A unique set of 21-nucleotide small RNAs may help tighten transposable element and gene expression via posttranscriptional gene silencing using a methylation state-activated isoform of DICER-LIKE4.
Abstract
Plant RNA silencing operates via RNA-directed DNA-methylation (RdDM) to repress transcription or by targeting mRNAs via posttranscriptional gene silencing (PTGS). These pathways rely on distinct Dicer-like (DCL) proteins that process double-stranded RNA (dsRNA) into small-interfering RNAs (siRNAs). Here, we explored the expression and subcellular localization of Arabidopsis thaliana DCL4. DCL4 expression predominates as a transcription start site isoform encoding a cytoplasmic protein, which also represents the ancestral form in plants. A longer DCL4 transcript isoform encoding a nuclear localization signal, DCL4NLS, is present in Arabidopsis, but DNA methylation normally suppresses its expression. Hypomethylation caused by mutation, developmental reprogramming, and biotic stress correlates with enhanced DCL4NLS expression, while hypermethylation of a DCL4 transgene causes a reduction in DCL4NLS expression. DCL4NLS functions in a noncanonical siRNA pathway, producing a unique set of 21-nucleotide-long “disiRNAs,” for DCL4NLS isoform-dependent siRNAs, through the nuclear RdDM dsRNA synthesis pathway. disiRNAs originate mostly from transposable elements (TEs) and TE-overlapping/proximal genes, load into the PTGS effector ARGONAUTE1 (AGO1), and display a subtle effect on transcript accumulation together with overlapping 24-nucleotide siRNAs. We propose that, via PTGS, disiRNAs could help to tighten the expression of epigenetically activated TEs and genes using the methylation-state-responsive DCL4NLS.
INTRODUCTION
In many eukaryotes, RNA silencing serves key roles in gene regulation and defense against invasive nucleic acids including transposable elements (TEs) and viruses. In the model plant Arabidopsis thaliana, posttranscriptional gene silencing (PTGS) targets mRNAs, whereas transcriptional gene silencing (TGS) can be directed on chromatin via RNA-directed DNA methylation (RdDM). PTGS and RdDM require different Dicer-like (DCL) RNase III endonucleases to cleave double-stranded RNA (dsRNA) precursors into small-interfering RNAs (siRNAs) or microRNAs (miRNAs) of discrete length (Borges and Martienssen, 2015; Bologna and Voinnet, 2014).
RdDM relies primarily on 24-nucleotide-long siRNAs produced by DCL3 from dsRNA synthesized by the concerted activities of RNA polymerase IV (PolIV) and RNA-dependent RNA polymerase 2 (RDR2). Twenty-four-nucleotide siRNAs then load into specific Argonaute (AGO) effectors, primarily AGO4, which are believed to bind RNA polymerase V transcripts and recruit the DNA methyltransferase DRM2 to methylate cytosines in any sequence context (CG, CHG, CHH) (Law and Jacobsen, 2010; Stroud et al., 2013). The de novo methylated CG and CHG states can be independently maintained by METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE2 (CMT2/CMT3), respectively, while maintenance of asymmetric CHH methylation relies on continued RdDM or CMT2 action (Stroud et al., 2014); additional repressive histone tail methylation reinforces TGS by further bolstering DNA methylation (Law and Jacobsen, 2010). Most 24-nucleotide siRNAs derive from TE- and repeat-associated loci. Constitutive loss of CHH methylation in RdDM mutants increases transcription at certain loci, yet causes minimal phenotypes, presumably because robust CG and CHG methylation maintains repression at most loci (Stroud et al., 2014), and/or because additional stimuli are necessary to alter gene expression. During pollen, ovule, and seed development, as well as in response to biotic stress, CHH methylation is reduced via active DNA demethylation and repression of methyltransferases. In these reproductive or infected tissues, TE-proximal genes involved in development or defense, respectively, are derepressed, leading to a broad hypothesis in the silencing field that dynamic epigenetic changes may enable developmental reprogramming and stress adaptation (Dowen et al., 2012; Yu et al., 2013; Ibarra et al., 2012; Gehring et al., 2009; Slotkin et al., 2009; Jullien et al., 2012).
PTGS, by contrast, targets expressed mRNAs for cleavage and/or translational repression (Chen, 2004; Aukerman and Sakai, 2003; Brodersen et al., 2008; Baumberger and Baulcombe, 2005) and is performed by several AGOs, including the main effector AGO1. PTGS is guided both by miRNAs, processed by DCL1 from RNA-PolII-dependent fold-back precursors, as well as 21-nucleotide trans-acting siRNAs (tasiRNAs) produced by DCL4 and its cofactor DRB4 (dsRNA BINDING PROTEIN4) following dsRNA amplification by RDR6 and SUPPRESSOR OF GENE SILENCING3 (SGS3) (Vazquez et al., 2004; Hiraguri et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Dunoyer et al., 2005; Adenot et al., 2006). DCL4 also processes evolutionarily young miRNAs with near-perfect complementarity and viral dsRNA (Rajagopalan et al., 2006; Bouché et al., 2006). DCL2 functions in these pathways as well, by producing distinct 22-nucleotide siRNAs from endogenous inverted repeat loci and RNA viruses (Henderson et al., 2006; Deleris et al., 2006; Wang et al., 2011).
TGS and PTGS are often considered to be genetically isolated pathways. However, divergent DCLs may substitute for one another to synthesize siRNAs from the same dsRNA templates. For example, in dcl3 mutants, RDR2-dependent dsRNA can be processed by DCL4 and DCL2, while in dcl4 mutants, DCL3 and DCL2 can process RDR6-dependent dsRNA (Gasciolli et al., 2005; Xie et al., 2005). In wild-type plants, DCL3 could access RDR6-synthesized dsRNA from a reactivated TE and promote RdDM (Marí-Ordóñez et al., 2013), and several additional variations of RDR6-induced RdDM have been described on TE and TAS (tasiRNA-producing) loci or during virus infection, suggesting that the DCL hierarchy can be modified in biologically relevant manners in wild-type plants (Bond and Baulcombe, 2015; Nuthikattu et al., 2013; Wu et al., 2012; Pontier et al., 2012; McCue et al., 2015). Thus, PTGS and TGS may be interconnected under some conditions.
Subcellular compartmentalization could work to isolate PTGS and TGS. Hence, nuclear proteins such as PolIV, RDR2, DCL3, and DRM2 regulate the TGS pathway (Pontes et al., 2006; Law and Jacobsen, 2010; Jullien et al., 2012), while AGO4 was shown to shuttle between the nucleus and cytoplasm (Ye et al., 2012). In contrast, PTGS components, including RDR6, SGS3, AGO1, and DRB4, are entirely or partly cytoplasmic, a logical state for a pathway that targets mature mRNAs (Kumakura et al., 2009; Derrien et al., 2012; Zhu et al., 2013; Martínez de Alba et al., 2015; Jouannet et al., 2012). Plant DCLs have all been assigned a nuclear localization based primarily on heterologous and/or overexpression studies (Xie et al., 2004; Hiraguri et al., 2005; Fang and Spector, 2007; Kumakura et al., 2009), while immunolocalization results presented by Hoffer et al. (2011) showed DCL4 antibody staining in both the nucleus and cytoplasm. Nuclear localization of Arabidopsis DCL4, in particular, poses a paradox because DCL4 operates on RDR6/SGS3 templates and on RNA viruses, which replicate in the cytoplasm (Laliberté and Sanfaçon, 2010; Pumplin and Voinnet, 2013). Here, we investigated aspects of DCL4 expression regulation and determined the localization of Arabidopsis DCL proteins using fluorescent reporter fusions. We provide evidence that the ancestral plant DCL4 is localized in the cytoplasm, while a DNA methylation-regulated DCL4 transcript isoform encodes a protein with a nuclear localization signal (NLS) that can process PolIV/RDR2-dependent dsRNA into 21-nucleotide siRNAs during silique development. We propose that this Arabidopsis nuclear DCL4 isoform tightens RNA silencing via PTGS when DNA methylation is reduced.
RESULTS
DNA Methylation Status Influences DCL4 TSS Usage
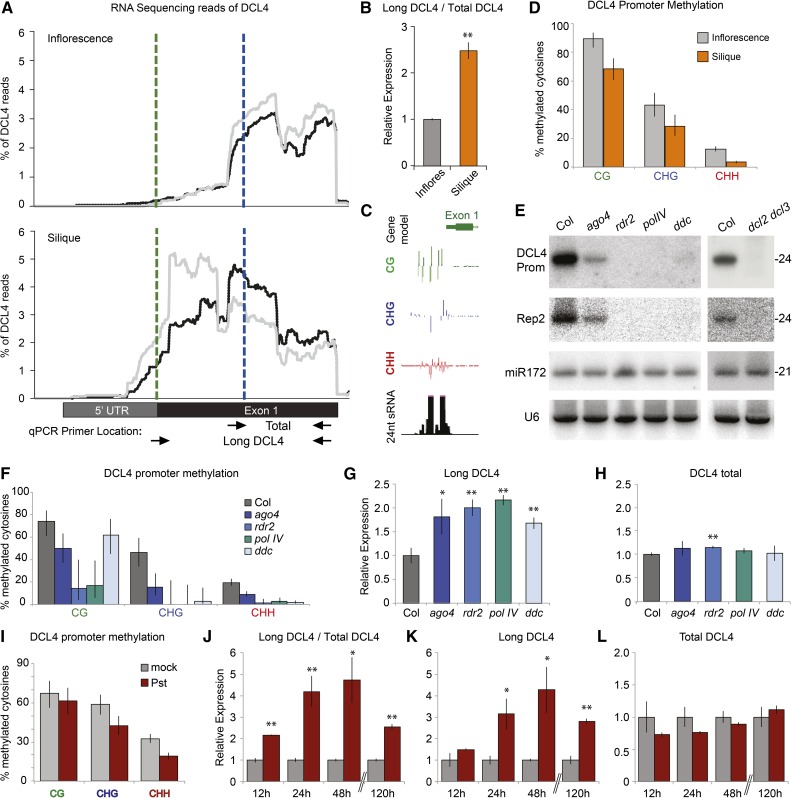
While investigating DCL4 expression in high-depth RNA-seq libraries (40 million paired-end reads) from inflorescence samples, we noted that the bulk of reads began mapping in the middle of the first exon of DCL4, while only a low proportion of reads mapped to the annotated 5′ end or overlapped with the predicted start codon (Figure 1A; Supplemental Figure 1). In RNA-seq libraries from silique samples, however, mapped reads included a higher proportion of sequences spanning the predicted first ATG of DCL4 (green line, Figure 1A). This observation suggested that DCL4 might be expressed as two isoforms in these different tissues. To verify this result, qRT-PCR was conducted using two primer sets to measure the relative level of total DCL4 (all mRNA forms) and longer DCL4 transcripts that include the upstream ATG. Expression was measured using the cycle threshold (Ct) value from the long DCL4 primer set and normalizing by subtracting the Ct value of total DCL4, in place of a housekeeping gene control. With this approach, siliques showed an increased proportion of long DCL4 transcripts normalized to total DCL4 transcript relative to inflorescence samples (Figure 1B).
Figure 1.
Influence of RdDM on DCL4 Transcript Isoform Expression.
(A) Distribution of RNA sequencing reads along DCL4 exon 1, as a percentage of total DCL4 reads from libraries of inflorescence (top) and silique tissues (bottom), each with two replicates (black and gray traces); ATGs marked with dashed green and blue lines. Results for entire DCL4 locus shown in Supplemental Figure 1. The locations of primers used for qRT-PCR are shown with arrows.
(B) Relative expression measured by qRT-PCR of long DCL4 normalized to total DCL4. Mean ± se of three independently harvested biological replicates, each with three technical replicates shown. Asterisks denote P value < 0.01 in two-tailed t test.
(C) Illustration from the UCSC genome browser of DCL4 exon 1, methylation status, and 24-nucleotide siRNA accumulation.
(D) Methylation analysis of the DCL4 promoter in inflorescences (n = 67 cloned sequences) and siliques (n = 70 cloned sequences), measured by sequencing of bisulfite-converted DNA (error bars show 95% Wilson score confidence intervals).
(E) RNA gel blot analysis of 24-nucleotide siRNAs arising from the DCL4 promoter in RdDM mutants; Rep2, miR172 and U6 shown as controls. Analysis performed by stripping and reprobing the same membranes.
(F) Methylation analysis of the DCL4 promoter region in wild-type and RdDM mutants (number of cloned sequences indicated in Supplemental Figure 6A, error bars indicate 95% Wilson score confidence intervals).
(G) and (H) Relative expression of long DCL4 (G) and DCL4 total measured by qRT-PCR (H), normalized to Actin2. For all qRT-PCR results, mean and se of three independently harvested biological replicates, each with three technical replicates are shown. *P value < 0.05 and **P value < 0.01 as determined by a two-tailed t test compared with Col.
(I) Methylation analysis as in (F), comparing mock infiltrated leaves (gray bars, n = 41 cloned sequences) to Pseudomonas syringae infected leaves (red bars, n = 44 cloned sequences), 5 d postinfiltration.
(J) Relative expression of long DCL4 normalized to total DCL4 measured by qRT-PCR. Samples from indicated time points after infiltration with P. syringae (red) or buffer control (gray). Mean and se of three independently harvested biological replicates, each with three technical replicates shown.
(K) and (L) Relative expression of long DCL4 (K) or total DCL4 (L), normalized to Actin2 with same samples used in (J). *P value < 0.05 and **P value < 0.01 as determined by t test compared with respective mock control.
A few studies in plants and mammals have suggested that promoter methylation changes might influence alternative mRNA isoform transcription (Du et al., 2014; Hoivik et al., 2013; Connolly et al., 2011), a possibility we therefore examined for DCL4 transcriptional start sites (TSSs). Analysis of the DCL4 locus revealed a discrete methylation patch affecting cytosines in CG, CHG, and CHH contexts immediately upstream of the coding sequence (Stroud et al., 2013) (Figure 1C; Supplemental Figure 2). Genome-wide DNA methylation levels vary dynamically throughout plant growth, particularly in siliques, in which the developing seed endosperm DNA becomes hypomethylated (Gehring et al., 2009; Ibarra et al., 2012). Accordingly, DCL4 promoter methylation was reduced in silique samples relative to inflorescences of wild-type plants (Figure 1D; Supplemental Figure 3). Furthermore, analysis of published methylation data from dissected embryo and endosperm samples (Pignatta et al., 2014) revealed that DCL4 promoter methylation is reduced in endosperm relative to embryos (Supplemental Figure 4). Finally, the long DCL4 transcript was enriched in dissected seeds and depleted from dissected silique valves, while total DCL4 expression was similar between the two tissue types (Supplemental Figure 5). These data suggest that DNA methylation influences DCL4 TSS usage in developing seeds, when the genome experiences global hypomethylation.
We found that the DCL4 promoter is associated with PolIV/RDR2/DCL3-dependent 24-nucleotide siRNAs, which together with asymmetric DNA methylation are hallmarks of an RdDM target (Figures 1C and 1E). Accordingly, the methylation level of the DCL4 promoter was reduced in RdDM mutants relative to wild-type Col-0 control plants (hereafter referred to as Col), including in single ago4, rdr2, and polIV and triple ddc (drm1 drm2 cmt3) mutants, particularly in the CHH and CHG contexts (Figure 1F; Supplemental Figure 6A). This promoter region does not share any similarity with TE sequences, and because the vast majority of siRNAs arising from this locus map uniquely to the DCL4 promoter, we conclude that this methylated region does not share high similarity with other sequences within the Arabidopsis genome. qRT-PCR analysis revealed that the levels of the long transcript of DCL4 consistently increased by ∼2-fold in all RdDM mutants tested, while total DCL4 expression remained largely unaffected, relative to wild-type plants (Figures 1G and 1H). Together, these data show that DCL4 TSS usage is influenced by the DNA methylation status of the plant via the RdDM pathway.
In addition to mutant backgrounds and specific developmental stages, the Arabidopsis genome undergoes dynamic, genome-wide DNA methylation changes upon infection with Pseudomonas syringae pv tomato strain DC3000 (Pst) (Dowen et al., 2012). We therefore investigated whether Pst infection has any effect on DCL4 expression and promoter methylation. In Pst-infected leaves, DCL4 promoter methylation decreased (Figure 1I; Supplemental Figure 6B), as observed in a previous genome-wide study (Dowen et al., 2012). Pst infection did not significantly alter total DCL4 expression; however, the long DCL4 transcript was strongly induced relative to mock-inoculated leaves (Figures 1J to 1L). These results from wild-type leaves experiencing biotic stress serve as a third independent example, in which increased expression of the long DCL4 TSS correlates with a decrease in promoter methylation. Taken together, induced expression of the long DCL4 isoform is thus a consistent outcome of promoter DNA hypomethylation.
The Major Arabidopsis DCL4 Isoform Is Cytoplasmic
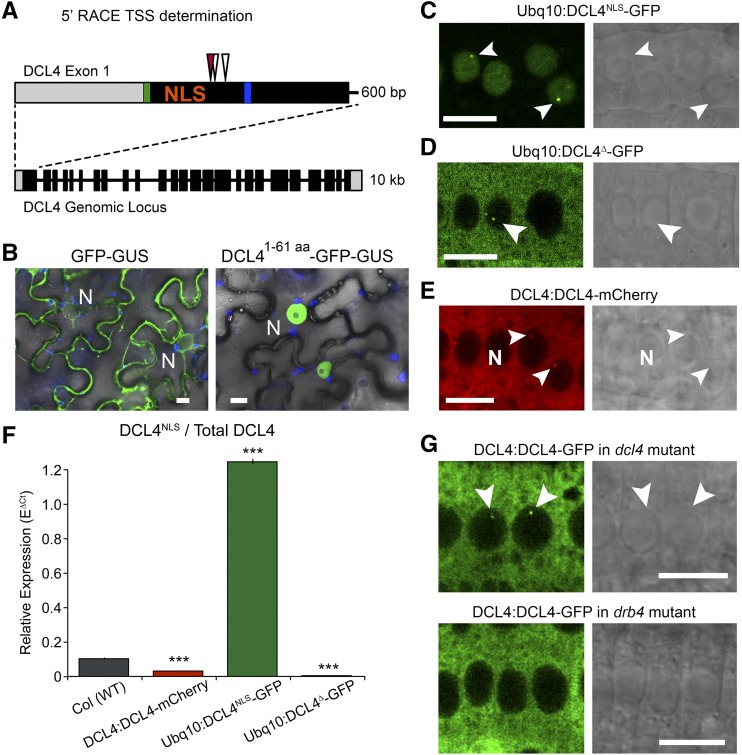
The 5′ RACE-PCR conducted on wild-type Col seedlings revealed that the DCL4 TSSs are within the first exon, consistent with the RNA-seq observation from inflorescences (Figure 1A and 2A; Supplemental Figure 7). This mRNA form bypasses the annotated first start codon and thus encodes a protein isoform beginning with a downstream ATG and truncated relative to the standard model (blue line, Figures 1A and 2A). The region of 61 amino acids encoded between the two alternate start codons includes an identifiable NLS (Kosugi et al., 2009) (Figure 2A), which is sufficient to confer nuclear localization to a GFP-GUS reporter fusion (Figure 2B). We therefore name the long isoform, which includes the upstream ATG and NLS sequence, “DCL4NLS” and refer to the shorter isoform dominantly expressed in inflorescences and seedlings as “DCL4∆.”
Figure 2.
DCL4∆ Isoform Predominates and Encodes a Cytoplasmic Protein.
(A) Illustration of DCL4 locus and detailed view of exon 1. Exons, blocks; introns, lines; UTR, gray; ATGs highlighted in green and blue. TSS determined by direct sequencing of bulk PCR products from nested RACE PCR are depicted with arrowheads: Col wild type (white) and ProDCL4:DCL4-mCherry (red).
(B) Images of GFP-GUS (left) or GFP-GUS fused to the first 61 amino acids of DCL4 (right) expressed from UBQ10 promoter in N. benthamiana leaves. Overlaid with chlorophyll autofluorescence (blue) and bright field (differential interference contrast).
(C) Confocal images of fluorescent signal and bright field (differential interference contrast; right) from roots of Arabidopsis plants transformed with Ubq10 promoter-driven GFP fusions to the genomic coding sequence of DCL4NLS,
(D) Genomic coding sequence of DCL4∆, corresponding to the TSS determined by RACE sequencing.
(E) DCL4 native promoter fused to the full DCL4 genomic coding sequence, including NLS, fused to mCherry.
(F) Relative expression by qRT-PCR of DCL4NLS normalized to total DCL4 in Col and transgenic seedlings. Mean ± se of three independently harvested biological replicates, each with three technical replicates shown. ***P value < 0.001 in t test relative to Col.
(G) Confocal images of ProDCL4:DCL4-GFP in roots of dcl4 (top) or drb4 mutant plants (bottom).
Images represent consistent results in at least three independent transgenic lines. Dicing bodies indicated with arrowheads. Bars = 10 μm.
To evaluate the localization of alternative TSS isoforms of DCL4, we expressed the two isoforms of DCL4 as genomic coding sequence fusions to GFP under the control of the constitutive Ubiquitin10 promoter in transgenic Arabidopsis plants. ProUbq10:DCL4NLS-GFP revealed exclusively nuclear signals in the roots of transgenic plants, demonstrating that the NLS confers efficient nuclear import (Figure 2C). ProUbq10:DCL4∆-GFP roots, by contrast, displayed cytoplasmic signals, with occasional nuclear foci in some cells at the root tip (Figure 2D). Interestingly, a transgene reporter encoding the entire DCL4 locus, including the NLS, under the control of the native DCL4 promoter (ProDCL4:DCL4-mCherry/GFP) produced cytoplasmic signals with some nuclear foci, the same localization pattern observed in ProUbq10:DCL4∆-GFP roots (Figures 2E and 2G). This result suggests that the native DCL4 promoter drives expression of the DCL4∆ isoform in roots, consistent with the RNA-seq results. Indeed, 5′ RACE sequencing of transgenic DCL4:DCL4-mCherry plants showed a TSS similar to that of nontransgenic plants (Figure 2A). Finally, qRT-PCR analysis of transgenic seedlings revealed that long DCL4NLS had a relative expression level close to 1 when normalized to total DCL4 transcripts in ProUbq10:DCL4NLS-GFP plants (Figure 2F). This result shows that expression of full-length DCL4 under the control of the Ubq10 promoter results in the accumulation of the DCL4NLS TSS isoform and does not give rise to significant levels of transcript encoding cytoplasmic DCL4, in agreement with the fluorescent signals observed in nuclei (Figure 2C). In contrast, the long TSS form showed a relative expression level below 0.1 compared with total DCL4 transcripts in Col and ProDCL4:DCL4-mCherry plants, and 0 in ProUbq10:DCL4∆-GFP plants. We conclude that endogenous Arabidopsis DCL4 is mainly expressed as an alternative shorter-than-annotated DCL4∆ mRNA isoform and that the endogenous DCL4 protein is therefore mostly cytoplasmic. These results show a distinctive behavior of DCL4 in comparison with DCL1, DCL2, and DCL3, which all localized to the nucleus when expressed from their respective native promoters or the 35S promoter, consistent with previous studies (Supplemental Figure 8).
DCL4∆ Localization in Nuclear Dicing Bodies Requires DRB4
ProDCL4:DCL4-mCherry/GFP, ProUbq10:DCL4NLS-GFP, and ProUbq10:DCL4∆-GFP transgenic roots all displayed signals in nucleoplasmic foci in some cells of the root tip (Figures 2C to 2E and 2G, arrowheads), resembling the previously described DCL1 dicing bodies (Song et al., 2007; Fang and Spector, 2007). Further studies of DCL4 and DRB4 in roots showed that these proposed dicing bodies are largely distinct from Coilin- and U2B′′-labeled Cajal bodies (Supplemental Figure 9).
DCL4∆ can localize to nuclear dicing bodies in roots despite lacking an NLS (Figure 2D), possibly through its interacting protein, DRB4. Indeed, DCL4 colocalizes with DRB4 in nuclear dicing bodies of native promoter-expressing plants (Supplemental Figure 10). ProDCL4:DCL4-GFP was transformed into drb4 mutants, and multiple roots of three independent transgenic lines were analyzed by microscopy for nuclear dicing body localization. Despite the consistent cytoplasmic localization throughout root tips, no dicing body signals were ever observed in drb4 mutant roots, in contrast to the easily detectable dicing body signals in dcl4 roots (Figure 2G). Therefore, at least two distinct mechanisms account for DCL4 nuclear localization: (1) as the purely nuclear DCL4NLS isoform, which enters through the traditional nuclear import machinery (Cook et al., 2007), or (2) via DRB4, which likely binds and imports a fraction of the predominant and cytoplasmic DCL4∆ isoform into dicing bodies in cells of the root tip.
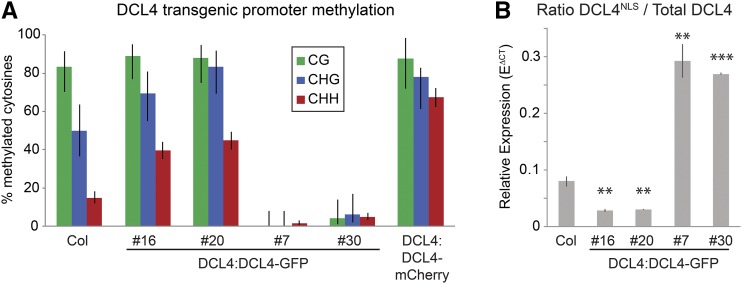
The DCL4 Promoter Methylation State Directly Influences TSS Usage
While analyzing expression in reporter fusion plants, we noted that the transgenic reporter ProDCL4:DCL4-mCherry expressed a significantly lower fraction of long transcript compared with wild-type plants (Figure 2F). Methylation analysis of the introduced transgenic promoter, using a specific primer that recognizes the artificial cloning junction between promoter and coding sequence, revealed a substantial level of hypermethylation (Figure 3A; Supplemental Figure 11). To test whether promoter methylation directly causes a change in isoform expression, we analyzed four independent transgenic lines expressing ProDCL4:DCL4-GFP. Two lines (#16 and #20) displayed hypermethylation on the introduced DCL4 promoter and expressed lower ratios of DCL4NLS transcript to total DCL4, relative to the nontransgenic control (Figure 3). By contrast, two different lines (#7 and #30) displayed extremely low methylation levels on the introduced promoter. These lines displayed a substantial increase in DCL4NLS ratio (>25% compared with <10% in the wild type), comparable with the increase observed in RdDM mutants. Interestingly, while all transgenic lines expressed transcript above the level of wild-type Col plants, hypermethylation correlated with higher expression of total DCL4 relative to that seen in hypomethylated lines (Supplemental Figure 12), showing that promoter methylation plays a role in repressing the DCL4NLS TSS isoform, as opposed to repressing DCL4∆, which is consistent with results from the RdDM mutants and bacterial infection (Figures 1F to 1L). These results therefore demonstrate that DCL4 promoter methylation likely plays a causative role in influencing TSS usage, and that the changes to TSS caused by genome-wide hypomethylation are not likely due to effects of other loci operating in trans. Nevertheless, the lack of promoter methylation does not cause a complete upstream shift in TSS usage, but rather it enhances the level of upstream TSS usage. Thus, additional factors must contribute to TSS specification, and even in hypomethylated conditions, DCL4∆ transcript isoform still predominates.
Figure 3.
Methylation Level Directly Influences DCL4NLS TSS Usage in Transgenic Plants.
(A) Methylation analysis of the transgenic DCL4 promoter in independent transgenic lines, including ProDCL4:DCL4-mCherry, whose expression is depicted in Figure 2F, compared with the endogenous promoter of wild-type Col plants, measured by sequencing of bisulfite-converted DNA. Graphs depict averages with error bars showing 95% Wilson score confidence intervals. Number of cloned sequences analyzed is indicated in Supplemental Figure 11.
(B) Relative expression by qRT-PCR of DCL4NLS normalized to total DCL4 in Col and transgenic seedlings. Mean ± se of three independently harvested biological replicates, each with three technical replicates shown. **P value < 0.01 and ***P value <0.001, as determined by a pairwise two-tailed t test compared with Col.
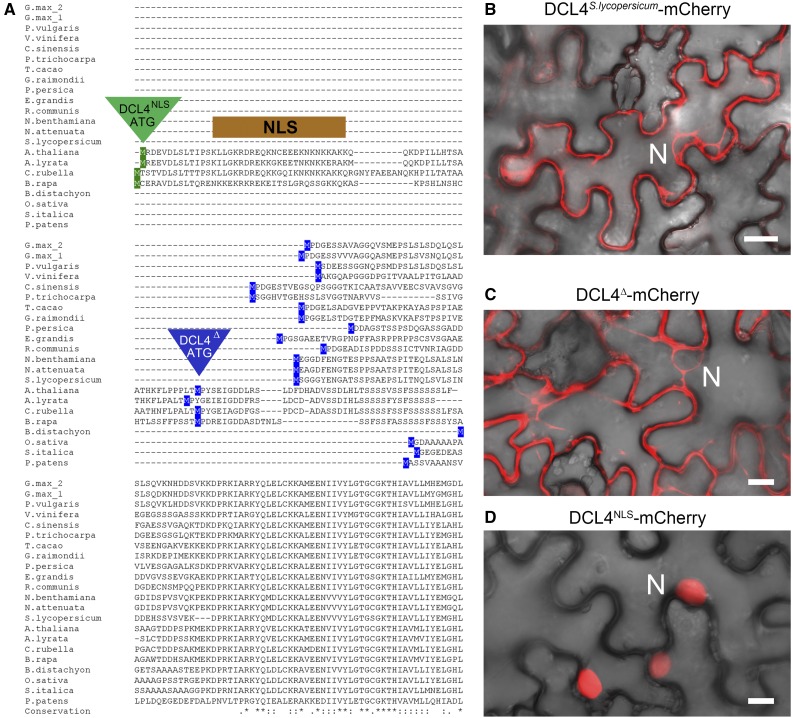
Cytoplasmic Localization of Plant DCL4 Homologs
In addition to the restricted expression pattern of DCL4NLS in Arabidopsis, amino acid sequence analyses conducted with DCL4 amino acid sequences from a range of plant families revealed that the NLS encoding N-terminal extension is limited to Brassicaceae homologs (Figure 4A). Interestingly, the promoter sequence of DCL4, which becomes methylated in the Arabidopsis genome, is not conserved in sequences upstream of the DCL4 coding sequence in Arabidopsis lyrata, Brassica rapa, nor Capsella rubella; however, it is not known whether the promoters of these DCL4 homologs are methylated. Non-Brassicaceae homologs, from dicots to the bryophyte Physcomitrella patens, encode a start codon more closely aligned to the DCL4∆ ATG of Arabidopsis and are devoid of predictable NLS sequences. DCL4 in non-Brassicaceae species is thus expected to localize to the cytoplasm. Accordingly, a full-length cDNA of tomato (Solanum lycopersicum) DCL4 fused to mCherry resulted in purely cytoplasmic signal in transient expression experiments in Nicotiana benthamiana leaves (Figure 4B). As expected, control mCherry fusions to DCL4∆ and DCL4NLS localized to the cytoplasm and nucleus, respectively (Figures 4C and 4D). Thus, DCL4NLS encodes a purely nuclear protein that arose during Brassicaceae evolution, while the mostly cytoplasmic DCL4∆ likely represents the ancestral protein form in plants.
Figure 4.
DCL4NLS Isoform Is Confined to Brassicaceae Homologs.
(A) Amino acid sequence alignment of DCL4 homologs from representative plant species, with alternative start codons and NLS from Arabidopsis highlighted.
(B) Confocal image of Pro35S:DCL4S.lycopersicum-mCherry expressed by transient expression in N. benthamiana leaves. Fluorescent signal (red) is overlaid with bright field (differential interference contrast).
(C) Confocal image of Pro35S:DCL4∆-mCherry.
(D) Pro35S:DCL4NLS-mCherry by transient expression in N. benthamiana leaves.
Nucleus is (N) indicated. Bars = 10 μm.
DCL4NLS and DCL4∆ Both Rescue DCL4 Functions in Endogenous PTGS
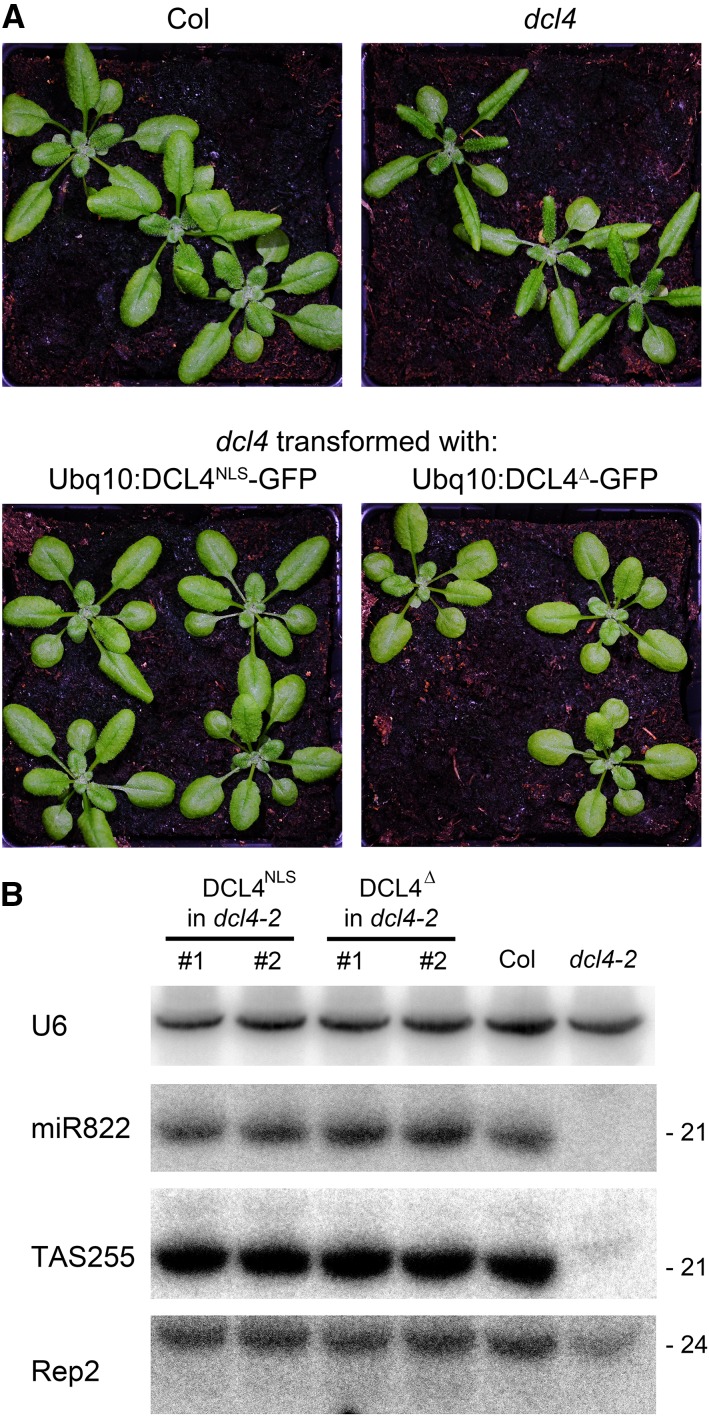
To determine the biological significance of alternate DCL4 localization, ProUbq10:DCL4NLS-GFP (nuclear), ProUbq10:DCL4∆-GFP (cytoplasmic), ProDCL4:DCL4-mCherry (cytoplasmic), and a Pro35S:DCL4-mCherry fusion (nuclear) were tested for complementation of the dcl4 mutant. All transgenes complemented the dcl4 leaf morphology defects and restored accumulation of tasiRNAs and of the DCL4-dependent miR822 (Figure 5; Supplemental Figure 13). Thus interestingly, both isoforms can rescue the known DCL4 functions in endogenous PTGS, possibly while localized in nuclear dicing bodies.
Figure 5.
DCL4 Isoforms Complement dcl4 Mutant Developmental Phenotype.
(A) Morphology of Col, dcl4-2 mutants, and complementation by ProUbq10:DCL4NLS-GFP and ProUbq10:DCL4∆-GFP. Plants shown were grown for 3 weeks in soil under stable growth chamber conditions with a 16-h-light/8-h-dark cycle.
(B) RNA gel blot hybridization showing complementation of tasiRNA255 and miR822 production by ProUbq10:DCL4NLS-GFP and ProUbq10:DCL4∆-GFP. U6 and Rep2 shown as controls.
A Unique Population of DCL4NLS-Dependent 21-Nucleotide siRNAs Accumulates in Siliques
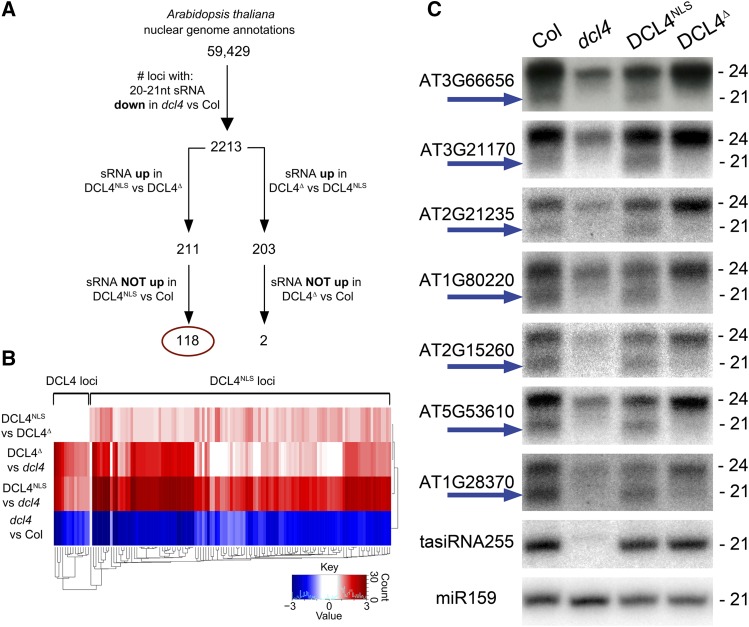
Analysis of siliques, which are naturally enriched in DCL4NLS (Figures 1A and 1B), may reveal an isoform-specific DCL4 function. Illumina HiSeq small RNA (sRNA) sequencing was thus conducted in siliques of wild-type Col, dcl4 mutants, and dcl4 mutants complemented with Ubq10 promoter-driven DCL4NLS or DCL4∆ to identify candidate loci for subsequent validation that might produce small RNAs from one specific isoform. Differential analysis of 20- to 21-nucleotide sRNAs between Col and dcl4 libraries identified 2213 DCL4-dependent sRNA loci (Figure 6A; Supplemental Data Set 1). Subsequently, those loci were tested for differential sRNA accumulation in DCL4∆- versus DCL4NLS-expressing plants. sRNA populations accumulating in transgenic lines above the levels of wild-type plants were discarded as probable artifacts due to DCL4 transgenic expression (Figure 6A). The vast majority of sRNAs were produced by both DCL4 isoforms, consistent with the results of complementation experiments shown in Figure 5. Of the 2213 loci, only two were specifically DCL4∆ dependent, whereas sRNA from 118 loci were DCL4NLS dependent (Figures 6A and 6B; Supplemental Data Set 2). Thus, ∼5% of DCL4-dependent loci were candidates to be produced by DCL4NLS, unraveling a more specific role for the NLS than the ∆ isoform. Selected loci identified by sequencing were investigated by RNA gel blot analysis. All probes tested revealed abundant 24-nucleotide species and additional populations of 21-nucleotide siRNAs in wild-type and DCL4NLS plants. The 21nt siRNAs were distinctively absent or their levels substantially reduced in dcl4 mutant and DCL4∆ plants (Figure 6C, arrow). The levels of 24-nucleotide species were also reduced in dcl4 mutants, consistent with previous reports (Pélissier et al., 2011), and in DCL4NLS- but not DCL4∆-expressing plants; although the mechanism underlying this phenomenon is unknown, we hypothesize that cytoplasmic DCL4 sequesters a DCL3 repressor away from the nucleus. We hereafter refer to the unique 21-nucleotide siRNAs as DCL4NLS isoform-dependent siRNAs or “disiRNAs.” Detection of disiRNAs in wild-type siliques and the finding that their production is complemented by DCL4NLS, but not by DCL4∆, confirms that DCL4NLS is expressed in wild-type plants and that nuclear DCL4 confers a unique molecular function, which is naturally present in Arabidopsis siliques.
Figure 6.
DCL4NLS Produces a Unique Set of siRNAs in Siliques.
(A) Flowchart of sequence analysis pipeline for library size-normalized 20- to 21-nucleotide sRNA reads, uncovering a population of candidate loci that produce sRNAs made by DCL4NLS. Loci with adjusted P value < 0.05 were considered as candidates.
(B) Heat map showing log(e)-fold differential accumulation of 20- to 21-nucleotide siRNAs in different pairwise library comparisons for a selection of loci.
(C) RNA gel blot detection of siRNAs in siliques of Col (wild type), dcl4 mutant plants, and dcl4 mutants expressing ProUbq10:DCL4NLS-GFP or ProUbq10:DCL4∆-GFP, revealing 24-nucleotide species and 21-nucleotide disiRNAs produced by DCL4NLS (arrow); analysis performed by stripping and reprobing the same membrane.
disiRNAs Arise from TE-Associated Loci in a Noncanonical RDR2/PolIV-Dependent Manner
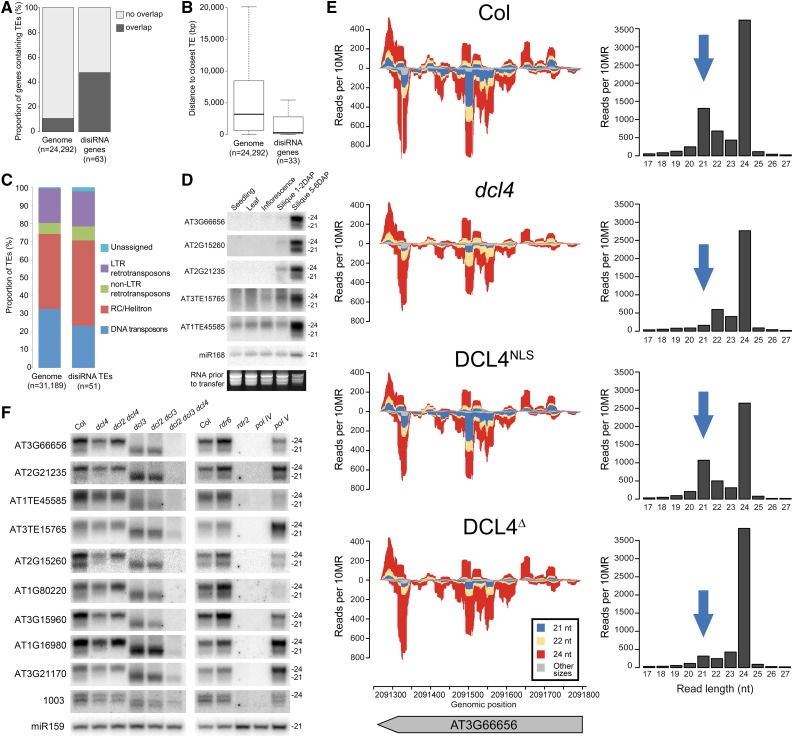
Loci that produce disiRNAs include both protein-coding genes (63 loci) and specific TE sequences (51 loci) (Supplemental Data Set 2). Interestingly, disiRNA-associated genes had a higher incidence of overlapping TEs compared with the entire genome (∼50% versus ∼10% of loci; Figure 7A). Genes without an overlapping TE had a substantially closer average distance to the nearest TE than the average of all Arabidopsis genes (Figure 7B), suggesting a key contribution of TEs to disiRNA production. disiRNA-associated TEs represent diverse families of both DNA and retrotransposons, in a proportion reflecting their abundance in the genome, suggesting a general process (Figure 7C; Supplemental Data Set 3). Further analyses showed that disiRNA accumulation predominates in siliques 5 to 6 d after pollination (DAP); disiRNA are below the detection limits of RNA gel blot analysis in seedlings, leaves, flowers, and young siliques 1 to 2 DAP (Figure 7D). Accordingly, disiRNA-associated mRNAs are primarily expressed in developing siliques, seeds, and pollen (Schmid et al., 2005) (Supplemental Figure 14). Genes in this population serve diverse functions, but are enriched in transcription factor annotation (Supplemental Figure 15) and include genes known to undergo silencing-mediated regulation in siliques, such as the TE-associated AGAMOUS-LIKE91 (AGL91) and AGL40 (Lu et al., 2012). Therefore, both the expression and silencing of disiRNA-associated loci appear to be developmental stage specific.
Figure 7.
disiRNAs Are Associated with TEs, and Biogenesis Requires RDR2 and PolIV.
(A) Genes that contain overlapping TEs or no overlapping TEs in genome and disiRNA loci. χ2 test P value < 2.2 e-16.
(B) Box plot analysis of average distance to nearest TE for genome versus disiRNA loci without an overlapping TE. Filled bars indicate median TE gene distance, and boxes indicate range between the first and third quartiles (Q1 and Q3, respectively) defining the interquartile range (IQR). Whiskers correspond to data between Q1 − 1.5 × IQR or Q3 + 1.5 IQR. Outliers are not shown. P value < 3e-05 by Wilcoxon rank test.
(C) Analysis of TE classes, by proportion, present in the Arabidopsis genome and those directly producing disiRNAs.
(D) RNA gel blot detection of disiRNAs in different tissues of Arabidopsis. miR168 and RNA staining is shown as a loading control. Analysis performed by stripping and reprobing the same membrane.
(E) Small RNA sequencing reads from both strands mapped onto an example locus, AT3G66656 (left); 24-nucleotide siRNAs in red, 22-nucleotide siRNAs green, and 21-nucleotide siRNAs blue. Histograms of sRNAs are shown at the right, with 21-nucleotide disiRNAs highlighted with a blue arrow.
(F) RNA gel blot detection of disiRNAs in siliques of dcl, rdr, and pol mutants reveals genetic basis for biogenesis. Analysis performed by stripping and reprobing the same membrane.
Mapping sRNA reads onto disiRNA-associated loci revealed remarkable overlapping populations of 21- and 24-nucleotide siRNAs covering parts or the entirety of corresponding coding regions, suggesting they shared the same dsRNA precursors (Figure 7E; Supplemental Figures 16 to 19). RNA gel blot analysis of RNA extracted from siliques harvested from a panel of mutant plants revealed, as expected, the strict DCL3 dependency of the 24-nucleotide species and confirmed the exclusive requirement for DCL4 for 21-nucleotide disiRNA accumulation (Figure 7F). Furthermore, disiRNAs overaccumulated in dcl3 mutants, consistent with the hierarchical DCL action on shared dsRNA substrates, and showing that in wild-type plants, DCL4NLS access to nuclear dsRNA substrates is somewhat limited by DCL3 activity. Some disiRNA loci also accumulated 22-nucleotide siRNA species in dcl4 and dcl3 mutants, which were lost in dcl2 dcl4 or dcl2 dcl3 double mutants (Figure 7F). Surprisingly, the rdr6 mutation did not reduce disiRNA accumulation, despite the established connection between RDR6 and DCL4 in mediating PTGS; however, mutants in RDR2 and PolIV, required to provide dsRNA templates to DCL3, lost both 24-nucleotide siRNA and 21-nucleotide disiRNA populations (Figure 7F). The polV mutation had variable outcomes, echoing its known effects on siRNA synthesis (Law and Jacobsen, 2010). As both small RNA sequencing and RNA gel blotting revealed discrete 24-nucleotide and 21-nucleotide siRNAs dependent, respectively, on DCL3 and DCL4, these RNA species are genuine and not likely due to degradation or experimental artifacts. Because both populations accumulate as overlapping populations, which require RDR2 and PolIV, but not RDR6, for biogenesis, we conclude that DCL3 and DCL4NLS can share the same population of dsRNA precursors in siliques. disiRNAs thus represent a small proportion of DCL4-dependent siRNAs, distinguished by a biogenesis mechanism in which the canonical RdDM components PolIV and RDR2 are connected to the downstream action of DCL4 through the nuclear variant.
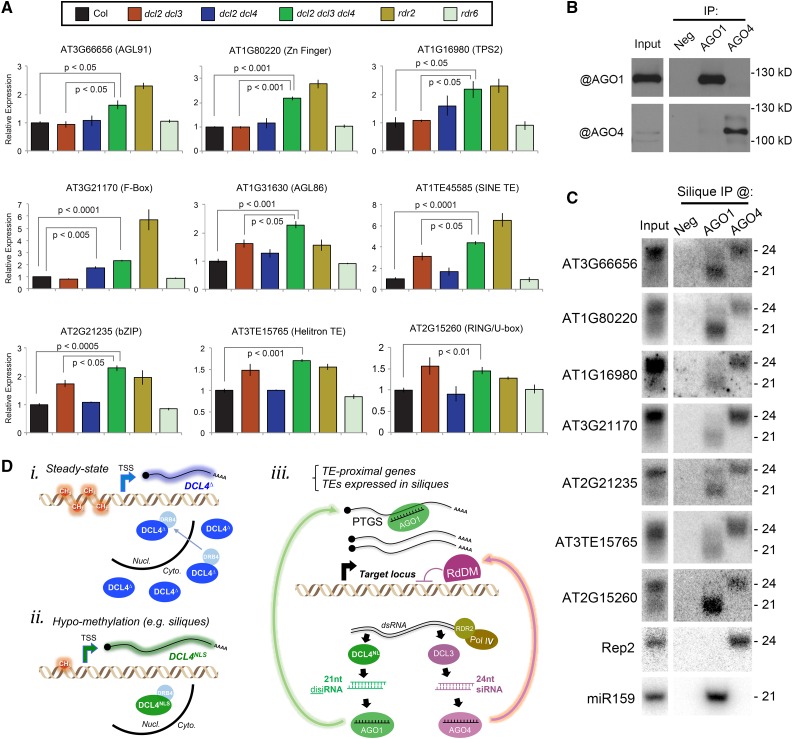
disiRNAs Load into AGO1 and Act with 24-Nucleotide siRNAs to Regulate Transcript Accumulation
To explore whether disiRNAs can function in silencing, we tested the expression of disiRNA-producing genes in siliques of wild-type Col as well as dcl2 dcl4, dcl2 dcl3, dcl2 dcl3, dcl4, rdr2, and rdr6 mutants, 5 d after hand-pollination to ensure optimal fertilization and synchronized development. The dcl2 mutation was included in these mutant combinations to exclude contributions from 22-nucleotide siRNAs found at some disiRNA loci in single dcl3 or dcl4 mutants (Figure 7F). The comparison between dcl2 dcl3 and dcl2 dcl3 dcl4 is most relevant to isolating a specific role for DCL4 and disiRNAs. Many disiRNA-associated gene and TE transcripts accumulated to significantly higher levels in dcl2 dcl3 dcl4 compared with dcl2 dcl3 or Col siliques, demonstrating that disiRNAs can mediate silencing on their own (Figure 8A). Because dcl2 dcl3 dcl4 mutants accumulate higher levels of these transcripts compared with dcl2 dcl4 mutants, the 24-nucleotide species produced coincidently with disiRNA are also sufficient to silence these loci. Transcripts from disiRNA-producing loci overaccumulated in the rdr2 but not in the rdr6 mutant, consistent with the genetics of disiRNA biogenesis (Figures 7F and 8A). We conclude that disiRNAs synthesized by nuclear DCL4 regulate TE and TE-proximal/overlapping gene expression to a slight but significant degree, in a cooperative or redundant manner with DCL3-dependent 24-nucleotide species.
Figure 8.
disiRNAs Act with 24-Nucleotide siRNAs to Regulate Transcript Accumulation.
(A) Relative accumulation of disiRNA-producing mRNA in hand-pollinated siliques of mutants. Mean ± se of three biological replicates shown, with P value derived from two-tailed t test.
(B) Protein gel blot analysis following immunoprecipitation from siliques by AGO1, AGO4, or no (Neg) antibodies. Immunoprecipitation (IP) samples and an Input control were analyzed by AGO1 or AGO4 antibodies (indicated with @).
(C) RNA gel blot detection of immunoprecipitated RNAs by AGO1, AGO4, or no primary antibody control (Neg), compared with Input. Blots were stripped and reprobed to analyze multiple loci.
(D) A model for DCL4NLS conditional expression and activity. (i) Steady state DCL4∆ expression gives rise to a cytoplasmic protein that can shuttle into nuclear dicing bodies via DRB4. (ii) Hypomethylation promotes usage of an upstream TSS, resulting in expression of the nuclear DCL4NLS isoform. Note that DCL4∆ is additionally expressed in siliques. (iii) Subset of TEs and TE-proximal genes are activated in siliques during a phase of genome-wide hypomethylation. RdDM, operated via AGO4-loaded 24-nucleotide siRNA, can mediate TGS at the DNA level, whereas the pool of preexisting mRNAs made from the activated loci is eliminated via PTGS mediated by DCL4NLS-dependent disiRNAs loaded in AGO1. Both 24-nucleotide siRNAs and 21-nucleotide disiRNAs share the same PolIV/RDR2-dependent dsRNA precursors.
The potential for cooperative regulation, rather than mere redundancy, could be revealed if 21-nucleotide disiRNAs and 24-nucleotide siRNAs are specifically loaded into different effectors of the PTGS and RdDM pathways. To test this idea, native AGO1 (PTGS) and AGO4 (RdDM) were immunoprecipitated from wild-type siliques (Figure 8B), and coprecipitated sRNA was tested by RNA gel blot analysis. This revealed loading of 21-nucleotide disiRNAs specifically in AGO1 and 24-nucleotide species in AGO4 (Figure 8C), consistent with the well-described sRNA size preference of these AGOs. While it is possible that disiRNAs load into additional AGOs, the well-established functions of AGO1 and its strong loading with disiRNAs (Figure 8C) lead us to conclude that disiRNAs have the necessary attributes to mediate PTGS and thus function distinctly from AGO4-loaded 24-nucleotide siRNAs.
DISCUSSION
Evolution of Dicer Localization and Function across Kingdoms
The amino acid sequence alignment and localization of tomato DCL4 presented here strongly suggest that DCL4 evolved in plants as a cytoplasmic enzyme that acquired an additional NLS-containing module during the emergence of the Brassicaceae. Previous conclusions that DCL4 is nuclear localized relied mostly on the NLS-containing annotated gene model of Arabidopsis, whereas experiments reported in this work show that a shorter transcript encoding cytoplasmic DCL4∆ dominates in Arabidopsis seedlings and flowers. Furthermore, immunolocalization results presented by Hoffer et al. (2011) were used to conclude that DCL4 localizes strictly in the nucleus; however, their data also show cytoplasmic localization, which was not discussed. Cytoplasmic localization would enable DCL4 to directly target RNA viruses, one of its primary and universal functions; additional experiments will be required to directly address the contribution of isoforms to antiviral defense and to clarify how nuclear DCL2 can perform its antiviral function.
This Brassicaceae-specific DCL4 elaboration is not without precedent, as Dicers have experienced multiple modifications during eukaryotic evolution. In monocot plants, gene duplication at the DCL3 locus spawned DCL3b/DCL5, which produces 24-nucleotide siRNAs from tasiRNA-like precursors (Song et al., 2012). Phylogenetic analyses also revealed that the DCL2 clade did not arise until the evolution of gymnosperms (Ma et al., 2015). Recent studies have further revealed examples of alternative Dicer localization that affect biological functions. Dicer of Schizosaccharomyces pombe, which directs heterochromatin formation, was shown to shuttle between the nucleus and cytoplasm through motifs that mediate nuclear export and retention (Emmerth et al., 2010). Caenorhabditis elegans Dicer can relocalize to the nucleus following ERK-mediated phosphorylation (Drake et al., 2014), while the nematode Trichinella spiralis evolved a Dicer with an N-terminal NLS, which can likely mediate RdDM to repress transposons (Sarkies et al., 2015). Finally, metazoan Dicer is cytoplasmic, but human Dicer encodes a noncanonical nuclear localization signal, normally masked in the folded protein, and was shown to shuttle into the nucleus where it represses spurious interferon response activation (White et al., 2014; Doyle et al., 2013; Burger and Gullerova, 2015).
The existence and specific role of DCL4NLS are clearly revealed by the sequencing and RNA gel blot results presented in Figures 6C and 7E, which demonstrate disiRNA species accumulating in wild-type and DCL4NLS-expressing dcl4 mutant plants, but not in dcl4 mutants or those expressing DCL4∆. The surprising noncanonical biogenesis pathway for disiRNA precursors is nonetheless logical, as PolIV and RDR2 produce dsRNA in the nucleus. Thus, DCL4NLS is probably able to compete with DCL3 for processing, while cytoplasmic DCL4∆ does not gain efficient access to these precursors. It is interesting to note that only a subset of PolIV/RDR2 dsRNAs give rise to disiRNAs in siliques, while in seedlings the expression of DCL4NLS does not seem to process PolIV/RDR2 products (Figures 5B and 7F; Supplemental Data Set 1), suggesting that additional factors are required for DCL4NLS to process RdDM dsRNA substrates. Somewhat puzzling is the observation that this study did not shed light on the location of miR822 and tasiRNA processing. These substrates can be processed by both DCL4 isoforms, suggesting that dsRNA precursors can shuttle between compartments or that they are produced in nuclear dicing bodies, where both DCL4 isoforms can localize.
An Unexplored Aspect of Epigenetic Regulation?
DNA methylation control of alternative TSS expression is a fascinating concept that is just starting to emerge: Only two such examples are available in metazoans (Hoivik et al., 2013; Connolly et al., 2011), and two imprinted genes were recently shown to undergo this type of regulation in rice (Oryza sativa) endosperm, although the functional significance of alternative TSS isoforms was not elucidated in this instance (Du et al., 2014). While expression studies on RdDM mutants did not detect profound gene expression changes (Reinders et al., 2009; Stroud et al., 2014), such studies would have unlikely revealed subtle changes in TSS usage such as DCL4∆ and DCL4NLS. Recent detailed studies in zebra fish uncovered ∼900 genes, in which the TSS shifted within 100 bp between maternal oocytes and zygotes, revealing that tissue-specific signals can direct subtle changes to promoter usage (Haberle et al., 2014). While DCL4 is not an imprinted gene (Gehring et al., 2011), the possibility remains that the NLS isoform is maternally expressed in the endosperm. We analyzed available endosperm-specific RNA-seq data; however, low read coverage over the DCL4 5′ region precluded any conclusions about whether this form is specifically expressed in the endosperm or imprinted.
Expression of the two TSS isoforms is likely due to the coexistence of two cis-element regions in DCL4, which guide distinct transcriptional starts. Our data in tissues, mutants, and biotic stress establish a correlation between methylation of the endogenous DCL4 promoter and alternative TSS usage, and our analysis of multiple independent transgenic lines reveals a causative role for promoter methylation in influencing TSS usage. We propose that this methylated region directly affects transcription factor binding, such that hypomethylation allows a transcription factor to bind an otherwise inaccessible, or poorly accessible, cis-element to initiate DCL4NLS transcription (Figure 8D). Hypermethylation of the promoter would thus prevent or decrease transcription factor binding to the DCL4NLS-specifying cis-element and would thus favor DCL4∆ expression. Based on the finding that DCL4NLS is never the major form expressed from the endogenous promoter, we further suggest that the transcription factor(s) is likely present in limited amounts and/or in discrete cell types. Deciphering the exact mechanisms and conditions that regulate DCL4 TSS usage would shed a key light on the epigenetic sensitivity of transcription initiation in plants.
Feedback Regulation and Epigenetic Control of RNA Silencing Genes
The methylation-dependent conditioning of DCL4NLS expression echoes the rheostatic mode of regulation at the DNA demethylase REPRESSOR OF SILENCING1 (ROS1) locus, which contains a methylated TE-derived fragment in its promoter. Unlike most loci, ROS1 expression increases when methylated and, conversely, decreases when unmethylated. ROS1 functions to remove DNA methylation, and its expression regulation represents an elegant mechanism to maintain DNA methylation homeostasis (Lei et al., 2015; Williams et al., 2015). This autoregulation mechanism differs from that of DCL4, which does not play a direct role in the DNA methylation pathway. Specifically, mRNA of both DCL4 isoforms accumulates to similar levels in dcl4-2 mutants as in wild-type plants, demonstrating that DCL4 does not regulate its own expression (Supplemental Figure 10). A recently evolved mouse Dicer isoform also exhibits rheostatic regulation, although the involvement of DNA methylation is not clear in this case: An MT-C retrotransposon inserted into the sixth intron of Dicer is highly transcribed in mouse oocytes, serving as an alternative promoter for the truncated and oocyte-specific DicerO isoform, which enables potent RNAi of coincidently induced transposons. Loss of DicerO causes sterility, highlighting the functional importance of this adaptation (Flemr et al., 2013).
We propose that the DCL4 locus in Arabidopsis functions in an analogous manner during seed development, whereby hypomethylation leads to induction of DCL4NLS, which produces a new population of disiRNAs arising from the PolIV/RDR2 portion of the RdDM pathway. disiRNAs then mediate PTGS via AGO1 to repress targets that themselves are induced during this period of hypomethylation (Supplemental Figure 14), thus contributing to transcriptome homeostasis (Figure 8D). This example of RdDM-to-PTGS interconnectivity represents the inverse of recent discoveries showing PTGS-to-RdDM connections (Bond and Baulcombe, 2015; Nuthikattu et al., 2013; Marí-Ordóñez et al., 2013; Wu et al., 2012; Pontier et al., 2012).
Tightening the Silencing of TEs and TE-Associated Genes
Genome-wide hypomethylation described in endosperm results from active DNA demethylation by DEMETER in the central cell before fertilization, and further repression of the MET1, DRM2, and CMT3 DNA methyltransferases in the endosperm after fertilization (Jullien et al., 2012; Ibarra et al., 2012). Interestingly, the methylation status of maternal-origin seed tissues is not known. In contrast to methyltransferase expression, the PolIV/RDR2 pathway is not downregulated by 5 DAP; rather, 24-nucleotide siRNAs accumulate to high levels (Mosher et al., 2009; Lu et al., 2012), although their exact role remains unclear. Some evidence suggests their movement from endosperm into embryo where they could reinforce TGS (Ibarra et al., 2012), while recent work demonstrated that an RDR2-dependent 24-nucleotide siRNA could mediate PTGS (Klein-Cosson et al., 2015). Nevertheless, the most parsimonious model is that endosperm 24-nucleotide siRNAs guide RdDM to silence gene expression as part of the normal endosperm developmental program, which requires the tightly controlled expression of several key MADS box transcription factors (Bemer et al., 2010; Lu et al., 2012). The simultaneous induction and repression of targets fits for genes whose function is required for only a brief developmental window, as is described for seed development (Belmonte et al., 2013; Bemer et al., 2010). Of the 63 disiRNA-producing genes identified here, seven are indeed AGL/MADS box genes, including AGL91 and AGL40, which were previously studied in the frame of seed size regulation in uneven tetraploid crosses leading to endosperm defects in Arabidopsis (Lu et al., 2012). Lu et al. (2012) pointed out that 24-nucleotide siRNA species found at the AGL91 and AGL40 loci coincided with an overlapping population of 21-nucleotide siRNAs, which, as shown here, are disiRNAs. While they can execute RNA silencing on their own in a dcl2 dcl3 mutant, we propose that AGO1-loaded disiRNA most likely function cooperatively with RdDM in wild-type siliques, to eliminate, via PTGS, the pool of preexisting transcripts produced by these loci (Figure 8D). The ensuing consolidation of silencing could contribute to subtly tighten the spatio-temporal expression window of endosperm-specific genes. We note that the mildly enhanced levels of disiRNA targets in dcl2 dcl3 dcl4 mutants relative to wild-type plants (∼2-fold or lower) is comparable to the often modest increase in accumulation of miRNA targets in plants mutated for components of the miRNA pathway (Todesco et al., 2010). Nevertheless, DCL4 does not play a major role in seed development under growth chamber conditions, as mutant plants, including dcl2 dcl3 dcl4, did not display any drastic impairment in seed development.
A second nonmutually exclusive function of disiRNAs might be to reduce the detrimental effects of epigenetically reactivated TEs during seed development. Although most disiRNAs are spawned from inert transposon remnants, their loading into the PTGS effector AGO1 suggests that they could trans-target active and potentially harmful TEs. Consistent with this idea, transposon expression is high in endosperm up to 3 to 4 DAP, but decreases coincidently with the burst of disiRNA and 24-nucleotide siRNAs (Figure 7D; Belmonte et al., 2013). In metazoan germlines, piwi-interacting RNAs silence TEs in trans through both PTGS and TGS mechanisms (Siomi et al., 2011), and in Arabidopsis, TEs can be repressed in the developing male germline by epigenetically activated siRNAs of vegetative origin (Slotkin et al., 2009). Production of these epigenetically activated siRNAs depends on both the miRNA pathway and DCL4 (Creasey et al., 2014; Slotkin et al., 2009). disiRNAs could thus be a functional equivalent, in seeds, of the pollen epigenetically activated siRNAs, and a defensive function against TEs might underpin the primary origin of these molecules.
A unique and crucial function for the DCL4NLS isoform is ultimately suggested by its conservation in all available Brassicaceae genomes. Whether these species also regulate DCL4 isoform expression through DNA methylation remains an open question. While the promoter sequence is not conserved between Arabidopsis and other Brassica species, it is possible that methylation on this locus was maintained throughout evolution of the nucleotide sequence. It is also possible that this isoform in Arabidopsis plays a more significant role in other tissues or under variable/adverse environments. For example, we used the hypomethylation induced by bacterial infection to support the connection between methylation state and isoform expression, and it is further possible that this regulation plays a role in plant disease responses. The continued evolution of Dicers indicates that grasping the full complexity of silencing mechanisms will require analyses of a large diversity of plant families. Finally, establishing the extent to which DNA methylation regulates protein isoform diversity may reveal an important and unappreciated epigenetic function.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Col-0 ecotype was used as the wild type in all studies and compared with previously described mutants dcl4-2, dcl2-1, dcl3-1, rdr2-1, rdr6-15, ago4-5, polIV (nrpd1a-3), polV (nrpd1b-11), ddc (drm1-2 drm2-2 cmt3-11), all in the Col background. Seedlings were grown on 0.5× MS medium without sucrose in a 12-h-light/12-h-dark growth chamber; expression analysis was performed on plants 10 to 14 d after germination. For inflorescence and silique tissue, plants were grown for 2 weeks in soil in a 12-h-light/12-h-dark cycle, then moved into a 16 h/8 h chamber. Growth chambers were equipped with 36-Watt fluorescent lights in a 2:1 ratio of 840 Cool White:Grolux with an average light intensity of 160 µmol m−2 s−1 and 60% relative humidity. Stable transgenic plants were established through the flower dip method (Clough and Bent, 1998) and selected for homozygous, single locus insertion by antibiotic resistance segregation. A minimum of three lines from independent transgenic events were analyzed for each reporter to ensure consistent results. Coilin and U2b′′ expressing plants were ordered from the NASC stock center. Transient expression in Nicotiana benthamiana was performed according to de Felippes and Weigel (2010), and the viral suppressor P38 was included for DCL4 expression experiments. For expression analysis in dissected siliques, 3- to 4-DAP siliques were harvested and anchored with tape under a dissecting microscope, and syringe tips were used to dissect and immediately freeze valve tissue. Septa, with attached seeds, were then removed to a new tube and frozen.
Plasmid Construction
All constructs reported here were created in the pB/K7m34GW vector by multisite Gateway recombination, as described by Karimi et al. (2005). Briefly, promoters and coding sequences were amplified from Arabidopsis genomic DNA with Phusion polymerase (Thermo Scientific) using primers listed in Supplemental Data Set 4 and recombined into the appropriate pDONR plasmid with Gateway BP Clonase (Invitrogen). DCL4 from tomato (Solanum lycopersicum) was amplified from cDNA synthesized as described below. Fluorescent reporters were similarly amplified, and all plasmids were sequenced. Three-way recombination was subsequently performed with Gateway LR Clonase to create the promoter:gene-reporter constructs used in this study.
Pseudomonas syringae Infection
Bacterial infection experiments were performed as described (Yu et al., 2013). Briefly, Pseudomonas syringae pv tomato strain DC3000 was grown in liquid culture to log phase (OD600 = 0.6), pelleted by centrifugation, and resuspended in 10 mM MgCl2 to a concentration of 107 colony-forming units per milliliter. Four leaves of 5- to 6-week old plants were syringe-infiltrated with the Pst solution or a buffer control and harvested after 12, 24, 48, and 120 h. Two plants were combined for each biological replicate, and three biological replicates were compared for each expression experiment, while two replicates were compared for methylation analysis. Pst infection experiments were conducted three times with similar results.
5′ RACE
RACE was performed according to Scotto-Lavino et al. (2006). RNA was extracted from Col and ProDCL4:DCL4-mCherry seedlings by TRIzol (Life Technologies), incubated with DNaseI (Thermo Scientific), and cleaned with an RNeasy mini kit (Qiagen). Following first-strand cDNA synthesis, a tailing reaction was performed and an adapter, which includes primer binding sites for oligos Outer and Inner, was added by PCR. Nested PCR was performed on cDNA first with primers Outer and DCL4R3, diluted 1/1000, and reamplified with primers Inner and DCL4R7. PCR products were purified on PCR Spin columns (Qiagen), and this population of products was sent directly for sequencing with internal primer DCL4R6. By avoiding subcloning and sequencing of individual PCR products, this approach yields the major 5′ cDNA end within the population.
Bisulfite Sequencing
DNA methylation was determined by extracting genomic DNA from the indicated tissues and genotypes using a DNeasy Plant Mini Kit (Qiagen) followed by bisulfite conversion with an EpiTect Bisulfite Kit (Qiagen). Converted DNA was amplified by PCR with primers designed specifically for bisulfite-treated DNA according to Henderson et al. (2010). PCR products were gel-purified and ligated into pGEM-T Easy (Promega), and single colonies were sent for sequencing. Percentage of methylated cytosines was determined by analyzing sequence data with the Kismeth web tool (Gruntman et al., 2008). Average methylation in a 168-nucleotide-long DCL4 promoter region was presented, with 95% Wilson score confidence intervals as error bars. Experiments were repeated at least twice with similar results.
Live-Cell and Immunofluorescence Imaging
All fluorescence data were acquired with a Zeiss 780 confocal laser scanning microscope using a 40× water immersion objective (LD C-Apochromat 40×/1.1). Images were analyzed and contrasted in Fiji (Schindelin et al., 2012) and assembled in Adobe Photoshop CS5 (Adobe). Fluorescent protein fusion-expressing roots and leaves were imaged immediately following excision from growing plants. Differential interference contrast (bright field) is included to show tissue/cellular details. Images shown in figures are representative of consistent results observed in multiple experiments.
cDNA Synthesis and qRT-PCR
RNA was extracted from frozen, ground tissue with TRIzol reagent (Invitrogen) from seedlings and leaves, while RNA from siliques and inflorescences was extracted with an RNeasy mini kit (Qiagen). RNA was treated with DNaseI and reverse transcribed with a Maxima First-Strand cDNA synthesis kit (Thermo Scientific). Quantitative RT-PCR was performed on a LightCycler480 II (Roche) in optical 384-well plates with a SYBR Fast qPCR Kit (KAPA Biosystems) and Ct values determined by 2nd derivative max on three technical replicates, defined as the same combination of cDNA and primers, per sample. Occasional individual technical outliers that differed from the other two technical replicates by more than one Ct value (usually a failed reaction) were deleted while calculating the average. ∆Ct was calculated by comparing genes of interest to Actin11 for siliques and GAPC and Actin2 for other tissues. For relative DCL4NLS measurements, ∆Ct was calculated by normalizing the Ct of DCL4NLS (F2 and R6) reactions to Ct of Total DCL4 (F1 and R6). Primer efficiency was determined for DCL4 primer sets as 1.93; for the remaining genes, efficiency of 2 was used to estimate RNA abundance. Data are presented as average of three biological replicates, defined as independent plants of similar genotype/tissue, with error bars indicating se of the mean. P values were calculated with a Student’s t test. Primers are listed in Supplemental Data Set 4. All qRT-PCR experiments were repeated at least three times with similar results.
Amino Acid Sequence Alignment and NLS Analysis
NLSs were predicted with NLS mapper (http://nls-mapper.iab.keio.ac.jp/) (Kosugi et al., 2009). Sequences of DCL4 homologs from representative plant species with available high-quality genome sequence were acquired through TBLASTN searches for genes with the highest predicted amino acid similarity to the Arabidopsis DCL4NLS sequence. Sequences are available in Supplemental File 1 and represent full-length coding annotations. Sequences from representative plant families were aligned with ClustalW.
RNA Gel Blot Analysis
Total RNA was extracted from frozen, ground tissue with TRIzol reagent (Invitrogen) and eluted in 50% formamide. Total RNA was separated on a 17.5% polyacrylamide-urea gel, electrotransferred to a Hybond-NX membrane (GE Healthcare), and cross-linked with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide-mediated chemical cross-linking, as previously described (Pall and Hamilton, 2008). All radiolabeled probes, except those noted below, were made by incubating gel-isolated PCR fragments with the Prime-A-Gene kit (Promega) in the presence of [α-32P]dCTP (Hartmann Analytic). Oligonucleotides complementary to U6, tasiRNA255, miR822, miR159, miR172, siRNA1003, and siRNA Rep2 were end labeled by incubation with T4 PNK (Thermo Scientific) in the presence of [γ-32P]dATP. Multiple sequences were probed on individual membranes by stripping twice with boiling 0.1% SDS and reprobing. Results shown are representative of at least three independent experiments. Raw data from RNA gel blotting are included in Supplemental Data Set 5.
Small RNA Sequencing
Total cellular RNA (10 µL, 200 to 300 ng/µL), extracted using TRIzol reagent (Invitrogen), was processed into sequencing libraries using adapted Illumina protocols and sequenced at Fasteris using the Illumina HiSeq sequencer. FASTQ file generation, demultiplexing, and adapter removal were done by Fasteris.
RNA Sequencing
Total RNA sequencing was done at the Zurich Functional Genomic Center. RNA was isolated from inflorescences and siliques using a Qiagen RNeasy kit. Ribosomal depletion and random hexamer paired-end (2 × 100 bp) stranded RNA sequencing were applied, producing around 2 × 40 million reads per library.
Bioinformatic Analysis
Small RNA reads were filtered to 15- to 35-nucleotide-long reads and matched against the Arabidopsis genome (TAIR10) using MUMmer v3.0 (Kurtz et al., 2004). Only reads with a perfect match over their entire length were analyzed further (16,267,721; 18,755,127; 27,059,402; and 21,670,047 reads for Col wild type, dcl4, DCL4NLS, and DCL4∆, respectively).
Differential Analysis
The number of 20- to 21-nucleotide reads matching nuclear genome annotations (TAIR10 genes and transposable elements as well as miRBase v19 miRNA precursor) were normalized (reads per 10 millions reads) and then compared between libraries using DESeq2 v1.2.10 (Love et al., 2014). Full analysis is shown in Supplemental Data Set 1. Loci with adjusted P value (which takes into account false discovery rate) lower than 0.05 were considered as significantly changed. This analysis was conducted to identify targets for further study by RNA gel blot analysis and qRT-PCR, and the lack of replicated samples limits the statistical power to detect genome-wide all loci that produce disiRNAs. Thus, the list of 118 candidates is likely an underestimate of the genome-wide disiRNA-producing loci.
Small RNA Profile Representation
Simple genomic position comparison was applied to retrieve sRNA read counts and positions corresponding to the selected loci. Those were then used to calculate the normalized read counts (reads per 10 millions reads) for each nucleotide and to produce a graphical representation using R. A heat map was constructed using the heatmap2 function from the gplot R package. For each locus, 20- to 21-nucleotide read count was calculated and library-size normalized. Those values were compared between libraries by calculating the log(e)-ratio of those value plus 1 (to avoid division by 0) and are shown in Supplemental Data Set 2. Loci in the heat map are shown hierarchically clustered by similarity based on Euclidian distance and single aggregation algorithm, while columns are hierarchically clustered by similarity based on Manhattan distance and Ward aggregation algorithm.
For RNA sequencing, reads from inflorescences and siliques were aligned on the Arabidopsis TAIR10 reference genome using TopHat (v2.0.11) (Kim et al., 2013) and Bowtie (v2.2.1.0) (Langmead et al., 2009). Reads corresponding to the DCL4 locus were retrieved by simple genomic position comparison.
Immunoprecipitation
Silique tissue, frozen in liquid nitrogen and ground with a mortar and pestle, was mixed 1:4 (volume ground tissue:volume buffer) with immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40) containing 2 μM proteasome inhibitor MG-132 and cOmplete protease inhibitor cocktail (Roche), and incubated for 25 min at 4°C on a rotating wheel. All steps were performed at 4°C and on ice. Cell debris was removed by centrifuging twice for 10 min at 8000g. The supernatant was precleared by incubating for 15 min with 50 μL of Protein A-agarose beads (Sigma-Aldrich). The beads were removed by centrifugation for 1 min at 10,000g, 100μL of supernatant was removed for the Input control, and the remaining sample was separated into three groups and incubated with previously established antibodies at 1/1000 dilution of anti-AGO1 (Qi et al., 2005) (Agrisera), 1/500 dilution of anti-AGO4 (Garcia et al., 2012) (Eurogentec), or no primary antibody for 45 min. Twenty-five microliters of Protein A-agarose beads was added, followed by incubation for 45 min. The beads were pelleted, washed three times in immunoprecipitation buffer, and resuspended in 100 μL PBS and 10μL for protein analysis and 90 μL for RNA analysis. RNA was extracted from precipitated complexes by the addition of 150 μL PBS + 1% SDS, 10 μL of Proteinase K and incubating 25 min at 37°C, followed by phenol extraction and precipitation in sodium acetate, ethanol, and glycogen and elution in 50% formamide. As a control, 100 μL of input sample was mixed with 1 mL of TRIzol reagent, and RNA was extracted and eluted in 50% formamide.
Protein Blot Analysis
Protein was resolved by SDS-PAGE, electrotransferred to Immobilon-P PVDF membrane (Millipore), and following 1 h blocking in PBS + 0.1% Tween 20 supplemented with 1% BSA, antibody incubations were performed overnight at 4°C with constant shaking. Primary antibodies were used with the following dilutions: AGO1 (1/8000) and AGO4 (1/4000). Following three washes in PBS + 0.1% Tween 20, membranes were incubated for 1 h in horseradish peroxidase-conjugated goat anti-rabbit antibodies then rinsed three times before detection with an ECL Western Blotting Detection Kit (GE Healthcare). Raw data from protein blotting are included in Supplemental Data Set 5.
Accession Numbers
Genes referred to in this study correspond to the following Arabidopsis Genome Initiative locus identifiers: DCL4, AT5G20320; DCL3, AT3G43920; DCL2, AT3G03300; DCL1, AT1G01040; DRB4, AT3G62800; RDR2, AT4G11130; PolIV, AT1G63020; PolV, AT2G40030; RDR6, AT3G49500; AGO4, AT2G27040; AGO1, AT1G48410; DRM1, AT5G15380; DRM2, AT5G14620; CMT3, AT1G69770; ACTIN2, AT3G18780; ACTIN11, AT3G12110; GAPC, AT3G04120; UBQ10, AT4G10590; ROS1, AT2G36490; MET1, AT5G49160; AGL91, AT3G66656; and AGL40, AT4G36590. Sequences of DCL4 homologs in other species correspond to the following NCBI references: S. lycopersicum, NM_001279281.2; Arabidopsis lyrata, XM_002873945.1; Brassica rapa, XM_009122569.2; Capsella rubella, XM_006289369.1. RNA and small RNA sequencing data are available from the NCBI Gene Expression Omnibus under the following reference numbers: GSE74731, GSM1930763, GSM1930764, GSM1930834, and GSM1930835.
Supplemental Data
Supplemental Figure 1. RNA-sequencing reads of DCL4.
Supplemental Figure 2. DCL4 methylation and small RNA accumulation.
Supplemental Figure 3. DCL4 promoter methylation bisulfite data.
Supplemental Figure 4. DCL4 promoter methylation in endosperm and embryo.
Supplemental Figure 5. DCL4 isoform expression in dissected seeds and valves.
Supplemental Figure 6. DCL4 promoter methylation bisulfite data.
Supplemental Figure 7. DCL4 TSS determined by 5′ RACE sequencing.
Supplemental Figure 8. Localization of DCL proteins.
Supplemental Figure 9. Colocalization analysis with Cajal body markers.
Supplemental Figure 10. Colocalization of DCL4 and DRB4 in nuclear bodies.
Supplemental Figure 11. DCL4 transgenic promoter methylation bisulfite data.
Supplemental Figure 12. DCL4 expression in independent transgenic lines.
Supplemental Figure 13. DCL4 isoform complementation analysis.
Supplemental Figure 14. Expression pattern of disiRNA-producing genes.
Supplemental Figure 15. GO annotation of disiRNA-producing genes.
Supplemental Figure 16. sRNA accumulation in siliques over AT3TE15765.
Supplemental Figure 17. sRNA accumulation in siliques over AT1G80220.
Supplemental Figure 18. sRNA accumulation in siliques over AT2G21235.
Supplemental Figure 19. sRNA accumulation in siliques over AT2G15260.
Supplemental Data Set 1. Differential analysis of silique small RNA sequencing.
Supplemental Data Set 2. Results of differential analysis of disiRNA-producing loci.
Supplemental Data Set 3. Comparison of disiRNA-producing TE loci.
Supplemental Data Set 4. Oligonucleotide sequences.
Supplemental Data Set 5. Raw scans of RNA and protein blotting data.
Supplemental File 1. Sequences used for amino acid alignment.
Supplementary Material
Acknowledgments
We thank the following people for contributions: A. Marí-Ordóñez for RNA sequencing libraries and assistance with DNA methylation analysis; A. Imboden for plant growth assistance; Alessandra Stürchler and Raphael Iselin for transgenic plant analysis; F. Jay for assistance with Pseudomonas syringae experiments; F. Brioudes for providing plasmids; J. van Eck for tomato material; N.-H. Chua for DRB4:DRB4-YFP-FLAG; ETH ScopeM for microscopy facilities; S. Zeeman for critical reading of the manuscript; and the Voinnet group for invaluable insight and comments. This research was supported by an Advanced Grant from the European Research Council (Frontiers in RNAi-II No. 323071), a core grant from ETH Zürich and a grant from the Swiss National Foundation (No. 310030B_152832), all attributed to O.V. N.P. was supported by an EMBO Long-Term Fellowship (ALTF 1487-2011) and Marie-Curie IIF Fellowship (Project 299789).
AUTHOR CONTRIBUTIONS
N.P. and O.V. designed the research. N.P., A.S., P.E.J., and S.O. performed experimental work and analyzed data. N.P. and N.G.B. contributed new biological material. N.P., A.S., and O.V. wrote the manuscript.
Glossary
- TE
transposable element
- PTGS
posttranscriptional gene silencing
- TGS
transcriptional gene silencing
- RdDM
RNA-directed DNA methylation
- dsRNA
double-stranded RNA
- siRNA
small-interfering RNA
- miRNA
microRNA
- tasiRNA
trans-acting siRNA
- NLS
nuclear localization signal
- TSS
transcriptional start site
- sRNA
small RNA
- disiRNA
DCL4NLS isoform-dependent siRNA
- DAP
days after pollination
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte M.F., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110: E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N.G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Lauressergues D., Gasciolli V., Vaucheret H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25: 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Burger K., Gullerova M. (2015). Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat. Rev. Mol. Cell Biol. 16: 417–430. [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Connolly D., Yang Z., Castaldi M., Simmons N., Oktay M.H., Coniglio S., Fazzari M.J., Verdier-Pinard P., Montagna C. (2011). Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression. Breast Cancer Res. 13: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. (2007). Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76: 647–671. [DOI] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes F.F., Weigel D. (2010). Transient assays for the analysis of miRNA processing and function. Methods Mol. Biol. 592: 255–264. [DOI] [PubMed] [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71. [DOI] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M., Badertscher L., Jaskiewicz L., Güttinger S., Jurado S., Hugenschmidt T., Kutay U., Filipowicz W. (2013). The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA 19: 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M., Furuta T., Suen K.M., Gonzalez G., Liu B., Kalia A., Ladbury J.E., Fire A.Z., Skeath J.B., Arur S. (2014). A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev. Cell 31: 614–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., Luo M., Zhang R., Finnegan E.J., Koltunow A.M. (2014). Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites. Plant J. 79: 232–242. [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360. [DOI] [PubMed] [Google Scholar]
- Emmerth S., Schober H., Gaidatzis D., Roloff T., Jacobeit K., Bühler M. (2010). Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev. Cell 18: 102–113. [DOI] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemr M., Malik R., Franke V., Nejepinska J., Sedlacek R., Vlahovicek K., Svoboda P. (2013). A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155: 807–816. [DOI] [PubMed] [Google Scholar]
- Garcia D., Garcia S., Pontier D., Marchais A., Renou J.P., Lagrange T., Voinnet O. (2012). Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol. Cell 48: 109–120. [DOI] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., et al. (2014). Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Chan S.R., Cao X., Johnson L., Jacobsen S.E. (2010). Accurate sodium bisulfite sequencing in plants. Epigenetics 5: 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T. (2005). Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57: 173–188. [DOI] [PubMed] [Google Scholar]
- Hoffer P., Ivashuta S., Pontes O., Vitins A., Pikaard C., Mroczka A., Wagner N., Voelker T. (2011). Posttranscriptional gene silencing in nuclei. Proc. Natl. Acad. Sci. USA 108: 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoivik E.A., Witsoe S.L., Bergheim I.R., Xu Y., Jakobsson I., Tengholm A., Doskeland S.O., Bakke M. (2013). DNA methylation of alternative promoters directs tissue specific expression of Epac2 isoforms. PLoS One 8: e67925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V., Moreno A.B., Elmayan T., Vaucheret H., Crespi M.D., Maizel A. (2012). Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 31: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P.E., Susaki D., Yelagandula R., Higashiyama T., Berger F. (2012). DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr. Biol. 22: 1825–1830. [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Cosson C., Chambrier P., Rogowsky P.M., Vernoud V. (2015). Regulation of a maize HD-ZIP IV transcription factor by a non-conventional RDR2-dependent small RNA. Plant J. 81: 747–758. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2009). Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 106: 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R., Watanabe Y. (2009). SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté J.F., Sanfaçon H. (2010). Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48: 69–91. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Zhang H., Julian R., Tang K., Xie S., Zhu J.K. (2015). Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Zhang C., Baulcombe D.C., Chen Z.J. (2012). Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 109: 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Hatlen A., Kelly L.J., Becher H., Wang W., Kovarik A., Leitch I.J., Leitch A.R. (2015). Angiosperms are unique among land plant lineages in the occurrence of key genes in the RNA-directed DNA methylation (RdDM) Pathway. Genome Biol. Evol. 7: 2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Moreno A.B., Gabriel M., Mallory A.C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M.D., Vaucheret H., Maizel A. (2015). In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 43: 2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K. (2015). ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G.S., Hamilton A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Pélissier T., Clavel M., Chaparro C., Pouch-Pélissier M.N., Vaucheret H., Deragon J.M. (2011). Double-stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV-dependent siRNA levels in Arabidopsis. RNA 17: 1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D., Erdmann R.M., Scheer E., Picard C.L., Bell G.W., Gehring M. (2014). Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92. [DOI] [PubMed] [Google Scholar]
- Pontier D., et al. (2012). NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell 48: 121–132. [DOI] [PubMed] [Google Scholar]
- Pumplin N., Voinnet O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11: 745–760. [DOI] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19: 421–428. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Wulff B.B., Mirouze M., Marí-Ordóñez A., Dapp M., Rozhon W., Bucher E., Theiler G., Paszkowski J. (2009). Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P., et al. (2015). Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol. 13: e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M.A. (2006). 5′ End cDNA amplification using classic RACE. Nat. Protoc. 1: 2555–2562. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., Aravin A.A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104: 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69: 462–474. [DOI] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Wang X.B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X., Gasciolli V., Vaucheret H., Ding S.W. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., Schlackow M., Kamieniarz-Gdula K., Proudfoot N.J., Gullerova M. (2014). Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct. Mol. Biol. 21: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.P., Pignatta D., Henikoff S., Gehring M. (2015). Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Mao L., Qi Y. (2012). Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Wang W., Iki T., Liu C., Wu Y., Ishikawa M., Zhou X., Qi Y. (2012). Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 46: 859–870. [DOI] [PubMed] [Google Scholar]
- Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., Abraham A.L., Penterman J., Fischer R.L., Voinnet O., Navarro L. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 110: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Jeong R.D., Lim G.H., Yu K., Wang C., Chandra-Shekara A.C., Navarre D., Klessig D.F., Kachroo A., Kachroo P. (2013). Double-stranded RNA-binding protein 4 is required for resistance signaling against viral and bacterial pathogens. Cell Reports 4: 1168–1184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.