Abstract
Fungi play important roles as decomposers, plant symbionts and pathogens in soils. The structure of fungal communities in the rhizosphere is the result of complex interactions among selection factors that may favour beneficial or detrimental relationships. Using culture-independent fungal community profiling, we have investigated the effects of nitrogen fertilizer dosage on fungal communities in soil and rhizosphere of field-grown sugarcane. The results show that the concentration of nitrogen fertilizer strongly modifies the composition but not the taxon richness of fungal communities in soil and rhizosphere. Increased nitrogen fertilizer dosage has a potential negative impact on carbon cycling in soil and promotes fungal genera with known pathogenic traits, uncovering a negative effect of intensive fertilization.
The rhizosphere, the interface between soil and roots, is a biologically active zone where roots and microorganisms interact1. The identity of microorganisms in the rhizosphere has great influence on plant vigour and growth with beneficial, neutral and detrimental microbial relationships2. Structural and functional diversity of rhizosphere microbial populations in natural and agricultural systems is affected by plant species, crop cultivar, phenology, soil type, agronomy practice and other environmental factors3,4,5,6.
It is estimated that global N fertilizer use will increase threefold by 2050 to meet the growing need for food7. The use of chemical fertilizers is often accompanied by inefficiences that result in pollution and soil degradation8. The type and quantity of N fertilizer affects physical, chemical and biochemical properties of soil9,10, as well as bacterial and arbuscular mycorrhizal fungal (AMF) communities in the rhizosphere11,12.
Increasing the dose of NPK fertilizer has been associated with an increased presence of bacteria and fungi in crop soils13. However, whether the dose of fertilizer modifies rhizosphere fungal communities is largely unknown, with the exception of AMF which diminish in taxon richness in maize roots in response to N fertilizer11. The objective of this study was to investigate how N fertilizer rates influence fungal communities in rhizosphere and soil of a commercial sugarcane crop system.
Results and Discussion
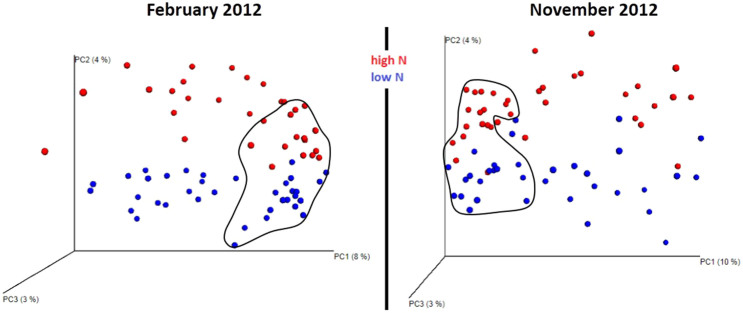
We sampled a total of 822 operational taxonomic units (OTUs) (February) and 820 OTUs (November) of fungi from 1135428 (February) and 1187048 (November) pyrosequence reads, respectively. Irrespective of collection time, the Chao1 metric showed no difference in species richness between low and high N treatment: in February 18.6 ± 8.83 (high N) and 17.2 ± 9.72 (low N); in November 19.3 ± 8.57 (high N) and 18.7 ± 9.63 (low N). Similar to the Chao1 metric, Simpson's index confirmed no difference in species richness: in February 0.768 ± 0.162 (high N) and 0.773 ± 0.125 (low N); in November 0.787 ± 0.126 (high N) and 0.782 ± 0.133 (low N). However, while fungal taxon richness did not differ between N-fertilizer doses, clear differences were detected in the fungal community composition in sugarcane rhizosphere and soil at each sampling time (Fig. 1).
Figure 1. Beta diversity metrics based on fungal ITS1 and ITS2 sequences reveal distinctly clustered rhizosphere and soil (soil data are circled in black) fungal communities structured by nitrogen fertilizer application.
Beta diversity fungal community clustering is observed for non-phylogenetic methods (binary Chord). The percentage of variation explained by the plotted principal coordinates is indicated on the axes.
Sequence-based community profiling is increasingly adopted to study plant root-associated bacterial and fungal communities14,15 as this approach circumvents culture bottlenecks. Species richness can be estimated from read counts of the internal transcribed spacer (ITS) region, but in the case of fungal communities these estimates may be biased due to the differing lengths of the ITS in different fungal species16, as shorter regions are preferentially amplified17. However, this bias does not significantly alter estimates of relative abundance (species evenness) of the dominant OTUs17,18 and is expected to be even further attenuated in comparisons of relative abundance within a community, e.g. between treatments. Our results (Fig. 1), based on the relative abundance of dominant OTUs and showing compositional dissimilarities between sugarcane soil and rhizosphere fungal communities associated with N fertilizer dose, are therefore unlikely to be significantly affected by this amplification bias, and detected community dissimilarities would reflect true biological variation.
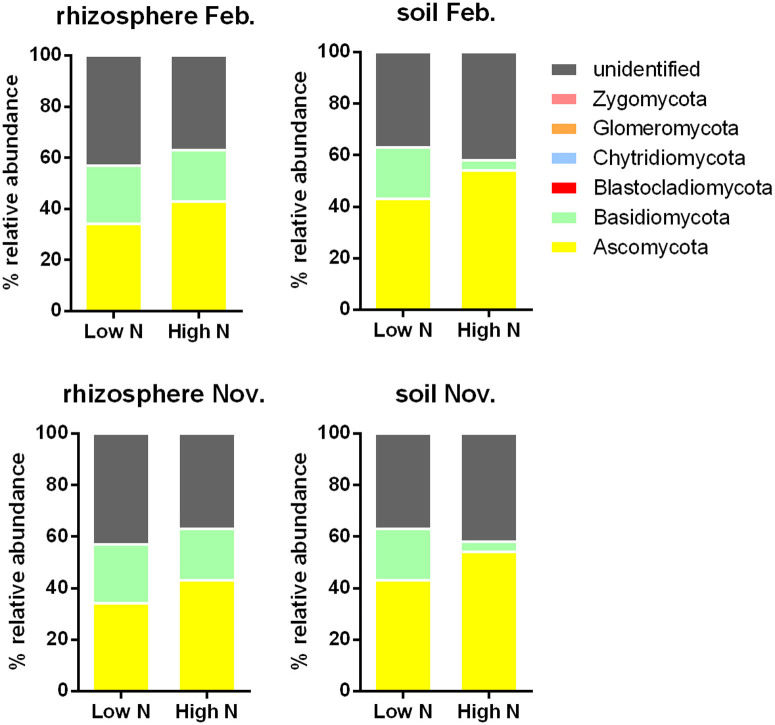
Interesting results are apparent even at the phylum level. Across all soil and rhizosphere samples collected in February and November, fungal taxonomic diversity involves mainly two phyla, Ascomycota and Basidiomycota (Fig. 2). The relative abundance of Ascomycota was generally higher in high N fertilizer dose conditions compared to low N, whereas for Basidiomycota it was lower. Consistent with this result, most saprotrophic microfungi are Ascomycota19 and their growth rate is correlated with N availability20. Basidiomycetes are widely recognised as lignin decomposers21 and thus important for carbon cycling in soil; in the same way, this beneficial function could be adversely affected by high N dose (Fig. 2). In agreement to our results, deleterious effects of mineral fertilizers on soil and plant function has been proposed because it negatively impacts on symbiotic relationships, including diazotrophic22 and AM symbioses23,24.
Figure 2. Effect of nitrogen fertilizer doses on fungal taxonomic diversity in sugarcane rhizosphere and soil.
The samples were collected at two time points (February and November 2012).
To identify the known fungal genera that were most-altered in relative abundance by N fertilizer doses, we compared the relative abundances of identified OTUs in rhizosphere and soil between low N and high N doses, using the ratio (low N/high N) as a means of evaluation (Tables 1 and 2). The data show that in all samples, the genera positively or negatively affected include groups known for their positive impact on soil and plant health (biocontrol, decomposers) or to the contrary, for their negative impact as plant pathogens. For example, irrespective of collection time, the relative abundance of OTUs corresponding to the ascomycete genus Clonostachys and to the basidiomycete genus Resinicium were amongst the most promoted in rhizosphere and soil by low N doses (Tables 1 and 2). Both genera contain species with known antagonistic effects against other microorganisms25,26 and are therefore of interest as potential biological control agents against pathogens. Fungal genera positive for plant health were also promoted by increased N dose, however to a lesser extent. There was a tendency for high N to substantially increase the proportion of pathogenic genera (Table 1 and 2). Whereas higher doses of NPK fertilizer have been associated with increased biomass of fungi in soils of crop systems13, this result indicates that in sugarcane rhizosphere and soil, increased N fertilizer also modifies the composition of the fungal communities and, by promoting pathogenic fungi, may have a negative impact on plant health.
Table 1. Fifteen fungal OTUs whose relative proportion in the community was increased or decreased the most between low and high N fertilizer dose. Ratio denotes the relative abundance of OTUs in low N compared to high N. The samples were from February 2012 collection.
| Rhizosphere | Soil | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OTU | Genus | Function | Ref | Ratio | OTU | Genus | Function | Ref | Ratio |
| 110 | Clonostachys | Biocontrol | 31 | 41.8 | 175 | Unidentified | n/a | 21 | |
| 178 | Agrocybe | Decomposer | 32 | 26.3 | 110 | Clonostachys | Biocontrol | 31 | 14.2 |
| 123 | Emericellopsis | Biocontrol | 33 | 26 | 82 | Calcarisporiella | n/a | 14 | |
| 205 | unidentified | n/a | 24.3 | 207 | Waitea | n/a | 13.3 | ||
| 183 | Clitopilus | Biocontrol | 34 | 23.7 | 199 | unidentified | n/a | 13 | |
| 212 | Resinicium | Decomposer, biocontrol | 26,35 | 16.2 | 43 | Epicoccum | Biocontrol | 36 | 11.3 |
| 145 | Kananascus | n/a | 11.8 | 253 | unidentified | n/a | 10.2 | ||
| 180 | Conocybe | Biocontrol | 37 | 11.2 | 250 | unidentified | n/a | 7.7 | |
| 126 | Sarocladium | Pathogen | 38 | 7.3 | 151 | Scedosporium | n/a | 6.7 | |
| 154 | Corynascus | Decomposer | 39 | 6 | 243 | unidentified | n/a | 6.6 | |
| 199 | unidentified | n/a | 5.5 | 180 | Conocybe | Biocontrol | 37 | 6.6 | |
| 196 | unidentified | n/a | 5.3 | 205 | unidentified | n/a | 5.7 | ||
| 204 | Ceratobasidium | Biocontrol | 40 | 5.3 | 216 | unidentified | n/a | 5.3 | |
| 1 | Aplosporella | n/a | 4.9 | 97 | Dactylella | Biocontrol | 41 | 5.3 | |
| 67 | Emericella | Pathogen | 42 | 4.6 | 76 | Spiromastix | n/a | 3.8 | |
| 136 | Nectria | Pathogen | 43 | 0.03 | 196 | unidentified | n/a | 0.04 | |
| 160 | Zopfiella | n/a | 0.03 | 89 | Scolecobasidium | n/a | 0.04 | ||
| 48 | Preussia | Biocontrol | 44 | 0.04 | 129 | Trichothecium | Pathogen | 45 | 0.05 |
| 161 | unidentified | n/a | 0.07 | 126 | Sarocladium | Pathogen | 38 | 0.07 | |
| 113 | Cordyceps | Biocontrol | 46 | 0.07 | 258 | Mortierella | Biocontrol | 47 | 0.09 |
| 4 | Capnodium | Pathogen | 48 | 0.15 | 200 | unidentified | n/a | 0.10 | |
| 194 | Coprinopsis | n/a | 0.17 | 197 | Psilocybe | n/a | 0.10 | ||
| 114 | Metarhizium | Biocontrol | 49 | 0.17 | 143 | Arthrinium | Biocontrol | 50 | 0.12 |
| 108 | unidentified | n/a | 0.18 | 187 | unidentified | n/a | 0.14 | ||
| 93 | Scytalidium | n/a | 0.21 | 186 | Vascellum | n/a | 0.20 | ||
| 44 | Exserohilum | Decomposer, biocontrol, pathogen | 51 | 0.21 | 64 | Thermoascus | Decomposer | 52 | 0.20 |
| 97 | Dactylella | Biocontrol | 41 | 0.22 | 122 | Acremonium | n/a | 0.20 | |
| 236 | Dioszegia | n/a | 0.23 | 236 | Dioszegia | n/a | 0.22 | ||
| 59 | Rhinocladiella | n/a | 0.25 | 111 | unidentified | n/a | 0.22 | ||
| 140 | Ophiocordyceps | Biocontrol | 53 | 0.27 | 191 | Pluteus | n/a | 0.24 | |
Table 2. Fifteen fungal OTUs whose relative proportion in the community was increased or decreased the most between low and high N fertilizer dose. Ratio denotes the relative abundance of OTUs in low N compared to high N. The samples were from November 2012 collection.
| Rhizosphere | Soil | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OTU | Genus | Function | Ref | Ratio | OTU | Genus | Function | Ref | Ratio |
| 183 | Clitopilus | Biocontrol | 34 | 122.9 | 178 | Bolbitius | n/a | 388.4 | |
| 208 | Resinicium | Decomposer, Biocontrol | 26,35 | 44.9 | 202 | Ceratobasidium | Biocontrol | 40 | 79.1 |
| 189 | Cyathus | Decomposer, Biocontrol | 54,55 | 34.5 | 179 | Conocybe | Biocontrol | 37 | 35.2 |
| 33 | unidentified | n/a | 29.2 | 103 | Clonostachys | Biocontrol | 31 | 24.6 | |
| 202 | Ceratobasidium | Biocontrol | 40 | 26.6 | 197 | unidentified | n/a | 17.8 | |
| 103 | Clonostachys | Biocontrol | 31 | 23.3 | 89 | Dactylella | Biocontrol | 41 | 16.2 |
| 211 | unidentified | n/a | 17.5 | 108 | Cordyceps | Biocontrol | 46 | 15.0 | |
| 230 | unidentified | n/a | 9.8 | 81 | Retroconis | n/a | 13.5 | ||
| 219 | unidentified | n/a | 6.0 | 83 | Xylogone | Biocontrol | 56 | 11.8 | |
| 204 | unidentified | n/a | 4.6 | 211 | unidentified | n/a | 11.5 | ||
| 182 | Gymnopilus | Decomposer | 57 | 4.5 | 187 | Marasmius | n/a | 9.5 | |
| 11 | Septoria | Pathogen | 58 | 4.5 | 239 | Spizellomyces | Decomposer | 59 | 8.8 |
| 218 | unidentified | n/a | 4.4 | 106 | Beauveria | Biocontrol | 60 | 7.1 | |
| 40 | Epicoccum | Biocontrol | 36 | 4.3 | 61 | Emericella | Biocontrol | 33 | 5.8 |
| 187 | Marasmius | n/a | 3.8 | 22 | Periconia | Pathogen | 61 | 5.0 | |
| 146 | Verticillium | Pathogen | 62 | 0.03 | 205 | Rhizoctonia | Pathogen | 63 | 0.02 |
| 186 | Unidentified | n/a | 0.04 | 177 | Agrocybe | Decomposer | 32 | 0.04 | |
| 125 | Sarocladium | Pathogen | 38 | 0.05 | 29 | unidentified | n/a | 0.08 | |
| 147 | Microascus | n/a | 0.07 | 196 | Lepista | n/a | 0.08 | ||
| 25 | Coniothyrium | Pathogen | 64 | 0.08 | 105 | unidentified | n/a | 0.08 | |
| 194 | unidentified | n/a | 0.09 | 192 | Coprinopsis | n/a | 0.11 | ||
| 156 | Cladorrhinum | Biocontrol | 65 | 0.09 | 62 | Eupenicillium | Biocontrol | 66 | 0.18 |
| 102 | Bionectria | Biocontrol | 67 | 0.10 | 147 | Microascus | n/a | 0.23 | |
| 50 | Cyphellophora | n/a | 0.10 | 92 | Meyerozyma | Biocontrol | 68 | 0.24 | |
| 68 | Talaromyces | Biocontrol | 69 | 0.13 | 12 | unidentified | n/a | 0.27 | |
| 34 | Phaeosphaeriopsis | Pathogen | 70 | 0.14 | 122 | Emericellopsis | Biocontrol | 33 | 0.27 |
| 90 | Dactylellina | Biocontrol | 71 | 0.16 | 194 | unidentified | n/a | 0.28 | |
| 245 | unidentified | n/a | 0.16 | 185 | Vascellum | n/a | 0.28 | ||
| 57 | unidentified | n/a | 0.17 | 121 | Acremonium | n/a | 0.29 | ||
| 45 | Preussia | Biocontrol | 44 | 0.17 | 167 | Nigrospora | Pathogen | 72 | 0.32 |
In summary, our findings add to understanding on how different doses of N influence fungal communities. The data show that the changes in relative abundance of fungal population in response to N doses are not restricted to AMF11 but span a wide range of fungal taxa, including genera known to influence plant health. Further research should elucidate the specific roles of these fungal taxa in sugarcane rhizosphere and soils, and on the heath of the plant. It is an attractive concept to manipulate the microbial community in the rhizosphere to reduce the need for agrochemicals, reduce disease incidence and improve crop performance27,28,29. To advance the ecological management of crop soils, understanding is needed of how beneficial microbial relationships can be fostered.
Methods
Sample collection
We sampled three individual plots within a 4-hectare field trial in the Burdekin region, Australia (near Ayr, S19°43.955′, E14°710.727′, 26 m above sea level). The soil is a silty-clay loam. Within each plot, half the sugarcane crops received an N supply rate of 200 kg N ha−1 y−1 (recommended ‘high’ N in the form of urea) while the other half received 40 kg N ha−1 y−1 (‘low’ N). From the three replicate plots, six bulk-soil and six roots with adhering soil (constituting the rhizosphere samples in our study) biological replicates were sampled at 0–10 cm depth from sugarcane receiving either N supply rate. Sampling was performed on a first and second ratoon crop of three sugarcane cultivars (Australian cultivars Q208 and Q186, and Brazilian cultivar SP79-2313) for a total of 144 root and bulk soil samples. Samples were immediately placed in a cool box for 2 days during transport to the laboratory and stored at −20°C for isolation of DNA. Sampling was carried out in February 2012 and November 2012 (4 and 3 months after fertilizer application, respectively) to assess the reproducibility of the observations.
DNA extraction and pyrosequencing
Total dsDNA was extracted from soil and rhizosphere samples using Mo Bio PowerSoil DNA isolation kits following manufacturer's instructions (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). To profile fungal communities, the fungal internal transcribed spacer (ITS) region was PCR-amplified from isolated soil and rhizosphere DNA using ITS1F (5'- CTTGGTCATTTAGAGGTAA-3') and ITS2R (5'-GCTGCGTTCTTCATCGATGC-3') primers modified on the 5' end to contain the 454 FLX Titanium Lib L adapters A and B, respectively. The forward and reverse primer contained a 10-base multiplex identifier (MID) barcode sequence between the primer target sequence and the adapter. A unique MID was used for each sample to identify sequencing reads to sample. PCR was performed using AmpliTaq Gold 360 master mix (Applied Biosystems). Thermocycling conditions were as follows: 95°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 45 s, 72°C for 60 s; 72°C for 7 min. Amplicons were purified using Ampure magnetic beads (Beckman Coulter), quantified using Picogreen (Invitrogen) fluorometry on the Quant Studio (Life Technologies) and normalized to 1 × 109 molecules/μL. Normalized samples were set up for qPCR (KAPA Biosystems kit) on the Quant Studio and then normalized and pooled for 454 sequencing. Sequencing was performed by the Australian Genome Research Facility Ltd.
Bioinformatics
The Quantitative Insights Into Microbial Ecology (QIIME) workflow was implemented for data analysis. Raw data were first de-multiplexed with a quality threshold of 150-bp minimum read length and minimum average quality score of 25. Further flanking regions were trimmed to extract ITS regions using ITSx 1.0.930. Quality filtered reads were clustered to pick operational taxonomic units (OTUs) using a closed reference-based method and then assigned taxonomy using the RDP classifier at a confidence level of 80% with reference to the UNITE database dated Nov 2012 (http://unite.ut.ee/repository.php). The resulting OTU table was used to estimate fungal diversity within (α-diversity) and between samples (β-diversity). Fungal species richness was represented using the Chao1 metric and Simpson's index. The binary Chord's metric was used to compare microbial communities based on their composition. Analyzed data were visualized by three-dimensional Principal Component Analysis (PCA).
Nucleotide sequence accession number
Pyrosequencing data were deposited in European Nucleotide Archive (ENA accession ERA372942).
Acknowledgments
We thank Jozef Visser for discussions and Michael Nuhn for depositing data in ENA. We are grateful to the ARC Centre of Excellence for Integrative Legume Research for access to research facilities. This project was carried out in part at Bioinformatics Resource Australia of EMBL Australia (BRAEMBL). This research was funded by a Queensland Co-Investment Fund award to MAR, PH and SS, and a James S. McDonnell Foundation grant to MAR. It was supported by access to facilities managed by Bioplatforms Australia and funded by the Australian Government National Collaborative Research Infrastructure Strategy and Education Investment Fund Super Science Initiative.
Footnotes
Author Contributions C.P.L. and Y.K.Y. have contributed equally. C.P.-L., T.G.A.L., P.H., M.A.R. and S.S. wrote the main manuscript text and C.P.-L., Y.K.Y., N.K. and N.R. performed the experiments. All authors reviewed the manuscript.
References
- Hinsinger P., Bengough A. G., Vetterlein D. & Young I. M. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152 (2009). [Google Scholar]
- Berendsen R. L., Pieterse C. M. J. & Bakker P. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012). [DOI] [PubMed] [Google Scholar]
- Grayston S. J., Wang S., Campbell C. D. & Edwards A. C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378 (1998). [Google Scholar]
- Marschner P., Yang C. H., Lieberei R. & Crowley D. E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33, 1437–1445 (2001). [Google Scholar]
- İnceoğlu Ö., Falcão Salles J. & van Elsas J. Soil and cultivar type shape the bacterial community in the potato rhizosphere. Microb. Ecol. 63, 460–470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupwayi N. Z., Rice W. A. & Clayton G. W. Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol. Biochem. 30, 1733–1741 (1998). [Google Scholar]
- Tilman D., Cassman K. G., Matson P. A., Naylor R. & Polasky S. Agricultural sustainability and intensive production practices. Nature 418, 671–677 (2002). [DOI] [PubMed] [Google Scholar]
- Cui Z. L. et al. Closing the N-use efficiency gap to achieve food and environmental security. Environ. Sci. Technol. 48, 5780–5787 (2014). [DOI] [PubMed] [Google Scholar]
- Schreiner O. The organic constituents of soils. U.S. Department of Agriculture, Bureau of Soils, Circular No. 74 - Washington, Government Printing Office, (1913) .https://archive.org/details/organicconstitue74schr (Date of access: December 01 2014)
- Paungfoo-Lonhienne C., Visser J., Lonhienne T. G. A. & Schmidt S. Past, present and future of organic nutrients. Plant Soil 359, 1–18 (2012). [Google Scholar]
- Toljander J. F., Santos-Gonzalez J. C., Tehler A. & Finlay R. D. Community analysis of arbuscular mycorrhizal fungi and bacteria in the maize mycorrhizosphere in a long-term fertilization trial. FEMS Microbiol. Ecol. 65, 323–338 (2008). [DOI] [PubMed] [Google Scholar]
- Verbruggen E. et al. Positive effects of organic farming on below-ground mutualists: large-scale comparison of mycorrhizal fungal communities in agricultural soils. New Phytol. 186, 968–979 (2010). [DOI] [PubMed] [Google Scholar]
- Aira M., Gomez-Brandon M., Lazcano C., Baath E. & Dominguez J. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281 (2010). [Google Scholar]
- Bulgarelli D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012). [DOI] [PubMed] [Google Scholar]
- Dumbrell A. J. et al. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 190, 794–804 (2011). [DOI] [PubMed] [Google Scholar]
- Bellemain E. et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A. et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME Journal 4, 642–647 (2010). [DOI] [PubMed] [Google Scholar]
- Amend A. S., Seifert K. A. & Bruns T. D. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Ecol. 19, 5555–5565 (2010). [DOI] [PubMed] [Google Scholar]
- Xiong J., Peng F., Sun H., Xue X. & Chu H. Divergent responses of soil fungi functional groups to short-term warming. Microb. Ecol. 68, 708–715 (2014). [DOI] [PubMed] [Google Scholar]
- Fontaine S. et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 43, 86–96 (2011). [Google Scholar]
- Hanson C., Allison S., Bradford M., Wallenstein M. & Treseder K. Fungal taxa target different carbon sources in forest soil. Ecosystems 11, 1157–1167 (2008). [Google Scholar]
- Laws M. T. & Graves W. R. Nitrogen inhibits nodulation and reversibly suppresses nitrogen fixation in nodules of Alnus maritima. J. Am. Soc. Hortic. Sci. 130, 496–499 (2005). [Google Scholar]
- Ryan M. H. & Graham J. H. Is there a role for arbuscular mycorrhizal fungi in production agriculture? Plant Soil 244, 263–271 (2002). [Google Scholar]
- Larsen J., Ravnskov S. & Sorensen J. in Mycorrhizae in crop production (eds C Hamel & C Planchette) .123–150 (The Haworth Press, 2007).
- ten Hoopen G. M. et al. Compatibility between Clonostachys isolates with a view to mixed inocula for biocontrol. Mycologia 102, 1204–1215 (2010). [DOI] [PubMed] [Google Scholar]
- Holmer L. & Stenlid J. Resinicium bicolor; a potential biological control agent for Heterobasidion annosum. European Journal of Forest Pathology 27, 159–172 (1997). [Google Scholar]
- Shen J. et al. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J. Exp. Bot. 64, 1181–1192 (2013). [DOI] [PubMed] [Google Scholar]
- Chaparro J. M., Sheflin A. M., Manter D. K. & Vivanco J. M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499 (2012). [Google Scholar]
- Bakker M. G., Manter D. K., Sheflin A. M., Weir T. L. & Vivanco J. M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360, 1–13 (2012). [Google Scholar]
- Bengtsson-Palme J. et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919 (2013). [Google Scholar]
- Cota L. V., Maffia L. A., Mizubuti E. S. G. & Macedo P. E. F. Biological control by Clonostachys rosea as a key component in the integrated management of strawberry gray mold. Biol. Control 50, 222–230 (2009). [Google Scholar]
- Philippoussis A., Zervakis G. & Diamantopoulou P. Bioconversion of agricultural lignocellulosic wastes through the cultivation of the edible mushrooms Agrocybe aegerita, Volvariella volvacea and Pleurotus spp. World J. Microbiol. Biotechnol. 17, 191–200 (2001). [Google Scholar]
- Fantini A. A. Genetics and antibiotic production of Emericellopsis species. Genetics 47, 161–177 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru S., Collins C. M., Hartley A. J., Bailey A. M. & Foster G. D. Establishing molecular tools for genetic manipulation of the pleuromutilin-producing fungus Clitopilus passeckerianus. Appl. Environ. Microbiol. 75, 7196–7204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J. H. & Jellison J. Calcium translocation, calcium oxalate accumulation and hyphal sheath morphology in the white-rot fungus Resinicum bicolor. Can. J. Bot. 73, 927–936 (1995). [Google Scholar]
- Larena I. et al. Biological control of postharvest brown rot (Monilinia spp.) of peaches by field applications of Epicoccum nigrum. Biol. Control 32, 305–310 (2005). [Google Scholar]
- Brady L. R., Benedict R. G., Tyler V. E., Stuntz D. E. & Malone M. H. Identification of Conocybe filaris as a toxic basidiomycete. Lloydia 38, 172–173 (1975). [PubMed] [Google Scholar]
- Tschen J. S. M., Chen L. L., Hsieh S. T. & Wu T. S. Isolation and phytotoxic effects of helvolic acid from plant pathogenic fungus Sarocladium oryzae. Bot. Bull. Acad. Sin. 38, 251–256 (1997). [Google Scholar]
- Tasca F. et al. A third generation glucose biosensor based on cellobiose dehydrogenase from Corynascus thermophilus and single-walled carbon nanotubes. Analyst 136, 2033–2036 (2011). [DOI] [PubMed] [Google Scholar]
- Mosquera-Espinosa A. T., Bayman P., Prado G. A., Gomez-Carabali A. & Otero J. T. The double life of Ceratobasidium: orchid mycorrhizal fungi and their potential for biocontrol of Rhizoctonia solani sheath blight of rice. Mycologia 105, 141–150 (2013). [DOI] [PubMed] [Google Scholar]
- Olatinwo R., Yin B., Becker J. O. & Borneman J. Suppression of the plant-parasitic nematode Heterodera schachtii by the fungus Dactylella oviparasitica. Phytopathology 96, 111–114 (2006). [DOI] [PubMed] [Google Scholar]
- Frisvad J. C. & Samson R. A. Emericella venezuelensis, a new species with stellate ascospores producing sterigmatocystin and aflatoxin B-1. Syst. Appl. Microbiol. 27, 672–680 (2004). [DOI] [PubMed] [Google Scholar]
- Wen F., White G. J., VanEtten H. D., Xiong Z. & Hawes M. C. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 151, 820–829 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapperson R. R., Kotiw M., Davis R. A. & Dearnaley J. D. The diversity and antimicrobial activity of Preussia sp. endophytes isolated from Australian dry rainforests. Curr. Microbiol. 68, 30–37 (2014). [DOI] [PubMed] [Google Scholar]
- Hamid M. I., Hussain M., Ghazanfar M. U., Raza M. & Liu X. Z. Trichothecium roseum causes fruit rot of tomato, orange, and apple in Pakistan. Plant Dis. 98, 1271–1271 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng P., Xia Y. L., Zhang S. W. & Wang C. S. Genetics of Cordyceps and related fungi. Appl. Microbiol. Biotechnol. 97, 2797–2804 (2013). [DOI] [PubMed] [Google Scholar]
- Al-Shammari T. A., Bahkali A. H., Elgorban A. M., El-Kahky M. T. & Al-Sum B. A. The use of Trichoderma longibrachiatum and Mortierella alpina against root-knot nematode, Meloidogyne javanica on tomato. J. Pure Appl. Microbiol. 7, 199–207 (2013). [Google Scholar]
- De Felice B. & Wilson R. R. Molecular characterization of a novel pathogen-responsive receptor kinase-like in Citrus limon. Tree Genet. Genomes 6, 47–56 (2010). [Google Scholar]
- Pattemore J. A. et al. The genome sequence of the biocontrol fungus Metarhizium anisopliae and comparative genomics of Metarhizium species. BMC Genomics 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos H. P., Braun G. H., Pupo M. T. & Said S. Antimicrobial activity from endophytic fungi Arthrinium state of Apiospora montagnei Sacc. and Papulaspora immersa. Braz. Arch. Biol. Technol. 53, 629–632 (2010). [Google Scholar]
- Haelewaters D. Exserohilum rostratum, the killing fungus . http://www.scilogs.com/life_off_the_edge/exserohilum-rostratum-the-killing-fungus/ (2012) (Date of access: December 01 2014).
- da Silva R. et al. Production of xylanase and CMCase on solid state fermentation in different residues by Thermoascus aurantiacus miehe. Braz. J. Microbiol. 36, 235–241 (2005). [Google Scholar]
- Evans H. C., Elliot S. L. & Hughes D. P. Hidden diversity behind the zombie-ant fungus Ophiocordyceps unilateralis: four new species described from carpenter ants in Minas Gerais, Brazil. PLoS One 6, e17024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anke T. & Oberwinkler F. The striatins-new antibiotics from the basidiomycete Cyathus striatus (Huds. ex Pers.) Willd. J. Antibiot 30, 221–225 (1977). [DOI] [PubMed] [Google Scholar]
- Blenis P. V. & Chow P. S. Evaluating fungi from wood and canola for their ability to decompose canola stubble. Can. J. Plant Pathol. 27, 259–267 (2005). [Google Scholar]
- Kang H. et al. Antifungal activities of Xylogone ganodermophthora KACC93082P against several plant pathogens. Phytopathology 103, 71–71 (2013). [Google Scholar]
- Hobbie E. A. et al. Fungal carbon sources in a pine forest: evidence from a C-13-labeled global change experiment. Fungal Ecology 10, 91–100 (2014). [DOI] [PubMed] [Google Scholar]
- Quaedvlieg W. et al. Sizing up Septoria. Stud. Mycol. 307–390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitz T. C. & Menge J. A. Is Spizellomyces punctatum a parasite or saprophyte of vesicular-arbuscular mycorrhizal fungi? Mycologia 76, 99–107 (1984). [Google Scholar]
- Ansari M. S., Ahmad N. & Hasan F. [Environmental Protection Strategies for Sustainable Development] Potential of Biopesticides in Sustainable Agriculture .[Malik A., & Grohmann E. (eds.)] [529–595] (Springer, London, 2012).
- Odvody S., Dunkle L. & Edmunds L. Characterization of the Periconia circinata population in a milo disease nursery on roots of Sorghum bicolor (L.) Moench. Phytopathology 67, 1485–1489 (1977). [Google Scholar]
- Barbara D. J. & Clewes E. Plant pathogenic Verticillium species: how many of them are there? Mol. Plant Pathol. 4, 297–305 (2003). [DOI] [PubMed] [Google Scholar]
- Taheri P., Gnanamanickam S. & Hofte M. Characterization, genetic structure, and pathogenicity of Rhizoctonia spp. associated with rice sheath diseases in India. Phytopathology 97, 373–383 (2007). [DOI] [PubMed] [Google Scholar]
- Gezahgne A., Cortinas M. N., Wingfield M. J. & Roux J. Characterisation of the Coniothyrium stem canker pathogen on Eucalyptus camaldulensis in Ethiopia. Australas. Plant Pathol. 34, 85–90 (2005). [Google Scholar]
- Lewis J. A., Fravel D. R. & Papavizas G. C. Cladorrhinum foecundissimum: A potential biological control agent for the reduction of Rhizoctonia solani Soil Biol. Biochem. 27, 863–869 (1995). [Google Scholar]
- Chen Y. & Ko W. Characterization of a fungicidal substance produced by Eupenicillium brefeldianum isolated from soil for plant disease control and its significance in nature. Bot. Stud. 55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaga P., Rai V. & Rai K. Bionectria ochroleuca NOTL33—an endophytic fungus from Nothapodytes foetida producing antimicrobial and free radical scavenging metabolites. Ann. Microbiol. 64, 275–285 (2014). [Google Scholar]
- Coda R. et al. Antifungal activity of Meyerozyma guilliermondii: identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food microbiol. 33, 243–251 (2013). [DOI] [PubMed] [Google Scholar]
- Marois J. J., Fravel D. R. & Papavizas G. C. Ability of Talaromyces flavus to occupy the rhizosphere and its interaction with Verticillium dahliae. Soil Biol. Biochem. 16, 387–390 (1984). [Google Scholar]
- Golzar H. & Wang C. First report of Phaeosphaeriopsis glaucopunctata as the cause of leaf spot and necrosis on Ruscus aculeatus in Australia. Australasian Plant Dis. Notes 7, 13–15 (2012). [Google Scholar]
- Yu H., Duan J., Wang B. & Jiang X. The function of snodprot in the Cerato-Platanin family from Dactylellina cionopaga in nematophagous fungi. Biosci. Biotechnol. Biochem. 76, 1835–1842 (2012). [DOI] [PubMed] [Google Scholar]
- Wu J. B., Zhang C. L., Mao P. P., Qian Y. S. & Wang H. Z. First report of leaf spot caused by Nigrospora oryzae on Dendrobium candidum in China. Plant Dis. 98, 996–996 (2014). [DOI] [PubMed] [Google Scholar]