Abstract
Background:
Data on non-small-cell lung cancer (NSCLC) patients with non-classic epidermal growth factor receptor (EGFR) mutations are scarce, especially in non-Asian populations. The purpose of this study was to evaluate prevalence, clinical characteristics and outcome on EGFR-TKI treatment according to type of EGFR mutation in a Dutch cohort of NSCLC patients.
Methods:
We retrospectively evaluated a cohort of 240 EGFR-mutated NSCLC patients. Data on demographics, clinical and tumour-related features, EGFR-TKI treatment and clinical outcome were collected and compared between patients with classic EGFR mutations, EGFR exon 20 insertions and other uncommon EGFR mutations.
Results:
Classic EGFR mutations were detected in 186 patients (77.5%) and non-classic EGFR mutations in 54 patients (22.5%); 23 patients with an exon 20 insertion (9.6%) and 31 patients with an uncommon EGFR mutation (12.9%). Median progression-free survival (PFS) and overall survival (OS) on EGFR-TKI treatment were 2.9 and 9.7 months, respectively, for patients with an EGFR exon 20 insertion, and 6.4 and 20.2 months, respectively, for patients with an uncommon EGFR mutation. Patients with a double uncommon EGFR mutation that included G719X/L861Q/S768I had longer PFS and OS on EGFR-TKI treatment compared with patients with a single G719X/L861Q/S768I EGFR mutation (both P=0.02).
Conclusions:
In our Dutch cohort, prevalence and genotype distribution of non-classic EGFR mutations were in accordance with previously reported data. The PFS and OS on EGFR-TKI treatment in patients with an uncommon EGFR mutation were shorter compared with patients with classic EGFR mutations, but varied among different uncommon EGFR mutations.
Keywords: NSCLC, classic EGFR mutation, non-classic EGFR mutation, EGFR-TKI, TKI, EGFR
Classic EGFR mutations
The discovery of mutations in the epidermal growth factor receptor (EGFR) gene as oncogenic driver in lung cancer patients has changed both the diagnostic process and treatment of such patients. The EGFR mutations are detected in ∼10% of Caucasian patients with non-squamous non-small-cell lung cancer (NSCLC) and in up to 50% of Asian NSCLC patients (Dearden et al, 2013). In addition to the higher prevalence in people from Asian descent, there is a higher prevalence of EGFR mutations in women, nonsmokers and adenocarcinoma patients (Barlesi et al, 2016). The vast majority of EGFR mutations comprise microdeletions in exon 19 (45–50%) and the Leu858Arg (L858R) substitution, resulting from a point mutation in exon 21 (40–45% Murray et al, 2008). These mutations are so-called sensitising EGFR mutations and hereafter referred to as ‘classic EGFR mutations' (Supplementary Figure 1). The beneficial effect of treatment with EGFR tyrosine kinase inhibitors (TKIs) in NSCLC patients who harbour a classic EGFR mutation in their tumour is well established (Lynch et al, 2004; Maemondo et al, 2010; Mitsudomi et al, 2010; Fukuoka et al, 2011; Zhou et al, 2011; Han et al, 2012; Rosell et al, 2012; Sequist et al, 2013; Wu et al, 2014). However, resistance is inevitable and median progression-free survival (PFS) on EGFR-TKI treatment for NSCLC patients with a classic EGFR mutation is 8.0–13.1 months (Mok et al, 2009; Maemondo et al, 2010; Mitsudomi et al, 2010; Fukuoka et al, 2011; Zhou et al, 2011; Han et al, 2012; Rosell et al, 2012; Sequist et al, 2013; Wu et al, 2014, 2015).
The T790M mutation
The T790M mutation is a distinct EGFR mutation that is located in exon 20. It interferes with binding of EGFR-TKI to EGFR, thereby prohibiting the inhibitory effect of these agents. Detection of the T790M mutation before EGFR-TKI treatment is rare (0.5% Yu et al, 2014), although the detection rate of pretreatment T790M is higher with more sensitive detection methods (Rosell et al, 2011). However, the T790M mutation is detected in ∼60% of EGFR-mutated NSCLC patients on or post treatment with an EGFR-TKI showing renewed tumour growth (Yu et al, 2013).
Non-classic EGFR mutations
The EGFR mutations other than the classic EGFR mutations and exon 20 T790M mutations are less prevalent (hereafter referred to as ‘non-classic EGFR-mutations') (Supplementary Figure 1). The most prevalent non-classic EGFR mutations are insertions or duplications in EGFR exon 20 (further referred to as ‘EGFR exon 20 insertions') that are detected in ∼2.2–5.0% of NSCLC patients (Wu et al, 2008a; Arcila et al, 2013; Oxnard et al, 2013; Beau-Faller et al, 2014). In the study of Arcila et al (2013), EGFR exon 20 insertions were furthermore mutually exclusive with mutations in other genes, such as KRAS and BRAF, except for PIK3CA and there were no associations with age, sex, race or stage. Patients with EGFR exon 20 insertions generally have a lower response rate to EGFR-TKI treatment and a poorer prognosis compared with NSCLC patients with classic EGFR mutations (Wu et al, 2008a; Oxnard et al, 2013). Other non-classic EGFR mutations include so-called uncommon mutations (Supplementary Figure 1), for example, in EGFR exon 18 (e.g., G719X; X=A, S or C), EGFR exon 20 (e.g., S768I) and EGFR exon 21 (e.g., L861Q). The proportion of uncommon EGFR mutations among EGFR-mutated NSCLC patients might be as high as 14%, but varies in different studies (Yokoyama et al, 2006; Zhang et al, 2007; Wu et al, 2008b, 2011; Hata et al, 2010; De Pas et al, 2011; Arcila et al, 2013; Kobayashi et al, 2013; Keam et al, 2014).
Multiple uncommon EGFR mutations or an uncommon EGFR mutation in combination with a classic EGFR mutation may co-exist in the same tumour. These so-called ‘double' (or complex, or compound) mutations are reported to occur in 6.6% of EGFR-mutated NSCLC patients (Hata et al, 2010).
Data on results of EGFR-TKI treatment in Caucasian patients with non-classic EGFR mutations are scarce as they are commonly reported in small series, whereas the larger series typically include patients of Asian descent. We therefore evaluated a cohort of Dutch (i.e., predominant Caucasian) EGFR-mutated NSCLC patients retrospectively. The purpose of this study was to evaluate the prevalence and genotype distribution of non-classic EGFR mutations in this cohort, as well as clinical characteristics and outcome on EGFR-TKI treatment.
MATERIALS AND METHODS
Patients
All NSCLC patients in whom an EGFR mutation was detected in the VU University Medical Center (VUmc) between May 2006 and November 2014 (N=240) were retrospectively evaluated. As the VUmc is a diagnostic referral centre, some patients were diagnosed at our centre, but follow-up and treatment were performed in other hospitals. Patients with missing data on follow-up were excluded from analysis of clinical characteristics and outcome on EGFR-TKI treatment. For all other patients, data on demographics, clinical and tumour-related features, treatments and clinical outcomes was extracted from the medical records.
Mutation analysis
All mutation analyses were part of the routine diagnostic procedures in VU University Medical Center, Amsterdam, The Netherlands. The molecular diagnostic modalities for EGFR mutation analysis included Sanger sequencing, HRM sequencing and cancer panel multiplexed targeted resequencing (Janmaat et al, 2006; Heideman et al, 2009; Sie et al, 2014). All assays are designed to identify deletions or insertions in EGFR exons 19 and 20, and hot spot mutations in EGFR exons 18 through 21.
For analytical purposes, deletions in EGFR exon 19 and the L858R point mutation in EGFR exon 21 are referred to as classic EGFR mutations. Among non-classic EGFR mutations, a distinction between exon 20 insertions and ‘uncommon EGFR-mutations' was made (Supplementary Figure 1). The post-treatment T790M mutations are not included in our analyses, nor are common EGFR polymorphisms. All alterations that were detected were checked in Alamut Visual version 2.7 (Interactive Biosoftware, Rouen, France), the mycancergenome database (www.mycancergenome.org; accessed 1 April 2016) and the Cosmic database (cancer.sanger.ac.uk/cosmic; accessed 23 April 2016).
Treatment and outcomes
Patients who were alive at closing date (26 November 2015) or who were lost to follow-up were censored at the last date of follow-up. The EGFR-TKI treatment included treatment with erlotinib, gefitinib or afatinib in patients with advanced-stage disease. Survival was calculated from date of diagnosis of advanced-stage (stage IIIB or IV) disease until date of death. Objective response rate (ORR) on EGFR-TKI treatment was calculated as the proportion of patients with complete or partial response according to Response Evaluation Criteria in Solid Tumours (RECIST) 1.1 (Eisenhauer et al, 2009). Disease control rate (DCR) on EGFR-TKI treatment was calculated as the proportion of patients with an objective response or stable disease (for at least 6 weeks) according to the RECIST 1.1 criteria (Eisenhauer et al, 2009). Progression-free survival on EGFR-TKI treatment was calculated as the time from first day of treatment until progression of disease or date of death (from any cause). Patients who had not progressed at data cutoff were censored at the last day of follow-up. Overall survival (OS) on EGFR-TKI treatment was either calculated as the time from the first day of EGFR-TKI treatment until date of death (from any cause), or patients were censored at last follow-up.
Statistical analyses
Comparison of categorical variables was performed with Pearson's χ2 test. Comparison of three or more continuous variables was performed with one-way ANOVA. The Kaplan–Meier method was used for survival analyses and the log rank test was used to test for significance. Two-sided P-values of ⩽0.05 were considered significant and confidence intervals (CIs) were calculated at a 95% CI. The SPSS for Windows (version 20; SPSS Inc., Chicago, IL, USA) was used for statistical analyses. The medical ethical committee of VU University Medical Center (Amsterdam, The Netherlands) approved the protocol.
Results
Classic EGFR mutations
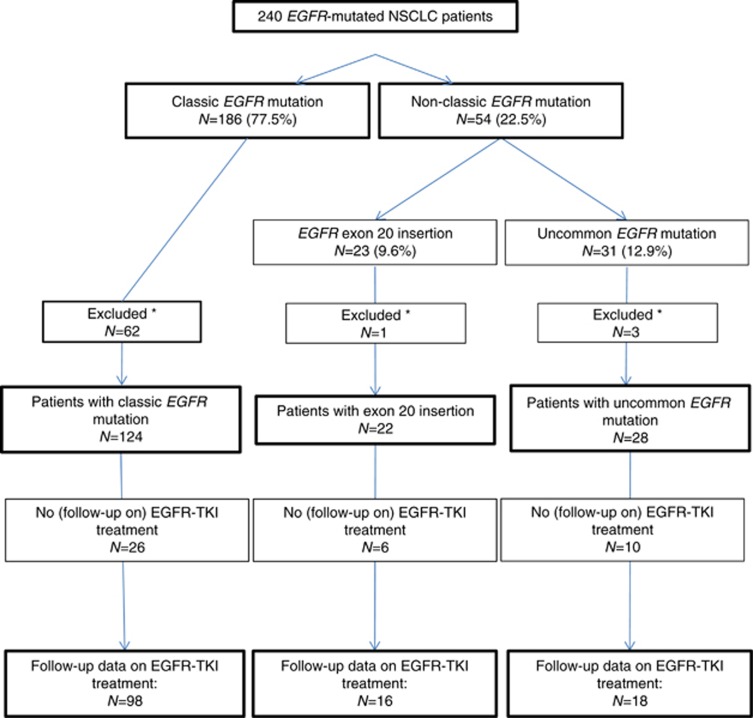
In 186 out of 240 patients (77.5%), a classic EGFR mutation was detected (Figure 1): 134 patients (72.0%) with an exon 19 deletion and 52 patients (28.0%) with an exon 21 L858R point mutation.
Figure 1.
Flowchart. *No treatment and follow-up in VUmc.
Sixty-two patients with a classic EGFR mutation were not treated at our centre and were excluded from further analysis. Clinical characteristics of the remaining 124 EGFR-mutated NSCLC patients are described in Table 1. Median follow-up was 31.6 months (95% CI, 26.1–27.3). The EGFR-TKI treatment was started in 111 patients (89.5%) (Supplementary Table 1). Clinical outcome on EGFR-TKI treatment of this group of patients is described in Table 2. Supplementary Tables provide more detailed data on start and/or progression on EGFR-TKI treatment (Supplementary Table 2), survival after EGFR-TKI treatment (Supplementary Table 3) and response setting (Supplementary Table 4).
Table 1. Patient characteristics.
| Patient characteristics | Classic EGFR mutation (N=124) | EGFR exon 20 insertion (N=22) | Uncommon EGFR mutation (N=28) | P-value | |||
|---|---|---|---|---|---|---|---|
| Median agea(years) | |
61.5 (range 29.5–83.0) |
61.0 (range 41.4–81.5) |
0.43 |
|||
| Frequency | (percentage) | Frequency | (percentage) | Frequency | (percentage) | ||
| Gender | |||||||
| Male | 29 | 23.4% | 8 | 36.4% | 9 | 32.1% | 0.34 |
| Female | 95 | 76.6% | 14 | 63.6% | 19 | 67.9% | |
| Ethnicity | |||||||
| Caucasian | 110 | 89.4% | 20 | 90.9% | 24 | 85.7% | 0.81 |
| Otherb | 13 | 10.6% | 2 | 9.1% | 4 | 14.3% | |
| Unknown | 1 | — | 0 | — | 0 | — | — |
| Smokingc | |||||||
| Current or previous smoker | 50 | 43.5% | 6 | 33.3% | 20 | 74.1% | 0.01 |
| Never smoker | 65 | 56.5% | 12 | 66.7% | 7 | 25.9% | |
| Unknown | 9 | — | 4 | — | 1 | — | — |
| Performance Status (PS) | |||||||
| PS 0–1 | 95 | 92.2% | 13 | 100.0% | 18 | 92.6% | 0.53 |
| PS >1 | 8 | 7.8% | 0 | 0.0% | 2 | 7.4% | |
| Unknown | 21 | — | 9 | — | 8 | — | — |
| Histology | |||||||
| Adenocarcinoma | 114 | 91.9% | 21 | 95.5% | 26 | 92.9% | 0.79 |
| Squamous cell carcinoma | 1 | 0.8% | 0 | 0.0% | 1 | 3.6% | |
| Adenosquamous carcinoma | 2 | 1.6% | 0 | 0.0% | 0 | 0.0% | |
| Large-cell neuroendocrine carcinoma | 4 | 3.2% | 0 | 0.0% | 0 | 0.0% | |
| Large-cell carcinoma | 3 | 2.4% | 1 | 4.5% | 1 | 3.6% | |
| Stagea | |||||||
| I–IIIA | 25 | 20.7% | 9 | 40.9% | 3 | 10.7% | 0.03 |
| IIIB–IV | 96 | 79.3% | 13 | 59.1% | 25 | 89.3% | |
| Unknown | 3 | — | 0 | — | 0 | — | — |
Abbreviation: EGFR=epidermal growth factor receptor.
At the time of first diagnosis of non-small-cell lung cancer (NSCLC).
Afro-American or Oriental.
Patients who smoked <100 cigarettes in a lifetime were considered as nonsmokers, patients who smoked within the last year before diagnosis were considered as current smokers and the remainder was considered as previous smoker.
Table 2. Median PFS, OS, ORR and DCR on EGFR-TKI treatment in advanced-stage NSCLC patients with classic EGFR mutations.
| N | Months | (95% CI) | P-value | |
|---|---|---|---|---|
|
Median PFS | ||||
| All patients | 98a | 12.2 | (10.8–13.5) | |
| Exon 19 | 75 | 12.6 | (11.2–14.1) | 0.26 |
| Exon 21 | 23 | 12.0 | (7.5–16.6) | |
|
Median OS | ||||
| All patients | 107b | 26.4 | (22.8–30.1) | |
| Exon 19 | 79 | 28.2 | (21.8–34.6) | 0.04 |
| Exon 21 | 28 | 21.0 | (20.4–21.6) | |
| N | % | P-value | ||
|
ORR | ||||
| All patients | 94c | 84.0% | ||
| Exon 19 | 70 | 84.3% | 0.91 | |
| Exon 21 | 24 | 83.3% | ||
|
DCR | ||||
| All patients | 94c | 95.7% | ||
| Exon 19 | 70 | 97.1% | 0.25 | |
| Exon 21 | 24 | 91.7% | ||
Abbreviations: CI=confidence interval; DCR=disease control rate; EGFR=epidermal growth factor receptor; NSCLC=non-small-cell lung cancer; ORR=objective response rate; OS=overall survival; PFS=progression-free survival; TKI=tyrosine kinase inhibitor.
For 13 patients, data on PFS on EGFR-TKI treatment were incomplete (Supplementary Table 2).
For 4 patients, data on OS on EGFR-TKI treatment were incomplete (Supplementary Table 3).
For 17, patients data on response on EGFR-TKI treatment were incomplete (Supplementary Table 4).
Non-classic EGFR mutations
A total of 54 patients (22.5%) harbouring a non-classic EGFR mutation were identified: 23 patients (9.6%) with an exon 20 insertion and 31 patients (12.9%) with an uncommon EGFR mutation in exons 18, 19, 20 and/or 21. In one patient, both an exon 20 insertion and an EGFR exon 20 V769L point mutation were detected. This patient was categorised in the EGFR exon 20 insertion group. All EGFR exon 20 insertions concerned insertions located on regions V769-N771 or H773-V774.
Of the group with uncommon EGFR mutations, 15 patients (6.3%) had a single uncommon EGFR mutation (Table 3) and 16 patients (6.7%) were identified with double uncommon EGFR mutations (Table 4). In three patients (1.3%) with a single uncommon EGFR mutation, a KRAS mutation was also detected (Table 3). In four patients (1.7%) with double EGFR mutations, one of these mutations concerned the classic EGFR mutation L858R on exon 21 (Table 4). There were two patients with a single G719X EGFR mutation and two patients with a single L861Q EGFR mutation (Table 3). Nine patients were identified with a double EGFR mutation that included G719X, L861Q and/or S768I (further referred to as ‘double G719X/L861Q/S768I' EGFR mutations; Table 4).
Table 3. Patients with single uncommon EGFR mutations.
|
EGFR
mutation |
Clinical characteristics |
EGFR-TKI treatment |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon | Codon | Protein | KRAS mutation | Sex | Histology | Smoking | Stagea | PSa | PFS | Response |
| 18 | c.2124G>T | p.K708N | No | F | Adeno | Current | No stage IV | Unknown | No TKI |
|
| 18 | c.2155G>A | p.G719S | No | M | Adeno | Previous | Stage IV | PS 1 | 1.6 | PD |
| 18 | c.2156G>C | p.G719A | No | F | Adeno | Previous | Stage IV | Unknown | 1.5 | NA |
| 18 | c.2161G>T | p.G721C | Yes | F | SqCC | Previous | Stage IV | PS 1 | No TKI |
|
| 19 | c.2232 C>G | p.I744M | No | M | Adeno | Current | Stage IV | Unknown | No TKI |
|
| 20 | c.2379 G>T | p.M793I | No | F | Adeno | Previous | Stage IV | PS 1 | 16.2 | SD |
| 20 | c.2327G>A | p.R776H | No | M | Adeno | Previous | Stage IV | PS 1 | 9.8 | PR |
| 20 | c.2327G>A | p.R776H | No | F | Adeno | Unknown | Stage IV | Unknown | No TKI |
|
| 20 | c.2327G>T | p.R776L | No | F | Adeno | Current | Stage IV | PS 1 | 2.6 | SD |
| 20 | c.2335_2336GG>TT | p.G779F | No | M | Adeno | Current | Stage IV | Unknown | No TKI |
|
| 21 | c.2582T>A | p.L861Q | No | M | Adeno | Current | Stage IV | PS 2 | 1.8 | PR |
| 21 | c.2513T>G | p.L838P | Yes | F | Adeno | Previous | Stage IV | PS 0 | 2.2 | SD |
| 21 | c.2495G>A | p.R832H | Yes | M | Adeno | Previous | Stage IV | PS 1 | No TKI |
|
| 19 | c.2231 2232ins18 | p.I744 K745insKIPVAI | No | F | Adeno | No clinical data |
||||
| 21 | c.2582T>A | p.L861Q | No | F | Adeno | No clinical data | ||||
Abbreviations: Adeno=adenocarcinoma; EGFR=epidermal growth factor receptor; F=female; M=male; NA=not available; PD=progressive disease; PFS=progression-free survival; PR=partial response; PS=performance status; SD=stable disease; SqCC=squamous cell carcinoma; TKI=tyrosine kinase inhibitor.
At the time of first diagnosis of non-small-cell lung cancer (NSCLC).
Table 4. Patients with double EGFR mutations (and at least one uncommon EGFR mutation).
|
EGFR
mutation |
Clinical characteristics |
EGFR-TKI treatment |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Classic/uncommon | Exon | Codon | Protein | Gender | Histology | Smoking | Stagea | PSa | PFS | Response |
| Double uncommon | 18+18 | c.2155G>A+c.2125G>A | p.G719S+p.E709K | F | Adeno | Nonsmoker | Stage IV | Unknown | 15.0 | PR |
| Double uncommon | 18+20 | c.2155G>T+c.2303G>T | p.G719C+p.S768I | F | Adeno | Previous | Stage IV | Unknown | 25.0 | NA |
| Double uncommon | 18+20 | c.2155G>A+c.2303G>T | p.G719S+p.S768I | F | Adeno | Previous | Stage IV | PS 0 | No TKI |
|
| Double uncommon | 18+20 | c.2155G>A+c.2327G>A | p.G719S+p.R776H | M | Adeno | Previous | Stage IV | PS 0 | 59.1 | PR |
| Double uncommon | 18+20 | c.2156G>C+c.2303G>T | p.G719A+p.S768I | F | Adeno | Previous | Stage IV | Unknown | 1.9 | PD |
| Double uncommon | 19+21 | c.2239-2253del15bp+c.2509G>T | p.del L747_T751+D837T | F | Adeno | Previous | Stage IV | PS 0 | 15.0 | CR |
| Double uncommon | 18+21 | c.2156G>C+c.2582T>A | p.G719A+p.L861Q | F | Adeno | Current | Stage IV | PS 1 | 2.1 | SD |
| Double uncommon | 20+20 | c.2303G>T+c.2305G>C | p.S768I+p.V769L | F | Adeno | Previous | Stage IV | PS 1 | 1.2 | NA |
| Double uncommon | 20+20 | c.2303G>T+c.2320G>A | p.S768I+p.V774M | F | Adeno | Nonsmoker | Stage IV | PS 1 | No TKI |
|
| Double uncommon | 21+21 | c.2497T>G+c.2504A>T | p.L833V+p.H835L | F | Adeno | Nonsmoker | Stage IV | PS 2 | 11.7 | PR |
| Double uncommon | 21+21 | c.2497T>G+c.2504A>T | p.L833V+p.H835L | F | Adeno | Nonsmoker | Stage IV | PS 1 | NA | NA |
| Double uncommon | 21+21 | c.2512C>G+c.2582T>A | p.L838V+p.L861Q | F | Large-cell | Nonsmoker | Stage IV | PS 1 | 6.4 | NA |
| Classic+uncommon | 21+21 | c.2573T>G+c.2618G>A | p.L858R+p.G873E | M | Adeno | Previous | Stage IV | PS 1 | NA | PR |
| Classic+uncommon | 21+21 | c.2573T>G+c.2612C>A | p.L858R+p.A871E | F | Adeno | Nonsmoker | Stage IV | PS 0 | 18.0 | PR |
| Classic+uncommon | 21+20 | c.2573T>G+c.2369C>T | p.L858R+p.T790M (pre-treatment T790M) | M | Adeno | Nonsmoker | Stage IV | PS 0 | 8.0 | SD |
| Classic+uncommon | 21+21 | c.2573T>G+c.2500G>T | p.L858R+p.V834L | F | Large-cell | No clinical characteristics | ||||
Abbreviations: Adeno=adenocarcinoma; CR=complete response; EGFR=epidermal growth factor receptor; F=female; Large-cell=large cell carcinoma; M=male; NA=not available; PD=progressive disease; PFS=progression-free survival; PR=partial response; PS=performance status; SD=stable disease; TKI=tyrosine kinase inhibitor.
At the time of first diagnosis of non-small-cell lung cancer (NSCLC).
Of the patients with non-classic EGFR-mutations, four were not treated in our centre and excluded from further analysis (i.e., one with an exon 20 insertion, one with L858R+V834L mutation, one with an exon 19 insertion and one with L861Q mutation).
Clinical characteristics of the remaining 22 patients with an EGFR exon 20 insertion and 28 patients with an uncommon EGFR mutation are described in Table 1. Median follow-up of these patients was 29.4 months (95% CI, 19.6–39.3). Baseline demographic characteristics were similar between the three groups, except for smoking (P<0.01).
EGFR-TKI treatment in patients with an EGFR exon 20 insertion
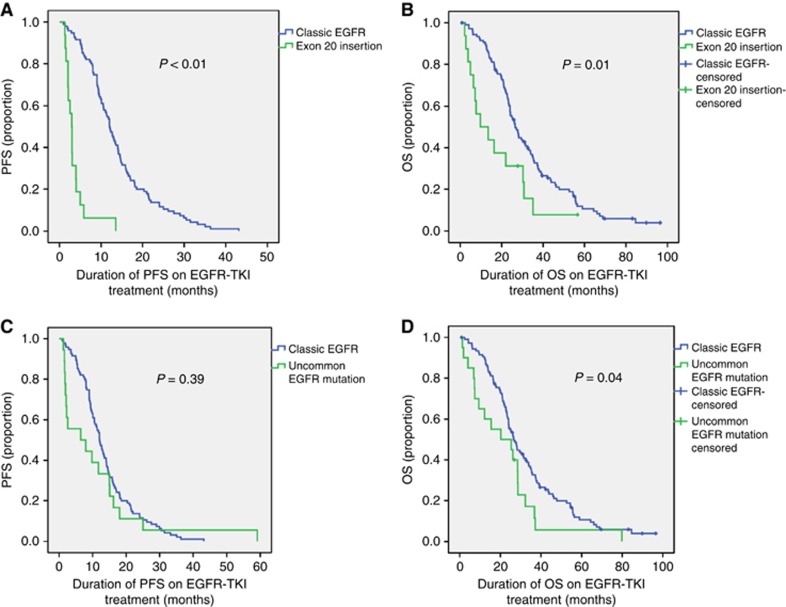
Sixteen patients with advanced-stage disease and an exon 20 insertion received EGFR-TKI treatment. Seven patients (43.8%) received EGFR-TKI as first-line treatment, but most patients received EGFR-TKI treatment as second-, third- or fourth-line treatment (Supplementary Table 5). Median PFS on EGFR-TKI treatment was 2.9 months (95% CI, 2.3–3.6). Median OS on EGFR-TKI treatment was 9.7 months (95% CI, 0.00–21.1). Both PFS and OS on EGFR-TKI treatment were significantly shorter in patients with an EGFR exon 20 insertion compared with patients with a classic EGFR mutation (P<0.01 and P=0.01, respectively; Figure 2A and B). The ORR was 0.0% and DCR was 56.3%.
Figure 2.
The PFS and OS on EGFR-TKI treatment in patients with a classic EGFR mutations vs EGFR exon 20 insertions or uncommon EGFR mutations. Difference between classic EGFR mutations vs EGFR exon 20 insertions in PFS (A) and OS (B) and between classic EGFR mutations and uncommon EGFR mutations in PFS (C) and OS (D). A full colour version of this figure is available at British Journal Of Cancer online.
EGFR-TKI treatment in patients with an uncommon EGFR exon 18, 19, 20 and 21 mutation
Twenty patients with an uncommon EGFR mutation received EGFR-TKI treatment. Sixteen patients (80%) received EGFR-TKI treatment as first-line and four patients (20%) as second-line treatment. For two patients, there was no registered date of progression. Median PFS on EGFR-TKI treatment for the remaining 18 patients (all advanced-stage disease) was 6.4 months (95% CI, 0.0–17.6). This was not significantly different compared with median PFS in patients with a classic EGFR mutation (P=0.39). Median OS on EGFR-TKI treatment in patients with an uncommon EGFR mutation was 20.2 months (95% CI, 0.0–41.7). This was significantly shorter compared with the median OS on EGFR-TKI treatment in patients with a classic EGFR mutation (P=0.04).
For 15 patients with uncommon EGFR mutations, data on response on EGFR-TKI treatment could be retrieved from the medical records: ORR was 53.3% and DCR was 86.7%.
Ten patients with single or double G719X/L861Q/S768I EGFR mutations were treated with an EGFR-TKI. Median PFS on EGFR-TKI treatment for patients with a double G719X/L861Q/S768I EGFR mutation (N=7) was 6.4 months (95% CI, 0.0–17.6), and this was significantly longer (P=0.02) than for patients with single-mutant status at these loci (N=3; 1.6 months (95% CI, 1.5–1.7)). Median OS on EGFR-TKI treatment for patients with a double G719X/L861Q/S768I EGFR mutation was 28.6 months (95% CI, 11.3–45.8), and 3.9 months (95% CI, 0.5–7.4) for those with a single G719X/L861Q/S768I EGFR mutation (P=0.02).
Discussion
Targeted agents are being developed rapidly and their clinical use is increasing in NSCLC patients. Considering the toxicities and costs of these drugs, their usage should be restricted to patients who truly benefit from them. In lung cancer, the efficiency of EGFR-TKIs is well known for classic EGFR mutations, but less data are available for patients with non-classic EGFR mutations. Moreover, most studies were performed in Asian populations. This study, among Dutch EGFR-mutated NSCLC-patients, adds to the current knowledge on non-classic EGFR mutations and the outcome on EGFR-TKI treatment in this subgroup of lung cancer patients.
In our cohort of 240 EGFR-mutated NSCLC patients, 54 patients (22.5%) were identified with a non-classic EGFR-mutation: 23 patients (9.6%) with an EGFR exon 20 insertion and 31 patients (12.9%) with an uncommon EGFR mutation. Previous studies on EGFR exon 20 insertions in predominantly non-Asian EGFR-mutated NSCLC patients reported a rate of 9%, 4.0% and 9.2% (Arcila et al, 2013; Oxnard et al, 2013; Beau-Faller et al, 2014), hence incidence of EGFR exon 20 insertions in our cohort is approximately in line with these studies. The incidence of uncommon EGFR mutations among non-Asian EGFR-mutated NSCLC patients varies between 5.9% and 20.4% (Pallis et al, 2007; De Pas et al, 2011; Beau-Faller et al, 2014; Stone et al, 2014; Arrieta et al, 2015; Lohinai et al, 2015), and this is also in accordance with results from our study. However, comparison to other studies is difficult, as there is a large variance in ethnicity of patients included, detection method of EGFR mutations and categorisation of non-classic EGFR mutations.
Interestingly, we detected a numerical difference in PFS and OS between patients with a classic EGFR mutation (exon 19 vs exon 21 mutation) that was significantly different for OS in favour of patients with an EGFR exon 19 deletion. Although originally it was thought that there was no difference between these two subtypes of classical EGFR mutations (Igawa et al, 2014), a meta-analysis detected a difference in PFS in favour of patients with an EGFR exon 19 deletion (Zhang et al, 2014). We did not detect a significant difference in PFS, but we did detect a difference in OS between these groups in accordance with a recent study (Rossi et al, 2016). Further investigation is warranted.
Several studies reported a higher prevalence of EGFR exon 20 insertions among women, nonsmokers and Asians (Huang et al, 2004; Kosaka et al, 2004; Shigematsu et al, 2005; Sasaki et al, 2007; Wu et al, 2008a), although another study did not find a significant difference in age, sex, ethnic origin or stage at diagnosis when compared with both patients with a classic EGFR mutation as in patients lacking a mutation (Arcila et al, 2013). Survival of NSCLC patients with an EGFR exon 20 insertion has generally been reported to be poor (Oxnard et al, 2013). Most exon 20 insertions are insensitive for treatment with both reversible and irreversible EGFR-TKIs (except for the EGFR exon 20 insertion A763_Y764insFQEA; Yasuda et al, 2013). This insensitivity is probably the reason of the poorer survival of this category of patients compared with NSCLC patients with classic EGFR mutations (Wu et al, 2008a; Lund-Iversen et al, 2012; Woo et al, 2014). In our cohort, PFS of patients with an EGFR exon 20 insertion on EGFR-TKI treatment was 2.9 months, comparable to the PFS of 1.5–2.0 months on erlotinib or gefitinib (Wu et al, 2008a; Jackman et al, 2009) and 2.7 months on afatinib (Yang et al, 2015b) that were reported previously. This suggests that these patients should preferably be treated with cytotoxic chemotherapy instead of first- and second-generation EGFR-TKIs. Recently, favourable results of a clinical study with AUY922 in NSCLC patients with an EGFR exon 20 insertion were reported and may hopefully provide a better treatment option for EGFR-mutated NSCLC patients with exon 20 insertion (Piotrowska et al, 2015).
Likewise, it has been reported that patients with uncommon EGFR mutations have lower EGFR-TKI sensitivity (Arrieta et al, 2015). We did not detect a statistically significant difference between patients with uncommon and classic EGFR mutations with respect to PFS, but considering the large numerical difference (6.4 vs 12.0 months, respectively), this is probably because of the small sample size and wide variation in PFS among patients with an uncommon EGFR mutation (Tables 3 and 4).
In our study, G719X/L816Q/S768I EGFR mutations are the most frequently detected among uncommon EGFR mutations, in line with previous reports (Mitsudomi and Yatabe, 2007; Shi et al, 2014). The G719X and the L861Q EGFR mutations were reported to have a shorter OS on gefitinib compared with classic EGFR mutations (Watanabe et al, 2014), although a recent study reported a PFS and OS of 13.8 and 26.9 months, respectively, for patients with a G719X EGFR mutation on first-line afatinib (Yang et al, 2015a). Chiu et al (2015) detected a statistically significant difference in median PFS on EGFR-TKI between patients (N=161) with single and double G719X/L816Q/S768I EGFR mutations. In our cohort, patients with double G719X/L816Q/S768I EGFR mutations not only had a statistically significant longer PFS on EGFR-TKI treatment compared with patients with single-mutant status at these loci, but also a longer OS. However, groups were small in our cohort, and hence interpretation should be with caution. Further investigation on the difference between single and double G719X, L816Q and/or S768I EGFR mutations is warranted.
It has been suggested that platinum-doublet treatment might be the best first-line treatment option for patients with (both single and double) G719X/L816Q/S768I EGFR mutations (Watanabe et al, 2014). However, taking into account the durable responses on EGFR-TKI treatment of several patients with a double EGFR mutation that included G719X, L861Q and/or S768I, in our opinion EGFR-TKI treatment could be considered as first-line treatment for these patients. In addition, high response rates of patients with G719X/L816Q/S768I EGFR mutations to first-line afatinib were recently reported (Yang et al, 2015a). Prospective trials are needed to elucidate this question.
Several limitations should be taken into account for this study. Because of the retrospective design, bias cannot be excluded. In addition, in a considerable part of the patients (especially in patients with an EGFR exon 20 insertion) data on performance score and smoking could not be retrieved from the medical records and the line of EGFR-TKI treatment (i.e., first-, second-line and so on) varied. Perhaps therefore, we detected a significant difference between the groups in smoking. In addition, in the early days of EGFR testing, clinical characteristics were taken into account. Therefore, there might have been a screening bias for women and nonsmokers. However, from 2012, all stage IV adenocarcinoma patients were tested for EGFR mutations, irrespective of gender, smoking status and race. A large molecular heterogeneity existed among patients with non-classic EGFR mutations. The results of the subgroup analyses should therefore be interpreted with caution. Furthermore, in routine pathology, solely tumour tissue is evaluated, and for most cases of our study no normal DNA was available to confirm the somatic origin of the mutations identified. The R776H mutation (detected in three patients in our cohort), for example, has both been reported as somatic and germline (Nagalakshmi et al, 2013; van Noesel et al, 2013). For one of our patients, analysis of normal DNA confirmed somatic nature (data not shown), but for the others because of absence of normal DNA a germline nature cannot be excluded.
To summarise, in this cohort of Dutch EGFR-mutated NSCLC patients, the prevalence and genotype distribution of non-classic EGFR mutations was in accordance with previously published studies among non-Asian, EGFR-mutated NSCLC patients. Outcome on EGFR-TKI treatment was poor for patients with EGFR exon 20 insertions and varied widely in patients with uncommon EGFR mutations. Further (prospective) studies on patients with non-classic EGFR mutations are warranted to hopefully improve prognosis of these patients.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
D Heideman has occasionally been member of the scientific advisory boards of Amgen and Pfizer.
Supplementary Material
References
- Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, Zakowski MF, Kris MG, Ladanyi M (2013) EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 12(2): 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta O, Cardona AF, Corrales L, Campos-Parra AD, Sanchez-Reyes R, Amieva-Rivera E, Rodriguez J, Vargas C, Carranza H, Otero J, Karachaliou N, Astudillo H, Rosell R (2015) The impact of common and rare EGFR mutations in response to EGFR tyrosine kinase inhibitors and platinum-based chemotherapy in patients with non-small cell lung cancer. Lung Cancer 87(2): 169–175. [DOI] [PubMed] [Google Scholar]
- Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I, Westeel V, Escande F, Monnet I, Lemoine A, Veillon R, Blons H, Audigier-Valette C, Bringuier PP, Lamy R, Beau-Faller M, Pujol JL, Sabourin JC, Penault-Llorca F, Denis MG, Lantuejoul S, Morin F, Tran Q, Missy P, Langlais A, Milleron B, Cadranel J, Soria JC, Zalcman G (2016) Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 387(10026): 1415–1426. [DOI] [PubMed] [Google Scholar]
- Beau-Faller M, Prim N, Ruppert AM, Nanni-Metellus I, Lacave R, Lacroix L, Escande F, Lizard S, Pretet JL, Rouquette I, de Crémoux P, Solassol J, de Fraipont F, Bieche I, Cayre A, Favre-Guillevin E, Tomasini P, Wislez M, Besse B, Legrain M, Voegeli AC, Baudrin L, Morin F, Zalcman G, Quoix E, Blons H, Cadranel J (2014) Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol 25(1): 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Yang CT, Shih JY, Huang MS, Su WC, Lai RS, Wang CC, Hsiao SH, Lin YC, Ho CL, Hsia TC, Wu MF, Lai CL, Lee KY, Lin CB, Yu-Wung YD, Chuang CY, Chang FK, Tsai CM, Perng RP, Chih-Hsin YJ (2015) Epidermal growth factor receptor tyrosine kinase inhibitor treatment response in advanced lung adenocarcinomas with G719X/L861Q/S768I mutations. J Thorac Oncol 10(5): 793–799. [DOI] [PubMed] [Google Scholar]
- De Pas T, Toffalorio F, Manzotti M, Fumagalli C, Spitaleri G, Catania C, Delmonte A, Giovannini M, Spaggiari L, De Braud F, Barberis M (2011) Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol 6(11): 1895–1901. [DOI] [PubMed] [Google Scholar]
- Dearden S, Stevens J, Wu YL, Blowers D (2013) Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 24(9): 2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45(2): 228–247. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu DT, Saijo N, Duffield EL, Rukazenkov Y, Speake G, Jiang H, Armour AA, To KF, Yang JC, Mok TS (2011) Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 29(21): 2866–2874. [DOI] [PubMed] [Google Scholar]
- Han JY, Park K, Kim SW, Lee DH, Kim HY, Kim HT, Ahn MJ, Yun T, Ahn JS, Suh C, Lee JS, Yoon SJ, Han JH, Lee JW, Jo SJ, Lee JS (2012) First-SIGNAL: first-line single-agent iressa versus gemcitabine and cisplatin trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol 30(10): 1122–1128. [DOI] [PubMed] [Google Scholar]
- Hata A, Yoshioka H, Fujita S, Kunimasa K, Kaji R, Imai Y, Tomii K, Iwasaku M, Nishiyama A, Ishida T, Katakami N (2010) Complex mutations in the epidermal growth factor receptor gene in non-small cell lung cancer. J Thorac Oncol 5(10): 1524–1528. [DOI] [PubMed] [Google Scholar]
- Heideman DA, Thunnissen FB, Doeleman M, Kramer D, Verheul HM, Smit EF, Postmus PE, Meijer CJ, Meijer GA, Snijders PJ (2009) A panel of high resolution melting (HRM) technology-based assays with direct sequencing possibility for effective mutation screening of EGFR and K-ras genes. Cell Oncol 31(5): 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, Wu YC, Chen YR, Tsai SF (2004) High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 10(24): 8195–8203. [DOI] [PubMed] [Google Scholar]
- Igawa S, Kasajima M, Ishihara M, Kimura M, Hiyoshi Y, Asakuma M, Otani S, Katono K, Sasaki J, Masuda N (2014) Comparison of the efficacy of gefitinib in patients with non-small cell lung cancer according to the type of epidermal growth factor receptor mutation. Oncology 87(4): 215–223. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE (2009) Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res 15(16): 5267–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmaat ML, Rodriguez JA, Gallegos-Ruiz M, Kruyt FA, Giaccone G (2006) Enhanced cytotoxicity induced by gefitinib and specific inhibitors of the Ras or phosphatidyl inositol-3 kinase pathways in non-small cell lung cancer cells. Int J Cancer 118(1): 209–214. [DOI] [PubMed] [Google Scholar]
- Keam B, Kim DW, Park JH, Lee JO, Kim TM, Lee SH, Chung DH, Heo DS (2014) Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol 19(4): 594–600. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Canepa HM, Bailey AS, Nakayama S, Yamaguchi N, Goldstein MA, Huberman MS, Costa DB (2013) Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol 8(1): 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T (2004) Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 64(24): 8919–8923. [DOI] [PubMed] [Google Scholar]
- Lohinai Z, Hoda MA, Fabian K, Ostoros G, Raso E, Barbai T, Timar J, Kovalszky I, Cserepes M, Rozsas A, Laszlo V, Grusch M, Berger W, Klepetko W, Moldvay J, Dome B, Hegedus B (2015) Distinct epidemiology and clinical consequence of classic versus rare EGFR mutations in lung adenocarcinoma. J Thorac Oncol 10(5): 738–746. [DOI] [PubMed] [Google Scholar]
- Lund-Iversen M, Kleinberg L, Fjellbirkeland L, Helland A, Brustugun OT (2012) Clinicopathological characteristics of 11 NSCLC patients with EGFR-exon 20 mutations. J Thorac Oncol 7(9): 1471–1473. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350(21): 2129–2139. [DOI] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362(25): 2380–2388. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11(2): 121–128. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Yatabe Y (2007) Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci 98(12): 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361(10): 947–957. [DOI] [PubMed] [Google Scholar]
- Murray S, Dahabreh IJ, Linardou H, Manoloukos M, Bafaloukos D, Kosmidis P (2008) Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol 3(8): 832–839. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi, Jamil K, Rani PU (2013) Association of EGFR gene polymorphism in head and neck cancer patients with tobacco and alcohol consuming habits. Biol Med 5: 69–77. [Google Scholar]
- Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, Jackman DM, Johnson BE, Janne PA (2013) Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 8(2): 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallis AG, Voutsina A, Kalikaki A, Souglakos J, Briasoulis E, Murray S, Koutsopoulos A, Tripaki M, Stathopoulos E, Mavroudis D, Georgoulias V (2007) 'Classical' but not 'other' mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer 97(11): 1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska Z, Costa DB, Huberman M, Oxnard GR, Gainor JF, Heist RS, Muzikansky A, Shaw AT, Niederst MJ, Fulton L, Engelman JA, Sequist LV (2015) Activity of AUY922 in NSCLC patients with EGFR exon 20 insertions. J Clin Oncol 33(Suppl): abstract 8015. [Google Scholar]
- Rosell R, Molina MA, Costa C, Simonetti S, Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P, Cardenal F, Isla D, Provencio M, Cobo M, Insa A, Garcia-Campelo R, Reguart N, Majem M, Viteri S, Carcereny E, Porta R, Massuti B, Queralt C, de Aguirre I, Sanchez JM, Sanchez-Ronco M, Mate JL, Ariza A, Benlloch S, Sanchez JJ, Bivona TG, Sawyers CL, Taron M (2011) Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 17(5): 1160–1168. [DOI] [PubMed] [Google Scholar]
- Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Munoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriguez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyj A, Queralt C, de Aquirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L Spanish Lung Cancer Group in collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13(3): 239–246. [DOI] [PubMed] [Google Scholar]
- Rossi S, D'Argento E, Basso M, Strippoli A, Dadduzio V, Cerchiaro E, Martini M, Cassano A, Barone C (2016) Different EGFR gene mutations in exon 18, 19 and 21 as prognostic and predictive markers in NSCLC: a single institution analysis. Mol Diagn Ther 20(1): 55–63. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Endo K, Takada M, Kawahara M, Kitahara N, Tanaka H, Okumura M, Matsumura A, Iuchi K, Kawaguchi T, Kawano O, Yukiue H, Yokoyama T, Yano M, Fujii Y (2007) EGFR exon 20 insertion mutation in Japanese lung cancer. Lung Cancer 58(3): 324–328. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, Massey D, Zazulina V, Shahidi M, Schuler M (2013) Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 31(27): 3327–3334. [DOI] [PubMed] [Google Scholar]
- Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang PC (2014) A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 9(2): 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF (2005) Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 97(5): 339–346. [DOI] [PubMed] [Google Scholar]
- Sie D, Snijders PJ, Meijer GA, Doeleman MW, van Moorsel MI, van Essen HF, Eijk PP, Grunberg K, van Grieken NC, Thunnissen E, Verheul HM, Smit EF, Ylstra B, Heideman DA (2014) Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell Oncol (Dordr) 37(5): 353–361. [DOI] [PubMed] [Google Scholar]
- Stone E, Allen HA, Saghaie T, Abbott A, Daniel R, Mead RS, Kohonen-Corish M, Plit M, Morgan L (2014) High proportion of rare and compound epidermal growth factor receptor mutations in an Australian population of non-squamous non-small-cell lung cancer. Intern Med J 44(12a): 1188–1192. [DOI] [PubMed] [Google Scholar]
- van Noesel J, van der Ven WH, van Os TA, Kunst PW, Weegenaar J, Reinten RJ, Kancha RK, Duyster J, van Noesel CJ (2013) Activating germline R776H mutation in the epidermal growth factor receptor associated with lung cancer with squamous differentiation. J Clin Oncol 31(10): e161–e164. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Minegishi Y, Yoshizawa H, Maemondo M, Inoue A, Sugawara S, Isobe H, Harada M, Ishii Y, Gemma A, Hagiwara K, Kobayashi K (2014) Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J Thorac Oncol 9(2): 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HS, Ahn HK, Lee HY, Park I, Kim YS, Hong J, Sym SJ, Park J, Lee JH, Shin DB, Cho EK (2014) Epidermal growth factor receptor (EGFR) exon 20 mutations in non-small-cell lung cancer and resistance to EGFR-tyrosine kinase inhibitors. Invest New Drugs 32(6): 1311–1315. [DOI] [PubMed] [Google Scholar]
- Wu JY, Wu SG, Yang CH, Gow CH, Chang YL, Yu CJ, Shih JY, Yang PC (2008. a) Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res 14(15): 4877–4882. [DOI] [PubMed] [Google Scholar]
- Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC (2011) Effectiveness of tyrosine kinase inhibitors on ‘uncommon' epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 17(11): 3812–3821. [DOI] [PubMed] [Google Scholar]
- Wu SG, Chang YL, Hsu YC, Wu JY, Yang CH, Yu CJ, Tsai MF, Shih JY, Yang PC (2008. b) Good response to gefitinib in lung adenocarcinoma of complex epidermal growth factor receptor (EGFR) mutations with the classical mutation pattern. Oncologist 13(12): 1276–1284. [DOI] [PubMed] [Google Scholar]
- Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, Li W, Hou M, Shi JH, Lee KY, Xu CR, Massey D, Kim M, Shi Y, Geater SL (2014) Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 15(2): 213–222. [DOI] [PubMed] [Google Scholar]
- Wu YL, Zhou C, Liam CK, Wu G, Liu X, Zhong Z, Lu S, Cheng Y, Han B, Chen L, Huang C, Qin S, Zhu Y, Pan H, Liang H, Li E, Jiang G, How SH, Fernando MC, Zhang Y, Xia F, Zuo Y (2015) First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 26(9): 1883–1889. [DOI] [PubMed] [Google Scholar]
- Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, Massey D, Zazulina V, Wu YL (2015. a) Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 16(7): 830–838. [DOI] [PubMed] [Google Scholar]
- Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, Lu S, Huang Y, Geater SL, Lee KY, Tsai CM, Gorbunova V, Hirsh V, Bennouna J, Orlov S, Mok T, Boyer M, Su WC, Lee KH, Kato T, Massey D, Shahidi M, Zazulina V, Sequist LV (2015. b) Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 16(2): 141–151. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, Huberman MS, Cohen DW, Nakayama S, Ishioka K, Yamaguchi N, Hanna M, Oxnard GR, Lathan CS, Moran T, Sequist LV, Chaft JE, Riely GJ, Arcila ME, Soo RA, Meyerson M, Eck MJ, Kobayashi SS, Costa DB (2013) Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 5(216): 216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Kondo M, Goto Y, Fukui T, Yoshioka H, Yokoi K, Osada H, Imaizumi K, Hasegawa Y, Shimokata K, Sekido Y (2006) EGFR point mutation in non-small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci 97(8): 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ (2014) Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol 25(2): 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ (2013) Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 19(8): 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GC, Lin JY, Wang Z, Zhou Q, Xu CR, Zhu JQ, Wang K, Yang XN, Chen G, Yang JJ, Huang YJ, Liao RQ, Wu YL (2007) Epidermal growth factor receptor double activating mutations involving both exons 19 and 21 exist in Chinese non-small cell lung cancer patients. Clin Oncol (R Coll Radiol) 19(7): 499–506. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sheng J, Kang S, Fang W, Yan Y, Hu Z, Hong S, Wu X, Qin T, Liang W, Zhang L (2014) Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: a meta-analysis. PLoS One 9(9): e107161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, Lu S, Zhang L, Hu C, Hu C, Luo Y, Chen L, Ye M, Huang J, Zhi X, Zhang Y, Xiu Q, Ma J, Zhang L, You C (2011) Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 12(8): 735–742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.