is a conserved motif in the promoter of cold-inducible genes (Mikkelsen and Thomashow, 2009; Maruyama et al., 2012). PRR5, PRR7, PRR9, and TOC1 were also reported to be involved in freezing tolerance (Nakamichi et al., 2009; Keily et al., 2013). Our results indicate that COR27 and COR28 work redundantly to regulate period length in the circadian clock (Figure 6) and suggest that COR27 and COR28 balance flowering and freezing tolerance via circadian clock regulation.
Low Temperature- and Blue Light-Regulated COR27 and COR28 Play Roles in the Circadian Clock
The circadian clock is an internal time-keeping system that coordinates daily and seasonal changes of environmental signals with biological processes. Light and temperature are two key input signals entraining circadian clock in higher plants (Song et al., 2015). Various photoreceptors act singularly or together to transduce the light signal into the clock. CRYs transduce blue light to the clock, while phytochromes transduce red/far-red light (Somers et al., 1998). UVR8 acts in mediating low-intensity UV-B light input to the clock (Feher et al., 2011). ZTL and FKF are circadian photoreceptors. They are clock genes and they can also sense blue light (Kim et al., 2007; Sawa et al., 2007). How CRYs, phytochromes, and UVR8 transduce light signals to the clock is largely unknown. Cold also acts as a clock input, as transcription of clock genes is damped at 4°C (Bieniawska et al., 2008), and low temperature-associated alternative splicing of CCA1 mediates clock responses to low temperatures. The low-temperature signal is transduced into the clock by the CCA1β isoform, whereas freezing tolerance is enhanced by the CCA1a isoform (Seo et al., 2012). CBF1 was shown to bind directly to the LUX promoter to regulate the transcription of LUX (Chow et al., 2014). Here, we showed that COR27 and COR28 worked redundantly to regulate period length in the circadian clock, while being regulated by blue light and low temperature. Furthermore, we showed that cold treatment affects COR27- and COR28-regulated transcription of PRR5, and transgenic plants overexpressing Pro35S:YFP-COR27 or Pro35S:GFP-COR28 showed a long-period phenotype in CL conditions with a 12-h-light/12-h-dark entrainment (Figures 6 and 7). By contrast, they showed an unrhythmic phenotype in CL conditions with 12 h 22°C/12 h 12° (Supplemental Figure 9), indicating that COR27 and COR28 might be involved in the temperature regulation of the circadian clock. Thus, there is the possibility that they mediate both light and low temperature inputs to the circadian clock.
In summary, here, we show that COR27 and COR28 are regulated by both light and low-temperature signals and that they are involved in the regulation of the circadian clock. We propose that they balance flowering and freezing tolerance in Arabidopsis. COR27 and COR28 bind to chromatin of TOC1 and PRR5 to repress their transcription, so that they affect the period length of the clock (Figure 9). COR27 and COR28 are not transcription factors, and they cannot bind to DNA themselves in vitro. However, they may act as transcriptional regulators, forming a complex with other DNA binding transcription factors that binds to the chromatin in vivo. To more precisely determine their mechanism of action, COR27- and COR28-interacting proteins need to be identified and further studied.
Figure 9.
A Hypothetical Model Depicting How Light- and Low Temperature-Regulated COR27 and COR28 Play Roles in the Circadian Clock.
The model hypothesizes that the transcription of COR27 and COR28 is induced by cold/low ambient temperature but repressed by blue light. Their proteins are stabilized in response to blue light, and they repress the transcription of TOC1 and PRR5 to affect the period length of the circadian clock. In the meantime, they are regulated by the circadian clock. CCA1 represses the transcription of COR27 and COR28 through direct promoter binding.
METHODS
Plant Materials and Growth Conditions
Except where indicated otherwise, the Columbia ecotype of Arabidopsis thaliana was used. T-DNA insertion mutants cor27-1 (CS834545), cor27-2 (SALK_042072), cor28-1 (CS812929), and cor28-2 (SALK_137155) were obtained from ABRC. The cor27 cor28 double mutant was prepared by crossing cor27-1 with cor28-2, and its identity was verified by genotyping and qRT-PCR. To produce cca1-1 lhy, the cca1-1 mutant in the Wassilewskija background (Green and Tobin, 1999) was backcrossed to Col-0 and was then crossed with T-DNA insertion mutant lhy in the Col-0 background (SALK_031092). For constitutive expression, full-length coding sequences of COR27 and COR28 were cloned into pEarly104 (Pro35S:YFP-COR27), pEarly203 (Pro35S:MYC-COR27), and pMDC43 (Pro35S:GFP-COR28) using the Gateway method. pEarly vectors and pMDC43 are from ABRC. The plasmid Pro35S:COR27-mcherry was prepared by cloning the COR27 cDNA into pCambia1300 vector (Cambia), which codes for the mCherry tag. ProCCR2:LUC reporter was constructed as reported before (Strayer et al., 2000). The ProCAB2:LUC reporter was reported before (Lu et al., 2011). Pro35S:YFP-COR27 and Pro35S:GFP-COR28 were transformed into Col-0 by the floral dip method (Clough and Bent, 1998). For every transformation, greater than 10 independent transgenic lines with a single copy of the transgene were generated. Phenotypes of transgenic plants were verified in at least three independent transgenic lines. Immunoblots were performed to verify overexpression of the transgenes. Pro35S:YFP-COR27 and Pro35S:MYC-COR27 (both in the cor27-1 mutant background) were generated by crossing transgenic lines expressing Pro35S:YFP-COR27 or Pro35S:MYC-COR27 in the wild type background with the cor27-1 mutant. Seeds were sterilized in 10% bleach, placed on Murashige and Skoog medium containing 0.8% agar and 1% sucrose, and stratified for 4 d at 4°C in the dark before being transferred to white light (70 μmol m−2 s−1) in a Percival growth chamber (Percival Scientific).
Light Conditions
Light conditions used were blue light (40 μmol m−2 s−1), red light (40 μmol m−2 s−1), white light (70 μmol m−2 s−1), LD (16 h light/ 8 h dark, white light intensity was 70 μmol m−2 s−1), MD (12 h light/12 h dark, white light intensity was 70 μmol m−2 s−1), SD (8 h light/16 h dark, white light intensity was 70 μmol m−2 s−1), and CL (white light intensity was 70 μmol m−2 s−1).
Analysis of Circadian Rhythms
Transgenic plants expressing the ProCCR2:LUC and ProCAB2:LUC reporters in the cor27-1, cor27-2, cor28-1, cor28-2, and cor27 cor28 backgrounds were prepared by the floral dip method (Clough and Bent, 1998). T2 seedlings were entrained for 6 d in LD conditions before transferred to CL for analysis of LUC activity using an Andor cool camera system. Bioluminescence rhythms of groups of ∼20 seedlings were analyzed using ImageJ software (Lu et al., 2011). The average of all the LUC activity of ∼20 seedlings were set as 1, and all the LUC activity data were normalized to the average to get the normalized LUC activity. For leaf movement analysis, seedlings were entrained for 10 d under a MD and then transferred to CL, and the vertical position of the primary leaves was monitored and analyzed (Lu et al., 2011). Seedlings were individually transferred to the wells of upright 24-well tissue culture plates and the positions of the primary leaves were recorded every 20 min for 9 d using a CCD camera (model LTC 0335) from Bosch. Leaf movement was assessed by measuring the vertical position of the primary leaves using the Image J software. The average of all the vertical positions of the primary leaf was set as 1, and all the vertical positions of the primary leaf were normalized to the average to get the normalized leaf position. Rhythm data of LUC activity and leaf movement were analyzed with BRASS software to get period and relative amplitude error estimates (available from http://www.amillar.org) using the fast Fourier transform nonlinear least square program (Millar et al., 1995; Plautz et al., 1997).
Analysis of Plant Freezing Tolerance
Analysis of freezing tolerance analysis was performed as described previously (Ding et al., 2015) with the following modifications. Arabidopsis plants were grown in LD conditions at 22°C on Murashige and Skoog plates containing 0.8% agar and 1% sucrose for 8 d. The seedlings were then used for the freezing assay in a freezing chamber (RuMED4001). After seedlings were pretreated in the freezing chamber at 4°C for 1 d, the freezing chamber was programmed to drop 2°C per hour to −8°C and was kept in −8°C for 2 h. After the freezing treatment, plants were put into darkness at 4°C for 12 h and then transferred to normal conditions for 3 d, at which time the survival rates were determined. Three independent experiments of three to six replicates each were conducted, and ∼40 to 50 seedlings were analyzed for each replicate.
mRNA Expression Analyses
mRNA expression analyses were performed as described previously (Ma et al., 2016). Total RNAs were isolated using the RNAiso Plus (Takara). cDNA was synthesized from 500 ng of total RNA using the PrimeScript RT reagent kit with genomic DNA Eraser (Takara). SYBR Premix Ex Tag (Takara) was used for the qPCR reactions, using the MX3000 system (Stratagene). Levels of ACT7 or IPP2 or APA1 mRNA expression were used as the internal controls. qRT-PCR data for each sample were normalized to the respective ACT7 or IPP2 or APA1 expression level. The cDNAs were amplified following denaturation, using the 40-cycle programs (95°C, 5 s; 60°C, 20 s per cycle). Biological replicates represent three independent experiments involving ∼30 seedlings per experiment. Three technical replicates were done for each experiment.
Protein Subcellular Localization
Protein subcellular localization experiments were performed as described previously (Y. Liu et al., 2013). GFP-derived and mCherry-derived fluorescence was analyzed using an Olympus BX53 microscope. Excitation/emission wavelengths were as follows: GFP (488 nm/505 to 575 nm) and mCherry (543 nm/560 to 615 nm).
ChIP Assays
ChIP assays were performed as described before (Ma et al., 2016). Seven-day-old LD-grown seedlings harboring Pro35S:YFP-COR27, Pro35S:GFP-COR28, or wild-type genes were used for ChIP. ChIP samples of COR27 and COR28 were collected at ZT15, whereas the sample for CCA1 ChIP was collected at ZT0. Anti-GFP or CCA1 antibodies were used for ChIP. Two grams of seedlings was harvested and treated with 1% formaldehyde (Sigma-Aldrich) under vacuum for 20 min. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M. The seedlings were rinsed with water, frozen in liquid nitrogen, and ground to a fine powder. This starting material was used to precipitate COR27, COR28, or CCA1.
Accession Numbers
Sequence data for genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: COR27 (AT5G42900), COR28 (AT4G33980), CO (AT5G15840), FT (AT1G65480), TSF (AT4g20370), FLC (AT5G10140), SOC1 (AT2G45660), CIB1 (AT4G34530), TOE1 (AT2G28550), SPL9 (AT2G42200), SPL15 (AT3G57920), CCA1 (At2G46830), LHY (At1G01060), PRR9 (At2G46790), PRR7 (At5G02810), PRR5 (At5G24470), TOC1 (At5G61380), GI (At1G22770), CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), ACT7 (AT5G09810), IPP2 (AT3G02780), and APA1 (AT1G11910). T-DNA insertion mutants were obtained from the ABRC: cor27-1 (CS834545), cor27-2 (SALK_042072), cor28-1 (CS812929), cor28-2 (SALK_137155), and lhy (SALK_031092).
Supplemental Data
Supplemental Figure 1. Red Light Represses Transcription of COR27 and COR28, and Low Temperature Does Not Affect COR27 and COR28 Stability.
Supplemental Figure 2. COR27 and COR28 Repress Their Own and Each Other’s Transcription.
Supplemental Figure 3. Isolation and Characterization of COR27 and COR28 T-DNA Insertional Mutants.
Supplemental Figure 4. Overexpression of COR27 and COR28 Leads to a Slightly Late-Flowering Phenotype.
Supplemental Figure 5. The cor27-1 Phenotype Was Fully Rescued by Introduction of Pro35S:MYC-COR27.
Supplemental Figure 6. COR27 Affects Freezing Tolerance.
Supplemental Figure 7. COR27 and COR28 Are Direct Targets of CCA1.
Supplemental Figure 8. The cor27 cor28 Double Mutation Lengthens the Free-Running Period of Central Oscillator Gene Expression.
Supplemental Figure 9. Transgenic Lines Expressing YFP-COR27 and GFP-COR28 Show an Unrhythmic Phenotype in a Temperature-Cycle Condition.
Supplemental Figure 10. COR27 and COR28 Affect the Transcription of PRR5 and TOC1 but Not PRR7 and PRR9.
Supplemental Figure 11. COR27 and COR28 Cannot Bind the Promoter of PRR5 and TOC1 Directly.
Supplemental Data Set 1. Oligonucleotide Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Shuhua Yang for experimental materials used in this study and Parker Johnson for proofreading the manuscript. This work was supported by the National Natural Science Foundation of China (31270285, 31322006, and 31400252) and the Hundred Talents Program of the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
X.L., S.X.L., and H.L. conceived the project. X.L. and D.M. performed most of the experiments. S.X.L. performed the leaf movement assay. X.H. and T.L. made some of the constructs. S.X.L., T.X., and E.M.T. contributed new reagents. D.M., X.L., and R.H. performed the freezing tolerance assays. X.L., D.M., and H.L. analyzed data. D.M. and H.L. wrote the manuscript.
Glossary
- EE
evening element
- LD
long-day
- SD
short-day
- MD
medium-day
- CL
constant light
References
- Amasino R.M., Michaels S.D. (2010). The timing of flowering. Plant Physiol. 154: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Bergonzi S., Albani M.C., Ver Loren van Themaat E., Nordström K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097. [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B.Y., Sanchez S.E., Breton G., Pruneda-Paz J.L., Krogan N.T., Kay S.A. (2014). Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Arabidopsis. Curr. Biol. 24: 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32: 278–289. [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Love J., Webb A.A. (2005). The plant clock shows its metal: circadian regulation of cytosolic free Ca2+. Trends Plant Sci. 10: 15–21. [DOI] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Shimizu H., Nohales M.A., Araki T., Kay S.A. (2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54. [DOI] [PubMed] [Google Scholar]
- Feher B., Kozma-Bognar L., Kevei E., Hajdu A., Binkert M., Davis S.J., Schafer E., Ulm R., Nagy F. (2011). Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana Plant J. 67: 37–48. [DOI] [PubMed] [Google Scholar]
- Foreman J., Johansson H., Hornitschek P., Josse E.M., Fankhauser C., Halliday K.J. (2011). Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65: 441–452. [DOI] [PubMed] [Google Scholar]
- Fowler S., Thomashow M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.D., et al. (2013). Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol. 9: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.M., Tobin E.M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96: 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K., McClung C.R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16: 598–610. [DOI] [PubMed] [Google Scholar]
- Hayama R., Coupland G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Borevitz J.O., Harmon F.G., Pruneda-Paz J.L., Schultz T.F., Yanovsky M.J., Liljegren S.J., Ecker J.R., Kay S.A. (2005). Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 138: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192. [DOI] [PubMed] [Google Scholar]
- Hicks K.A., Millar A.J., Carré I.A., Somers D.E., Straume M., Meeks-Wagner D.R., Kay S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792. [DOI] [PubMed] [Google Scholar]
- Hsu P.Y., Harmer S.L. (2014). Global profiling of the circadian transcriptome using microarrays. Methods Mol. Biol. 1158: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H., Jones H.J., Foreman J., Hemsted J.R., Stewart K., Grima R., Halliday K.J. (2014). Arabidopsis cell expansion is controlled by a photothermal switch. Nat. Commun. 5: 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keily J., MacGregor D.R., Smith R.W., Millar A.J., Halliday K.J., Penfield S. (2013). Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. 76: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang Q., Liu Y., Zhao X., Imaizumi T., Somers D.E., Tobin E.M., Lin C. (2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. USA 110: 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Li K., Liu H., Lin C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Liu H., Knowles S.M., Li J., Ma L., Tobin E.M., Lin C. (2011). A role for protein kinase casein kinase2 α-subunits in the Arabidopsis circadian clock. Plant Physiol. 157: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- Maruyama K., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Kim W.Y., Somers D.E., Kay S.A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570. [DOI] [PubMed] [Google Scholar]
- McClung C.R. (2014). Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Thomashow M.F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60: 328–339. [DOI] [PubMed] [Google Scholar]
- Millar A.J., Carré I.A., Strayer C.A., Chua N.H., Kay S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 (suppl.): S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Fukushima A., Kusano M., Sakakibara H., Mizuno T., Saito K. (2009). Linkage between circadian clock and tricarboxylic acid cycle in Arabidopsis. Plant Signal. Behav. 4: 660–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K., Ishiura M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972. [DOI] [PubMed] [Google Scholar]
- Onouchi H., Igeño M.I., Périlleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Kaneko M., Hall J.C., Kay S.A. (1997). Independent photoreceptive circadian clocks throughout Drosophila. Science 278: 1632–1635. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M., Andrés F., Coupland G. (2014). Flowering responses to seasonal cues: what’s new? Curr. Opin. Plant Biol. 21: 120–127. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sanchez S.E., Yanovsky M.J. (2013). Time for a change. eLife 2: e00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Park M.J., Lim M.H., Kim S.G., Lee M., Baldwin I.T., Park C.M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Imaizumi T. (2015). Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- Thomas B. (2006). Light signals and flowering. J. Exp. Bot. 57: 3387–3393. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang J.W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65: 4723–4730. [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312. [DOI] [PubMed] [Google Scholar]
- Zhou C.M., Zhang T.Q., Wang X., Yu S., Lian H., Tang H., Feng Z.Y., Zozomova-Lihová J., Wang J.W. (2013). Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.