Epidermal phyB non-cell-autonomously promotes the degradation of endodermal PIFs to inhibit hypocotyl negative gravitropism.
Abstract
Seedling hypocotyls display negative gravitropism in the dark but agravitropism in the light. The Arabidopsis thaliana pif quadruple mutant (pifQ), which lacks four PHYTOCHROME-INTERACTING FACTORS (PIFs), is agravitropic in the dark. Endodermis-specific expression of PIF1 rescues gravitropism in pifQ mutant seedlings. Since phytochromes induce light responses by inhibiting PIFs and the COP1-SPA ubiquitin E3 ligase complex in the nucleus, we asked whether phyB can cell autonomously inhibit hypocotyl negative gravitropism in the endodermis. We found that while epidermis-specific expression of PHYB rescues hypocotyl negative gravitropism and all other phyB mutant phenotypes, endodermis-specific expression of PHYB does not. Epidermal phyB induces the phosphorylation and degradation of endodermal PIFs in response to red light. This induces a global gene expression pattern similar to that induced by red light treatment of seedlings expressing PHYB under the control of its own endogenous promoter. Our results imply that epidermal phyB generates an unidentified mobile signal that travels to the endodermis where it promotes PIF degradation and inhibits hypocotyl negative gravitropism.
INTRODUCTION
Phytochromes are red and far-red light photoreceptors that regulate light responses like seed germination, hypocotyl negative gravitropism, seedling photomorphogenesis, shade avoidance, and leaf senescence (Franklin and Quail, 2010; Wang and Wang, 2015). Light-activated phytochromes promote light responses via at least two pathways. Light-activated phytochromes enter the nucleus, interact with SUPPRESSOR OF PHYA-105 (SPA) proteins, and inhibit the ubiquitin E3 ligase activity of the CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)-SPA complex by disrupting the interaction between COP1 and SPA proteins (Zheng et al., 2013; Sheerin et al., 2015; Menon et al., 2016). The COP1-SPA complex promotes the degradation of positive light signaling components like ELONGATED HYPOCOTYL5 (HY5), LONG HYPOCOTYL IN FAR-RED, LONG AFTER FAR-RED LIGHT1, PHY RAPIDLY REGULATED1 (PAR1), and PAR2 (Osterlund et al., 2000; Seo et al., 2003; Duek et al., 2004; Zhou et al., 2014). This means that light responses are induced by activation of phytochromes to inhibit the COP1-SPA complex, thereby stabilizing these positive factors.
Light-activated phytochromes also enter the nucleus and interact with a group of bHLH transcription factors called PHYTOCHROME INTERACTING FACTORs (PIFs) (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999; Leivar and Quail, 2011; Jeong and Choi, 2013). Activated phytochromes inhibit PIFs both by promoting their degradation and by interfering with PIF binding to PIF-target promoters (Bauer et al., 2004; Park et al., 2004, 2012; Nozue et al., 2007; Lorrain et al., 2008). In general, PIFs repress light responses by regulating the transcription of various light-related genes (Leivar et al., 2008, 2009; Shin et al., 2009). PIFs bind hundreds or even thousands of sites across the genome, activating or repressing many target genes, including those related to cell wall biosynthesis and maintenance as well as auxin, gibberellin, and brassinosteroid biosynthesis and signaling (Oh et al., 2009, 2012; Hornitschek et al., 2012; Zhang et al., 2013; Pfeiffer et al., 2014). Since PIFs repress light responses, when light-activated phytochromes inhibit PIFs, light responses are induced by reversal of the PIF-induced gene expression changes. Although these two pathways are the most well characterized, phytochromes likely regulate light responses via other pathways, as other phytochrome-interacting proteins that do not fit either pathway have been reported (Fankhauser et al., 1999; Bae and Choi, 2008; Yasui et al., 2012; Choi et al., 2014; Huang et al., 2016).
Phytochromes are ubiquitously expressed in plants. Early spectroscopic and immunocytochemical analyses showed wide distribution of phytochromes (predominantly phyA) in etiolated seedlings of oat (Avena sativa) and other plant seedlings (Briggs and Siegelman, 1965; Pratt and Coleman, 1971). Later, GUS and GFP reporters corroborated the ubiquitous expression of PHYA and PHYB in Arabidopsis thaliana leaves and roots, as well as other locations including the epidermis, cortex, endodermis, and vascular tissues (Somers and Quail, 1995; Goosey et al., 1997; Salisbury et al., 2007). PHYA1 is expressed throughout tobacco (Nicotiana tabacum) seedlings with strongest expression in the hook, root tips, and vascular tissues (Adam et al., 1994). Rice (Oryza sativa) PHYA is expressed throughout etiolated seedlings with strongest expression in the vascular bundles of the coleoptile and all parts of immature leaves. However, rice PHYB is expressed only weakly in the nodes and internodes of mature plants without any cell-type specificity (Baba-Kasai et al., 2014). In contrast to the phytochromes, the tissue-specific expression patterns of the PIFs are less well characterized. Still, PIFs seem also to be ubiquitously expressed in Arabidopsis seedlings; whole transgenic PIF1pro, PIF3pro, PIF4pro, and PIF5pro-GUS reporter seedlings stain strongly for GUS (Zhang et al., 2013). Considering this likely ubiquitous expression of both phytochromes and PIFs, we were curious whether phytochromes always act cell autonomously in all organs and tissues.
There are some indications that sites of light perception by phytochromes may be distinguishable from the sites of corresponding light responses. In cucumber (Cucumis sativus) seedlings, for example, hypocotyl elongation is inhibited more effectively by shining red light on the cotyledons rather than on the hypocotyls (Black and Shuttleworth, 1974). In Sinapis alba, internode elongation is promoted more effectively by shining end-of-day far-red light (EOD-FR) or light with a low red:far-red ratio (R:FR) on the leaves rather than on the internodes (Casal and Smith, 1988). This suggests phytochromes in the cotyledons and leaves may generate mobile signals that regulate hypocotyl and internode elongation from a distance. In cotyledons and leaves, the phytochromes in the mesophyll cells appear to perceive light and generate interorgan mobile signals, as the removal of phytochrome chromophores in mesophyll cells by CHLOROPHYLL A/B BINDING PROTEIN3 (CAB3) promoter-driven biliverdin IXa reductase (BVR) leads to elongated hypocotyls under both red and far-red light (Warnasooriya and Montgomery, 2009). Both strong phyB expression in mesophyll cells and weak phyB expression in the epidermis of enhancer trap lines rescues phyB mutant phenotypes for flowering time, hypocotyl length, and cotyledon size (Endo et al., 2005). Phytochromes can generate mobile signals that move between cells within an organ: A red light microbeam spotted onto cotyledons induces anthocyanin production and CAB promoter-driven luciferase expression both within and outside the irradiated area (Nick et al., 1993; Bischoff et al., 1997). Mesophyll phyB but not vascular phyB delays flowering by repressing the expression of FLOWERING LOCUS T in vascular tissues (Endo et al., 2005). In other developmental stages, both stomata-specific and vascular tissue-specific expression of PHYB promote stomatal development (Casson and Hetherington, 2014), while endosperm phyB promotes seed germination by inhibiting the biosynthesis of the antigermination hormone abscisic acid (ABA) and its subsequent transport into the embryo (Lee et al., 2012; Kang et al., 2015). Together, these studies suggest light-activated phytochromes generate signals capable of moving to surrounding cells and organs to induce light responses at a distance.
Interorgan phytochrome signaling for hypocotyl and petiole elongation is partly mediated by auxin. Arabidopsis enhancer trap reporter lines with insertions near the auxin-inducible GRETCHEN HAGEN3 or SMALL AUXIN UPREGULATED genes show hypocotyl reporter expression after far-red light irradiation of their cotyledons but not their hypocotyls (Tanaka et al., 2002). Consistent with this result, transcriptome analyses of microsamples from the cotyledons and shoot apex show cotyledon-specific EOD-FR induces auxin- and PIF7-dependent expression of shade genes autonomously in the cotyledons themselves and non-autonomously in the shoot apex (Nito et al., 2015). Low R:FR irradiation of entire plants increases auxin levels by inducing the expression of auxin biosynthetic genes in the leaf lamina. This increased auxin is then transported basipetally to promote petiole elongation (de Wit et al., 2015). Similar non-autonomous interorgan signaling between cotyledons and hypocotyls mediated by auxin has also been observed in Brassica rapa (Procko et al., 2014). Shoot phyB may also promote lateral root formation by enhancing auxin transport (Salisbury et al., 2007). Together, phytochromes in cotyledons and leaves perceive light and generate signals that move downward to regulate light responses in hypocotyls and roots. However, it should be noted that not all phytochrome signals move basipetally. Acropetal induction of light responses within a cotyledon has been observed, as has shoot apex to cotyledon induction and lateral induction from leaf mesophyll cells to vascular tissues (Bischoff et al., 1997; Endo et al., 2005; Nito et al., 2015). Recent reports showing that brassinosteroid (BR) and gibberellic acid (GA) stabilize PIFs independent of light suggest that phytochromes may also regulate light responses non-cell-autonomously by suppressing BR and GA biosynthesis or signaling (Bernardo-García et al., 2014; Li et al., 2016).
Hypocotyl negative gravitropism and its inhibition by phytochromes may provide a good model system for examining the cellular autonomy of phytochrome functions. Gravity detection depends on the sedimentation of amyloplasts in the endodermis (Kiss et al., 1997; Hashiguchi et al., 2013; Toyota et al., 2013). This leads to asymmetric auxin distribution across a hypocotyl and subsequent asymmetric hypocotyl growth against the gravity vector (Brauner and Diemer, 1971; Li et al., 1991; Rakusová et al., 2011). In the dark, PIFs maintain starch-filled amyloplasts in the endodermis, allowing amyloplast sedimentation along the gravity vector. pif quadruple (pifQ) mutant seedlings lack PIF1, PIF3, PIF4, and PIF5 (Shin et al., 2009; Kim et al., 2011). In the dark, pifQ mutant seedlings have etioplast-like plastids instead of starch-filled amyloplasts in their endodermis. This makes them agravitropic, even in the dark. Endodermis-specific expression of PIF1 under the control of the SCR promoter fully rescues the agravitropic phenotype of the pifQ mutant in the dark without rescuing the dark germination or short hypocotyl phenotypes (Kim et al., 2011). This finding suggests that endodermal PIFs promote hypocotyl negative gravitropism in the dark. Importantly, red light still inhibits hypocotyl negative gravitropism of this endodermal PIF1 line (Kim et al., 2011). Together, these results indicate that phytochromes inhibit hypocotyl negative gravitropism by inhibiting endodermal PIFs. It remains unclear, though, whether this inhibition of endodermal PIFs by endodermal phytochromes is cell autonomous.
To determine whether phytochrome B’s inhibition of hypocotyl negative gravitropism is cell autonomous, we generated and characterized transgenic plants expressing PHYB under the control of tissue-specific promoters. We found that epidermal but not endodermal phyB rescues the various phyB mutant phenotypes, including hypocotyl negative gravitropism. To our surprise, but consistent with the rescue experiments, we found epidermal phyB promotes the degradation of endodermal PIFs. Our results indicate epidermal phyB generates a mobile signal that moves from the epidermis to the endodermis to promote the degradation of endodermal PIFs.
RESULTS
Generation of Transgenic Tissue-Specific phyB Lines
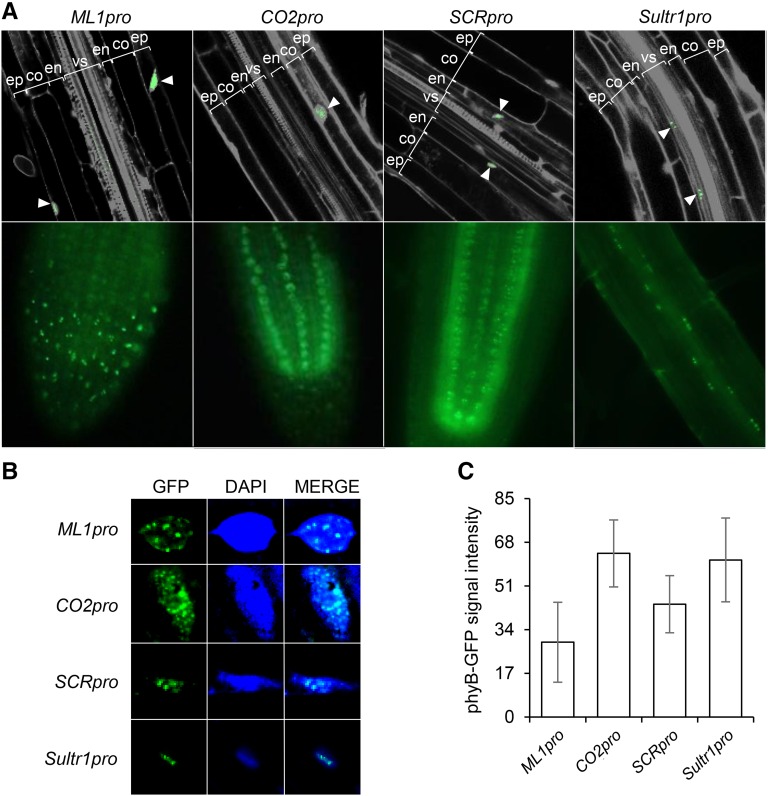
To determine whether phyB regulates hypocotyl negative gravitropism tissue specifically, we generated transgenic plants expressing Arabidopsis PHYB-GFP under the control of several tissue-specific promoters: the MERISTEM LAYER1 (ML1) promoter (ML1pro) for the epidermis, the CO2 promoter (CO2pro) for the cortex, the SCARECROW (SCR) promoter (SCRpro) for the endodermis, and the SULFATE TRANSPORTER 1;3 (SULTR1;3) promoter (Sultr1pro) for the phloem (Di Laurenzio et al., 1996; Sessions et al., 1999; Yoshimoto et al., 2003; Heidstra et al., 2004). We introduced each of these PHYB-GFP transgenes into the phyB-9 mutant to generate at least two independent homozygous lines for each tissue-specific promoter. We then examined the GFP signal in each transgenic plant grown in the light. As expected, we observed GFP signal and, therefore, phyB expression only in the epidermis for ML1pro and in the endodermis for SCRpro (Figure 1A). For CO2pro and Sultr1pro, however, we observed GFP signals mainly in cortex and vascular tissues, respectively, but also occasionally in the epidermis. This nonspecific expression of CO2pro and Sultr1pro was more prominent in white light-grown seedlings.
Figure 1.
Transgenic Arabidopsis Expressing phyB-GFP under the Control of Tissue-Specific Promoters.
(A) Confocal (upper panel) and epifluorescence (lower panel) microscopic images showing tissue-specific phyB-GFP expression (green; white arrowheads) in the roots of 3-d-old red-light-grown transgenic seedlings. PI (gray) was used to counterstain the cell walls and nuclei. ep, epidermis; co, cortex; en, endodermis; vs, vascular structure.
(B) Confocal images of roots from 3-d-old red-light-grown transgenic seedlings showing nuclear body formation by tissue-specific phyB-GFPs (green). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue).
(C) Per cell tissue-specific phyB-GFP signal intensities were quantified from confocal images of individual cells from 3-d-old red-light-grown seedlings of each tissue-specific line. Error bars indicate sd (n ≥ 100).
Since active phyB forms nuclear bodies (Kircher et al., 1999; Yamaguchi et al., 1999; Chen et al., 2003), we confirmed the presence of nuclear bodies in all our transgenic lines as evidence of functional tissue-specific phyB expression (Figure 1B). We next measured the amount of phyB protein per cell in red light-grown transgenic seedlings by quantifying GFP fluorescence intensity from confocal images. We observed similar per cell GFP signal intensities across all the lines and tissues except the ML1pro line, which showed slightly lower intensities (Figure 1C). Together, these results indicate our epidermis- and endodermis-specific PHYB lines produce functional, nuclear body-forming phyB in the expected tissue.
Epidermal phyB Promotes Light Responses
We next asked which light responses are regulated by tissue-specific phyBs. We examined seed germination, hypocotyl elongation, hypocotyl negative gravitropism, and shade avoidance. Although we examined and recorded the phenotypes for all of our lines, we excluded the CO2pro and Sultr1pro lines because of their leaky expression, drawing conclusions only from the ML1pro and SCRpro lines.
Since phyB promotes seed germination, wild-type seeds show high germination frequencies in response to a pulse of red light, but phyB mutant seeds do not. We therefore exposed the tissue-specific PHYB lines to a red-light pulse and examined seed germination. ML1pro and CO2pro showed germination frequencies of 80% or more in response to a red-light pulse, but SCRpro and Sultr1pro showed virtually no seed germination (Figures 2A and 2B). However, the SCRpro and Sultr1pro seeds are capable of germination because all lines showed more than 80% germination under continuous white light conditions. We detected phyB-GFP signal in the endodermis of imbibed SCRpro seeds and in the epidermis of imbibed ML1pro seeds (Figure 2C). From the results of a seed coat bedding assay, we can infer phyB promotes seed germination partly by inhibiting ABA biosynthesis in the endosperm (Lee et al., 2012). We noted ML1pro, although weaker than PHYBpro, is expressed in the endosperm, while SCRpro is not (Figure 2D). Thus, these results suggest phyB in the epidermis or endosperm but not the endodermis can promote seed germination.
Figure 2.
Epidermal phyB Promotes Seed Germination in Response to Red Light.
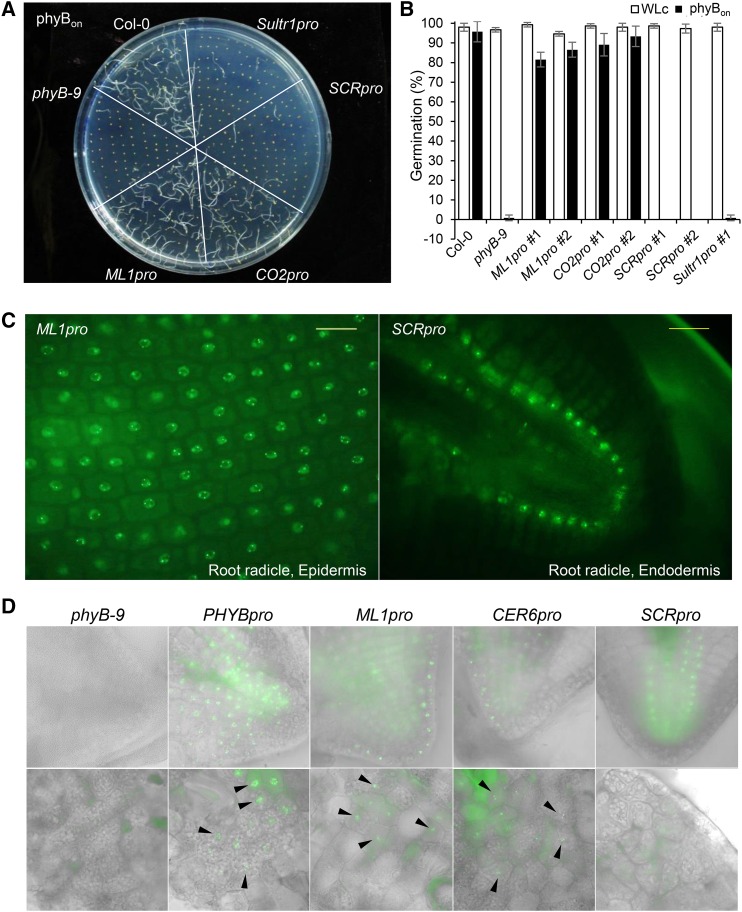
(A) Visualization of germination frequencies for tissue-specific phyB lines under conditions that activate phyB. Seeds were surface-sterilized, treated with far-red light (2.56 μmol m−2 s−1) for 5 min followed by red light (11.4 μmol m−2 s−1) for 5 min, and then incubated in the dark for 4 d (phyBon).
(B) Germination frequencies for each tissue-specific phyB line. White bars indicate the continuous white light (WLc) condition, and black bars indicate the phyBon condition. Error bars indicate sd (n = 3 biological replicates, 50 seeds each).
(C) Epifluorescence images showing the expression of phyB-GFP in the epidermis (ML1pro) and endodermis (SCRpro) of imbibed seeds. Seeds were imbibed for 21 h before their seed coats were removed for epifluorescence imaging. Bars = 20 μm.
(D) Epifluorescence images showing the expression of phyB-GFP in the endosperm of imbibed seeds. Seeds were imbibed for 21 h before their seed coats were removed for epifluorescence imaging. Bright-field images in gray scale were merged with the GFP channel. The black arrowheads indicate phyB-GFP in the endosperm.
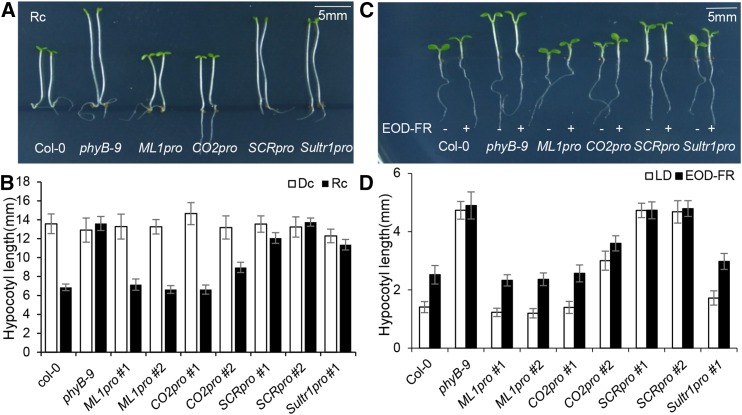
Since phyB inhibits hypocotyl elongation in continuous red light, wild-type seedlings have shorter hypocotyls than phyB mutant seedlings. We therefore exposed the tissue-specific PHYB lines to red light and measured the resulting hypocotyl lengths. While ML1pro and CO2pro seedlings produced short hypocotyls like wild-type, SCRpro, and Sultr1pro seedlings produced long hypocotyls like phyB mutant seedlings (Figures 3A and 3B). In the dark, however, all lines produced equally long hypocotyls. phyB also inhibits hypocotyl elongation in a non-shade light condition. This is why, compared with the constitutively long hypocotyls of phyB mutant seedlings, wild-type hypocotyls grow longer in response to simulated shade (EOD-FR light). We found in a simulated shade experiment that ML1pro, CO2pro, and Sultr1pro seedlings produced long hypocotyls like those of wild-type seedlings. SCRpro seedlings, by contrast, produced constitutively long hypocotyls regardless of the light stimulus, like phyB mutant seedlings (Figures 3C and 3D). The shorter hypocotyls of Sultr1pro seedlings under non-shaded white light may be caused by the leaky epidermal phyB-GFP expression from Sultr1pro under white light. Together, these results indicate that epidermal but not endodermal phyB inhibits hypocotyl elongation in response to red light.
Figure 3.
Epidermal phyB Inhibits Hypocotyl Elongation in Response to Red Light.
(A) Epidermal phyB inhibits hypocotyl elongation. Seedlings were grown for 4 d under continuous red light (Rc; 11.4 μmol m−2 s−1) or in the dark (Dc). Bar = 5 mm.
(B) Hypocotyl lengths for the various tissue-specific phyB lines. White bars dark-grown seedlings (Dc), and black bars indicate red light-grown seedlings (Rc). Error bars indicate sd (n = 60 seedlings).
(C) Epidermal phyB restores EOD-FR responses. Seedlings were grown either in long days for 6 d (LD) or in long days for 2 d followed by 4 d of EOD-FR. Pairs of seedlings are shown for each tissue-specific line. The left seedling of each pair was long-day grown (−) and the right was EOD-FR treated (+). Bar = 5 mm.
(D) Hypocotyl lengths for each tissue-specific phyB line. White bars indicate long-day-grown seedlings, and black bars indicate EOD-FR-treated seedlings. Error bars indicate sd (n = 60 seedlings).
Epidermal phyB Non-Cell-Autonomously Inhibits Hypocotyl Negative Gravitropism
Epidermal phyB’s ability to promote various light responses that may also require nonepidermal tissues suggests epidermal phyB may generate signals that travel to and coordinate the action of other tissues and organs. Among light responses, the site of action for hypocotyl negative gravitropism is well characterized. phyB inhibits hypocotyl negative gravitropism at least in part by reducing the size of endodermal amyloplast starch granules, thus interfering with gravity sensing. The finding that endodermis-specific expression of PIF1 in pifQ mutant seedlings fully rescues both endodermal amyloplast starch levels and hypocotyl negative gravitropism (Shin et al., 2009; Kim et al., 2011) suggests that phyB inhibits negative gravitropism by inhibiting endodermal PIFs. The fact that endodermis-specific expression of a dominant-negative PIF3 in wild-type seedlings disrupts hypocotyl negative gravitropism in the dark also supports this hypothesis (Kim et al., 2016). Since the epidermis and endodermis are quite distinct, hypocotyl negative gravitropism provides a good model system for investigating the potential non-cell-autonomous functions of phyB.
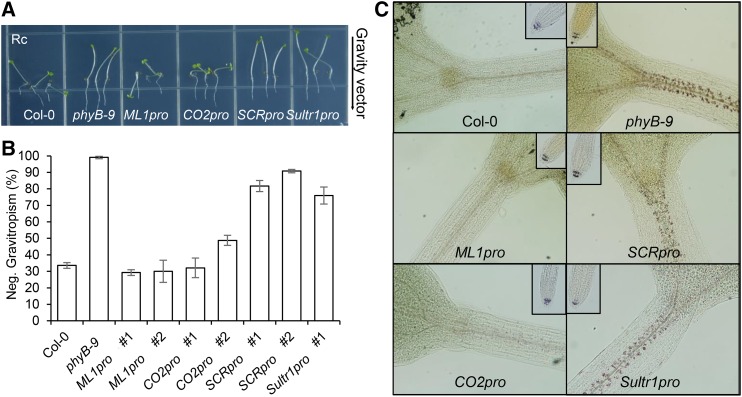
We measured hypocotyl negative gravitropism in the tissue-specific PHYB lines, counting hypocotyls growing within ±45 degrees of vertical as negatively gravitropic. While wild-type hypocotyls grew in random directions under red light, phyB mutant hypocotyls showed vertical growth. This result indicates that endogenous phyB inhibits hypocotyl negative gravitropism in red light. ML1pro and CO2pro seedlings grew randomly under red light like wild-type seedlings, but SCRpro and Sultr1pro lines grew vertically like phyB mutants (Figure 4A). Roughly 30% of wild-type, ML1pro, and CO2pro seedlings showed negative gravitropism in red light compared with more than 80% of phyB mutant, SCRpro, and Sultr1pro seedlings (Figure 4B). Thus, epidermal phyB inhibits hypocotyl negative gravitropism but endodermal phyB does not.
Figure 4.
Epidermal phyB Non-Cell-Autonomously Inhibits Hypocotyl Negative Gravitropism in Response to Red Light.
(A) Inhibition of hypocotyl negative gravitropism by ML1pro-driven epidermal phyB. Seedlings from each line after growing on vertical plates for 4 d in continuous red light (Rc).
(B) Hypocotyl negative gravitropism was quantified by counting seedlings growing within 45° of vertical. Error bars indicate sd (n = 3, 80 seedlings each).
(C) Epidermal phyB induces degradation endodermal amyloplast starch granules. Four-day-old red-light-grown seedlings were stained with Lugol’s iodine to reveal the amyloplast starch granules (brown spots). Small inset images show amyloplast starch granules in the columellar cells of root tips.
Endogenous phyB inhibits hypocotyl negative gravitropism at least in part by reducing the size of endodermal amyloplast starch granules. Consistent with this, Lugol’s iodine only weakly stained endodermal amyloplasts of red light-grown wild-type seedlings but strongly stained those of red light-grown phyB mutant seedlings. Lugol’s iodine also only weakly stained red light-grown ML1pro and CO2pro seedlings, but strongly stained SCRpro and Sultr1pro seedlings (Figure 4C). This staining profile was consistent with the hypocotyl negative gravitropic phenotypes of these lines under red light illumination. In contrast to the endodermal amyloplasts, Lugol’s iodine strongly stained the root columella amyloplasts in all genotypes, including the wild type (Figure 4C). Together, these results indicate that epidermal phyB non-cell-autonomously inhibits hypocotyl negative gravitropism by decreasing the size of endodermal amyloplast starch granules, whereas endodermal phyB cannot. The fact that endodermal PIF1 rescues the pifQ mutant seedling defects in starch-filled endodermal amyloplasts and hypocotyl negative gravitropism strengthens the case for epidermal phyB’s non-cell-autonomous inhibition of endodermal PIFs.
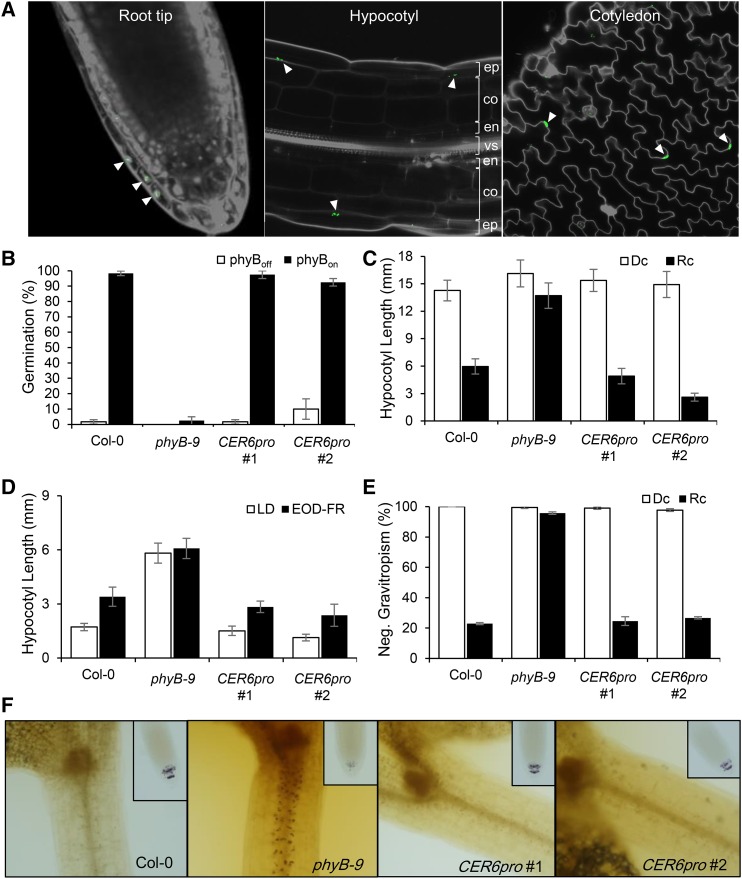
CER6 Promoter-Driven Epidermal phyB Promotes Light Responses
To confirm that epidermal phyB can indeed promote light responses in the epidermis, we generated another epidermis-specific PHYB transgenic line using the CER6 promoter (CER6pro). CER6 encodes ketoacyl coA synthase 6, which is necessary for wax biosynthesis. Like ML1, CER6 expression is limited to the epidermis and its promoter is used regularly to drive epidermis-specific expression (Hooker et al., 2002). As expected, we were able to detect CER6pro-driven phyB-GFP only in the outermost cell layer (Figure 5A). We therefore proceeded to examine various light responses in CER6pro seedlings. Like ML1pro and the wild type, more than 80% of CER6pro seeds germinated in response to a red-light pulse (Figure 5B). Similarly, CER6pro seedling hypocotyl elongation was inhibited under continuous red light (Figure 5C) and stimulated in response to shade (Figure 5D). In addition, CER6pro seedling hypocotyls grew in random orientations under red light (Figure 5E) and showed only weak staining by Lugol’s iodine (Figure 5F). Together, these results confirm that epidermal phyB (ML1pro or CER6pro) can promote various light responses.
Figure 5.
Epidermal phyB (CER6pro) Also Promotes Light Responses.
(A) Confocal images showing epidermis-specific phyB expression (white arrowheads) in the root tips, hypocotyls, and cotyledons of 3-d-old red-light-grown CER6pro seedlings. PI (gray) was used to counterstain the cell walls and nuclei. ep, epidermis; co, cortex; en, endodermis; vs, vascular structure.
(B) Seed germination frequencies of the CER6pro lines. Seeds were surface-sterilized and either treated with far-red light (2.56 μmol m−2 s−1) for 5 min followed by red light (11.4 μmol m−2 s−1) for 5 min followed by 4 d of incubation in the dark (phyBon; black bars) or treated with far-red light (2.56 μmol m−2 s−1) for 5 min followed by 4 d of incubation in the dark (phyBoff; white bars). Error bars indicate sd (n = 3 biological replicates, 50 seeds each).
(C) Hypocotyl lengths for the tissue-specific phyB lines. White bars indicate hypocotyl length of dark-grown seedlings (Dc), and black bars indicate hypocotyl length of red-light-grown seedlings (Rc). Error bars indicate sd (n = 60 seedlings).
(D) Hypocotyl lengths were measured after seedlings were grown either in long days for 6 d (LD) or in long days for 2 d followed by 4 d of EOD-FR treatment. White bars indicate hypocotyl length of long-day-grown seedlings, and black bars indicate hypocotyl length of EOD-FR-treated seedlings. Error bars indicate sd (n = 60 seedlings).
(E) Hypocotyl negative gravitropism was quantified by counting seedlings growing within 45° of vertical. Error bars indicate sd (n = 3, 80 seedlings each).
(F) Epidermal phyB induces degradation of endodermal amyloplast starch granules. Four-day-old red-light-grown seedlings were stained with Lugol’s iodine to reveal the amyloplast starch granules (brown spots). Small inset images show amyloplast starch granules in the columellar cells of root tips.
Epidermal phyB Non-Cell-Autonomously Promotes the Degradation of Endodermal PIFs
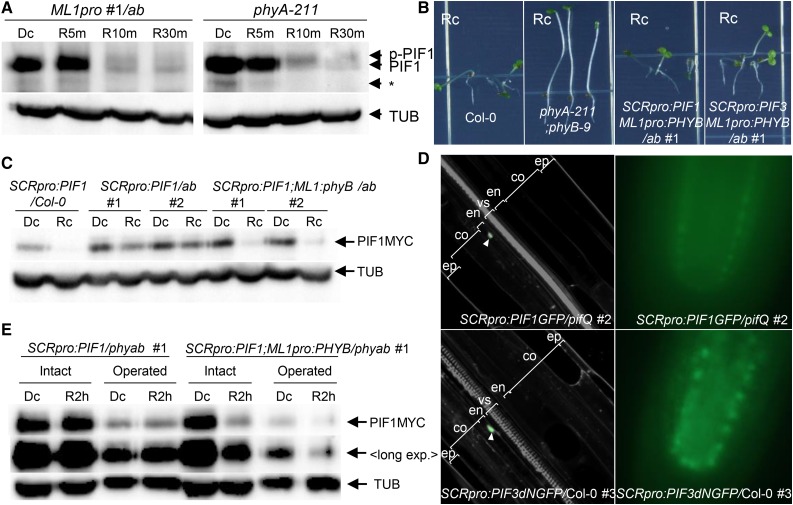
Phytochromes induce light responses in part by promoting the degradation of PIFs. Since epidermal phyB is capable of inducing light responses in the endodermis, we next asked whether it induces PIF degradation by comparing PIF1 levels in phyA-211 and ML1pro:PHYB/phyA-211;phyB-9 (ML1pro:PHYB/ab) seedlings. Indeed, endogenous PIF1 protein levels fell rapidly by similar amounts when dark-grown phyA and ML1pro:PHYB/ab mutant seedlings were transferred to red light (Figure 6A). The fact that epidermis-specific PHYB expression and endogenous PHYB expression induced similar levels of PIF1 degradation suggested that epidermal phyB may promote the degradation of PIF1 both in the epidermis and elsewhere.
Figure 6.
Epidermal phyB Non-Cell-Autonomously Promotes the Degradation of Endodermal PIFs.
(A) Epidermal phyB induces endogenous PIF1 degradation. Dark-grown seedlings were transferred to red light (9 μmol m−2 s−1) for 5, 10, or 30 min (R5m, R10m, or R30m) before being sampled to measure PIF1 levels using an anti-PIF1 antibody. PIF1 indicates endogenous PIF1, while p-PIF1 indicates the slower-migrating phosphorylated form. TUB indicates α-tubulin. The asterisk indicates nonspecific bands.
(B) Epidermal phyB inhibits hypocotyl negative gravitropism in lines expressing PIFs specifically in the endodermis. These images were taken after allowing the seedlings to grow on vertical plates for 4 d in continuous red light (Rc).
(C) Epidermal phyB induces degradation of endodermal PIF1. Seedlings were grown either in the dark (Dc) or in continuous red light (Rc) for 4 d before detection of MYC-tagged endodermal PIF1 (PIF1MYC) with an anti-MYC antibody.
(D) Confocal (left) and epifluorescence (right) microscopic images showing endodermis-specific GFP signal (green; white arrowheads) of 3-d-old dark-grown SCRpro:PIF3dNGFP/Col-0 seedlings counterstained with PI (gray). PIF3dN lacks the N-terminal 300 amino acids of wild-type PIF3. ep, epidermis; co, cortex; en, endodermis; vs, vascular structure.
(E) Epidermal phyB induces degradation of endodermal PIF1 in seedlings lacking cotyledons and the shoot apex. Four-day-old dark-grown seedlings were subjected to cotyledon and shoot apex removal (Operated) under a green safety light before being transferred back to either the dark (Dc) or to red light for 2 h (R2 h). MYC-tagged PIF1 was detected using an anti-MYC antibody. “Long exp.” indicates higher exposure.
To determine whether epidermal phyB non-cell-autonomously promotes PIF1 degradation, we generated transgenic lines expressing MYC-tagged PIF1 in the endodermis under the control of the SCR promoter in either the phyA-211 phyB-9 (ab) double mutant background (SCRpro:PIF1/ab) or the ML1pro:PHYB/ab background (SCRpro:PIF1;ML1pro:PHYB/ab). In the dark, both of these lines showed normal vertical growth. In red light, SCRpro:PIF1/ab mutant seedlings showed vertical growth, but the SCRpro:PIF1;ML1pro:PHYB/ab seedlings showed random growth orientations (Figure 6B; Supplemental Figure 1). This loss of hypocotyl negative gravitropism suggests that epidermal phyB inhibits endodermal PIFs in red light. Indeed, we were able to observe a strong reduction in endodermal PIF1 in the presence of epidermal phyB (SCRpro:PIF1;ML1pro:PHYB/ab) but not in its absence (SCRpro:PIF1/ab) (Figure 6C). The weak residual degradation of endodermal PIF1 in SCRpro:PIF1/ab seedlings is likely caused by other phytochromes such as phyD (Bauer et al., 2004). This degradation of endodermal PIF1 by epidermal phyB could not be attributed to movement of endodermal PIF1 to the epidermis; endodermis-specific expression of PIF1-GFP in the pifQ mutant background remained localized to the endodermis, as did endodermis-specific expression of a nondegradable PIF3dN-GFP in the wild-type background (Figure 6D). The SCR promoter reportedly produces additional expression in the epidermis of the shoot apical meristem (Wysocka-Diller et al., 2000), suggesting a potential overlap between ML1pro-driven phyB and SCRpro-driven PIF1 in the epidermis of shoot apex. However, we still observed PIF1 degradation in seedlings lacking cotyledons and the shoot apex (Figure 6E). Together, these results indicate epidermal phyB non-cell-autonomously promotes the degradation of endodermal PIF1s in response to red light.
The degradation of PIFs induced by red light is preceded by phosphorylation. We therefore asked whether epidermal phyB induces endodermal PIF1 phosphorylation and degradation in response to red light. Unfortunately, we were unable to reliably detect red light-induced shifts in the migration of MYC-tagged PIF1 protein bands. In contrast to the situation with PIF1, we were able to reliably detect shifts in MYC-tagged PIF3 protein bands. Thus, we generated a new transgenic line with endodermis-specific expression of MYC-tagged PIF3 in the ML1pro:PHYB/ab background (SCRpro:PIF3;ML1pro:PHYB/ab). This line, like SCRpro:PIF1/ML1pro:PHYB/ab, grew in random directions under red light (Figure 6B; Supplemental Figure 1). This result is consistent with epidermal phyB inhibiting endodermal PIF3, as it does PIF1.
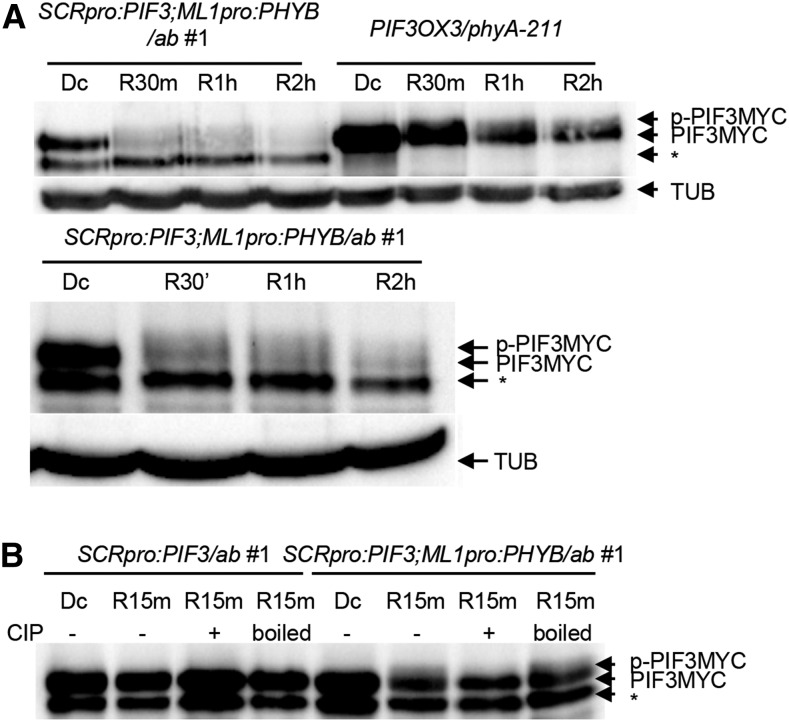
We next measured endodermal PIF3 degradation during the dark-to-light transition. Like endodermal PIF1, endodermal PIF3 is rapidly degraded when dark-grown seedlings are transferred to red light (Figure 7A). In fact, red light induced the degradation of endodermal PIF3 by epidermal phyB just as rapidly as globally expressed PIF3 was degraded by endogenous phyB. In other words, the physical separation of endodermal PIF3 and epidermal phyB by the cortex did not noticeably delay endodermal PIF3 degradation. We next asked whether epidermal phyB induces the phosphorylation of endodermal PIF3 in response to red light. According to a previous report, phosphorylated PIFs migrate more slowly on a gel than their unphosphorylated forms. We observed these slow-migrating endogenous PIF1 bands in lysates from both ML1pro:PHYB/ab and phyA mutant seedlings transferred to red light (Figure 6A). We also noticed similar slow-migrating PIF3 bands in lysates from the SCRpro:PIF3;ML1pro:PHYB/ab line and PIF3OX3/phyA-211 seedlings (Figure 7A), suggesting epidermal phyB induces the phosphorylation of endodermal PIF3. In the absence of epidermal phyB (SCRpro:PIF3/ab), red light did not shift the migration of endodermal PIF3, nor was its migration affected by phosphatase treatment (Figure 7B). Phosphatase treatment converted the slow-migrating PIF3 bands from SCRpro:PIF3;ML1pro:PHYB/ab seedlings to their faster-migrating counterparts, but treatment with heat-inactivated phosphatase did not (Figure 7B). These experiments confirm that the slow-migrating endodermal PIF3 band induced by epidermal phyB is indeed the phosphorylated form. Together, these results indicate epidermal phyB non-cell-autonomously induces the phosphorylation and degradation of endodermal PIF3.
Figure 7.
Epidermal phyB Non-Cell-Autonomously Promotes the Phosphorylation and Degradation of Endodermal PIF3.
(A) Epidermal phyB induces the degradation of endodermal PIF3. Dark-grown seedlings were transferred to red light (9 μmol m−2 s−1) for 30 min, 1 h, or 2 h (R30, R1 h, or R2 h) before being sampled to measure endodermal MYC-tagged PIF3 levels using an anti-MYC antibody. PIF3MYC indicates endodermal MYC-tagged PIF3, while p-PIF3MYC indicates the slower-migrating phosphorylated form. TUB indicates α-tubulin. The asterisk indicates nonspecific bands. The lower panel is an independent experiment with a longer exposure.
(B) Epidermal phyB induces phosphorylation of endodermal PIF3. Dark-grown seedlings were transferred to red light for 15 min before sampling. Precipitated PIF3MYC proteins were treated with either CIP or boiled CIP (boiled). MYC-tagged PIF3 was detected using an anti-MYC antibody. The asterisk indicates nonspecific bands.
Epidermal phyB and Endogenous Global phyB Induce Similar Gene Expression Patterns in Response to Red Light
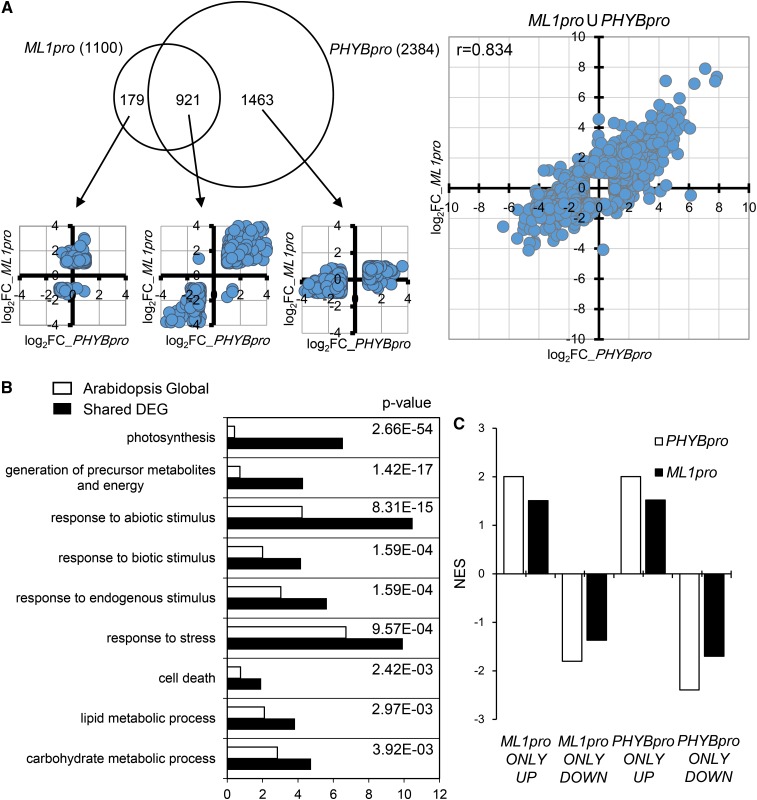
Since epidermal phyB induces degradation of PIFs and rescues all phyB mutant phenotypes, it is likely epidermal phyB induces changes in gene expression similar to those induced by global phyB. To confirm this, we compared the transcriptomes of phyB mutants with either an epidermis-specific phyB rescue (ML1pro:PHYB/phyB-9) or a global phyB rescue (PHYBpro:PHYB/phyB-9), all under red light. If we set the threshold for differentially expressed genes (DEGs) at 2-fold or higher, the comparison of the phyB mutant with a global phyB rescue produced 2384 DEGs, but the comparison with an epidermal phyB rescue produced only 1100 DEGs (Figure 8A; Supplemental Data Set 1). Of these, 921 were shared DEGs, comprising 84% of the epidermal phyB DEGs (921 of 1100) but only 39% of the global phyB DEGs (921 of 2384). All of these shared DEGs fell in the first and third dot plot quadrants and produced a correlation coefficient of 0.917 (Figure 8A), indicating that epidermal and global phyB regulated these genes in the same direction. Using Gene Ontology analysis, we found a strong enrichment for photosynthesis-related genes among these shared DEGs (Figure 8B). This suggests that photosynthesis is an important biological process regulated by both global and epidermal phyB.
Figure 8.
Epidermal and Global phyB Induce Similar Changes in Gene Expression in Response to Red Light.
(A) A Venn diagram comparing DEGs induced by epidermal and global phyB. Transcriptome analyses were performed by comparing 4-d-old red light-grown ML1pro (ML1pro:PHYB-GFP/phyB-9) seedlings or PHYBpro (PHYBpro:PHYB-GFP/phyB-9) seedlings with phyB-9 mutant seedlings. DEGs are genes with ≥2-fold changes in expression and false discovery rate ≤ 0.05. Dot plots comparing log2 fold changes for the indicated DEGs from the epidermal and global phyB transcriptomes. The x axis indicates log2 expression fold differences between PHYBpro seedlings and phyB-9 seedlings, while the y axis indicates log2 expression fold differences between ML1pro seedlings and phyB-9 seedlings.
(B) Gene Ontology analysis showing an enrichment for photosynthesis genes and other biological processes among the shared DEGs. The top nine enriched biological processes are ranked by P values that compare the enrichment in the whole transcriptome (white bars) to the shared DEGs (black bars).
(C) Gene set enrichment analysis showing enrichment of nonshared DEGs in the other group’s transcriptome. Statistically significant enrichments (false discovery rate ≤ 0.15) are presented as NES (normalized enrichment scores). ML1pro and PHYBpro indicate the corresponding transcriptomes. UP and DOWN indicate gene sets comprising up- or downregulated genes among the nonshared epidermal phyB and global phyB DEGs.
Although epidermal and global phyB shared many DEGs, these shared DEGs were asymmetrically distributed (i.e., 84% of epidermal phyB DEGs versus 39% of global phyB DEGs). This asymmetry may indicate that epidermal phyB alters the expression of a subset of genes altered by global phyB. Alternatively, a similar pattern would arise if epidermal and global phyB regulate similar genes, but epidermal phyB was expressed more weakly than global phyB. When we performed a dot plot analysis, we found that the non-shared DEGs fell mainly in the first and third quadrants (1340 of 1463 global phyB-specific DEGs [proportion test, P = 0.00017] and 150 of 179 epidermal phyB-specific DEGs [proportion test, P = 0.0014]) (Figure 8A). These DEGs in the first and third quadrants show a correlation coefficient of 0.834 (Figure 8A). This indicates the non-shared DEGs, as with the shared DEGs, were regulated in the same direction by epidermal and global phyB. When we performed a gene set enrichment analysis (Subramanian et al., 2005), we found that the up- and downregulated non-shared DEGs were enriched in genes up- and downregulated, respectively, in the other group’s transcriptome (Figure 8C). This suggests that the non-shared DEGs were classified as “non-shared” mainly because they showed a lower fold change in one transcriptome versus the other, primarily in that of epidermal phyB. Roughly 70% of the shared DEGs also showed a lower fold change in the epidermal phyB transcriptome than in the global phyB transcriptome. The fact that epidermal and global phyB regulated similar sets of genes likely explains the nearly identical regulation of light responses induced by epidermal and global phyB expression.
DISCUSSION
phyB induces red light responses in part by inhibiting PIF transcription factors and the COP1-SPA ubiquitin E3 ligase complex in the nucleus. This mechanism implies a cell autonomous function for phyB. The fact that phytochromes are ubiquitously expressed coupled with the penetration of light into plant tissues strengthens the case for a cell autonomous function for phytochromes in regulating light responses (Somers and Quail, 1995; Goosey et al., 1997). However, some studies suggest that shade (low R:FR or EOD-FR) regulates hypocotyl, petiole, and internode elongation non-cell-autonomously by increasing auxin biosynthesis in the leaves (Casal and Smith, 1988; Tanaka et al., 2002; Procko et al., 2014; de Wit et al., 2015) and that red light promotes seed germination by decreasing ABA biosynthesis in the endosperm (Lee et al., 2012). Thus, the extent to which phyB-induced mobile signals may regulate light responses remains unclear.
Here, we characterized various light responses including hypocotyl negative gravitropism using tissue-specific PHYB lines. We found that epidermal and not endodermal phyB rescues all phyB mutant phenotypes. Epidermal phyB promotes seed germination in response to a red light pulse, inhibits hypocotyl negative gravitropism by reducing amyloplast starch in the endodermis, inhibits hypocotyl elongation under continuous red light, and suppresses shade responses in the light. Importantly, endodermal phyB does not even rescue the phenotype that is known to depend on the endodermis (i.e., hypocotyl negative gravitropism). Epidermal phyB’s ability to promote light responses is not an artifact of high PHYB-GFP mRNA expression levels as the ML1pro line, which expresses PHYB-GFP at levels even lower than the SCRpro line, also promotes light responses (Supplemental Figure 2). Although it is possible that differential chromophore availability may block the function of endodermal phyB, we observed nuclear body formation with all tissue-specific phyBs in their respective tissues. This suggests phyBs are assembled properly with chromophore in all tissues (Chen et al., 2003). Others have reported phyB expression in the mesophyll, another “external” not “internal” tissue layer, also rescues phyB mutant phenotypes (Endo et al., 2005). In addition, enhancer trap lines expressing phyB in the vascular tissues, another “internal” location, does not rescue the phyB mutant hypocotyl elongation defect (Endo et al., 2005). All of these results, including the lack of rescue in our endodermal phyB line, suggest that phyBs in these inner tissue layers do not play critical roles in phyB-mediated light responses. Our finding that epidermal phyB regulates hypocotyl elongation and other light responses is consistent with the notion that the epidermis restricts stem growth (Kutschera and Niklas, 2007) and that epidermal BR and auxin signaling are sufficient to regulate growth (Savaldi-Goldstein et al., 2007; Procko et al., 2016).
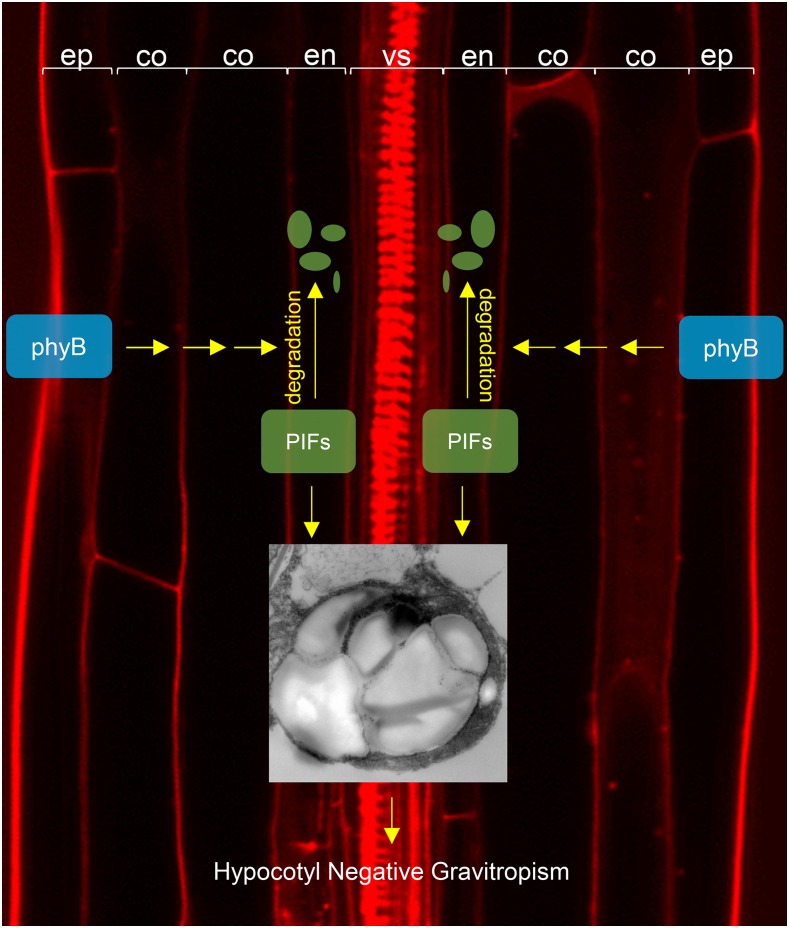
Our data indicate that phyB generates mobile signals that inhibit hypocotyl negative gravitropism. Red light disrupts hypocotyl negative gravitropism in the presence of epidermal phyB but not in the presence of endodermal phyB. As in wild-type seedlings, this disruption of hypocotyl negative gravitropism by red light in the presence of epidermal but not endodermal phyB is accompanied by a dramatic reduction in endodermal amyloplast starch. This result supports a model in which epidermal phyB generates mobile signals that travel to the endodermis to decrease amyloplast starch, thereby inhibiting hypocotyl negative gravitropism (Figure 9). Tissue-specific expression of PHOTOTROPIN1 (PHOT1) under the control of the ML1 or SCR promoters induces similar global dephosphorylation of NON-PHOTOTROPIC HYPOCOTYL3 (Preuten et al., 2013). This implies that mobile signals coordinate light responses downstream of other photoreceptors as well.
Figure 9.
Model Depicting the Non-Cell-Autonomous Function of phyB in the Inhibition of Hypocotyl Negative Gravitropism.
Epidermal phyB inhibits hypocotyl negative gravitropism at least in part by promoting the degradation of endodermal PIFs in response to red light. This loss of endodermal PIFs results in a reduction in the size of endodermal starch granules. The physical separation of the epidermis and endodermis by the layers of the cortex suggests that epidermal phyB generates mobile signals that travel to the endodermis to promote PIF degradation. These mobile signals have not yet been identified.
We found epidermal and global phyB activate similar signaling pathways to rescue phyB mutant phenotypes. Our data indicate that both epidermal and global phyB promote PIF degradation in response to red light. It is not yet known whether mesophyll-specific expression of phyB affects PIF stability in red light, but the fact that several PIF target genes (e.g., PIL2) are DEGs in the CAB3pro-driven BVR transcriptome suggests as much (Oh et al., 2013). In addition, epidermal and global phyB induce many shared DEGs in response to red light, indicating that they regulate gene expression in similar ways. Although epidermal and global phyB do induce non-shared DEGs, most of these genes were regulated in the same direction by epidermal and global phyB and were classified as non-shared only because they fell below the 2-fold change threshold in one transcriptome or the other. The epidermal phyB transcriptome shows a tendency toward weaker fold changes in gene expression than the global phyB transcriptome. This may be due either to lower levels of phyB in the tissue of interest or simply to its expression in a limited area. Regardless, these data support our conclusion that epidermal and global phyB activate similar signaling pathways.
A recent characterization of epidermal phyA, also under the control of the ML1 promoter (Kirchenbauer et al., 2016), indicates that its ability to induce light responses is distinct from that of epidermal phyB. While we found that epidermal phyB fully rescues the long-hypocotyl phenotype of phyB mutants in response to red light, epidermal phyA shows only a partial rescue of the long-hypocotyl phenotype of phyA mutants in response to far-red light (Kirchenbauer et al., 2016). The long hypocotyls of phyA mutant seedlings are further shortened when mesophyll-specific and phloem-specific expression of PHYA are added. The cotyledon-expansion phenotype of phyA mutant seedlings, too, is partially rescued by epidermal phyA and shows progressive improvement with the addition of mesophyll and phloem PHYA expression. Epidermal phyA does not rescue the phototropism and flowering phenotypes of phyA mutants. By contrast, our current results show that epidermal phyB fully rescues all phyB mutant phenotypes including seed germination, hypocotyl negative gravitropism, hypocotyl length, and shade responses. Furthermore, epidermal phyB promotes the degradation of endodermal PIFs non-cell-autonomously in response to red light, whereas epidermal phyA promotes the degradation of PIF1 only cell-autonomously in response to far-red light (Kirchenbauer et al., 2016). The similar levels of endogenous PIF1 degradation induced by epidermal phyB and endogenous phyB suggest that epidermal phyB promotes the degradation of PIFs via both cell autonomous and non-cell-autonomous mechanisms. The differences between epidermal phyA and epidermal phyB that underlie their differing abilities to non-cell-autonomously induce light responses remain unclear. Although far-red light deeply penetrates plant tissues, chloroplasts absorb and thus limit the penetration of red light. Since phyB responds to red light, it may have evolved the ability to coordinate light responses in deeper layers by generating signals that migrate inward from the red light-exposed outer cell layers such as the epidermis and mesophyll (Figure 9). By contrast, phyA responds to deeply penetrating far-red light, and perhaps it evolved to function cell-autonomously.
Our results raise an unexpected question concerning phyB signaling: Must phyB interact with PIFs to promote the phosphorylation and degradation of PIFs? phyB interacts with PIFs via its APB domain and promotes their phosphorylation and degradation in response to red light (Khanna et al., 2004; Al-Sady et al., 2006). Mutations in the APB domain abolish both phyB’s ability to interact with PIFs and its ability to induce their degradation (Al-Sady et al., 2006; Lorrain et al., 2008; Shen et al., 2008). This suggests that PIF degradation requires phyB and PIFs to physically interact. It is unclear, though, whether phytochromes promote PIF degradation only by physically interacting with them. Our data show that epidermal phyB can promote the phosphorylation and degradation of endodermal PIFs, suggesting that physical interaction may not be an absolute requirement.
The degradation of endodermal PIFs we observed is unlikely to be caused by overlapping expression from the ML1 and SCR promoters. Consistent with published reports of strict tissue specificity for these promoters, we were able to see GFP signal only in the epidermis for ML1pro and in the endodermis for SCRpro. However, another group reported some expression from the SCR promoter in the epidermis of the shoot apical meristem (Wysocka-Diller et al., 2000). Since epidermal phyB induces the degradation of endodermal PIF3 to the same extent as endogenous phyB induces the degradation of global PIF3, it is unlikely that the phenotypes we observed can be attributed to the localized degradation of SCRpro-driven PIF3 in a few cells in the shoot apex. To further rule out this possibility, we showed that epidermal phyB induces the degradation of endodermal PIF1 even when the cotyledons and shoot apex are removed before red light treatment. If PIF degradation occurs mainly in the shoot apex, specific expression of BVR in the meristematic regions should mimic phyB mutant phenotypes, which it does not (Warnasooriya and Montgomery, 2009). Enhancer trap lines expressing PHYB in shoot apex also do not rescue phyB mutant phenotypes (Endo et al., 2005). Our results thus argue against a localized degradation endodermal PIFs in the epidermis of the shoot apex.
Since proteins larger than the size limit of plasmodesmata can reportedly be transported to neighboring cells, it is possible epidermal phyB and endodermal PIFs move into and interact within the same cells (Wolf et al., 1989; Nakajima et al., 2001; Kim et al., 2003; Zhou et al., 2013; Han et al., 2014). HY5, a bZIP transcription factor, is one example. HY5 expressed in shoots moves to the roots to induce root expression of NITRATE TRANSPORTER 2.1 (Chen et al., 2016). However, such transport is unlikely in the case of phyB-GFP and PIF-GFP, as we were unable to observe any phyB-GFP signal outside the epidermis in the ML1pro:PHYB line (Figure 1) or any PIF-GFP signal outside the endodermis in either the SCRpro:PIF1-GFP line or the nondegradable SCRpro:PIF3dN-GFP line (Figure 6D). Although we cannot completely rule out the transfer of a small fraction of phyB and/or PIF molecules to neighboring tissues for degradation, our observation that endogenous phyB and epidermal phyB induce similar levels of PIF degradation (Figure 6A) argues against this possibility. Our results indicate that phyB is capable of promoting the degradation of PIFs without direct physical interaction (Figure 9). However, it is possible that despite the lack of detectable phyB-GFP signal outside the epidermis (ML1pro), an undetectable level of extraepidermal phyB-GFP is sufficient to induce light responses. A similar hypothesis was offered as a possible explanation for how low levels of ectopic PHOT1-GFP expression under the control of the CUP-SHAPED COTYLEDON3 promoter can induce phototropism (Sullivan et al., 2016).
Although we still do not know the identity of the mobile signals generated by epidermal phyB, plant hormones are appealing candidates. Light antagonizes the action of growth-promoting hormones like auxin, BR, and GA by decreasing hormone levels or by repressing hormonal signaling (Jeong et al., 2007; Halliday et al., 2009; Lau and Deng, 2010; Kim et al., 2014; Kurepin and Pharis, 2014). However, it remains unclear where phyB would regulate hormone signaling. Interestingly, ML1-driven epidermal expression of CPD, a BR biosynthetic gene, or BRI1, a BR receptor, rescues the cpd or bri1 mutant phenotypes, respectively. ATHB8-driven expression of CPD or BRI1 in internal tissues does not rescue the mutant phenotypes, suggesting that epidermal BR and its downstream signaling pathways are important promoters of growth (Savaldi-Goldstein et al., 2007). Similarly, ML1-driven epidermal expression of axr3-1, a nondegradable form of INDOLEACETIC ACID-INDUCED PROTEIN17 that represses auxin signaling, is sufficient to repress auxin signaling. This suggests that the epidermis is an important site for auxin signaling (Procko et al., 2016). By contrast, bioactive GAs accumulate in the root endodermis. Endodermis-specific expression of gai-1, a nondegradable form of GAI that represses GA signaling, is sufficient to repress the GA signal required for root elongation (Ubeda-Tomás et al., 2009). In addition, BR stabilizes PIF4 protein by preventing its phosphorylation by BRASSINOSTEROID-INSENSITIVE2 protein kinase (Bernardo-García et al., 2014). GA stabilizes four PIFs (i.e., PIF1, 3, 4, and 5) by degrading DELLA proteins and preventing their interaction with the PIFs (Li et al., 2016). This stabilization of the PIFs by BR and GA is light independent, suggesting that hormone regulation in one tissue may affect PIF protein levels in other tissues. Future experiments will clarify whether any of these plant hormones is the mobile phyB signal that induces light responses by promoting PIF degradation in distant tissues.
METHODS
Plant Materials and Growth
Arabidopsis thaliana plants were grown in a growth room with a 16-h-light/8-h-dark cycle at 22 to 24°C for general growth and seed harvesting (fluorescent light bulb [FL40EX-D], 100 μmol m−2 s−1). To generate the tissue-specific PHYB lines, the ML1, CO2, SCR, Sultr1;3, CER6, and PHYB promoters, as well as the PHYB coding region, were PCR-amplified using primer sets detailed in Supplemental Table 1 and cloned into pCAMBIA1300 (Di Laurenzio et al., 1996; Sessions et al., 1999; Yoshimoto et al., 2003; Heidstra et al., 2004) using the indicated restriction sites. These cloned vectors were transformed by floral dipping into the phyB-9 single mutant background, and homozygous lines were established for analysis. ML1pro:PHYB/phyA-211/phyB-9 line was generated by crossing ML1pro:PHYB/phyB-9 #1 line with the phyA-211 phyB-9 double mutant followed by appropriate selection. To generate the endodermal PIF lines, the SCR promoter and the PIF1 and PIF3 cDNAs were PCR-amplified with primer sets detailed in Supplemental Table 1 and cloned into a pBI121 vector in which the GUS gene was replaced with a HTM tag (9xHis-6xMYC) (Park et al., 2004). The cloned vectors were then transformed into Col-0, phyA-211;phyB-9, and ML1pro:PHYB/phyA-211;phyB-9 (ML1pro/ab #1) followed by selection of homozygous lines for analysis. PIF3OX3/phyA-211, SCRpro:PIF1-GFP/pifQ, and SCRpro:PIF3dN were previously described (Kim et al., 2011, 2016; Park et al., 2012).
Fluorescence Microscopic Imaging
Seedlings were mounted with distilled water and observed using either an epifluorescence microscope (Olympus BX-21) or a confocal microscope (Zeiss LSM 710). Cell walls and nuclei were visualized by staining with propidium iodide (PI; 30 μM in distilled water). In some cases, 4′,6-diamidino-2-phenylindole was also used to visualize nuclei. To quantify phyB-GFP intensities, all fluorescence images were taken at the same laser intensity and the same digital gain settings. phyB-GFP signal intensities were quantified using ImageJ with mean gray value options.
Seed Germination and Seedling Light Responses
The germination assays were performed as previously described. Seeds were surface sterilized and plated on 1/100 MS agar (1/100 MS, 0.8% phytoagar, and 0.05% MES, pH 5.7). The plates were irradiated with far-red light (2.56 μmol m−2 s−1) for 5 min (phyBoff) or far-red light followed by red light (11.4 μmol m−2 s−1) for 5 min (phyBon). After incubation for 4 d in the dark at 22°C, germination frequencies were determined by counting the seeds with protruding radicles.
For seedling light responses (i.e., hypocotyl negative gravitropism, hypocotyl elongation, and EOD-FR responses), surface sterilized seeds were plated on MS agar (0.5× MS, 0.8% phytoagar, and 0.05% MES, pH 5.7), imbibed for 3 d at 4°C in the dark, and irradiated with white light (100 μmol m−2 s−1) for 6 h for the induction of germination. For the hypocotyl negative gravitropism assay, plates were incubated vertically for 4 d under continuous red light (11.4 μmol m−2 s−1). The frequency of hypocotyl negative gravitropism was quantified by counting seedlings growing within 45° of vertical. For the hypocotyl length measurements, plates were incubated horizontally for 4 d either in the dark or under continuous red light (11.4 μmol m−2 s−1). Hypocotyl lengths of ≥45 seedlings were measured per sample. For EOD-FR responses, plates were incubated horizontally for 2 d in a 16-h-light/8-h-dark cycle and grown 4 d more with or without a 5-min far-red light (2.56 μmol m−2 s−1) pulse at the end of each light cycle. Hypocotyl lengths of ≥60 seedlings were measured per sample. Endodermal amyloplasts were stained with I2-KI solution as previously described (Kim et al., 2011).
CIP Treatment and Immunoblotting
The calf intestine alkaline phosphatase (CIP) treatment experiment was performed as previously described. For CIP treatments, seedlings were grown in the dark for 4 d and transferred to red light for 15 min. Sampled seedlings were ground in liquid nitrogen and further homogenized in a denaturing buffer (100 mM NaH2PO4, pH 8.0, 10 mM Tris-Cl, 8 M urea, 1 mM PMSF, and 1× complete protease inhibitor cocktail [Roche]). After centrifugation at 20,676g for 10 min at 4°C, PIF3 was purified from the supernatants using Ni-NTA beads (Qiagen). The pellets were then washed twice with PBS (8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, and 0.24 g L−1 KH2PO4, pH 7.4) and once with CIP buffer (NEB buffer 3: 100 mM NaCl, 50 mM Tris/HCl, 10 mM MgCl2, and 1 mM DTT, pH 7.9). The resuspended pellets were then treated for 15 min at 37°C with either no enzyme, 100 units of CIP (NEB), or a comparable amount of boiled CIP. Following CIP treatment, the reaction mixtures were boiled in 2× SDS sample buffer (45 mM Tris/HCl, pH 6.8, 10% v/v glycerol, 1% w/v SDS, 0.05% w/v bromophenol blue, and 50 mM DTT) and subjected to immunoblot analysis with an anti-MYC antibody. TUBULIN was detected with anti-α-tubulin antibody (T-5168; Sigma-Aldrich). MYC was detected with c-Myc (A-14) antibody (sc-789; Santa Cruz). Endogenous PIF1 was detected with previously reported PIF1 antibody (Lee et al., 2014).
Immunoblotting was performed as previously described. For the immunoblots in Figure 6E, the cotyledons and shoot apex of 4-d-old dark grown seedlings were cut with scissors below the apical hook under a green safety light. Cut seedlings were then transferred either to the dark or to red light (11.4 μmol m−2 s−1) for 2 h before being sampled for immunoblotting analysis.
Microarray Analysis
Seedlings were grown for 96 h in continuous red light (11.4 μmol m−2 s−1) before sampling. The Agilent Arabidopsis Genome 44k chip was used for microarray analyses. The analysis was performed using the LIMMA package from the Bioconductor R project. Background correction was performed using the normexp method as it is implemented in LIMMA. The background-corrected intensity data were normalized using the Lowess method to remove bias from the arrays. DEGs were defined as genes with a false discovery rate below 5% that showed a 2-fold or greater difference in expression (Supplemental Data Set 1).
Gene Expression Analysis
Seedlings were grown under continuous red light (11.4 μmol m−2 s−1) for 4 d and harvested for RNA extraction. Total RNAs were isolated using the Spectrum plant total RNA kit (Sigma-Aldrich) according to the manufacturer’s protocol and converted to cDNA using MMLV-RTase (Promega). The transcript levels were determined by real-time PCR using specific primer sets (Supplemental Table 1) and normalized with respect to the expression levels of PP2A.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHYB (AT2G18790), PHYA (AT1G09570), PIF1 (AT2G20180), PIF3 (AT1G09530), ML1 (AT4G21750), CO2 (AT1G62500), SCR (AT3G54220), Sultr1;3 (AT1G22150), CER6 (AT1G68530), and PP2A (AT1G13320). Expression data are available at the NCBI Gene Expression Omnibus under accession number GSE87591.
Supplemental Data
Supplemental Figure 1. Epidermal phyB inhibits hypocotyl negative gravitropism in SCRpro:PIF1;ML1pro:PHYB/ab and SCRpro:PIF3;ML1pro:PHYB/ab.
Supplemental Figure 2. ML1pro expresses less PHYB-GFP mRNA than SCRpro, but still promotes light responses.
Supplemental Table 1. List of primers.
Supplemental Data Set1. Genes regulated by epidermal and global phyB.
Supplementary Material
Acknowledgments
We thank Akira Nagatani at Kyoto University for sharing data. This work was supported in part by grants from the National Research Foundation of Korea (2015R1A2A1A05001091 and 2011-0031955) and the Rural Development Administration (SSAC-PJ011073) to G.C.
AUTHOR CONTRIBUTIONS
J.K., K.S., E.P., K.K., G.B., and G.C. designed the experiments. J.K., K.S., E.P., K.K., and G.B. performed the experiments. J.K. and G.C. wrote the article.
Glossary
- R:FR
red:far-red ratio
- EOD-FR
end-of-day far-red light
- ABA
abscisic acid
- BR
brassinosteroid
- GA
gibberellic acid
- DEG
differentially expressed gene
- PI
propidium iodide
- CIP
calf intestine alkaline phosphatase
References
- Adam E., Szell M., Szekeres M., Schaefer E., Nagy F. (1994). The developmental and tissue-specific expression of tobacco phytochrome-a genes. Plant J. 6: 283–293. [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Baba-Kasai A., Hara N., Takano M. (2014). Tissue-specific and light-dependent regulation of phytochrome gene expression in rice. Plant Cell Environ. 37: 2654–2666. [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311. [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.M., Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F., Millar A.J., Kay S.A., Furuya M. (1997). Phytochrome-induced intercellular signalling activates cab:luciferase gene expression. Plant J. 12: 839–849. [Google Scholar]
- Black M., Shuttleworth J.E. (1974). The role of the cotyledons in the photocontrol of hypocotyl extension in Cucumis sativus L. Planta 117: 57–66. [DOI] [PubMed] [Google Scholar]
- Brauner L., Diemer R. (1971). [The influence of the geotropic induction on the content and the distribution of auxin in the hypocotyls of Helianthus and on their sensitivity to the growth substance]. Planta 97: 337–353. [DOI] [PubMed] [Google Scholar]
- Briggs W.R., Siegelman H.W. (1965). Distribution of phytochrome in etiolated seedlings. Plant Physiol. 40: 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Smith H. (1988). Persistent effects of changes in phytochrome status on internode growth in light-grown mustard: Occurrence, kinetics and locus of perception. Planta 175: 214–220. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. (2014). phytochrome B Is required for light-mediated systemic control of stomatal development. Curr. Biol. 24: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Schwab R., Chory J. (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA 100: 14493–14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N.P., Fu X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26: 640–646. [DOI] [PubMed] [Google Scholar]
- Choi H., Jeong S., Kim D.S., Na H.J., Ryu J.S., Lee S.S., Nam H.G., Lim P.O., Woo H.R. (2014). The homeodomain-leucine zipper ATHB23, a phytochrome B-interacting protein, is important for phytochrome B-mediated red light signaling. Physiol. Plant. 150: 308–320. [DOI] [PubMed] [Google Scholar]
- de Wit M., Ljung K., Fankhauser C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208: 198–209. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301. [DOI] [PubMed] [Google Scholar]
- Endo M., Nakamura S., Araki T., Mochizuki N., Nagatani A. (2005). Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosey L., Palecanda L., Sharrock R.A. (1997). Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday K.J., Martinez-Garcia J.F., Josse E.M. (2009). Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 1: a001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Kumar D., Chen H., Wu S., Kim J.Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65: 1737–1749. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y., Tasaka M., Morita M.T. (2013). Mechanism of higher plant gravity sensing. Am. J. Bot. 100: 91–100. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18: 1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang H., Yoo C.Y., Bindbeutel R., Goldsworthy J., Tielking A., Alvarez S., Naldrett M.J., Evans B.S., Chen M., Nusinow D.A. (2016). PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife 5: e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., Lee S., Kim S.L., Hwang I., An G. (2007). Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ. 30: 590–599. [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yim S., Choi H., Kim A., Lee K.P., Lopez-Molina L., Martinoia E., Lee Y. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6: 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Jeong Y.J., Corvalán C., Fujioka S., Cho S., Park T., Choe S. (2014). Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 77: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Yuan Z., Jackson D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362. [DOI] [PubMed] [Google Scholar]
- Kim K., Shin J., Lee S.H., Kweon H.S., Maloof J.N., Choi G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 108: 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Jeong J., Kim J., Lee N., Kim M.E., Lee S., Chang Kim S., Choi G. (2016). PIF1 regulates plastid development by repressing photosynthetic genes in the endodermis. Mol. Plant 9: 1415–1427. [DOI] [PubMed] [Google Scholar]
- Kirchenbauer D., Viczián A., Ádám É., Hegedűs Z., Klose C., Leppert M., Hiltbrunner A., Kircher S., Schäfer E., Nagy F. (2016). Characterization of photomorphogenic responses and signaling cascades controlled by phytochrome-A expressed in different tissues. New Phytol. 211: 584–598. [DOI] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J.Z., Guisinger M.M., Miller A.J., Stackhouse K.S. (1997). Reduced gravitropism in hypocotyls of starch-deficient mutants of Arabidopsis. Plant Cell Physiol. 38: 518–525. [DOI] [PubMed] [Google Scholar]
- Kurepin L.V., Pharis R.P. (2014). Light signaling and the phytohormonal regulation of shoot growth. Plant Sci. 229: 280–289. [DOI] [PubMed] [Google Scholar]
- Kutschera U., Niklas K.J. (2007). The epidermal-growth-control theory of stem elongation: an old and a new perspective. J. Plant Physiol. 164: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577. [DOI] [PubMed] [Google Scholar]
- Lee N., Kang H., Lee D., Choi G. (2014). A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. Plant J. 78: 282–293. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26: 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Yu R., Fan L.M., Wei N., Chen H., Deng X.W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hagen G., Guilfoyle T.J. (1991). An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell 3: 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Menon C., Sheerin D.J., Hiltbrunner A. (2016). SPA proteins: SPAnning the gap between visible light and gene expression. Planta 244: 297–312. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Nick P., Ehmann B., Furuya M., Schafer E. (1993). Cell communication, stochastic cell responses, and anthocyanin pattern in mustard cotyledons. Plant Cell 5: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K., Kajiyama T., Unten-Kobayashi J., Fujii A., Mochizuki N., Kambara H., Nagatani A. (2015). Spatial regulation of the gene expression response to shade in Arabidopsis seedlings. Plant Cell Physiol. 56: 1306–1319. [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Warnasooriya S.N., Montgomery B.L. (2013). Downstream effectors of light- and phytochrome-dependent regulation of hypocotyl elongation in Arabidopsis thaliana. Plant Mol. Biol. 81: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt L.H., Coleman R.A. (1971). Immunocytochemical localization of phytochrome. Proc. Natl. Acad. Sci. USA 68: 2431–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T., Hohm T., Bergmann S., Fankhauser C. (2013). Defining the site of light perception and initiation of phototropism in Arabidopsis. Curr. Biol. 23: 1934–1938. [DOI] [PubMed] [Google Scholar]
- Procko C., Crenshaw C.M., Ljung K., Noel J.P., Chory J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Burko Y., Jaillais Y., Ljung K., Long J.A., Chory J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30: 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H., Gallego-Bartolomé J., Vanstraelen M., Robert H.S., Alabadí D., Blázquez M.A., Benková E., Friml J. (2011). Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67: 817–826. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Nagatani A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10: 859–868. [DOI] [PubMed] [Google Scholar]
- Salisbury F.J., Hall A., Grierson C.S., Halliday K.J. (2007). Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 50: 429–438. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263. [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Quail P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7: 413–427. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S., Takemiya A., Kharshiing E., Cloix C., Shimazaki K.I., Christie J.M. (2016). Functional characterisation of Arabidopsis phototropin 1 in the hypocotyl apex. Plant J., doi/10.1111/tpj.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nakamura S., Mochizuki N., Nagatani A. (2002). Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol. 43: 1171–1181. [DOI] [PubMed] [Google Scholar]
- Toyota M., Ikeda N., Sawai-Toyota S., Kato T., Gilroy S., Tasaka M., Morita M.T. (2013). Amyloplast displacement is necessary for gravisensing in Arabidopsis shoots as revealed by a centrifuge microscope. Plant J. 76: 648–660. [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T.S., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang H. (2015). Phytochrome signaling: time to tighten up the loose ends. Mol. Plant 8: 540–551. [DOI] [PubMed] [Google Scholar]
- Warnasooriya S.N., Montgomery B.L. (2009). Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol. 149: 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom C.M., Beachy R.N., Lucas W.J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Mukougawa K., Uemoto M., Yokofuji A., Suzuri R., Nishitani A., Kohchi T. (2012). The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24: 3248–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Saito K., Yamaya T., Takahashi H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., Lee J.Y., Lee J.Y. (2013). Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 25: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Song M., Yang Q., Su L., Hou P., Guo L., Zheng X., Xi Y., Meng F., Xiao Y., Yang L., Yang J. (2014). Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 164: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.