Abstract
Fruit softening is mainly associated with cell wall structural modifications, and members of the xyloglucan endotransglucosylase/hydrolase (XTH) family are key enzymes involved in cleaving and re-joining xyloglucan in the cell wall. In this work, we isolated a new XTH gene, DkXTH8, from persimmon fruit. Transcriptional profiling revealed that DkXTH8 peaked during dramatic fruit softening, and expression of DkXTH8 was stimulated by propylene and abscisic acid but suppressed by gibberellic acid and 1-MCP. Transient expression assays in onion epidermal cells indicated direct localization of DkXTH8 to the cell wall via its signal peptide. When expressed in vitro, the recombinant DkXTH8 protein exhibited strict xyloglucan endotransglycosylase activity, whereas no xyloglucan endohydrolase activity was observed. Furthermore, overexpression of DkXTH8 resulted in increased leaf senescence coupled with higher electrolyte leakage in Arabidopsis and faster fruit ripening and softening rates in tomato. Most importantly, transgenic plants overexpressing DkXTH8 displayed more irregular and twisted cells due to cell wall restructuring, resulting in wider interstitial spaces with less compact cells. We suggest that DkXTH8 expression causes cells to be easily destroyed, increases membrane permeability and cell peroxidation, and accelerates leaf senescence and fruit softening in transgenic plants.
Fruit softening occurs primarily through modifications to the cell wall as the result of cell wall polymer degradation catalyzed by diverse enzymes such as cellulase, polygalacturonase, β-galactosidase, pectate lyase, and xyloglucan endotransglycosylase/hydrolase (XTH)1,2,3. Indeed, the depolymerization and solubilization of pectic and hemicellulosic polysaccharides in the cell wall have been demonstrated to be the major processes in fruit softening4. Xyloglucan, the major hemicellulose in the primary cell wall of dicotyledonous plants, comprises a network with cellulose microfibrils to provide strength to the cell wall5,6, with xyloglucan endotransglucosylases/hydrolases (XTHs) functioning in xyloglucan metabolism through xyloglucan endotransglycosylase (XET) and/or xyloglucan endohydrolase (XEH) activities7,8. XET activity results in the transfer of one xyloglucan molecule to another, whereas XEH activity hydrolyzes one xyloglucan molecule from the polymer9,10.
Enzymes exhibiting XTH activity belong to a multigene family11 with at least 33 genes isolated from Arabidopsis thaliana12 and 25 genes from tomato8. Expression of XTH genes is regulated by developmental and environmental stimuli13, such as darkness, touch, cold/heat-shock14,15, and by many hormones, such as ethylene16, abscisic acid (ABA)6, gibberellic acid (GA3)17, and auxins18.
XTHs have generally been thought to play important roles in fruit ripening and softening through activities that loosen the cell wall and break down the cellulose-xyloglucan matrix19,20,21,22. XET activity was found to peak at the stage of fruit ripening in apple and kiwifruit, and this activity was suggested to be responsible for fruit softening23. However, the SlXTH1 gene of tomato, which was found to be mainly expressed during fruit fast growth24,25, was demonstrated to be involved in maintaining fruit firmness after storage26. Persimmon DkXTH1 and DkXTH2 have very distinct transcriptional patterns during various physiological stages, and the encoded XET enzymes exhibited diverse enzymatic characteristics, which suggested to play different roles in fruit ripening and softening27.
Persimmon (Diospyros kaki L.) is not only an important economic crop but also displays evident changes in texture during ripening, making this species a good model for studying fruit softening28,29. In our previous studies, seven XTH genes (DkXTH1–7) were amplified from persimmon, and these genes were proposed to be involved in fruit development, ripening or softening6,27. However, there is a lack of direct genetic evidence for these activities, and additional genes should be identified to provide a better understanding of the roles of specific genes in fruit. Accordingly, in this study, we identified a new XTH gene, DkXTH8, from persimmon fruit and analyzed its patterns of expression in different tissues and in response to several hormones (propylene, ABA, GA3 and 1-MCP (1-methylcyclopropene)). Furthermore, the subcellular localization of DkXTH8 was examined, and the enzymatic characteristics of the recombinant DkXTH8 protein were also investigated. Most importantly, we generated transgenic Arabidopsis and tomato overexpressing DkXTH8, and leaf senescence and fruit softening were evaluated. Lastly, microscopic structures were observed in transgenic plants to explore changes in the cell wall.
Results
Cloning and phylogenetic analysis of DkXTH8
A new full-length sequence named DkXTH8 was amplified from persimmon (Diospyros kaki L. cv Fuping jianshi); the sequence has been deposited in GenBank under accession number KF318888. The DkXTH8 cDNA is 1088 bp long, with an open reading frame (ORF) spanning nucleotides 130 to 996. The deduced protein is 288 amino acids long, with a predicted molecular weight of 32.53 kDa and a pI of 8.97. DkXTH8 shares 50–70% amino acid homology with DkXTH1–7, which were previously amplified from persimmon. Moreover, DkXTH8 is predicted to contain an N-terminal signal peptide, with a cleavage site between residues 25 and 26.
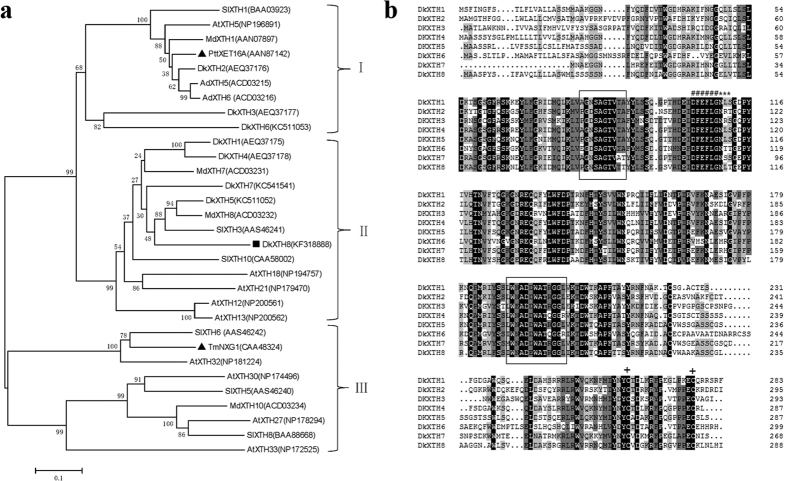
A phylogenetic tree was generated using the deduced amino acid sequences of DkXTH8 and another 30 XTHs from various plant species (Fig. 1a), with the XTHs classified into three groups, as reported by Campbell and Braam11. DkXTH2, DkXTH3 and DkXTH6 belong to group I, together with PttXET16A, a strict XET enzyme30. DkXTH8 as well as DkXTH1, DkXTH4, DkXTH5 and DkXTH7 belong to group II and is closely related to the apple protein MdXTH8 and tomato protein SlXTH3. TmNXG1 of group III is a strict XEH enzyme according to Baumann et al.31.
Figure 1. Phylogenetic tree and alignment of deduced amino acid sequences of XTHs.
(a) Phylogenetic tree of XTHs. The phylogenetic tree was constructed by the Neighbour-Joining method (1000 trials) with bootstrap using MEGA 5.1 software. DkXTH8 is set as bold (square). PttXET16A and TmNXG1 (triangle) were the first XET and XEH with three-dimensional structures, respectively. The GenBank accession numbers are indicated in the figure. (b) Alignment of predicted DkXTHs proteins. The conserved regions are framed boxes. Putative catalytic domain, N-glycosylation site, and two cysteines are marked with “#,” “*,” and “+,” respectively.
A multiple alignment was generated to assess relationships among persimmon DkXTH1-8 (Fig. 1b). All DkXTHs possess the conserved motif DEIDFEFLG, which is attributed to the putative active site, and a nearby potential N-linked glycosylation (N-X-S/T) site. Moreover, as typical characteristics of glycosyl-hydrolase family 16 enzymes, DkXTHs have two conserved central domains, and two cysteine residues are located in the C terminal region, suggesting that DkXTH8 shares common features with XTHs from other plants.
Physiological characterization during persimmon fruit storage
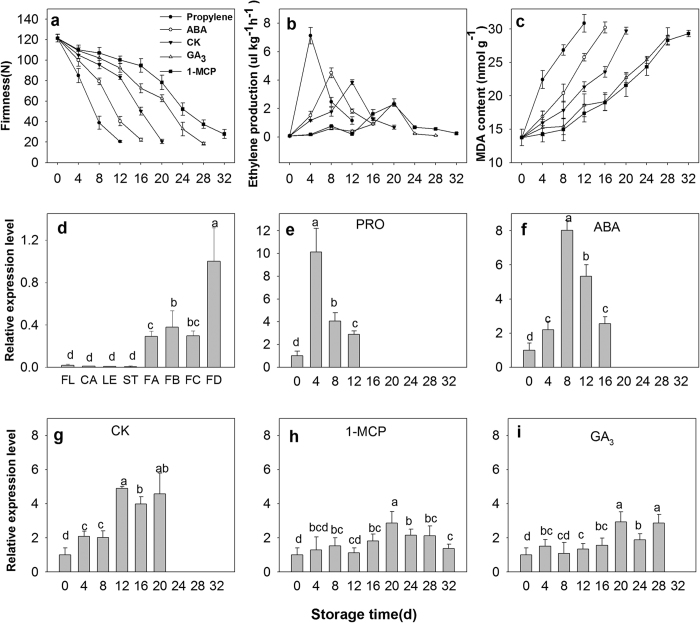
To analyze postharvest softening and senescence, uniform persimmon fruits free from visible defects and with 70–80% surface yellow coloration were harvested. After treatment (propylene, ABA, GA3 and 1-MCP), fruits were stored at room temperature and randomly collected every 4 days for physiological characterization. When testing firmness (Fig. 2a), the control fruit (“Fuping jianshi” fruit without any treatment, CK) were obviously softened at 12 days after harvest; firmness was 122 N at harvest time, declining to 21 N on day 20. However, the firmness of the fruits treated with propylene and ABA (“Fuping jianshi” fruit treated with propylene and ABA, respectively) decreased more quickly than that of the CK fruit, showing a higher rate of softening. In detail, the CK fruit firmness was 75% and 51% firmer than the propylene and ABA fruits at 12 days of storage, respectively. In contrast, the firmness of the GA3 and 1-MCP fruits (“Fuping jianshi” fruits treated with GA3 and 1-MCP, respectively) declined more slowly than that of the CK fruit, showing a lower rate of softening. Specifically, the GA3 and 1-MCP fruit firmness was 67% and 74% firmer than the CK fruit at 20 days of storage, respectively.
Figure 2. Physiological characterization of persimmon and expression pattern of DkXTH8.
Firmness (a), ethylene production (b) and MDA content (c) of persimmon fruits during storage. ‘Propylene’ ‘1-MCP’ ‘ABA’ and ‘GA3’ indicated Fuping Jianshi fruit treated with propylene (5000 μl L−1, 24 h), 1-MCP (500 nL L−1, 24 h), ABA (50 mg L−1, 2 min) and GA3 (60 mg L−1, 2 min), respectively. The fruit without any treatment was served as the ‘CK’. (d) Expression pattern of DkXTH8 in various tissues of persimmon. ‘FL’ ‘CA’ ‘LE’ and ‘ST’ are indicated the flowers, calyces, leaves and stems, respectively. ‘FA’ ‘FB’ ‘FC’ and ‘FD’ are indicated fruits harvested at 20, 60, 100 and 140 days after full bloom, respectively. Expression of DkXTH8 at ‘FD’ was used as the control with a nominal value of 1. Expression pattern of DkXTH8 in ‘Propylene’(e), ‘ABA’(f), ‘CK’(g), ‘1-MCP’(h) and ‘GA3’(i) persimmon fruits. Expression of DkXTH8 at 0 day was used as the control with a nominal value of 1. Vertical bars indicate the standard error of three replicate assays. Columns with different letters at each time point are significantly different (LSD, P < 0.05).
The ethylene production of the fruits was measured during storage (Fig. 2b). Ethylene production was stimulated by propylene and ABA, and the maximal values in the propylene- (4 days) and ABA-treated fruits (8 days) was 46% and 16% higher than that in the CK fruit (12 days), respectively. Conversely, the ethylene production was inhibited in GA3 and 1-MCP fruits. In detail, the maximal ethylene production in GA3 and 1-MCP fruit (20 days) was only 63% and 61% of that in CK fruit, respectively.
After harvest, the malonaldehyde (MDA) content rose consistently in all of the tested fruits (Fig. 2c). In CK fruits, the MDA content was 13.8 nmol g−1 at harvest time and increased up to 29.8 nmol g−1 at the end of storage. Whereas, the MDA contents of the propylene and ABA fruits remained higher than that of the CK fruit, revealing accelerated MDA accumulation in the treated fruits. In contrast, the MDA content of the fruits treated with GA3 and 1-MCP remained at low levels, and the value at 20 days was only 76% and 72% of that in the CK fruit, respectively.
Expression of DkXTH8 in different persimmon tissues and during fruit storage
Leaves, flowers, calyces, stems and fruits were analyzed to examine the expression pattern of DkXTH8 in different tissues (Fig. 2d). DkXTH8 transcripts were notably detectable in fruits, though very little expression was found in other tissues. Moreover, fruits collected at 140 days after full bloom shown evidently higher DkXTH8 expression levels than fruits harvested at 20, 60 or 100 days after full bloom.
To analyze the association of DkXTH8 with fruit softening, the levels of expression were measured in propylene-, ABA-, GA3- and 1-MCP-treated fruits (Fig. 2e–i). After harvest, DkXTH8 transcripts increased rapidly, peaking at 12 days after storage in CK fruit. Moreover, a similar expression pattern was observed in the propylene and ABA fruits, peaking at 4 or 8 days after storage, respectively. The expression pattern of DkXTH8 appeared parallel to ethylene production, and both of them peaked during dramatic fruit softening. Additionally, the maximal vales of DkXTH8 expression in the propylene- and ABA-treated fruits were 52% and 39% higher than that in the CK fruit, revealing the synergistic effect of propylene and ABA on DkXTH8 expression. In contrast, the GA3 and 1-MCP fruits exhibited lower levels of DkXTH8 expression, with respective maximal values of only 60% and 58% of CK fruit.
Direct localization of DkXTH8 to the cell wall via its signal peptide
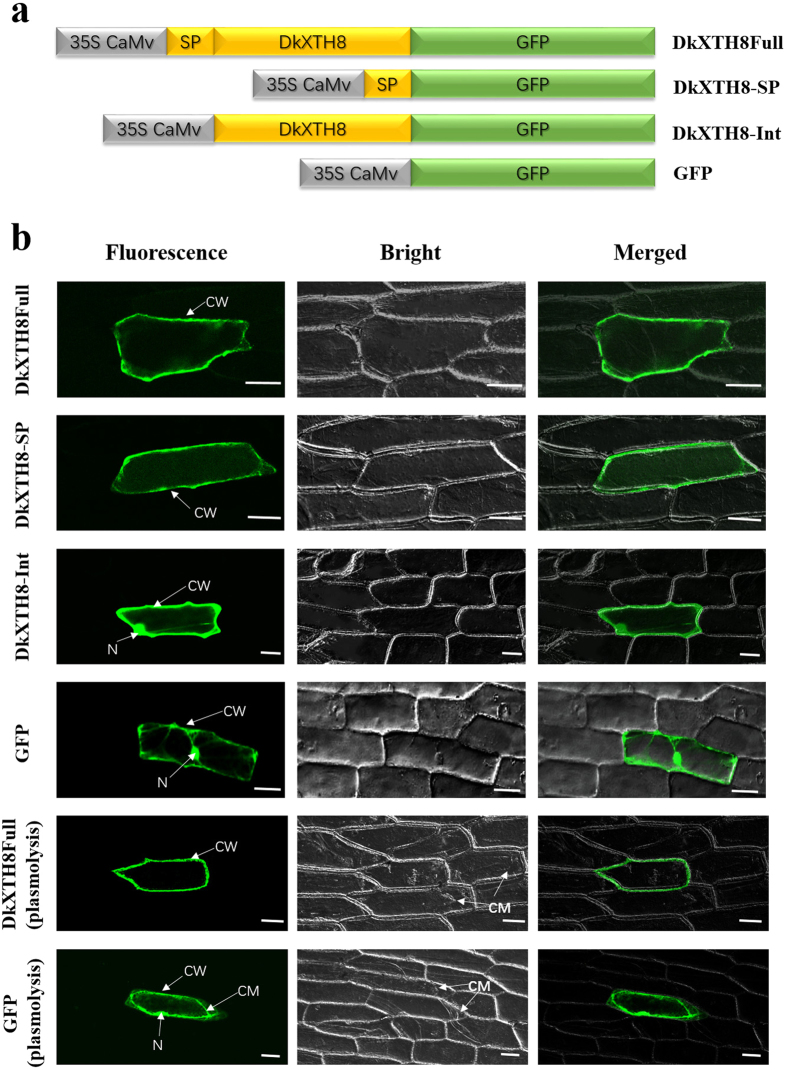
The ORF of DkXTH8 (DkXTH8Full), the signal peptide of DkXTH8 (DkXTH8-SP), and the ORF sequence of DkXTH8 without the signal peptide (DkXTH8-Int) were amplified. A schematic diagram of the vector construction is shown in Fig. 3a. The subcellular localization of DkXTH8 was analyzed by bombarding plasmids into onion epidermal cells. Three types of results were observed: “Fluorescent”, “Bright” and “Merged” (see Fig. 3b). DkXTH8Full protein was detected in cell walls by monitoring the plasmolyzed and non-plasmolyzed cells; this was different from the GFP control, for which protein was found throughout the cell. In control plasmolyzed cells, the fluorescence protein was obviously found in both the cell wall and plasma membrane, however, DkXTH8Full protein was only observed in cell walls. Besides, DkXTH8-SP protein was specifically localized to the cell wall. Nevertheless, in the absence of the signal peptide, DkXTH8-Int was localized throughout the cell, suggesting that its N-terminal signal peptide targets DkXTH8 to the cell wall.
Figure 3. Subcellular localization of DkXTH8.
(a) Diagram of DkXTH8 constructs fused to GFP. (b) “Fluorescent”, “Bright” and “Merged” images of subcellular localization of DkXTH8 and GFP control. Plasmolysis was induced by 400 mM sucrose. CW, cell wall; N, nucleus; CM, cell membrane. Scale bar = 50 μm.
The recombinant DkXTH8 protein possesses strict XET activity
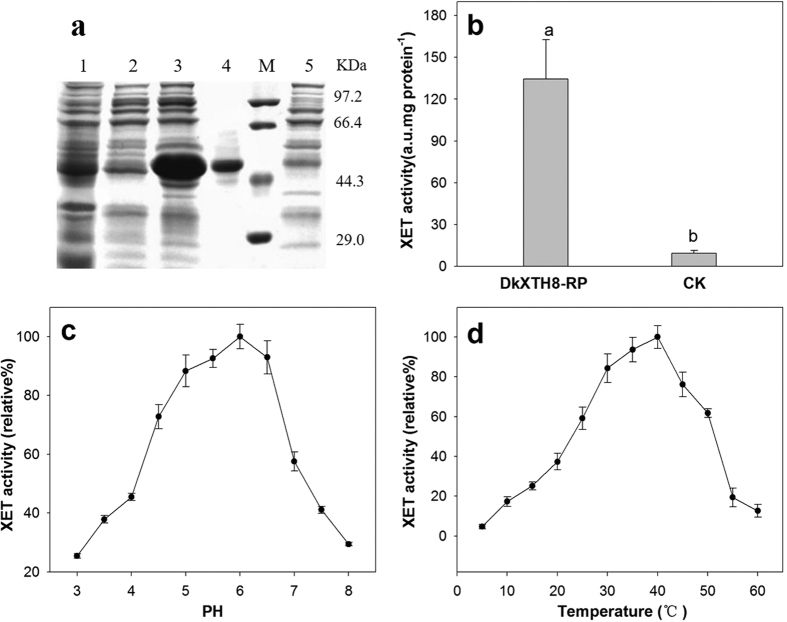
The recombinant DkXTH8 protein (DkXTH8-RP) was expressed in bacteria to investigate its enzymatic properties. To promote correct protein folding, the protein was induced at low-speed shaking and at a low temperature. However, only a small proportion of the protein was soluble, with most of the recombinant protein present in the insoluble fraction (Fig. 4a). After concentration and purification using a Ni-NTA resin column, the soluble recombinant protein was used for assessing enzyme activity. Obvious XET activity was detected for DkXTH8-RP in comparison with the blank control, suggesting that the purified recombined DkXTH8 protein was an active enzyme (Fig. 4b). The XEH activity of DkXTH8-RP was also measured by a viscometric assay using Trichoderma reesei cellulose as a positive control. Unlike Trichoderma reesei cellulose, which could reduce the viscosity of xyloglucan via hydrolytic action, DkXTH8-RP did not cause any evident decrease in viscosity of xyloglucan after a set reaction time. These results indicate that DkXTH8-RP possesses strict XET activity, with no XEH activity.
Figure 4. Expression and activity of recombinant DkXTH8 proteins.
(a) Proteins were separated on SDS–polyacrylamide gels and stained with Coomassie Blue. Lane 1, total soluble protein (DkXTH8); lane 2, unbound protein; lane 3, total insoluble protein (DkXTH8); lane 4, purified protein (DkXTH8); M, protein marks (Takara, Dalian, China); and lane 5, pET-32a control protein. (b) In vitro XET assay of recombinant DkXTH8 proteins. The XET assay was performed by colorimetric method as described in Section 4.7. The empty vector pET-32a was used as the control. (c) The pH–rate profile of recombinant DkXTH8 proteins. (d) The temperature profile of recombinant DkXTH8 proteins. Vertical bars indicate standard errors of three replicates.
To examine the pH profile of DkXTH8-RP, XET activity was tested over the pH range of 3–8 (Fig. 4c). A bell-shaped pH profile was found and the XET activity declined sharply when the pH decreased from 5 to 4, as a common feature of XET enzymes32. The XET activity of DkXTH8-RP was also tested over the temperature range from 5 to 60 °C, and the optimum temperature for the enzyme was found to be in the range 30–40 °C (Fig. 4d).
Overexpression of DkXTH8 in Arabidopsis promotes dark-induced leaf senescence
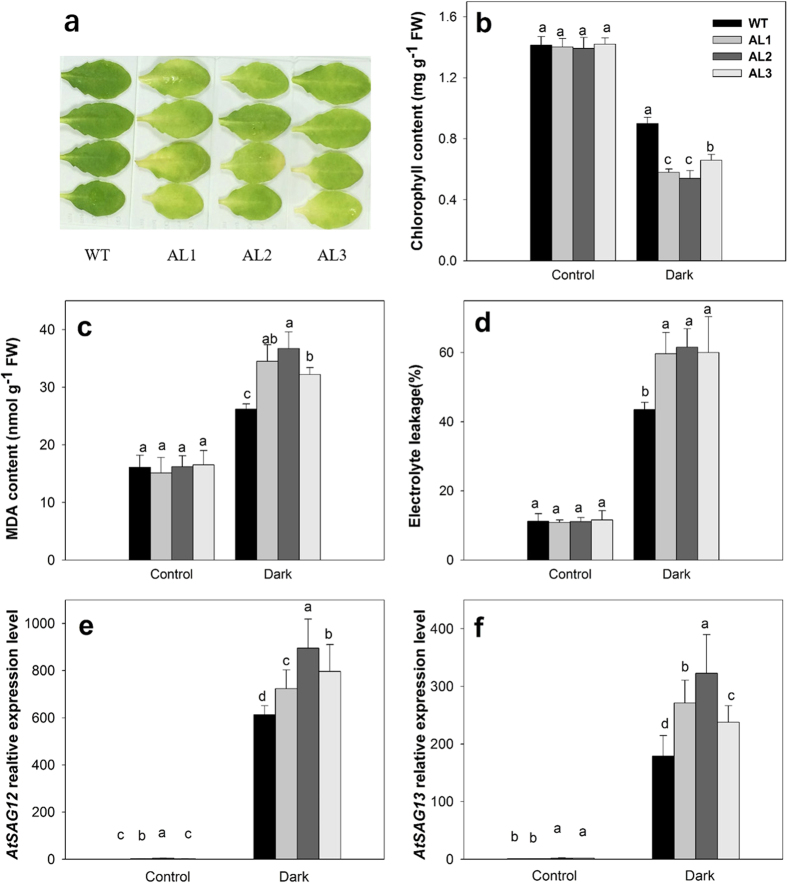
To verify whether DkXTH8 is involved in plant senescence, transgenic Arabidopsis lines (AL1, AL2, AL3) overexpressing DkXTH8 were generated (Fig. S1b). After stored in the dark for four days, detached leaves of transgenic Arabidopsis became more visibly yellow than the leaves of wild type (WT, Fig. 5a). Compared with the control, the chlorophyll content declined in both WT and transgenic Arabidopsis leaves after storage in the dark. However, the transgenic Arabidopsis leaves contained less chlorophyll than WT (Fig. 5b), suggesting accelerated senescence in DkXTH8-overexpressing Arabidopsis. Furthermore, the MDA content and electrolyte leakage were measured in detached leaves to indicate the degree of cell peroxidation (Fig. 5c,d). The transgenic Arabidopsis leaves showed higher levels of electrolyte leakage and MDA content than WT, indicating more lipid peroxidation in the transgenic plant cells. Two senescence associated genes, AtSAG12 and AtSAG13, were induced rapidly in dark stored leaves (Fig. 5e,f). While, both AtSAG12 and AtSAG13 exhibited higher expression levels in transgenic plants than that in WT, indicating critical leaf senescence in DkXTH8-overexpressing Arabidopsis.
Figure 5. Dark-induced leaves senescence of WT and DkXTH8-overexpressing Arabidopsis.
(a) Visual appearance of detached leaves of WT and DkXTH8-overexpressing Arabidopsis after four days in dark. (b) Chlorophyll content. (c) MDA content. (d) Electrolyte leakage. (e) Relative expression level of AtSAG12 gene in WT and DkXTH8-transgenic Arabidopsis leaves. (f) Relative expression level of AtSAG13 gene in WT and DkXTH8-transgenic Arabidopsis leaves. Detached leaves stored under growth conditions served as the control. Vertical bars indicate the standard error of three replicate assays. Columns with different letters at each time point are significantly different (LSD, P < 0.05).
Overexpression of DkXTH8 in tomato promotes fruit ripening and softening
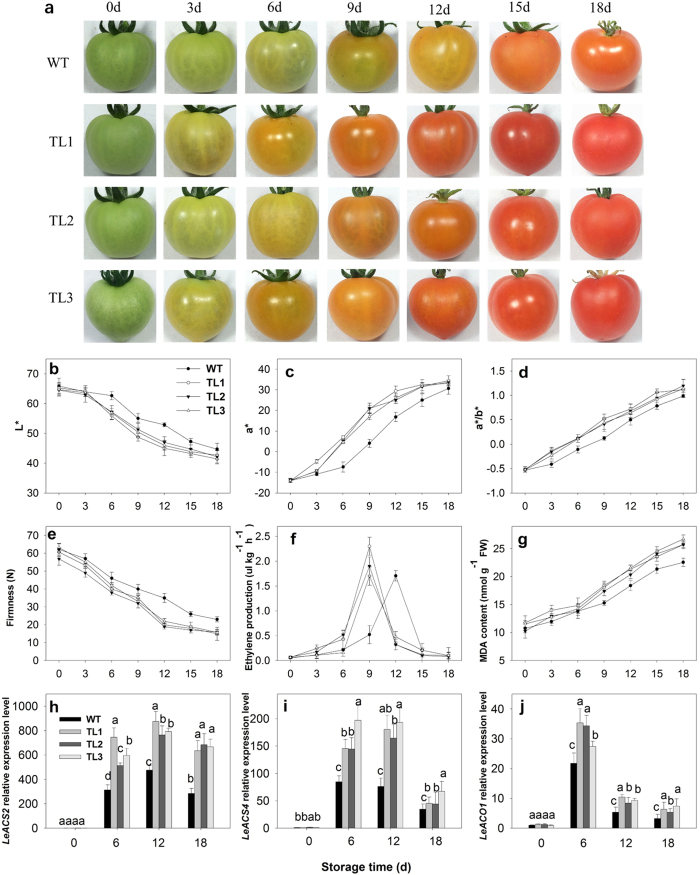
DkXTH8-transgenic tomato lines (TL1, TL2, TL3) were generated to explore whether DkXTH8 is related to fruit ripening and softening (Fig. S1c). Tomato fruits were collected at the mature green stage and stored at room temperature. Samples were randomly collected every 3 days, as shown in Fig. 6a. After harvesting, the fruits began to turn yellow and then red; however, the transgenic tomato fruits exhibited accelerated color change compared with the WT fruits. As a representation of color, L*, a* and a*/b* values were measured to indicate tomato fruit maturity (Fig. 6b–d). In the WT fruits, the level of L* declined constantly during storage. A marked decline was detected from 6 to 9 days, at which time the fruit turned from green to yellow. In contrast, the three transgenic tomato lines displayed a faster decrease in L*, indicating rapid color change. Similarly, the values of a* and a*/b* increased after storage though more rapidly in the transgenic fruits than in the WT fruits. The values of a* and a*/b* were 76–81% and 71–77% higher, respectively, in the transgenic fruits than in the WT fruits after 9 days of storage.
Figure 6. Phenotype and physiological parameters of WT and DkXTH8-overexpressing tomato.
(a) Fruit phenotype. Tomato fruits of WT and DkXTH8-transgenic lines were collected at the mature green period and stored at room temperature. Samples were randomly collected every 3 days. (b–d) Changes in L*, a* and a*/b* colour parameters of tomato fruit. (e–g) Changes in firmness, ethylene production and MDA contents of fruit. (h–j) Relative expression levels of LeACS2, LeACS4 and LeACO1 genes in fruit. Vertical bars indicate the standard error of three replicate assays.
When evaluating firmness, the transgenic fruits decreased faster than WT (Fig. 6e), with the firmness of the transgenic fruits only 75–88% and 54–63% of that in WT at 9 and 12 days, respectively. In addition, the maximal values of ethylene production by the transgenic fruits were higher than that in WT, and the peak appeared three days earlier (Fig. 6f). Moreover, the MDA content rose constantly after the fruits were harvested, 12–16% higher in the transgenic fruits than that in WT at the end of storage (Fig. 6g).
Ethylene synthesis related genes were also assessed to indicate the degree of fruit ripening and softening in WT and DkXTH8-overexpressed tomatoes. Both ACS and ACO genes were up regulated during fruit storage, however, relative higher expression levels were found in transgenic tomato fruits (Fig. 6h–j). In DkXTH8-overexpressed fruits, the expression levels of LeACS2 and LeACO1 were 39–58% and 21–38% higher than that in WT, respectively (6 days after storage, p < 0.05).
Microscopic observation of WT and DkXTH8-transgenic plants
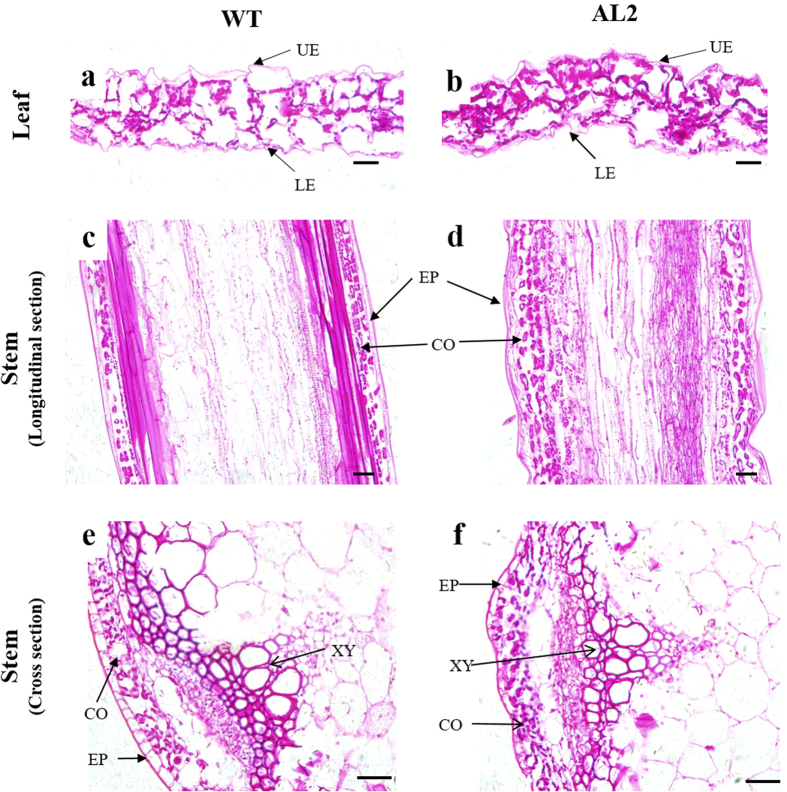
To assess whether the differences in the leaf senescence rate and fruit softening were due to changes in cell wall structure, the microscopic structures of WT and DkXTH8-transgenic plants were compared. The stems and fifth–sixth leaves of Arabidopsis plants were collected at four weeks after sowing. Compared with WT, the leaf sections of the transgenic Arabidopsis plants showed more irregularity, especially in the lower and upper epidermis layers, exhibiting a winding shape (Fig. 7a,b). Similar observations were found in stem sections. A longitudinal section of the stem from the DkXTH8-transgenic plants exhibited an irregular shape with a slightly wave-like border in the epidermis (Fig. 7c,d). Similarly, the epidermis and cortex contained more irregularly shaped cells in stem cross sections from the transgenic Arabidopsis plants (Fig. 7e,f). In particular, the cells of the xylem were rounded and smooth in WT but showed an angular and irregular shape in the transgenic plants.
Figure 7. Microscopic observation of WT and DkXTH8-overexpressing Arabidopsis.
Leaf section of WT (a) and DkXTH8-transgenic Arabidopsis (b). Longitudinal section of the stem from WT (c) and DkXTH8-transgenic Arabidopsis (d). Cross section of the stem from WT (e) and DkXTH8-transgenic Arabidopsis (f). Scale bar = 20 μm. UE, upper epidermis; LE, lower epidermis; EP, epidermis; CO, cortex; XY, xylem.
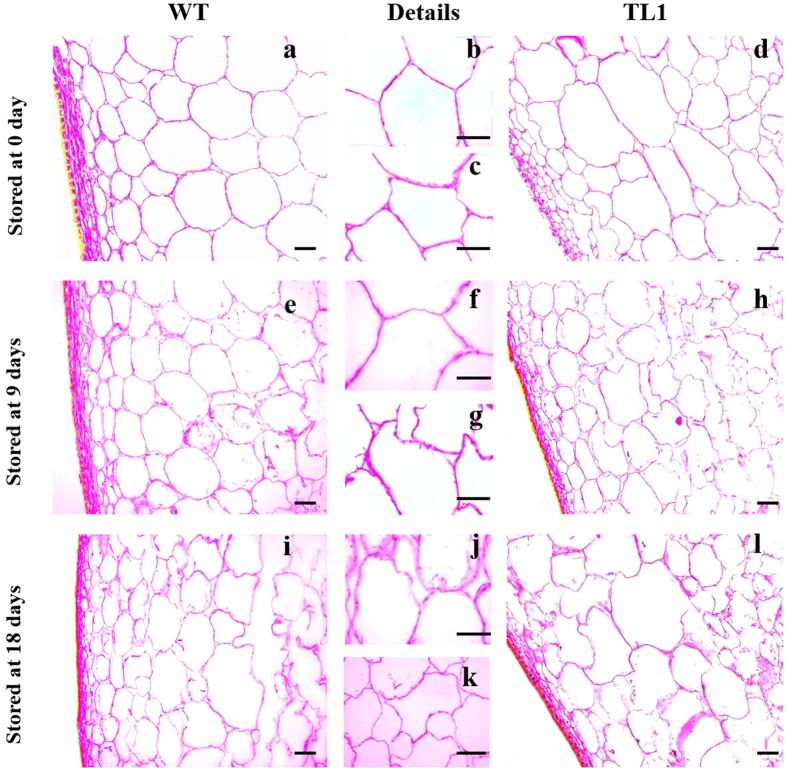
Microscopic observations of WT and DkXTH8-transgenic tomato were carried out using fruits stored for 0, 9 and 18 days. At harvest time (0 days), the cells from WT fruits were rounded and smooth with a uniform size (Fig. 8a,b). In contrast, the cells from the transgenic fruits were more angular and irregular with multiple sizes, resulting in a wider interstitial space and less compact cells (Fig. 8c,d). At the middle of the storage period (9 days), the majority of cells from the WT fruits retained their integrity, and only a few cells were degraded (Fig. 8e,f). However, more than half of the cells from the transgenic fruits were degraded, suggesting a higher rate of fruit softening (Fig. 8g,h). At the end of storage (18 days), nearly all cells from the transgenic fruits were degraded (Fig. 8k,l). Although most of the cells from the WT fruits were destroyed, the third-fourth layer cells under the peel retained integrity (Fig. 8i,j).
Figure 8. Microscopic observation of WT and DkXTH8-overexpressing tomato.
Microscopic observation of WT (a,b) and DkXTH8-overexpressing tomato fruit (c,d) stored at 0 day; Microscopic observation of WT (e,f) and DkXTH8-overexpressing tomato fruit (g,h) stored at 9 days; Microscopic observation of WT (i,j) and DkXTH8-overexpressing tomato fruit (k,l) stored at 18 days. Scale bar = 50 μm.
Discussion
Previous works have reported that XTHs are encoded by a large multigene family9,10. Individual XTHs exhibit multiple expression patterns and diverse responses to hormonal or environmental stimuli, which may account for their unique roles in fruit33,34. In previous studies, we isolated seven XTH genes from persimmon, and all of these genes were found to play important roles in fruit development, ripening or softening6,27. In the present study, a new XTH gene from persimmon was identified: DkXTH8. Phylogenetic analysis revealed that DkXTH8 belongs to group II (Fig. 1a), different from PttXET16A and TmNXG1, strict XET and XEH enzymes, respectively30,31. Sequence analysis indicated that DkXTH8 shares 50–70% homology with DkXTH1–7 and contains the conserved regions of glycosyl hydrolase family 16 genes (Fig. 1b), indicating that this new gene possesses the common structural features of XTHs11.
Propylene and ABA treatments of persimmon fruit resulted in a higher climacteric ethylene peak, lower firmness and an increased MDA content compared to CK fruit (Fig. 2a–c). More importantly, expression level of DkXTH8 was effectively stimulated and appeared to parallel the fruit softening rate (Fig. 2e–i). This feature is consistent with previous work of rose RbXTH1 and RbXTH2, which play important roles in senescence16. In contrast, exogenous GA3 and 1-MCP inhibited ethylene production and effectively suppressed DkXTH8 expression, which appeared to result in higher firmness of persimmon fruit. Similar results have been reported for papaya CTR135, cherimoya AcXET1-336, and apple MdXTH10 and MdXTH1122, which have been demonstrated to be involved in fruit softening. Interestingly, DkXTH8 was notably detected in mature persimmon fruit but scarcely in other tissues or unripe fruit (Fig. 2d). Overall, the results suggest that DkXTH8 is a fruit ripening-specific gene that most likely operates in conjunction with ethylene during postharvest fruit softening.
Isoenzymes of XTHs possess distinct enzymatic properties37,38, with specialized functions in cell wall modification10,39. In persimmon, DkXTH1 and DkXTH2 exhibit different affinities for small acceptor molecules, and the former might participate in cell wall assembly, whereas the latter is likely involved in cell wall restructuring27. The kinetic properties of the recombinant DkXTH8 protein (DkXTH8-RP) were investigated, with DkXTH8-RP showing significant XET activity without any detectable XEH activity (Fig. 4b). These results are similar to the reports of recombinant SlXTH5 protein from tomato40,8 and AtXTH14 and AtXTH26 from Arabidopsis41. DkXTH8-RP exhibited a bell-shaped pH profile (Fig. 4c), and the optimum temperature for the enzyme was in the range 30–40 °C (Fig. 4d), as a common feature of XET enzymes42. In addition, the DkXTH8 protein was directly localized to the cell wall via its signal peptide (Fig. 3). ZmXTH1, a cell wall-bound maize protein, has been shown to affect the cell wall structure and composition in transgenic Arabidopsis43. The PeXTH gene from Populus euphratica caused anatomical and physiological alterations in transgenic tobacco and was localized to the endoplasmic reticulum and cell wall44. Therefore, DkXTH8 is suggested to act as an XET enzyme that is directly localized to the cell wall and is involved in cell wall modification.
The relationship between DkXTH8 and leaf senescence was investigated in transgenic Arabidopsis. Leaf senescence was detected based on the loss of chlorophyll45, which was accompanied by an increase in lipid peroxidation and membrane permeability46. In our study, leaf senescence was promoted in DkXTH8-transgenic Arabidopsis, coupled with higher chlorophyll degradation, electrolyte leakage and MDA content (Fig. 5). Meanwhile, both AtSAG12 and AtSAG13 shown higher expression levels in transgenic plants than that in WT, which expression were strictly associated with senescence. Wagstaff et al. suggested that decreased expression of lettuce LsXTH altered the leaf biophysical structure and increased the leaf strength, leading to an extended shelf-life of transgenic plants47. Both the leaf and stem cells of DkXTH8-transgenic Arabidopsis showed more irregular and twisted shapes, resulting in a wider interstitial space and less compact cells compared with WT (Fig. 7). Similar results have been reported in maize: ZmXTH1 was demonstrated to affect cell wall structure in transgenic Arabidopsis, with a wider middle lamella region that resulted in a widening of the space between cells43. These results raise the possibility that overexpression of DkXTH8 affected the structure of the cell wall, resulting in a wider interstitial space and less compact cells. Notably, these changes in shape caused the cells to be easily destroyed and also increased lipid peroxidation and membrane permeability, exacerbating leaf senescence.
To confirm whether DkXTH8 is involved in fruit ripening and softening, further verification was performed in DkXTH8-transgenic tomato. Compared to WT, the DkXTH8-transgenic tomato fruit exhibited accelerated color change, decreased firmness and increased MDA content (Fig. 6). The expression levels of LeACS2, LeACS4 and LeACO1 displayed higher values in transgenic fruits accompanied by an earlier and higher ethylene peak. This is the first direct genetic evidence for the promotion of fruit ripening and softening by XTHs. Overexpression of tomato SlXTH1 was demonstrated to reduce the softening of transgenic fruit, and the author suggested that XET was involved in maintaining the structural integrity of the cell wall26. However, SlXTH1 was transiently detected at high levels during the early stage of fruit development, with little expression during fruit ripening or softening in tomato24,25. DkXTH1, which was found to be largely expressed in fast-growing persimmon fruit, was demonstrated to contain fruit firmness by participating in cell wall assembly27. While, DkXTH2, a gene expressed mainly in ripening persimmon fruit, likely promotes fruit softening via restructuring of the cell wall27. Thus, we suggest that the fruit ripening-specific gene DkXTH8 may promote transgenic tomato fruit softening through involvement in cell wall restructuring.
In agreement with the results in Arabidopsis, DkXTH8-transgenic tomato fruit exhibited more irregular and twisted cells, leading to wider interstitial spaces and less compact cells compared with WT (Fig. 8). Furthermore, accelerated cell degradation was found in the postharvest transgenic fruit, resulting in a higher degree of fruit softening. Altogether, the results suggest that overexpression of DkXTH8 altered cell shape in the transgenic fruit by acting in cell wall restructuring, which resulted in wider interstitial spaces and less compact cells. These changes in shape caused the cells to be easily destroyed, intensifying fruit softening.
In conclusion, a new xyloglucan endotransglucosylase/hydrolase, DkXTH8, was identified from persimmon. This gene presented the highest expression levels during fruit ripening and softening. The recombinant DkXTH8 protein showed strict XET activity and no XEH activity. Overexpression of DkXTH8 caused more irregular and twisted cells, with wider interstitial spaces and less compact cells via its involvement in cell wall restructuring, which resulted in accelerated leaf senescence in Arabidopsis and fruit softening in tomato.
Methods
Plant materials and treatments
Persimmon materials were obtained from a commercial orchard in Fuping County, Shaanxi Province, China. The propylene treatment was performed by placing fruits in a 360-L chamber and exposing them to 5000 μL L−1 propylene for 24 h. The 1-MCP treatment was performed by exposing fruits to 500 nL L−1 1-MCP (EthylBloc®, Dow Chemical Co., Shanghai, China, a.i. 0.14%) for 24 h. The GA3 treatment was carried out by immersing fruits in 60 mg L−1 GA3 for 2 min and the ABA treatment by immersing fruits in 50 mg L−1 ABA for 2 min. Untreated fruits served as the control (‘CK’). After treatment, each group was divided randomly into three subgroups, and all fruits were stored at 25 ± 1 °C.
For DkXTH8 functional analyses, Arabidopsis thaliana ecotype ‘Columbia’ and Solanum lycopersicum Mill. cultivar ‘Micro-Tom’ were used.
RNA extraction and isolation of the full-length DkXTH8 cDNA
Total RNA was isolated from frozen persimmon tissues using the hot borate method27, and TransZol Up Plus RNA Kit (Transgen Biotech, Beijing, China) was used to extract total RNA from Arabidopsis and tomato. First-strand cDNA was obtained using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, Japan). Based on the degenerate primers designed by Zhu et al.6, a new conserved XTH gene region was amplified using material from persimmon fruit as a template. Subsequently, 3′- and 5′-rapid amplification of cDNA ends polymerase chain reactions (PCR) were performed as described in Han et al.27, and the full-length cDNA of XTH was then amplified based on the 3′-end and 5′-end fragments. The primer sequences are listed in Table 1.
Table 1. Oligonucleotide sequences for primers used in this study.
| Gene name | Prime sequences (5′–3′) | Purpose |
|---|---|---|
| DkXTH8 | Outer: ATTCCGCCGCACCCATCTCA | 5′RACE |
| Inner: TCATCGGCTGGCGAAGGTAG | ||
| Outer: GAACGGGAGCAGCAGTTTC | 3′RACE | |
| F: ACTGCTACTGCTGGCTTCATG | Full-length cDNA clone | |
| R: ATCTGATTCCGCCGCACCCATC | ||
| F: TCCTCCAACTTTAACCAGG | RT-qPCR | |
| R: ATCAATCTTGCCGAACAG | ||
| F: GCTCTAGAATGGCGGCTTCTCCATATT | DkXTH8Full | |
| R:GGGGTACCTATATGGAGATTTAATTTGC | ||
| F: GCTCTAGAATGGCGGCTTCTCCATATT | DkXTH8-SP | |
| R:GGGGTACCGGAGGAAGAAGAGGAAAGC | ||
| F: GCTCTAGAAACTTTAACCAGGATTTTAA | DkXTH8-Int | |
| R:GGGGTACCTATATGGAGATTTAATTTGC | ||
| F: CGGGATCCAACTTTAACCAGGATTTT | Recombinant protein expression | |
| R: CCCAAGCTTTATATGGAGATTTAATTTGC | ||
| F: GCTCTAGAATGGCGGCTTCTCCATATTCC | Generation of transgenic | |
| R: CGGGATCCTTATATATGGAGATTTAATTTGC | plants | |
| DkACTIN | F:TGCTCTTCCAGCCATCACTCATT | RT-qPCR |
| R:ATTTCCTTGCTCATCCGGTCAG | ||
| AtACTIN2 | F: TTGTGCTGGATTCTGGTGATGGT | RT-qPCR |
| R: CCGCTCTGCTGTTGTGGTGAA | ||
| AtSAG12 | F: GGATGTCCCGGTTAATGATG | RT-qPCR |
| R: TCCACTTTCTCCCCATTTTG | ||
| AtSAG13 | F: GCTGTGGTGGAGGAACTAGC | RT-qPCR |
| R: CCACATTGTTGACGAGGATG | ||
| LeUBI3 | F: CTACAACATCCAGAAGG | RT-qPCR |
| R: TGCAACACAGCGAGCTTAACC | ||
| LeACS2 | F: CCTCACCATTAGTTCGTTAAGACT | RT-qPCR |
| R: CCTCACCATTAGTTCGTTAAGACT | ||
| LeACS4 | F: GCAAGGATTCGGATGTTTATGGATGC | RT-qPCR |
| R: TGCTCGCACTACGAGCGAGGAATTG | ||
| LeACO1 | F: ACACGAATGTCACTAGCCTCAT | RT-qPCR |
| R: TCCATTGCCTTCATTGCTTCAA |
aLetters “F” and “R” indicate the forward and reverse primers, respectively.
Sequence analysis and bioinformatic methods
The full-length XTH sequence was confirmed using the BLAST program in GenBank, and ORF Finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used for ORF and protein predictions. The PeptideMass program (http://us.expasy.org/tools/peptidemass.html) was employed to calculate the molecular weight and theoretical isoelectric point (pI) of the putative protein, and SignalP (http://www.cbs.dtu.dk/services/SignalP/) was used for N-terminal signal peptide prediction. The deduced amino acid sequences were initially aligned and compared using the DNAMAN program, and a phylogenetic tree was generated based on the Neighbor-Joining method (1000 bootstrap replicates) using MEGA 5.1 software.
Fruit firmness, ethylene production and MDA content determination
To measure fruit firmness, three small slices of skin were removed from 120° intervals around the equatorial axis of a fruit. A pressure tester equipped with a 5- or 1-mm diameter probe was used for persimmon and tomato fruits, respectively. For each time point, six fruits were tested for replications.
Six fruits from each treatment subgroup were placed in an airtight chamber for 1 h at room temperature, and 1 mL of gas was collected three times using a syringe. The ethylene concentration was then quantified using a GC-14A gas chromatograph (Shimadzu, Kyoto, Japan), as described by Zhu et al.6 The respiration rate was measured using a CO2 infrared gas analyzer (TEL7001; GE Telaire, CA, USA) according to Han et al.27.
The MDA content was measured and calculated according to Hou et al.28.
Quantitative real-time (RT)- qPCR expression analysis
First-strand cDNA was synthesized using the methods described above. RT-qPCR was then carried out using an iCycler iQ5 (Bio-Rad, Hercules, CA, USA) as described by Han et al.27. DkACTIN, AtACTIN2 and LeUBI3 were used as an internal control for persimmon, Arabidopsis and tomato, respectively. Standard curves were generated to ensure amplification efficiencies between the primers for the housekeeping genes and studied genes. The gene relative expression level was calculated using the comparative CT (2−△△CT) method48. Three biological replicates were performed for all of the samples. The specific primer sequences used for qRT-PCR are listed in Table 1.
Subcellular localization
Subcellular localization of DkXTH8 was investigated in onion epidermal cells with a biolistic PDS-1000/He particle delivery system (Bio-Rad), as described by Han et al.27. The ORF of DkXTH8 (DkXTH8Full), the signal peptide of DkXTH8 (DkXTH8-SP), and the ORF sequence of DkXTH8 without the signal peptide (DkXTH8-Int) were amplified using the specific primers listed in Table 1. After confirmation, the sequences were digested with XbaI and KpnI and then inserted into the pBI 221-GFP vector. After bombardment and cultivation, the onion epidermal cells were imaged using a confocal laser-scanning microscope. A Nikon A1 confocal microscope system operating on a Ti-E inverted microscope and equipped with a Plan Apo 10x microscope objective (Nikon, Tokyo, Japan) was used for this study. The confocal settings were excitation at 488 nm and emission wavelength was 525 nm. Images were obtained with 1x zoom and recorded at high resolution (2 comps 12 bit) using 2-fold line averaging. When indicated, cells were plasmolyzed in 400 mM sucrose for 15 min.
Production and purification of recombinant XTH proteins and enzyme activity analysis
The coding region of DkXTH8 without the signal peptide sequence was amplified using combinations of specific primers (Table 1). After confirmation, the products were digested with BamHI and HindIII and ligated into the pET-32a vector. The recombinant plasmid DkXTH8- pET-32a was introduced into Escherichia coli BL21 via the heat-shock method, and expression of the recombinant protein was induced with 0.100 mM isopropyl β-d-thiogalactopyranoside at 16 °C for 68 h with shaking at 20 revs−1. After sonication, the bacterial cell lysate was centrifuged for 10 minutes at 10,000 × g. Subsequently, the crude proteins in the supernatant were concentrated and dissolved in binding-wash buffer (40 mM Tris-HCl, 0.5 M NaCl, 20 mM imidazole, 10% glycerol, pH 7.9) and purified on a nickel-nitrilotriacetic acid (Ni-NTA) resin column. The pET-32a vector alone was used as the blank control. After analysis by SDS-polyacrylamide gel electrophoresis, DkXTH8-RP was concentrated and dialyzed in citrate/phosphate buffer (pH 5.5) for determination of XET/XEH activity according to Han et al.27.
Generation of transgenic Arabidopsis and tomato plants overexpressing DkXTH8
The coding region of DkXTH8 was amplified by PCR using the primers listed in Table 1. After digestion with the corresponding restriction enzymes (underlined in the primers), the resulting PCR products were inserted into the BamHI- and XbaI-digested binary vector 35 S:pVBG2307 (Fig. S1a). The recombinant plasmid DkXTH8-pVBG2307 was then introduced into Agrobacterium tumefaciens GV3101. A. thaliana ecotype ‘Columbia’ was transformed via the floral dip method49, and T3 homozygous transgenic lines (AL1, AL2 and AL3) were generated. Cultivar Micro-Tom was transformed via Agrobacterium-mediated leaf transformation as described by Hou et al.28, and T1 transgenic lines (TL1, TL2 and TL3) were used.
Dark-induced Arabidopsis leaf senescence
The fifth–sixth leaves of 4-week-old Arabidopsis plants were detached according to Hou et al.28. The leaves were floated on water in 9-mm-diameter Petri dishes and stored for up to 4 d in the dark at 22 °C to promote senescence. Detached leaves stored under growth conditions (16 h of light at 22 °C and 8 h of dark at 18 °C) served as the control.
Measurements of the chlorophyll content and relative electrolyte leakage
The chlorophyll concentration was determined spectrophotometrically as described by Wellburn50.
For the relative electrolyte leakage measurement, collected leaves were cut into discs (5 mm in diameter). After vacuum-infiltration of deionized water for 30 min, 10 discs were incubated in 5 mL ddH2O for 2 h. The initial conductivities (C1) of the solutions were measured using a conductivity meter. The samples were then boiled for 15 min and cooled to room temperature for recording the final conductivities (C2). The relative electrolyte leakage is expressed as C1/C2.
Storage of transgenic tomato fruits
Tomato fruits of WT and DkXTH8-transgenic lines were collected at the mature green period with no obvious color change. For postharvest softening and senescence analyses, fruits from each group were divided randomly into three subgroups, and all groups were stored at 25 ± 1 °C and 85–95% relative humidity. To investigate fruit color, a chroma meter CR-400 (Konica Minolta, Osaka, Japan) equipped with an 8-mm-diameter measuring area in the head was used.
Microscopic observation of WT and DkXTH8-transgenic plants
The fifth–sixth mature leaves (0.5 × 0.5 cm) and stems (0.5 cm) at 1–2 cm height above the ground were obtained from Arabidopsis WT and DkXTH8-transgenic line AL2 at four weeks after sowing. Tomato fruit materials (approximately 2 mm3) were collected from WT and DkXTH8-transgenic line TL1, as described in 4.11. The samples were immediately fixed in FAA solution (2% formaldehyde, 5% acetic acid, and 63% ethanol), placed under vacuum for 1 h and then processed as follows: dehydration in 70%, 85%, 95% and 100% ethanol (1 h each step); vitrified with a gradient from 100% ethanol to 100% xylene; infiltrated and embedded in paraffin. The polymerized samples were cut into 8-μm thick sections using a microtome (Leica RM2016, Germany) and then mounted onto microscopic slides. After staining with safranin, the samples were examined by light microscopy and imaged by confocal laser-scanning microscopy (T2; Olympus, Tokyo, Japan).
Statistical analysis
Data were evaluated by analysis of variance (ANOVA) using SPSS Statistics 22.0 software, and the means were compared by Fisher’s least significant difference (LSD) test. P values below 0.05 were considered statistically significant (P < 0.05). The data are expressed as the mean ± standard error.
Additional Information
How to cite this article: Han, Y. et al. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Sci. Rep. 6, 39155; doi: 10.1038/srep39155 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by National Key Research and Development Program (2016YFD0400102). We thank Prof. Zhenhui Gong (Northwest A&F University, China) for providing the pVBG2307 vector.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.Han and J.R. conceived and designed research. Y.Han and Q.B. conducted experiments. Y.Han, Q.B. and H.L. analyzed data. Y.Hou, M.J. and S.H. contributed new reagents or analytical tools. Y.Han and Q.B. wrote the manuscript. The work has not been submitted elsewhere for publication, and all the authors listed have approved the manuscript that is enclosed.
References
- Cosgrove D. J. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6, 850–861, doi: 10.1038/nrm1746 (2005). [DOI] [PubMed] [Google Scholar]
- Figueroa C. R. et al. Softening rate of the Chilean strawberry (Fragaria chiloensis) fruit reflects the expression of polygalacturonase and pectate lyase genes. Postharvest Biology and Technology 49, 210–220, doi: 10.1016/j.postharvbio.2008.01.018 (2008). [DOI] [Google Scholar]
- Payasi A., Mishra N. N., Chaves A. L. S. & Singh R. Biochemistry of fruit softening: an overview. Physiol. Mol. Biol. Plants 15, 103–113, doi: 10.1007/s12298-009-0012-z (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell D. A. & Harpster M. H. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Molecular Biology 47, 311–340, doi: 10.1023/a:1010656104304 (2001). [DOI] [PubMed] [Google Scholar]
- Schroder R., Atkinson R. G., Langenkamper G. & Redgwell R. J. Biochemical and molecular characterisation of xyloglucan endotransglycosylase from ripe kiwifruit. Planta 204, 242–251, doi: 10.1007/s004250050253 (1998). [DOI] [PubMed] [Google Scholar]
- Zhu Q. et al. Identification of xyloglucan endotransglucosylase/hydrolase genes (XTHs) and their expression in persimmon fruit as influenced by 1-methylcyclopropene and gibberellic acid during storage at ambient temperature. Food chemistry 138, 471–477, doi: 10.1016/j.foodchem.2012.09.141 (2013). [DOI] [PubMed] [Google Scholar]
- Nishitani K. In International Review of Cytology - a Survey of Cell Biology, Vol 173 Vol. 173 International Review of Cytology-a Survey of Cell Biology (ed. Jeon K. W.) 157–206 (1997). [DOI] [PubMed] [Google Scholar]
- Saladie M., Rose J. K., Cosgrove D. J. & Catala C. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. The Plant journal: for cell and molecular biology 47, 282–295, doi: 10.1111/j.1365-313X.2006.02784.x (2006). [DOI] [PubMed] [Google Scholar]
- Rose J. K. C., Braam J., Fry S. C. & Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant and Cell Physiology 43, 1421–1435, doi: 10.1093/pcp/pcf171 (2002). [DOI] [PubMed] [Google Scholar]
- Eklof J. M. & Brumer H. The XTH Gene Family: An Update on Enzyme Structure, Function, and Phylogeny in Xyloglucan Remodeling. Plant Physiology 153, 456–466, doi: 10.1104/pp.110.156844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P. & Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends in Plant Science 4, 361–366, doi: 10.1016/s1360-1385(99)01468-5 (1999). [DOI] [PubMed] [Google Scholar]
- Yokoyama R. & Nishitani K. Functional diversity of xyloglucan-related proteins and its implications in the cell wall dynamics in plants. Plant Biology 2, 598–604, doi: 10.1055/s-2000-16643 (2000). [DOI] [Google Scholar]
- Fry S. C. Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytologist 161, 641–675, doi: 10.1111/j.1469-8137.2003.00980.x (2004). [DOI] [PubMed] [Google Scholar]
- Xu W., Campbell P., Vargheese A. K. & Braam J. The Arabidopsis XET-related gene family: Environmental and hormonal regulation of expression. Plant Journal 9, 879–889, doi: 10.1046/j.1365-313X.1996.9060879.x (1996). [DOI] [PubMed] [Google Scholar]
- Xu W. et al. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7, 1555–1567 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. P., Tripathi S. K., Nath P. & Sane A. P. Petal abscission in rose is associated with the differential expression of two ethylene-responsive xyloglucan endotransglucosylase/hydrolase genes, RbXTH1 and RbXTH2. J. Exp. Bot. 62, 5091–5103, doi: 10.1093/jxb/err209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-B., Lu S.-M., Zhang J.-F., Liu S. & Lu Y.-T. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226, 1547–1560, doi: 10.1007/s00425-007-0591-2 (2007). [DOI] [PubMed] [Google Scholar]
- Potter I. & Fry S. C. Xyloglucan endotransglycosylase activity in pea internodes - effects of applied gibberellic-acid. Plant Physiology 103, 235–241, doi: 10.1104/pp.103.1.235 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H.-L. et al. Differential expression and regulation of longan XET genes in relation to fruit growth. Plant Science 174, 32–37, doi: 10.1016/j.plantsci.2007.09.008 (2008). [DOI] [Google Scholar]
- Ishimaru M. & Kobayashi S. Expression of a xyloglucan endo-transglycosylase gene is closely related to grape berry softening. Plant Science 162, 621–628, doi: 10.1016/s0168-9452(01)00608-2 (2002). [DOI] [Google Scholar]
- Ishimaru M., Smith D. L., Gross K. C. & Kobayashi S. Expression of three expansin genes during development and maturation of Kyoho grape berries. J. Plant Physiol. 164, 1675–1682, doi: 10.1016/j.jplph.2006.07.017 (2007). [DOI] [PubMed] [Google Scholar]
- Munoz-Bertomeu J., Miedes E. & Lorences E. P. Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. Journal of plant physiology 170, 1194–1201, doi: 10.1016/j.jplph.2013.03.015 (2013). [DOI] [PubMed] [Google Scholar]
- Percy A. E. et al. Xyloglucan endotransglycosylase activity during fruit development and ripening of apple and kiwifruit. Physiol. Plant. 96, 43–50 (1996). [Google Scholar]
- Miedes E. et al. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 168, 196–203, doi: 10.1016/j.jplph.2010.06.029 (2011). [DOI] [PubMed] [Google Scholar]
- Ohba T., Takahashi S. & Asada K. Alteration of fruit characteristics in transgenic tomatoes with modified expression of a xyloglucan endotransglucosylase/hydrolase gene. Plant Biotechnology 28, 25–32, doi: 10.5511/plantbiotechnology.10.0922a (2011). [DOI] [Google Scholar]
- Miedes E., Herbers K., Sonnewald U. & Lorences E. P. Overexpression of a cell wall enzyme reduces xyloglucan depolymerization and softening of transgenic tomato fruits. Journal of agricultural and food chemistry 58, 5708–5713, doi: 10.1021/jf100242z (2010). [DOI] [PubMed] [Google Scholar]
- Han Y. et al. Analysis of Xyloglucan Endotransglycosylase/Hydrolase (XTH) Genes and Diverse Roles of Isoenzymes during Persimmon Fruit Development and Postharvest Softening. Plos One 10, doi: 10.1371/journal.pone.0123668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. et al. The Persimmon 9-lipoxygenase Gene DkLOX3 Plays Positive Roles in Both Promoting Senescence and Enhancing Tolerance to Abiotic Stress. Frontiers in Plant Science 6, doi: 10.3389/fpls.2015.01073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J. et al. Cloning and expression of lipoxygenase genes and enzyme activity in ripening persimmon fruit in response to GA and ABA treatments. Postharvest Biology and Technology 92, 54–61, doi: 10.1016/j.postharvbio.2014.01.015 (2014). [DOI] [Google Scholar]
- Johansson P. et al. Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16, 874–886, doi: 10.1105/tpc.020065 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M. J. et al. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 19, 1947–1963, doi: 10.1105/tpc.107.051391 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas A. M. et al. Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula x tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochemical Journal 390, 105–113, doi: 10.1042/bj20041749 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha C. M. et al. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiology and Biochemistry 70, 433–444, doi: 10.1016/j.plaphy.2013.06.008 (2013). [DOI] [PubMed] [Google Scholar]
- Han Y. et al. Isolation and Characterization of Two Persimmon Xyloglucan Endotransglycosylase/Hydrolase (XTH) Genes That Have Divergent Functions in Cell Wall Modification and Fruit Postharvest Softening. Frontiers in Plant Science 7, doi: 10.3389/fpls.2016.00624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. et al. Molecular cloning, characterizing, and expression analysis of CTR1 genes in harvested papaya fruit. European Food Research and Technology 238, 503–513, doi: 10.1007/s00217-013-2131-6 (2014). [DOI] [Google Scholar]
- Li C.-r. et al. 1-MCP delayed softening and affected expression of XET and EXP genes in harvested cherimoya fruit. Postharvest Biology and Technology 52, 254–259, doi: 10.1016/j.postharvbio.2008.12.009 (2009). [DOI] [Google Scholar]
- Tabuchi A., Mori H., Kamisaka S. & Hoson T. A new type of endo-xyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant and Cell Physiology 42, 154–161, doi: 10.1093/pcp/pce016 (2001). [DOI] [PubMed] [Google Scholar]
- Steele N. M. & Fry S. C. Differences in catalytic properties between native isoenzymes of xyloglucan endotransglycosylase (XET). Phytochemistry 54, 667–680, doi: 10.1016/s0031-9422(00)00203-x (2000). [DOI] [PubMed] [Google Scholar]
- Sulova Z., Baran R. & Farkas V. Divergent modes of action on xyloglucan of two isoenzymes of xyloglucan endo-transglycosylase from Tropaeolum majus. Plant Physiology and Biochemistry 41, 431–437, doi: 10.1016/s0981-9428(03)00050-0 (2003). [DOI] [Google Scholar]
- Atkinson R. G., Johnston S. L., Yauk Y.-K., Sharma N. N. & Schroder R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biology and Technology 51, 149–157, doi: 10.1016/j.postharvbio.2008.06.014 (2009). [DOI] [Google Scholar]
- Maris A., Suslov D., Fry S. C., Verbelen J.-P. & Vissenberg K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60, 3959–3972, doi: 10.1093/jxb/erp229 (2009). [DOI] [PubMed] [Google Scholar]
- Bollok M. et al. Production of poplar xyloglucan endotransglycosylase using the methylotrophic yeast Pichia pastoris. Applied Biochemistry and Biotechnology 126, 61–77, doi: 10.1007/s12010-005-0006-4 (2005). [DOI] [PubMed] [Google Scholar]
- Genovesi V. et al. ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. Journal of experimental botany 59, 875–889, doi: 10.1093/jxb/ern013 (2008). [DOI] [PubMed] [Google Scholar]
- Han Y. et al. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J. Exp. Bot. 64, 4225–4238, doi: 10.1093/jxb/ert229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B. F., Noh Y. S., Himelblau E. & Amasino R. M. Molecular aspects of leaf senescence. Trends in Plant Science 5, 278–282, doi: 10.1016/s1360-1385(00)01655-1 (2000). [DOI] [PubMed] [Google Scholar]
- Strother S. The role of free radicals in leaf senescence. Gerontology 34, 151–156 (1988). [DOI] [PubMed] [Google Scholar]
- Wagstaff C. et al. Modification of cell wall properties in lettuce improves shelf life. J. Exp. Bot. 61, 1239–1248, doi: 10.1093/jxb/erq038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, doi: 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743, doi: 10.1046/j.1365-313x.1998.00343.x (1998). [DOI] [PubMed] [Google Scholar]
- Wellburn A. R. The spectral determination of chlorophyll-a and chlorophhyll-b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313 (1994). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.