Abstract
The cellular origin of gliomas remains a topic of controversy in cancer research. Advances in neurobiology, molecular genetics, and functional genomics have ushered new insights through exploiting the development of more sophisticated tools to address this question. Diverse distinct cell populations in the adult brain have been reported to give rise to gliomas, although how these studies relate physiologically to mechanisms of spontaneous tumour formation via accumulation of tumour-initiating mutations within a single cell are less well developed. Recent studies in animal models indicate that the lineage of the tumour-initiating cell may contribute to the biological and genomic phenotype of glioblastoma. These results suggest that the cell of origin may not only serve as a source of diversity for these tumours, but may also provide new avenues for improved diagnostics and therapeutic targeting that may prolong the lives of patients.
Keywords: cell of origin, tumour-initiating cell, glioma, glioblastoma, GBM, molecular subtypes, mouse models
Gliomas are the most common primary malignancies in the central nervous system (CNS). This heterogeneous group of tumours is characterised by their resemblance to glia that perform a variety of important functions including support to neurons. The World Health Organisation (WHO) classifies astrocytic gliomas based on histologic criteria from low-grade lesions (grades I–II) to high-grade (grades III–IV) malignancies. Glioblastoma multiforme (GBM) is a highly aggressive grade IV tumour that is the most prevalent and presents with the poorest prognosis, with a median survival of 15 months from the time of diagnosis. Typical histological features of GBM include regions of necrosis, microvascular proliferation, abundant mitoses, and pleiomorphic cells (Wen and Kesari, 2008). These highly infiltrative tumours are resistant to conventional therapies, including chemotherapy and radiation, and surgical intervention fails to remove the entire tumour. The consequence is eventual tumour recurrence and death (Stupp et al, 2005).
Genetic changes and molecular classification
In recent years, extensive molecular profiling has afforded an increasing understanding of the genomic landscape of malignant gliomas. Large-scale sequencing analyses have provided new data on molecular changes in gene expression, copy number, somatic mutations, and epigenetic signatures in these tumours (TCGA, 2008; Brennan et al, 2013). These studies validate the most commonly mutated gene signatures in human GBM that most frequently include a subset of the following oncogenes and tumour suppressors: TP53, PTEN, NF1, EGFR, ERBB2, RB1, PIK3R1, and PIK3CA (TCGA, 2008). This diverse group of cancer-associated genes represents a core set of pathways that are commonly deregulated in GBM, including growth factor signalling (receptor tyrosine kinase (RTK)/phosphatidylinositide 3-kinase (PI3K)/Ras), p53, and Rb signalling pathways. These lead to aberrant signalling in proliferation, cell cycle regulation, senescence, and apoptosis, underscoring the importance of such pathways in the development of malignant gliomas (TCGA, 2008; Brennan et al, 2013). These GBM-associated genes are altered in a variety of ways, including gene amplification and deletion or mutation, and are present in more than three-quarters of patients. Isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) mutations have also emerged as important drivers of a more rare subset of gliomas, particularly low-grade gliomas and secondary GBM (Verhaak et al, 2010).
Based on signature genome-wide gene expression changes, coupled with somatic mutations and copy number changes, GBM is commonly classified into four subtypes: proneural, mesenchymal, classical, and neural subtypes (Verhaak et al, 2010; Brennan et al, 2013). In addition to exhibiting characteristic mutational and gene expression profiles, mutations in NF1 are prevalent in the mesenchymal subtype, EGFR in the classical subtype, and PGDRFA and IDH1 in the proneural subtype. These subtypes are also found to bear resemblance with gene expression profiles of normal brain cells, with proneural tumours enriched for the oligodendrocyte development signature. Mesenchymal tumours are enriched for the cultured astroglia signature, although it remains unclear whether cultured astroglia represent any specific stem or progenitor cells in vivo. More recent analyses have been expanded to include other types of gliomas, such as grade II and III astrocytomas, oligodendrogliomas, or oligoastrocytomas. These glioma subgroups are based on newer criteria, such as the presence of IDH1 and TERT promoter mutations and chromosome 1p/19q co-deletion, as well as telomere alterations and DNA methylation (Eckel-Passow et al, 2015; Ceccarelli et al, 2016). However, the reproducibility, clinical relevance, and functional basis of these subclasses remain to be established.
On the other hand, Glioblastoma multiforme is a highly heterogeneous tumour consisting of mutant cells with varying morphologies, levels of aneuploidy, and expression of different cell markers. The cellular and genetic heterogeneity is well illustrated using recently developed techniques involving single-cell sequencing analysis that has demonstrated that single GBM cells, even within tumour cells from the same patient, are not genetic phenocopies. Single cells are found to exhibit variable expression of transcriptional programs in cellular processes such as hypoxia, cell cycle, and immune signalling, underscoring the inherent heterogeneity and complexity of these tumours (Patel et al, 2014). Moreover, genomic analysis reveals divergence of presumed mutational and epigenetic drivers between therapy-naive primary and recurrent GBM, as well as between low- and high-grade recurrent lesions, showing the versatility and evolutionary plasticity of these tumour cells (Johnson et al, 2014; Kim et al, 2015). These advances have shed light on the evolving genome of GBM, and speak to dynamic changes in the genomic make-up of tumour cells during malignant progression and in response to different therapies.
Genetically engineered mouse models
A variety of animal models have been developed that incorporate signature mutations found in human patients. Genetically engineered mouse models (GEMMs) are powerful tools in investigating the natural history of glioma development and also as preclinical models. The general approach involves loss of function of driver tumour suppressor genes, such as Cdkn2a (Ink4A/Arf), Nf1, Trp53, Pten, and Rb1 and/or the expression of driver oncogenes, such as mutant epidermal growth factor receptor (e.g., EGFR VIII) and Ras (K-Ras, H-Ras), Akt, and platelet-derived growth factor (PDGF) ligands (Kegelman et al, 2014). Such mutations are introduced as conventional or conditional knockouts and transgenic models. Modifications include the RCAS (replication-competent ASLV long terminal repeat with splice acceptor) system that employs Rous sarcoma virus-based vectors that are then injected into mice expressing the TVA receptor under the control of cell type-specific promoters, and MADM (mosaic analysis with double markers), a cre-based genetic mosaic model (Liu et al, 2011; Kegelman et al, 2014). Adenoviral or lentiviral vectors expressing cre or any gene of interest, or cultured cells first transduced with virus, can also be directly injected into mice. The use of conditional knockouts target initiating mutations into discrete cell populations using specific promoters that limit the extent of cre-mediated recombination in restricted cell types. Inducible cre systems further provide temporal and spatial control of gene targeting that prevents the developmental phenotype and early lethality complications associated with conventional knockouts. Such methods have produced a plethora of mouse models that develop gliomas with a range of histologic types, penetrance, and latencies, and which recapitulate in varying degrees the pathological hallmarks of human gliomas (Janbazian et al, 2014). Variations caused by differences in genetic mutations, cell of origin, and timing of initiation make it imperative that physiologic relevance and resemblance to human disease be established whenever animal models are used to study glioma pathophysiology.
Cells of origin of GBM
The cell of origin refers to the normal cell that acquires the initial cancer-promoting genetic hit(s). The identity of tumour-initiating cells in different organ cancers is an area of intense study. To date, many examples exist demonstrating the potential of immature, proliferating cells in animal models of leukaemia, colon, breast, lung, and other cancers (Visvader, 2011).
In GBM, addressing this question requires an understanding of the hierarchy of cell lineages in the CNS. In the past several decades, advances in developmental neurobiology have identified various cellular compartments that allow targeting of discrete cell populations. In the adult mammalian brain, the two main neurogenic regions are in the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus in the hippocampal formation. These adult neural stem cells (NSCs) are quiescent glial fibrillary acidic protein-positive (Gfap+) cells that exhibit unlimited self-renewal and multipotentiality. These give rise to proliferative progenitor cells that have more limited self-renewal and are fated to differentiate into different cell types. Whereas Gfap+ cells in the SVZ and SGZ identify quiescent neural stem cells, Nestin is widely expressed in both stem and progenitor cells; other markers that have been reported include Glast, Sox2, Tlx, Gli1, EGFR, PlexinB2, and Dlx1 (Doetsch et al, 1999; Canoll and Goldman, 2008; Mich et al, 2014). The bHLH transcription factor Ascl1 has recently been shown to mark a population of adult bipotential progenitor cells that can give rise to both the neuronal and oligodendrocyte lineage, as well as give rise to neurospheres in vitro (Kim et al, 2007; Mich et al, 2014). Neuronal progenitor cells in the SVZ give rise to new neurons that migrate through the rostral migratory stream and into the olfactory bulb, whereas those in the SGZ locally migrate into the granular cell layer. On the other hand, unipotent progenitors such as oligodendrocyte progenitor cells (OPCs) are broadly arrayed in the adult brain, where they comprise ∼5% of CNS cells. These OPCs express lineage-specific proteins including NG2, PDGFRα, and Olig1, and typically give rise to mature oligodendrocytes in both the developing and adult brain (Nishiyama et al, 2009).
The identification of cell lineage markers has provided necessary tools for the interrogation of the cell of origin. Both stereotactic and genetic approaches have been used, and inducible cre lines have allowed cell type-specific targeting of initiating mutations at adult ages (Figure 1). Earlier models target mutations using cell type-specific promoters but in embryonic or early postnatal cells, which is not ideal as GBM is an adult disease, with a median age of 64 years at the time of diagnosis (Stupp et al, 2005; Wen and Kesari, 2008). Direct injection of cre recombinase-expressing adenovirus into the SVZ of adult mutant mice containing conditional tumour suppressor alleles of Nf1, Trp53, and Pten has been shown to induce GBM formation (Alcantara Llaguno et al, 2009; Jacques et al, 2010). Mutant mice with the Nestin-creERT2 transgene, which contains the Nestin promoter/enhancer with the intron 2 regulatory element that limits its expression in NSCs but not other cell types such as endothelial cells, when bred to contain the same tumour suppressor mutations and induced at adult ages (4–8 weeks of age), develop GBM (Alcantara Llaguno et al, 2009). On the other hand, the use of Gfap as a marker for quiescent NSCs is complicated by its expression in some mature astrocytes, particularly in the dentate gyrus and striatum. Gfap-creERTM transgene-mediated loss of Pten, Trp53, and/or Rb induces high-grade glioma formation, where a high proportion of tumours can be found in close association with proliferative niches in the SVZ-RMS and SGZ. This would be consistent with a scenario wherein the tumours originate from the NSC-targeted cells even though cre expression in stem cells was significantly lower than in non-neurogenic regions (Chow et al, 2011). The contribution of lineage-restricted progenitors is determined using the Ascl1-creERTM transgene that targets bipotential progenitors, whereby expression is observed in both adult neural and oligodendrocyte progenitors. Ascl1-creERTM mice carrying Nf1, Trp53, and/or Pten mutations develop GBM, as do NG2-creERTM mice, where adult oligodendrocyte progenitor cells alone are targeted (Galvao et al, 2014; Alcantara Llaguno et al, 2015).
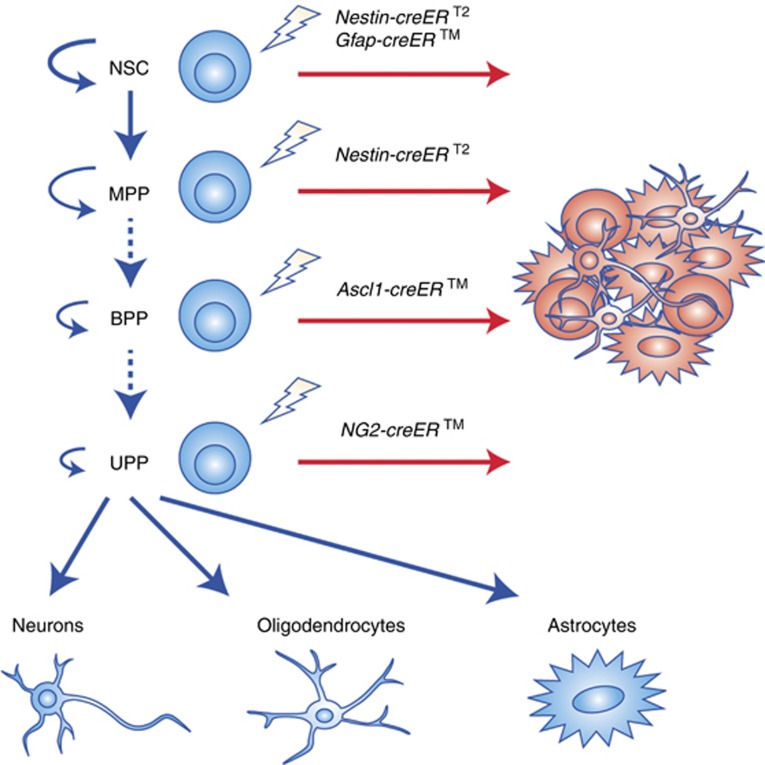
Figure 1.
Lineage hierarchy of tumour-initiating cells in GBM. Neural stem cells (NSC) are activated to give rise to progenitor cells that exhibit varying levels of potentiality: multipotent progenitors (MPPs) that give rise to all CNS cell types; bipotential progenitors (BPP), such as Ascl1 progenitors, that identify both adult oligodendrocyte progenitor cells (OPCs) and neural progenitor cells (NPCs); and unipotent progenitors (UPP) that differentiate into oligodendrocytes, neurons, or astrocytes. Dashed lines indicate that the hierarchical relationship between different progenitors is still unclear. Experiments using different inducible cre driver lines in conjunction with known driver mutations employed to target different adult brain cell types are shown. Models using Nestin-creERT2, which is expressed in neural stem cells as well as multipotent progenitors, can give rise to GBM. The Gfap-creERTM, which is expressed in SVZ neural stem cells as well as a subset of mature astrocytes, develop tumours near the neurogenic niches. The Ascl1-creERTM model, which targets NPCs and OPCs, gives rise to two GBM subtypes. One of these subtypes appears similar to GBM that develop using NG2-creERTM. Such animal models in which cre recombination is temporally controlled in the adult stem and progenitor populations have been shown to induce GBM formation, although with different molecular subtypes.
On the other hand, the role of post-mitotic, fully differentiated CNS cells in GBM formation is still controversial. Whether astrocytes are tumour-initiating cells is debatable because markers for astrocytes, such as Gfap and Glast, are also expressed in neural stem cells (Chow et al, 2011), although reports using cultured proliferative, early postnatal astrocytes (or possibly astrocyte progenitors) may not replicate the characteristics of adult astrocytes in vivo (Bachoo et al, 2002). There is evidence, however, that direct targeting of Gfap+ parenchymal astrocytes in adult mice does not easily form glioma (Alcantara Llaguno et al, 2009; Jacques et al, 2010). Meanwhile, differentiated neurons have been reported to transform into tumours in the experimental setting (Friedmann-Morvinski et al, 2012); however, it is not clear whether the targeted cells are exclusively post-mitotic neurons. Confirmation of these findings using temporally controlled and more specific lineage-restricted drivers as well as interrogation of the molecular profiles of these probable differentiated cell type-initiated tumours would be important in addressing this question.
Whether specific cells of origin are susceptible to certain mutations is not settled but most data from animal models suggest that stem cells and restricted progenitors are widely susceptible to a variety of mutations (Visvader, 2011). It is possible that specific cell types may exhibit preferential vulnerability to certain mutations, and that certain cell of origin-mutation combinations lead to specific tumour types or subtypes (Jacques et al, 2010).
Cell of origin as a driver of intertumoural diversity
The advent of advanced genomics has greatly increased our understanding of the molecular underpinnings of glioblastoma. Although GBMs from human patients contain different types of driver mutations that may represent different subtypes, the contribution of the cell of origin is not known. It is interesting to note that the TCGA molecular subtypes are enriched for lineage markers characteristic of distinct CNS cell types, such as proneural tumours exhibiting enrichment for the oligodendrocytic progenitor signature, suggesting that the molecular and/or epigenetic signature of the tumour-initiating cell is maintained during tumourigenesis (Verhaak et al, 2010; Levine et al, 2015).
What are the consequences of driving the same initiating mutations in different cells of origin? In mouse models, tumour suppressor deletions in different stem and progenitor lineages all lead to histologic GBM. However, these same GBMs are found to be molecularly distinct and separable based on the cell of origin. Hence, GBM driven by Nestin-creERT2, Ascl1-creERTM, and NG2-creERTM exhibit lineage-specific signatures in the context of Nf1, Trp53, and Pten loss (Alcantara Llaguno et al, 2015). Moreover, direct comparison of GBM from the Ascl1-creERTM model, where both adult neural and oligodendrocyte progenitors are targeted, demonstrate two subtypes of GBM, showing different predilection sites, marker expression, tumour boundaries, and gene expression profiles. Analysis of the mouse model subtype tumours also show similarities to the gene expression signatures of their normal progenitor cell counterparts. These observations suggest that the cell of origin can be an important determinant of tumour phenotype and genotype in GBM, and thus plays an important role in its malignant behaviour.
These studies are reminiscent of findings reported for leukaemia and medulloblastoma. For example, leukaemic stem cells (LSCs) derived from haematopoeitic stem cells transduced with mixed lineage leukaemia (MLL) rearrangements express high levels of Evi-1 that has been correlated with adverse clinical outcome, are hypermethylated, and are more resistant to chemotherapeutic agents, whereas LSCs from granulocyte-macrophage progenitors exhibit low Evi-1 expression and global methylation, and are less resistant to the same pharmacologic agents (Krivtsov et al, 2013). In medulloblastoma, molecular subtypes reflect distinct developmental origins. Sonic hedgehog (Shh) subtype medulloblastoma arising through aberrant Shh signalling (most often through inactivating mutations in Patched) in committed granule neuron precursor cells form within cerebellar hemispheres, whereas Wnt subtype medullobastoma arises from progenitor cells of the dorsal brainstem in response to activating mutations in the Wnt effector pathway, and tend to infiltrate the brainstem (Gibson et al, 2010).
These studies from GBM and other model systems are consistent with the cell of origin as a source of intertumoural diversity, whereby mutations in different cell types form different molecular subtypes (Figure 2). However, these subtypes can also be influenced by genetic mutations, epigenetic changes, and the microenvironment. These influences are not mutually exclusive, however, and more likely than not, intertumoural heterogeneity is a function of the combined effects of these several factors. Moreover, the relationship between cell lineage and biological phenotype suggests that mechanisms operative in normal cells may contribute to tumourigenesis. Signalling pathways such as Wnt and Notch, which are developmentally regulated in specific cell types, and especially stem and progenitor cells, may play important roles in tumourigenesis (Visvader, 2011).
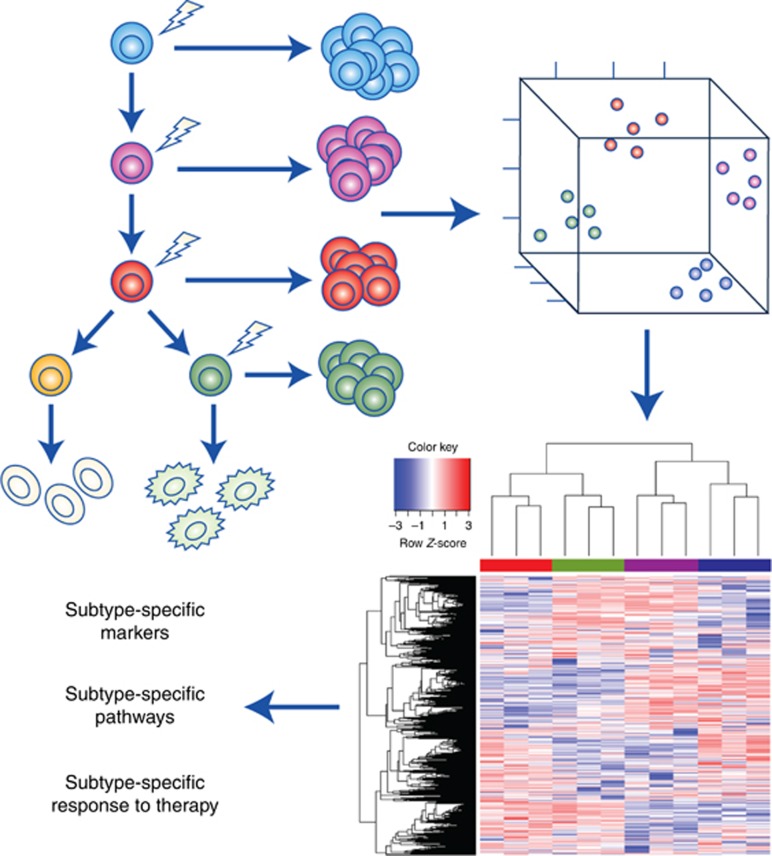
Figure 2.
Lineage-based functional subtyping of GBM. (Upper left panel) Cell of origin model of intertumoural diversity. Mouse models developed from targeting of tumour-initiating mutations into different cells of origin give rise to different GBM subtypes (represented by different colours). The GBM subtypes from different mouse models are molecularly separable, as hypothetically shown via locally linear embedding (LLE) dimension reduction analysis (upper right panel), and gene expression profiling (lower right panel). Studies on the molecular subtypes of GBMs that express distinct biomarkers, activate or turn off specific signalling pathways, and exhibit differential response to different treatment modalities may pave the way for improved diagnostics and therapeutics.
On the other hand, although it has not been experimentally proven for GBM, the lineage of the tumour-initiating cell may also directly influence the phenotype of the ensuing cancer stem cell that drives malignant behaviour and tumour progression that may also play a role in intratumoural heterogeneity.
Clinical consequences
Molecular subtyping of tumours, whether based on the lineage of the cell of origin, signature mutations, or a combination, provide important tools that can result in more refined diagnosis of brain tumours. Currently, the definitive clinical diagnosis for GBM is based on histopathologic examination by a neuropathologist. However, the subjective nature of this classic method makes it susceptible to bias and human error, in addition to relying on correlative and incomplete information. The incremental use of biomarkers and precision genomics will greatly supplement traditional histopathology in identifying gliomas. Molecular diagnostics will greatly advance the field, and facilitate the sharing of clinical and genomic information between academic and health centres. On the other hand, classification of these tumours into molecular subtypes using epigenetic, genetic, and gene expression profiling will further improve prognostication and lead to better stratification of patients for guided therapy (Figure 2). For example, the classical and mesenchymal subtypes have reportedly not only been associated with slightly better response to aggressive therapies, but also portend poorer prognosis compared with other subtypes (Verhaak et al, 2010; Ceccarelli et al, 2016). The CpG island methylator phenotype (G-CIMP), on the other hand, has been shown to represent a small subgroup of proneural gliomas that are associated with lower-grade lesions, IDH1 mutations, and better prognosis (Noushmehr et al, 2010). Furthermore, although single-cell sequencing analyses have demonstrated the expected intra-tumoural genetic heterogeneity of GBM cells at pre- and post-treatment conditions, studies in leukaemias suggest that evolving tumour cells retain the genetic and epigenetic signatures of their cells of origin (Levine et al, 2015). The maintenance of these lineage identifiers in tumour cells can potentially be used to identify specific tumour subtypes and track the evolution of individual cells. With more sophisticated technology, this may allow early detection of incipient tumours, as well as pave the way for preventative therapies for patients at high risk of developing brain tumours, such as neurofibromatosis type 1 (NF1) patients.
On the other hand, the identity of the cell of origin informing GBM subtype suggests that there may be subtype-specific pathways that may be open to therapeutic exploitation in different subsets of patients. Improved stratification will prevent unnecessary delay in delivery of appropriate treatments to which patients will most likely respond. Molecular diagnosis and subtype-specific treatments in CNS tumours is well under way in medulloblastomas (Taylor et al, 2012) and the glioblastoma field should not be far behind.
Finally, the use of experimental systems that address the cell of origin has led to the development of needed preclinical models that can be used to test candidate therapies. Physiologically relevant animal models that mirror human GBM at the molecular and cellular level are being used in both high-throughput screens and specific inhibitor studies in conjunction with patient-derived xenografts in drug development pipelines. Animal models provide large cohorts of in vitro and in vivo testing material that can be used to narrow down the most promising agents for subsequent testing in more limited patient samples. These molecularly defined subtype-specific mouse models can also provide unique insight into therapeutic response and resistance. Although mouse tumours do not always exhibit 100% equivalence with human GBM, these tools can be harnessed not only for drug validation but also inform target pathways that otherwise may not be uncovered by using only human tumour cells. The hope is that a deeper and more integrated understanding of the developmental origins, molecular underpinnings, and clinical behaviour of gliomas will pave the way for better therapeutic approaches that will benefit these patients.
Acknowledgments
We thank the members of the Parada lab for helpful discussion. This work was supported in part by Children's Tumor Foundation Young Investigator Award and NIH T32 Postdoctoral Trainee support (2T32CA124334-06; PI: Jerry Shay) to SRAL. LFP is an American Cancer Society Research Professor and recipient of NIH R01 Grant CA131313-01A1 and Cancer Prevention and Research Institute of Texas Grant RP100782.
The authors declare no conflict of interest.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF (2009) Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15(1): 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Wang Z, Sun D, Chen J, Xu J, Kim E, Hatanpaa KJ, Raisanen JM, Burns DK, Johnson JE, Parada LF (2015) Adult lineage-restricted CNS Progenitors specify distinct glioblastoma subtypes. Cancer Cell 28(4): 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA (2002) Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1(3): 269–277. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O'Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L (2013) The somatic genomic landscape of glioblastoma. Cell 155(2): 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoll P, Goldman JE (2008) The interface between glial progenitors and gliomas. Acta Neuropathol 116(5): 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG Jr., Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Noushmehr H, Iavarone A, Verhaak RG (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164(3): 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ (2011) Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19(3): 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97(6): 703–716. [DOI] [PubMed] [Google Scholar]
- Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O'Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372(26): 2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM (2012) Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338(6110): 1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao RP, Kasina A, McNeill RS, Harbin JE, Foreman O, Verhaak RG, Nishiyama A, Miller CR, Zong H (2014) Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proc Natl Acad Sci USA 111(40): E4214–E4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, Finkelstein D, Pounds S, Weiss A, Patay Z, Scoggins M, Ogg R, Pei Y, Yang ZJ, Brun S, Lee Y, Zindy F, Lindsey JC, Taketo MM, Boop FA, Sanford RA, Gajjar A, Clifford SC, Roussel MF, McKinnon PJ, Gutmann DH, Ellison DW, Wechsler-Reya R, Gilbertson RJ (2010) Subtypes of medulloblastoma have distinct developmental origins. Nature 468(7327): 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques TS, Swales A, Brzozowski MJ, Henriquez NV, Linehan JM, Mirzadeh Z, O' Malley C, Naumann H, Alvarez-Buylla A, Brandner S (2010) Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J 29(1): 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbazian L, Karamchandani J, Das S (2014) Mouse models of glioblastoma: lessons learned and questions to be answered. J Neurooncol 118(1): 1–8. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJ, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF (2014) Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343(6167): 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegelman TP, Hu B, Emdad L, Das SK, Sarkar D, Fisher PB (2014) In vivo modeling of malignant glioma: the road to effective therapy. Adv Cancer Res 121: 261–330. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Leung CT, Reed RR, Johnson JE (2007) In vivo analysis of Ascl1 defined progenitors reveals distinct developmental dynamics during adult neurogenesis and gliogenesis. J Neurosci 27(47): 12764–12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, Nam SH, Kim BS, Johnson MD, Kong DS, Seol HJ, Lee JI, Joo KM, Yoon Y, Park WY, Lee J, Park PJ, Nam DH (2015) Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 28(3): 318–328. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJ, Delwel R, Dohner K, Bullinger L, Kung AL, Melnick AM, Armstrong SA (2013) Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia 27(4): 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe'er D, Nolan GP (2015) Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162(1): 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H (2011) Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146(2): 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, Morrison SJ (2014) Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife 3: e02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, Zhu X (2009) Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci 10(1): 9–22. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17(5): 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344(6190): 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10): 987–996. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM (2012) Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123(4): 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216): 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1): 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE (2011) Cells of origin in cancer. Nature 469(7330): 314–322. [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359(5): 492–507. [DOI] [PubMed] [Google Scholar]