Abstract
Cystic fibrosis is often associated with intestinal inflammation due to several factors, including altered gut microbiota composition. In this study, we analyzed the fecal microbiota among patients with cystic fibrosis of 10–22 years of age, and compared the findings with age-matched healthy subjects. The participating patients included 14 homozygotes and 14 heterozygotes with the delF508 mutation, and 2 heterozygotes presenting non-delF508 mutations. We used PCR-DGGE and qPCR to analyze the presence of bacteria, archaea and sulfate-reducing bacteria. Overall, our findings confirmed disruption of the cystic fibrosis gut microbiota. Principal component analysis of the qPCR data revealed no differences between homozygotes and heterozygotes, while both groups were distinct from healthy subjects who showed higher biodiversity. Archaea were under the detection limit in all homozygotes subjects, whereas methanogens were detected in 62% of both cystic fibrosis heterozygotes and healthy subjects. Our qPCR results revealed a low frequency of sulfate-reducing bacteria in the homozygote (13%) and heterozygote (13%) patients with cystic fibrosis compared with healthy subjects (87.5%). This is a pioneer study showing that patients with cystic fibrosis exhibit significant reduction of H2-consuming microorganisms, which could increase hydrogen accumulation in the colon and the expulsion of this gas through non-microbial routes.
Keywords: cystic fibrosis; gut microbiota; methanogen archaea; sulfate-reducing bacteria, DGGE-PCR; qPCR
A different balance of hydrogenotrophic bacteria in cystic fibrosis intestinal microbiota, compared with healthy subjects, could increase hydrogen accumulation in the colon.
INTRODUCTION
Cystic fibrosis (CF) frequently involves the gastrointestinal tract, with one of the most common complications being chronic inflammation (Li and Somerset 2014). Decreased bacterial diversity is associated with overgrowth of particular groups of resident bacteria that can induce inflammatory responses, which could be a factor leading to intestinal inflammation in CF. Several mechanisms linking microbial dysbiosis to gut inflammation in inflammatory bowel diseases have been proposed. An altered composition of the microbial communities might cause an immune response in the gastrointestinal tract due to the inflammatory potential of some commensal bacteria such as Escherichia coli and Enterococcus faecalis (Kim et al.2005). Functional changes in the gut microbiota, such as the increase of stress oxidative pathways (Morgan et al.2012), the decrease of butyrate production (Takaishi et al.2008) or an increase of sulfate-reducing bacteria (SRB) generating toxic H2S, might also be implicated in intestinal inflammation (Pitcher 2000). However, for several gastrointestinal diseases, it is still not clear if the reduction of species richness is a cause or a downstream consequence of the inflammatory process (Mosca, Leclerc and Hugot 2016). Concerning CF, recent studies have examined the intestinal microbiota composition using both culture-dependent and culture-independent techniques (Duytschaever et al.2011, 2013; Scanlan et al.2012; Schippa et al.2013; Bruzzese et al.2014). Results of these studies commonly describe reductions of members of the Bacteroides-Prevotella group, Bifidobacterium spp. and Clostridium cluster XIVa. However, the literature includes some discrepancies, for example, the complexity of CF gut microbiota is sometimes described as reduced and sometimes as similar compared to that of healthy subjects. Such discrepancies are likely because the studies use different methodology and because the investigated patient cohorts are heterogeneous in terms of age, genotype, antibiotic treatments, inflammation status, etc. To the best of our knowledge, there are no studies showing a clear relationship between reduced microbiota complexity and inflammation in CF; in one study (Bruzzese et al.2014), the authors attempted to correlate bacterial biodiversity with fecal calprotectin (CLP) values but no association was found.
CF disease is characterized by an altered intraluminal environment in which many factors can affect the gut microbiota composition and biodiversity. Among these, alterations in intestinal motility, pH, production of digestive enzymes and mucus are mostly responsible for CF-associated gastrointestinal complications (De Lisle and Borowitz 2013). The excessive mucus production, together with its high viscosity (Garcia, Yang and Quinton 2009), not only could create niches in which bacterial colonization is altered, but also contribute to the onset of some gastrointestinal symptoms such as obstruction and malabsorption (Freudenberg et al. 2008; Houwen et al.2010). Malabsorption and inadequate nutrient intake represent the primary causes of malnutrition in patients with CF (Stephenson et al.2013). In this context, the gut microbiota could be one of the factors affecting nutrient metabolism and energy balance (Li and Somerset 2014).
Gut microbiota is directly involved in the anaerobic fermentation of undigested dietary components that reach the colon, and of mucin glycoproteins that constitute the mucus gel covering the intestinal epithelium. These complex metabolic reactions produce a variety of final products, mainly short-chain fatty acids (SCFAs). The type and quantity of SCFAs depend on the metabolic functions of the resident bacterial communities and on the gut environmental conditions as determined by pH, nutrient availability, transit time, etc. Besides butyrate, acetate and propionate, which are the major end products of fermentation, a lot of less represented SCFAs can result from substrate fermentations and bacterial cross-feeding, including valerate, hexanoate and octanoate. Secondary products of several microbial fermentations are gases such as CO2 and H2. Since high hydrogen levels can inhibit normal fermentative reactions (Carbonero, Benefiel and Gaskins 2012), mechanisms involved in H2 disposal are required for proper function of the intestinal ecosystem. A low H2 partial pressure is maintained in the colon by both H2 expulsion as flatus or breath and microbial hydrogenotrophic activity (Nakamura et al.2010). Therefore, microorganisms able to use hydrogen such as methanogenic archaea, acetogenic bacteria and SRB may play an important role in nutrient digestion and absorption through cross-feeding interactions.
In this study, we sought to determine whether the gut microbiota disruption in CF may involve modification of the archaea and SRB populations in response to the altered intestinal environment. We used PCR-DGGE and qPCR to investigate the gut bacterial and archaeal communities in 30 subjects with CF as well as in age-matched healthy controls. In addition, we evaluated potential correlations between gut microbiota composition and key clinical parameters as host genetic background, gut inflammation and status of lung function.
MATERIALS AND METHODS
Study subjects
For this study, we enrolled 30 clinically stable young adults with CF, who did not require antibiotics for at least 2 weeks and who did not manifest acute intestinal or extraintestinal diseases. We registered the patients’ sex, age, body mass index (BMI) centile, CF genotype and Pseudomonas aeruginosa colonization, based on sputum cultures. Host genotype was classified as either mild or severe depending on the occurrence of at least one mild mutation or two severe mutations, respectively, according to the class of mutations (Zielenski 2000) and their related functional consequences (Castellani et al.2008). Antibiotic burden in the last 12 months for patients with P. aeruginosa colonization consisted in four cycles of intravenous antibiotic therapy. One or two additional cycles of therapy were necessary in case of pulmonary exacerbation. Patients showing no P. aeruginosa colonization received on average two cycles of intravenous antibiotic therapy for pulmonary exacerbation.
The forced expiratory volume in 1 s (FEV1) and CLP levels were measured in order to evaluate lung function and intestinal inflammation, respectively. Patients’ mean age was 14.54 years (range, 10–22 years). To investigate intestinal microbiota composition and inflammation, fecal samples were collected from these subjects. We also collected fecal samples from eight healthy volunteers of the same age (mean age, 14.38 years). Participants older than 18 years provided their written informed consent to participate in this study. Moreover, we obtained written informed consent from the parents of each enrolled child. The study protocol was approved by the Ethical Committee of University Federico II of Naples.
Intestinal inflammation and lung function
To assess the level of intestinal inflammation, we measured fecal CLP production using an enzyme-linked immunosorbent assay (Calprest Eurospital SpA, Trieste, Italy). Normal inflammation was defined as a fecal CLP level of 0–50 μg/g of stool, intermediate inflammation corresponded to 50–100 μg CLP/g stool and severe intestinal inflammation was indicated by greater than 100 μg CLP/g stool (Fagerberg et al. 2003). CLP levels were not assessed in healthy volunteers. FEV1 was measured as described by Corey et al. (1997). At least three maximum expiratory maneuvers were performed during each test and the best result was recorded.
Bacterial DNA extraction and PCR-DGGE
We performed DNA extraction and PCR-DGGE of the dominant bacterial communities and the Bacteroides-Prevotella group as previously described (Bruzzese et al.2014). Our analysis of the Clostridium coccoides group utilized previously reported primers and thermal protocol (Maukonen et al.2006). All DGGE gels were run in an INGENYphor 2×2 System (INGENYphor, Goes, the Netherlands) using 1× buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0). DGGE gels were digitally processed via a multistep procedure using Fingerprinting II SW (Bio-Rad Laboratories, Hercules, CA, USA). After profile normalization, dendrograms were constructed using Pearson's correlation coefficient and the unweighted-pair group method. Discriminating bands, i.e. bands whose intensity varied significantly between patients with CF and healthy controls, were excised, re-amplified, and sequenced (BMR Genomics, Padova, Italy). Next, the blastn algorithm was used to compare the nucleotide sequences with those deposited in GenBank (http://www.ncbi.nlm.nih.gov/) and in the Ribosomal Database Project (Maidak et al.1994). We calculated the Shannon Wiener Diversity (H) Index of the biodiversity using the previously reported formula (Deng et al.2012). We also attempted to perform PCR-DGGE analysis of archaea and SRB communities by using group-specific primers as described (Geets et al.2006; Federici et al.2015). Unfortunately, PCR amplification of methanogenic 16S rRNA genes resulted in bands that were all identified as Methanobrevibacter smithii, while for SRB no amplicons were obtained from fecal DNA of patients with CF.
qPCR
We used qPCR to quantify Escherichia coli, Bifidobacterium spp., Bifidobacterium adolescentis, B. catenulatum, Eubacterium rectale, the Bacteroides-Prevotella group, Bacteroides uniformis, Ba. vulgatus and Faecalibacterium prausnitzii as previously described (Bruzzese et al.2014). Table 1 lists the remaining qPCR primers. For the detection of hydrogenotrophic groups, primers targeting key functional genes, namely the methyl-coenzyme M reductase α-subunit (mcrA), dissimilatory (bi)sulfite reductase α-subunit (dsrA), butyryl-CoA: acetate CoA-transferase (BcoAT) and acetyl-CoA synthase β-subunit (acsB) were used to quantify methanogens, SRB, butyrate-producing bacteria and acetogens, respectively. All qPCR reactions were performed in a LightCycler® 480 Instrument II (Roche Life Science, Mannheim, Germany). Standard curves were generated using reference DNA, and thermal conditions were set up to achieve optimal amplification efficiency. Results were expressed as gene copy number per gram of wet fecal sample.
Table 1.
qPCR primers used in this study.
| Species/group | Target gene | Reference |
|---|---|---|
| Dialister invisus | 16S rRNA | Kumar et al. (2003) |
| Collinsella aerofaciens | 16S rRNA | Kassinen et al. (2007) |
| Ruminococcaceae | 16S rRNA | Garcia-Mazcorro et al. (2012) |
| Blautia spp. | 16S rRNA | Suchodolski et al. (2012) |
| Clostridium coccoides group | 16S rRNA | Rinttilä et al. (2004) |
| Ruminococcus gnavus | 16S rRNA | Joossens et al. (2011) |
| Methanobrevibacter smithii | 16S rRNA | Dridi et al.2009) |
| Methanosphaera stadtmanae | 16S rRNA | Dridi et al. (2009) |
| Methanobrevibacter | 16S rRNA | Skillman et al. (2004) |
| Total archaea | 16S rRNA | Yu et al. (2005) |
| SRB | dsrA | Kondo et al. (2004) |
| Acetogens | acsB | Gagen et al. (2010) |
| Methanogens | mcrA | Denman, Tomkins and McSweeney (2007) |
| Butyrate-producing bacteria | BcoAT | Louis and Flint (2007) |
dsrA = dissimilatory (bi)sulfite reductase gene, α subunit; acsB = acetyl-CoA synthase gene, β subunit; mcrA = methyl-coenzyme M reductase gene, α subunit; BcoAT = butyryl-CoA: acetate CoA transferase gene.
Statistical analysis
Principal component (PC) analysis was performed using the PRINCOMP procedure of SAS release 8.0 (SAS Inst. Inc., Cary, NC, USA). The CLP concentration and FEV1 data of patients with CF were analyzed by ANOVA using the GLM procedure of SAS. The applied statistical model included the fixed effects of genetic mutation (heterozygote or homozygote) and the random effect of the patient. Spearman's rank correlation was calculated by separately considering the two groups (healthy and CF) using the CORR procedure of SAS. To investigate the relationship between CLP level, FEV1data and bacterial population in patients with CF, we performed a stepwise regression analysis using the REG procedure of SAS. Statistical significance was established using a conventional P level of 0.05.
RESULTS AND DISCUSSION
Study population
Of the included patients, 14 (46.7%) were homozygous and 14 (46.7%) were heterozygous for the delF508 mutation, which is the most common polymorphism of the CF transmembrane conductance regulator gene. The remaining two subjects (6.6%) displayed a mild genotype, which was not correlated with a mild inflammation status since CLP value was greater than 100 μg CLP/g stool for both individuals. The BMI centile, ranging from 10th to 75th centile, indicated that all enrolled patients had a good nutritional status. In the patients with CF, we measured fecal CLP concentration, which is reported as a good marker of intestinal inflammation (Hradsky et al.2014; Dhaliwal et al.2015). Severe inflammation was found in 19 subjects (63%), intermediate inflammation in 7 subjects (23%) and mild inflammation in 4 subjects (13%). Analysis by unpaired t test revealed no correlation between genotype and intestinal inflammation: mean of 213.9 μg/g ±145.2 μg/g in homozygous subjects versus 231.3 μg/g ± 299.2 μg/g among heterozygous subjects. FEV1 (% predicted) ranged from 32 to 99 (mean value, 75.71±13.82). No correlation was found between genotype and FEV1 data: mean (% predicted) of 73 ± 17.35 in homozygous subjects versus 78 ± 9.9 (% predicted) among heterozygous subjects. FEV1 values indicated a mild pulmonary involvement in 23 patients, a moderate involvement in 5 patients and a severe pulmonary involvement in 2 patients. FEV1 is widely used as marker of lung function throughout the course of CF lung disease and it is considered the best clinical predictor of poor outcome. Pseudomonas aeruginosa lung colonization was detected in 11 patients (36%).
PCR-DGGE
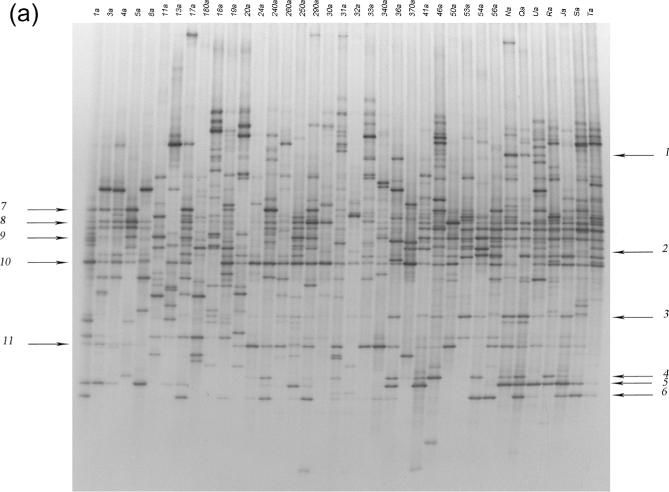
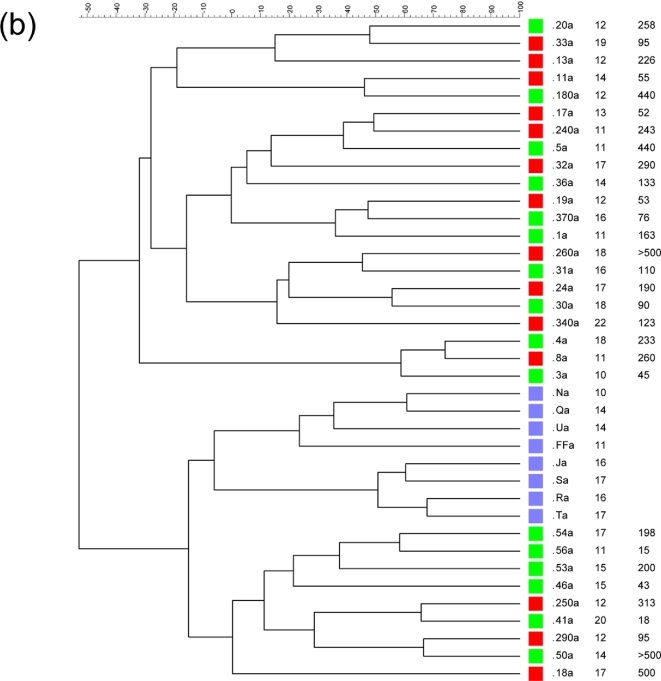
PCR-DGGE analysis of amplicons obtained using universal primers revealed a more complex profile in healthy subjects than in the majority of patients with CF (Fig. 1a). The main Shannon index values obtained for healthy subjects and patients with CF were 1.17 and 0.94, respectively (P = 0.007). This difference in biodiversity was lower than that previously reported among younger subjects (Bruzzese et al.2014). Figure 1b presents the dendrogram produced using software analysis. Healthy profiles formed two distinct subclusters: one containing subjects aged 10–14 years and the other including subjects from 16–17 years of age. In contrast, the clusterization of patients with CF was apparently unrelated to age or CLP level, and we did not find distinctiveness related to homozygote and heterozygote (delF508 and non-delF508) profiles (Fig. 1b). Notably, the frequent use of antibiotic therapy to treat pulmonary exacerbation could make individuals cluster distinctly regardless of host age or genotype. The effects on gut microbiota composition due to antibiotic treatments can be, in some cases, persistent (Jernberg et al. 2005, 2007; Jakobsson et al.2010). Moreover, reduced transit time, occasionally resulting in intestinal constipation, may exert a great impact on the relative proportions of specific bacterial populations.
Figure 1.
(a) DGGE profiles obtained using universal primers designed for the 16S rRNA gene. CF samples are indicated by a number, while samples from healthy subjects are indicated by capital letters. (b) Dendrogram constructed from analysis of DGGE gels using Pearson's correlation coefficient and the unweighted-pair group method. Red squares indicate homozygotes, green squares heterozygotes and light blue squares healthy subjects. Next to each patient's identification code, their age and CLP value are reported.
Figure 1.
(Continued).
Our results differ from previous findings that genetic background (homozygous-delF508, heterozygous-delF508 and non-delF508) was correlated with differences in gut microbiota composition (Schippa et al.2013). These discrepancies could be related to the analysis of different regions of the 16S rRNA gene between studies. Indeed, the quality of PCR-DGGE data reportedly depends on the number and the resolution of the amplicons in the gel (Yu and Morrison 2004). We chose the V3 region for PCR-DGGE analysis since it is considered a reliable target for gut microbiome studies (Huys, Vanhoutte and Vandamme 2008) although longer fragments, as those corresponding to V6–V8 regions, can allow a more accurate taxonomic affiliation of DNA bands. Yu and Morrison (2004) recommended a different set of primers compared to that used by Schippa et al. (2013) for the V6–V8 regions, in order to improve the biodiversity indices. In our present study, further software analysis enabled identification of the discriminant bands of fingerprints in decreasing order as related to Escherichia coli/Shigella, Blautia spp., Faecalibacterium prausnitzii, Collinsella aerofaciens, Dialister invisus, Eubacterium rectale and Bifidobacterium adolescentis (Table 2).
Table 2.
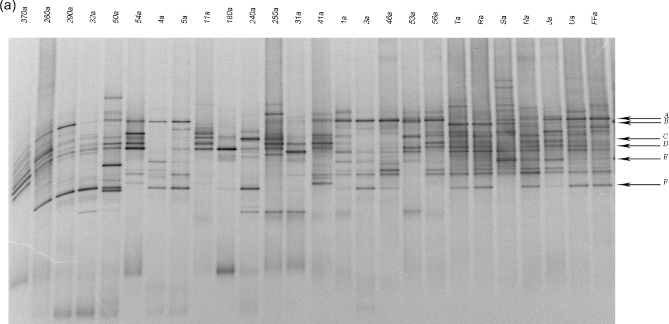
Sequence analysis of the bands indicated in Fig 1a (numerical ID) and 2a (capital letters ID).
| Band ID | % Blast similarity | Nearest species | Accession number | Presence in profiles of healthy controls (%) | Presence in profiles of CF patients (%) |
|---|---|---|---|---|---|
| 1 | 100 | Bifidobacterium adolescentis | KP256215.1 | 62.5 | 23.3 |
| 2 | 100 | Faecalibacterium prausnitzii | AJ270469 | 100 | 60 |
| 3 | 99 | Collinsella aerofaciens | NR113316.1 | 100 | 30 |
| 4 | 100 | Bifidobacterium catenulatum | KP256217.1 | 50 | 26.7 |
| 100 | Bifidobacterium pseudocatenulatum | AP012330.1 | |||
| 5 | 100 | Bifidobacterium adolescentis | KP256215.1 | 87.5 | 30 |
| 6 | 100 | Collinsella aerofaciens | NR113316.1 | 75 | 33.3 |
| 7 | 99 | Eubacterium rectale | AY804151 | 87.5 | 53.3 |
| 8 | 100 | Collinsella aerofaciens | NR113316.1 | 50 | 33.3 |
| 9 | 100 | Bifidobacterium coccoides | NR104700.1 | 100 | 63.3 |
| 100 | Blautia luti | NR041960.1 | |||
| 10 | 99 | Escherichia coli | LN867523.1 | 50 | 90 |
| 9 | Shigella flexneri | NR026331.1 | |||
| 99 | Escherichia fergusonii | NE074902.1 | |||
| 11 | 99 | Dorea invisus | NR113355.1 | 75 | 50 |
| A | 100 | Eubacterium rectale | NR 074634.1 | 100 | 66.7 |
| B | 95 | Uncultured Roseburia | JX 230356.1 | 87.5 | 16.7 |
| C | 99 | Blautia glucerasea | NR 113231.1 | 100 | 73.3 |
| D | 97 | Ruminococcus gnavus | AB 910745.1 | 100 | 62.5 |
| E | 98 | Blautia luti | NR 041960.1 | 88 | 33.0 |
| F | 99 | Dorea longicatena | NR 028883.1 | 100 | 44.3 |
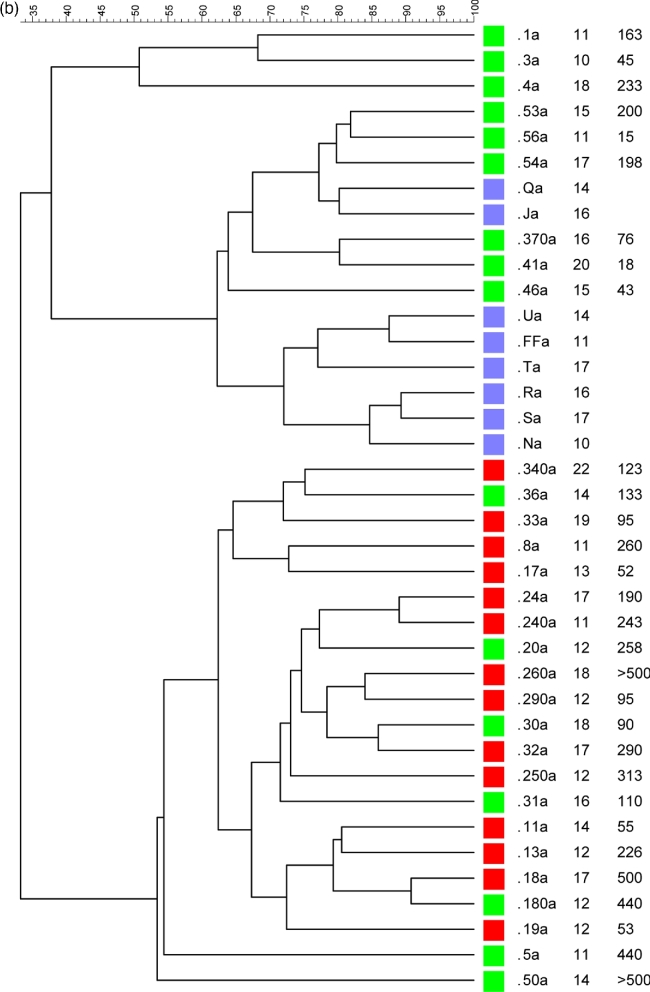
The majority of discriminant bands belonged to the Clostridium coccoides group, which is consistent with prior reports (Duytschaever et al.2013; Schippa et al.2013). Thus, we analyzed this population in detail using PCR-DGGE. Ruminococcus gnavus, Blautia luti, E. rectale and Bl. glucerasea were more frequently identified in healthy controls than in patients with CF, while Dorea longicatena was indistinctly present in both groups (Fig. 2a). The fecal microbiota clusters did not correlate with the CLP level, FEV1 data, lung colonization or age. Healthy subjects and nine heterozygotes (delF508 and non-delF508) formed one group, while the remaining seven heterozygotes formed a separate cluster together with homozygotes (Fig. 2b).
Figure 2.
(a) DGGE profiles obtained using Cl. coccoides group-specific primers. CF samples are indicated by a number, while healthy subjects are indicated by capital letters. (b) Dendrogram constructed from analysis of DGGE gels using Pearson's correlation coefficient and the unweighted-pair group method. Red squares indicate homozygotes, green squares heterozygotes and light blue squares healthy subjects. Next to each patient's identification code, their age and CLP value are reported.
Figure 2.
(Continued).
We previously found that Bacteroides uniformis presence was remarkably discriminant for healthy profiles (Bruzzese et al.2014); thus, we also evaluated the composition of the Bacteroides-Prevotella group using PCR-DGGE. A prior study reported that healthy subjects showed higher numbers of organisms from the Bacteroides-Prevotella group (obtained by count plates) compared to their siblings with CF (Duytschaever et al.2011). However, the PCR-DGGE profiles did not reveal particular differences in terms of the number of bands, and the intensity of Ba. uniformis-related amplicons did not discriminate healthy subjects from patients with CF.
Depletion of butyrate-producing bacteria such as F. prausnitzii, E. rectale and Roseburia spp. may have negative effects on the supply of butyrate to gut epithelial cells and also on the luminal pH owing to their ability to utilize lactate. Most butyrate producers within the gut use the butyryl-CoA:acetate CoA-transferase route which depends on acetate (Barcenilla et al. 2000). Acetate is in turn produced by acetogens such as some species belonging to the genera Ruminococcus and Blautia (Bernalier et al.1996). Other important acetate producers are members of Bifidobacterium and Bacteroides genera (Rajilić-Stojanović and de Vos 2014). Since the concentration of SCFAs in the intestinal lumen depends on the balance of production, microbial utilization and mucosal uptake, reduction of key bacteria groups has important effects on gut ecology and host mucosa.
qPCR
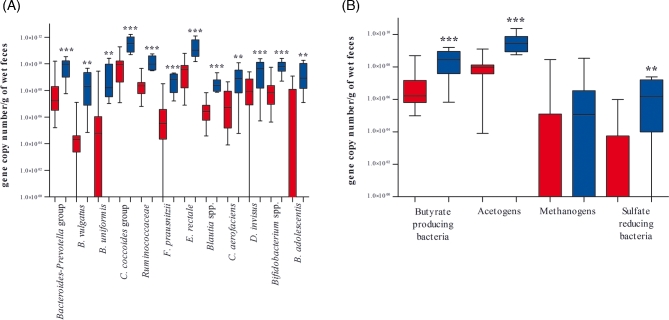
To confirm the results of qualitative analysis, we performed qPCR targeting the different bacterial genera or species that emerged as discriminant in PCR-DGGE analysis (Fig. 3A). Compared with healthy controls, patients with CF showed significantly reduced contents of D. invisus (P = 0.0001), B. adolescentis (P = 0.0011), Bifidobacterium spp. (P < 0.0001) and C. aerofaciens (P = 0.0025). Our results concerning Bifidobacterium spp. are consistent with previous findings (Scanlan et al.2012; Duytschaever et al.2013; Schippa et al.2013). A search of the literature revealed no information concerning B. adolescentis in patients with CF, but B. adolescentis, C. aerofaciens and D. invisus are constituents of the human gut microbiota (Lyra et al.2009; Qin et al.2010; Agans et al.2011; Hakansson and Molin 2011) and their abundance is reportedly reduced in the microbiota of patients with Crohn's disease (Suchodolski et al.2012).
Figure 3.
(A) Box plot of qPCR data (expressed as gene copy number per gram of wet feces) from patients with CF (red) and healthy controls (blue). The boxes show the first and third quartiles, with a horizontal line inside to represent the median. The whiskers represent the maximum and minimum data values, without outliers. ∗∗P < 0.050 and ∗∗∗P < 0.001. (A) qPCR data of species/groups of bacteria significantly different between CF and control individuals. (B) qPCR results for BcoAT, acs, mcr and dsr functional genes.
Enterobacteriaceae (Debyser et al.2016), particularly Es. coli (Hoffman et al.2014), were more abundant in CF fecal samples, although according to our data this difference did not reach statistical significance (P = 0.6278). We performed further quantitative analyses targeting bacteria belonging to the Cl. coccoides group, specifically F. prausnitzii, E. rectale, Blautia spp. and the Ruminococcaceae family. All of these groups were significantly reduced among adolescents with CF (P ≤ 0.0002). Persistent use of broad-spectrum antibiotics, frequent in patients with CF, can reduce bacterial diversity promoting or inhibiting specific indigenous taxa (Modi, Collins and Relman 2014). For example, a marked decrease of butyrate-producing bacteria, especially F. prausnitzii, and of Bifidobacterium spp. was reported after 4 days of amoxicillin-clavulanic acid administration (Young and Schmidt 2004). Reduced abundance of Cl. coccoides group in CF gut microbiota could be due to an altered intestinal environment since these populations are extremely sensitive to pH variations (Flint et al.2007). Moreover, butyrate-producing bacteria, and in particular Roseburia intestinalis and E. rectale, were able to colonize mucins in an in vitro study (Van den Abbeele et al.2013). So it is possible that the very viscous mucus present in the gastrointestinal tract of patients with CF could entrap these strong colonizers removing them from the intestinal lumen. On the other hand, R. gnavus abundance did not statistically differ (P = 0.4225) between patients with CF and the control group. These results differ from the findings of a recent study that applied a metaproteomic approach to analyze CF fecal samples, and reported reduced proteins taxonomically attributed to butyrate-producing bacteria and increased proteins associated with R. gnavus and Clostridia species (Debyser et al.2016). Further investigations are required to explain these discrepancies.
Eubacterium rectale and F. prausnitzii are butyrate-producing bacteria that are considered important markers of inflammatory status (Sokol et al.2008; Gupta et al. 2014). Reductions of this bacterial group could substantially impact enterocyte nutrition and the maintenance of intestinal health (Flint et al.2007). With regard to the Bacteroides-Prevotella group, Ba. vulgatus and Ba. uniformis were found to be significantly reduced in patients with CF compared to healthy subjects (P = 0.0002, P = 0.0015 and P = 0.0025, respectively). These modifications of the Bacteroides-Prevotella group contents were unrelated to age, since we previously found the same results among children with CF (Bruzzese et al.2014). A previous study used culture-based counts to compare the gut microbiota of children with CF to that of their healthy siblings, and found a borderline significant difference in Bacteroides-Prevotella presence (P = 0.07) (Duytschaever et al.2011). This discrepancy could be due to the difference in methodological approaches, and further investigation into this subject is needed.
We additionally quantified butyrate-producing bacteria, acetogens, methanogenic archaea and SRB by qPCR using primers targeting the functional genes BcoAT, acs, mcr and dsr (Fig. 3B). The BcoAT gene has been demonstrated to be prevalent in the human gut (Louis et al. 2004). The acs gene encodes for an enzyme exclusive of the acetyl-CoA pathway (Ragsdale 1991), mcr is highly conserved among methanogens (Luton et al.2002) and dsr is common to all SRB (Christophersen, Morrison and Conlon 2011). Thus, amplification of these genes represents a suitable approach for specific detection of these metabolic groups.
Compared to healthy subjects, patients with CF exhibited significantly decreased butyrate-producing bacteria and acetogens (P = 0.0002 and P = 0.0003, respectively). The observed reduction of butyrate-producing bacteria is consistent with previous findings (Duytschaever et al.2013; Schippa et al.2013), while no prior study has investigated acetogens presence in CF disease. SRB were detected in eight CF subjects (26.7%) with equal distribution among homozygotes and heterozygotes, compared to in 87.5% of healthy subjects. In subjects with detectable SRB, the amounts ranged from 104–107 gene copy number per gram of wet feces in healthy subjects and 104–105 gene copy number per gram of wet feces in patients with CF. Overall, SRB prevalence and abundance were significantly reduced in patients with CF (P = 0.0011), which is in contrast to other bowel diseases (Gibson, Cummings and Macfarlane 1991; Loubinoux et al.2002). Prior studies have described H2S involvement in retaining both proinflammatory and anti-inflammatory properties depending on the specific work (Singh and Lin 2015), and thus its precise role in intestinal inflammation remains unclear.
Archaea were detected in 62.5% of healthy subjects, compared to in 33.3% of patients with CF, all of which were heterozygotes. Within the subgroup of heterozygotes with CF, the prevalence of archaea was 62.5%, the same proportion as in healthy individuals. Archaea were present in both delF508 and non-delF508 heterozygote patients with CF. Methanogens were less abundant in CF fecal samples than in healthy controls, although this difference did not reach statistical significance (P = 0.2572). In contrast, a statistically significant difference was found between homozygotes and healthy subjects (P < 0.05), and between homozygotes and heterozygotes (P < 0.01). Methanobrevibacter spp. were the predominant genera in both healthy subjects and patients with CF, with amounts comparable with the total archaea content. As expected, Methanobrevibacter smithii was the most abundant methanogen, with quantification data comparable with the amounts of Methanobrevibacter spp., ranging from 104 to 109 gene copy number per gram of wet feces in both healthy and CF subjects. Methanosphaera stadtmanae was detected in 40% of healthy subjects and 20% of patients with CF, and the abundance of this species ranged from 104 to 105 gene copy number per gram of wet feces. Our results regarding M. stadtmanae prevalence and abundance for both groups are in accordance with those previously described in healthy subjects (Mihajlovski et al.2010; Dridi et al.2012).
It can be speculated that the reduced colonization of SRB and archaea in CF subjects (mainly in homozygotes) may be due to disease-related changes in the intestinal environment that interfere with these microorganisms’ ability to thrive. The reduction of SRB in CF subjects could be due to an altered lumen environment in which the availability of SCFAs and proteins is somewhat reduced. This hypothesis is based on the ability of SRB to use not only molecular hydrogen but also a wide range of substrates such as SCFAs or proteins as electron donors. In the human colon, the predominant SRB are those able to oxidize organic acids, mainly lactate and proteins (Newton et al.1998) whereas the H2/CO2 utilizers are less represented (Gibson, Macfarlane and Cummings 1988). In addition, one possible explanation for the reductions of both methanogens and SRB could be an alteration of the pH gradient from the right to left colon that enables distribution of these bacterial groups in accordance with their physiological requirements in healthy subjects (Nakamura et al.2010). Unfortunately, no detailed data are available regarding colon pH in patients with CF to support our hypothesis. Further investigations are required to elucidate the causes and effects of the unbalanced hydrogenotrophic populations in CF microbiota.
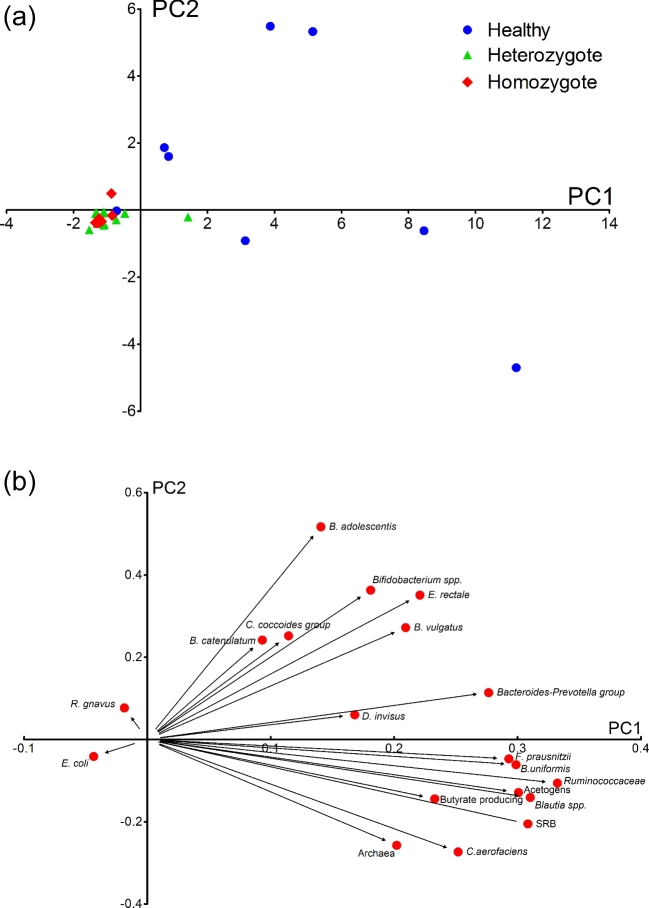
Figure 4a shows the results of PCA of qPCR data, and Fig. 4b shows the eigenvalues between all considered variables. The first three PC explained 66% of the total variation: 42% for PC1, 13% for PC2 and 11% for PC3. PC1 could efficiently separate the samples according to CF disease (Fig. 4a). Ordination analysis showed no discrimination between heterozygotes and homozygotes, and indicated higher interindividual biodiversity among healthy subjects compared with the microbiota of patients with CF. The abundance fluctuations of Ba. uniformis, Ruminococcaceae, F. prausnitzii, Blautia spp., acetogens and SRB were the predominant variables producing a positive impact on PC1 (Fig. 4b). The CF disease seemed to be the most important factor affecting this grouping since ANOVA revealed no correlation between bacterial population levels and lung colonization, FEV1 data, CLP concentration or genetic background (P = 0.7). Furthermore, stepwise regression analysis did not identify any group/bacterial species as being associated with CLP concentration, showing no link between intestinal inflammation and key bacterial groups in patients with CF.
Figure 4.
(a) Principal component analysis (PCA) plots of the qPCR dataset, indicating healthy subjects (blue dots), heterozygote patients with CF (green triangles) and homozygote patients with CF (red rhombus). (b) Graphical representation of the eigenvectors between qPCR data and PC1 and PC2, explaining 55% of the total variation (42% and 13% for PC1 and PC2, respectively).
Spearman's correlation analysis identified fewer correlations in healthy subjects (see Fig. S1A, Supporting Information) compared to among CF samples (see Fig. S1B, Supporting Information), possibly due to there being fewer subjects in the control group. In patients with CF, Ruminococcaceae and Blautia spp. were positively correlated with E. rectale and F. prausnitzii. Increased numbers of these butyrate-producing bacteria were correlated to higher abundance of Ba. uniformis and/or Ba. vulgatus. One possible metabolic interaction could be acetate production by members of the Bacteroides-Prevotella group and Blautia genera, which is then metabolized by butyrate-producing bacteria (Chassard and Bernalier-Donadille 2006). Bifidobacterium adolescentis was positively correlated with F. prausnitzii, confirming previous reports that this species was correlated to butyrate-producing bacteria (Belenguer et al.2006). This analysis also revealed a positive correlation of D. invisus with members of the Cl. coccoides group, Ba. vulgatus, Ba. uniformis and Ba. catenulatum. Archaea were negatively correlated with the Bacteroides-Prevotella group and positively correlated with Blautia spp. Previous findings have shown a negative correlation between Bacteroides and Methanobrevibacter (Hoffman et al.2014), as well as positive correlations between Methanobacteriales and Bifidobacterium spp., members of Lachnospiraceae, Dialister spp. and Blautia spp. (Vanderhaeghen, Lacroix and Schwab 2015). The presently observed low prevalence of SRB in CF patient samples prevent detailed analysis of their possible correlations with other groups of bacteria.
It is well recognized that a balance between different functional groups is crucial for maintaining efficient degradation of organic matter in the gastrointestinal tract. Disruption of this balance can affect bacterial community stability, leading to impaired host health. In our present study, patients with CF exhibited reduced abundance of several dominant bacterial groups, including C. coccoides, Bacteroides-Prevotella group and the Bifidobacterium genera. Patients with CF also showed strong modifications in the abundance of several functional groups, particularly acetogens, methanogens and SRB. We did not identify associations among bacterial groups, CLP values, lung colonization and genetic backgrounds. It is possible that the intestinal environment in CF disease leads to very strong selection of the more resilient enteric bacteria, as well as archaea and SRB, which showed low prevalence in homozygote patients with CF.
Considering the many CF-associated factors that could influence the gut microbiota composition and function, a major limitation of this study is represented by the limited number of recruited patients. A further limitation consists in the use of fecal samples in place of mucosal specimens. The gut bacteria community composition changes along the gastrointestinal tract and microbial communities living in the gut lumen actually differs from those adhering to the intestinal mucosa. Since mucosa-associated bacteria are in close contact with the epithelial tissues they could interact with the host more strongly than luminal microorganisms. Moreover, mucosal samples as those obtained by biopsies could be more representative of the inflammatory processes eventually occurring in the gut. To address some of these limitations, a larger number of patients with CF should be enrolled to increase the power of future studies. Clinical and nutritional parameters evaluation, together with the use of metagenomic approaches, could improve our understanding of the ecological relationships between gut microorganisms and their impact on CF disease.
CONCLUSION
The present work even if with some limitations represents the pioneer study investigating the balance of hydrogenotrophic bacteria in CF fecal microbiota. Our results confirm that the gut microbiota composition of patients with CF is significantly altered compared with that of healthy controls, although no correlation was found with either gut inflammation or host genotype. The reduction of species belonging to Clostridium coccoides group, Bacteroides-Prevotella group and Bifidobacterium genera could influence the metabolic potential of the microbiota in CF. Decrease of key butyrate-producers such as Faecalibacterium prausnitzii and Eubacteriumrectale or important functional bacteria groups as Blautia spp., E. rectale Ruminococcaceae and Bacteroides can have important effects on gut ecology and host cell metabolism. Hydrogenotrophic microorganisms, which resulted to be significantly less abundant in CF compared with controls, can have important ecological effects on the gut microbiota influencing the hydrogen economy of the colon. Our results warrant further investigation to highlight the link between these microbiota differences and disease states.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work was partially funded by Cystic Fibrosis Foundation Therapeutics, Inc. with a trial formally registered as a clinical trial at https://clinicaltrials.gov/ NCT01956916.
Conflict of interest. None declared.
REFERENCES
- Agans R, Rigsbee L, Kenche H et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol 2011;77:404–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin JC et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microb 2000;66:1654–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder AG et al. Two routes of metabolic cross-feeding between. Society 2006;72:3593–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernalier A, Rochet V, Leclerc M et al. Diversity of H2/CO2-utilizing acetogenic bacteria from feces of non-methane-producing humans. Curr Microbiol 1996;33:94–9 [DOI] [PubMed] [Google Scholar]
- Bruzzese E, Callegari ML, Raia V et al. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: A randomised clinical trial. PLoS One 2014;9:e87796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroentero 2012;9:504–18 [DOI] [PubMed] [Google Scholar]
- Castellani C, Cuppens H, Macek M et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros 2008;7:179–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett 2006;254:116–22 [DOI] [PubMed] [Google Scholar]
- Christophersen CT, Morrison M, Conlon MA. Overestimation of the abundance of sulfate-reducing bacteria in human feces by quantitative PCR targeting the Desulfovibrio 16S rRNA gene. Appl Environ Microb 2011;77:3544–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey M, Edwards L, Levison H et al. Longitudinal analysis of pulmonary function decline in patients with cystic fibrosis. J Pediatr 1997;131:809–14 [DOI] [PubMed] [Google Scholar]
- De Lisle RC, Borowitz D. The cystic fibrosis intestine. Cold Spring Harb Perspect Med 2013;3:a009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debyser G, Mesuere B, Clement L et al. Faecal proteomics: a tool to investigate dysbiosis and inflammation in patients with cystic fibrosis. J Cyst Fibros 2016;15:242–50 [DOI] [PubMed] [Google Scholar]
- Deng B, Shen CH, Shan XH et al. PCR-DGGE analysis on microbial communities in pit mud of cellars used for different periods of time. J Inst Brew 2012;118:120–6 [Google Scholar]
- Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 2007;62:313–22 [DOI] [PubMed] [Google Scholar]
- Dhaliwal J, Leach S, Katz T et al. Intestinal inflammation and impact on growth in children with cystic fibrosis. J Pediatr Gastr Nutr 2015;60:521–6 [DOI] [PubMed] [Google Scholar]
- Dridi B, Henry M, El Khéchine A et al. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 2009;4:e7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Henry M, Richet H et al. Age-related prevalence of Methanomassiliicoccus luminyensis in the human gut microbiome. APMIS 2012;120:773–7 [DOI] [PubMed] [Google Scholar]
- Duytschaever G, Huys G, Bekaert M et al. Cross-sectional and longitudinal comparisons of the predominant fecal microbiota compositions of a group of pediatric patients with cystic fibrosis and their healthy siblings. Appl Environ Microb 2011;77:8015–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duytschaever G, Huys G, Bekaert M et al. Dysbiosis of bifidobacteria and Clostridium cluster XIVa in the cystic fibrosis fecal microbiota. J Cyst Fibros 2013;12:206–15 [DOI] [PubMed] [Google Scholar]
- Fagerberg UL, Lööf L, Merzoug RD et al. Fecal calprotectin levels in healthy children studied with an improved assay. J Pediatr Gastr Nutr 2003;37:468–72 [DOI] [PubMed] [Google Scholar]
- Federici S, Miragoli F, Pisacane V et al. Archaeal microbiota population in piglet feces shifts in response to weaning: Methanobrevibacter smithii is replaced with Methanobrevibacter boviskoreani. FEMS Microbiol Lett 2015;362:1–7 [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP et al. Interactions and competition within the microbial community of the human colon: links between diet and health: minireview. Environ Microbiol 2007;9:1101–11 [DOI] [PubMed] [Google Scholar]
- Freudenberg F, Broderick AL, Yu BB et al. Pathophysiological basis of liver disease in cystic fibrosis employing a DeltaF508 mouse model. Am J Physiol Gastr L 2008;294:G1411–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagen EJ, Denman SE, Padmanabha J et al. Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl Environ Microbiol 2010;76:7785–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MAS, Yang N, Quinton PM. Normal mouse intestinal mucus release requires cystic fibrosis transmembrane regulator-dependent bicarbonate secretion. J Clin Invest 2009;119:2613–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mazcorro JF, Suchodolski JS, Jones KR et al. Effect of the proton pump inhibitor omeprazole on the gastrointestinal bacterial microbiota of healthy dogs. FEMS Microbiol Ecol 2012;80:624–36 [DOI] [PubMed] [Google Scholar]
- Geets J, Borremans B, Diels L et al. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J Microbiol Methods 2006;66:194–205 [DOI] [PubMed] [Google Scholar]
- Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Lett 1991;86:103–11 [Google Scholar]
- Gibson GR, Macfarlane GT, Cummings JH. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol 1988;65:103–11 [DOI] [PubMed] [Google Scholar]
- Gupta A, Kang S, Wagner J et al. Analysis of mucosal microbiota in inflammatory bowel disease using a custom phylogenetic microarray. Austin J Gastroenterol 2014;1:1020 [Google Scholar]
- Hakansson A, Molin G. Gut microbiota and inflammation. Nutrients 2011;3:637–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, Pope CE, Hayden HS et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis 2014;58:396–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwen RH, van der Doef HP, Sermet I et al. Defining DIOS and constipation in cystic fibrosis with a multicentre study on the incidence, characteristics, and treatment of DIOS. J Pediatr Gastr Nutr 2010;50:38–42 [DOI] [PubMed] [Google Scholar]
- Hradsky O, Ohem J, Mitrova K et al. Fecal calprotectin levels in children is more tightly associated with histological than with macroscopic endoscopy findings. Clin Lab 2014;60:1993–2000 [DOI] [PubMed] [Google Scholar]
- Huys G, Vanhoutte T, Vandamme P. Application of sequence-dependent electrophoresis fingerprinting in exploring biodiversity and population dynamics of human intestinal microbiota: what can be revealed?. Interdiscip Perspect Infect Dis 2008;2008:597603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 2010;5:e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007;1:56–66 [DOI] [PubMed] [Google Scholar]
- Jernberg C, Å Sullivan, Edlund C et al. Monitoring of antibiotic-induced alterations in the human intestinal microflora and detection of probiotic strains by use of terminal restriction fragment length polymorphism. Appl Environ Microb 2005;71:501–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M, Huys G, Cnockaert M et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 2011;60:631–7 [DOI] [PubMed] [Google Scholar]
- Kassinen A, Krogius-Kurikka L, Mäkivuokko H et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007;133:24–33 [DOI] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005;128:891–906 [DOI] [PubMed] [Google Scholar]
- Kondo R, Nedwell DB, Purdy KJ et al. Detection and enumeration of sulphate-reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol J 2004;21:145–57 [Google Scholar]
- Kumar PS, Griffen AL, Barton JA et al. New bacterial species associated with chronic periodontitis. J Dent Res 2003;82:338–44 [DOI] [PubMed] [Google Scholar]
- Li L, Somerset S. The clinical significance of the gut microbiota in cystic fibrosis and the potential for dietary therapies. Clin Nutr 2014;33:571–80 [DOI] [PubMed] [Google Scholar]
- Loubinoux J, Bronowicki JP, Pereira IAC et al. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol 2002;40:107–12 [DOI] [PubMed] [Google Scholar]
- Louis P, Duncan SH, McCrae SI et al. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 2004;186:2099–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Development of a semiquantitative degenerate real-time PCR-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microb 2007;73:2009–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luton PE, Wayne JM, Sharp RJ et al. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 2002;148:3521–30 [DOI] [PubMed] [Google Scholar]
- Lyra A, Rinttilä T, Nikkilä J et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol 2009;15:5936–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak BL, Larsen N, McCaughey MJ et al. The ribosomal database project. Nucleic Acids Res 1994;22:3485–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maukonen J, Satokari R, Mättö J et al. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol 2006;55:625–33 [DOI] [PubMed] [Google Scholar]
- Mihajlovski A, Doré J, Levenez F et al. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ Microbiol Rep 2010;2:272–80 [DOI] [PubMed] [Google Scholar]
- Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest 2014;124:4212–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem?. Front Microbiol 2016;7:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lin HC, McSweeney CS et al. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol 2010;1:363–95 [DOI] [PubMed] [Google Scholar]
- Newton DF, Cummings JH, Macfarlane S et al. Growth of a human intestinal Desulfovibrio desulfuricans in continuous cultures containing defined populations of saccharolytic and amino acid fermenting bacteria. J Appl Microbiol 1998;85:372–80 [DOI] [PubMed] [Google Scholar]
- Pitcher MCL. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 2000;46:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J et al. A human gut microbial gene catalog established by metagenomic sequencing. Nature 2010;464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale SW. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit Rev Biochem Mol 1991;26:261–300 [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014;38:996–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinttilä T, Kassinen A, Malinen E et al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 2004;97:1166–77 [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Buckling A, Kong W et al. Gut dysbiosis in cystic fibrosis. J Cyst Fibros 2012;11:454–5 [DOI] [PubMed] [Google Scholar]
- Schippa S, Iebba V, Santangelo F et al. Cystic fibrosis transmembrane conductance regulator (CFTR) allelic variants relate to shifts in faecal microbiota of cystic fibrosis patients. PLoS One 2013;8:e61176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Lin H. Hydrogen sulfide in physiology and diseases of the digestive tract. Microorganisms 2015;3:866–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skillman LC, Evans PN, Naylor GE et al. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 2004;10:277–85 [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. P Natl Acad Sci USA 2008;105:16731–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson AL, Mannik LA, Walsh S et al. Longitudinal trends in nutritional status and the relation between lung function and BMl in cystic fibrosis: a population-based cohort. Am J Clin Nutr 2013;97:872–7 [DOI] [PubMed] [Google Scholar]
- Suchodolski JS, Markel ME, Garcia-Mazcorro JF et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 2012;7:e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi H, Matsuki T, Nakazawa A et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol 2008;298:463–72 [DOI] [PubMed] [Google Scholar]
- Van den Abbeele P, Belzer C, Goossens M et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 2013;7:949–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen S, Lacroix C, Schwab C. Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol Lett 2015;362 10.1093/femsec/fnv092 [DOI] [PubMed] [Google Scholar]
- Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol 2004;42:1203–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Lee C, Kim J et al. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 2005;89:670–9 [DOI] [PubMed] [Google Scholar]
- Yu Z, Morrison M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Environ Microb 2004;70:4800–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration 2000;67:117–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.