Abstract
Children with autism spectrum disorders (ASD) often suffer gastrointestinal problems consistent with imbalances in the gut microbial population. Treatment with antibiotics or pro/prebiotics has been postulated to regulate microbiota and improve gut symptoms, but there is a lack of evidence for such approaches, especially for prebiotics. This study assessed the influence of a prebiotic galactooligosaccharide (B-GOS) on gut microbial ecology and metabolic function using faecal samples from autistic and non-autistic children in an in vitro gut model system. Bacteriology was analysed using flow cytometry combined with fluorescence in situ hybridization and metabolic activity by HPLC and 1H-NMR. Consistent with previous studies, the microbiota of children with ASD contained a higher number of Clostridium spp. and a lower number of bifidobacteria compared with non-autistic children. B-GOS administration significantly increased bifidobacterial populations in each compartment of the models, both with autistic and non-autistic-derived samples, and lactobacilli in the final vessel of non-autistic models. In addition, changes in other bacterial population have been seen in particular for Clostridium, Rosburia, Bacteroides, Atopobium, Faecalibacterium prausnitzii, Sutterella spp. and Veillonellaceae. Furthermore, the addition of B-GOS to the models significantly altered short-chain fatty acid production in both groups, and increased ethanol and lactate in autistic children.
Keywords: autism, gut microbiota, B-GOS, prebiotics, in vitro fermentation, SCFAs
B-GOS affects microbiota of autistic and non-autistic children.
INTRODUCTION
Autism typically develops in childhood, and it is considered as ‘a systemic spectrum disorder with multiple development trajectories with an incidence four times higher in males than in females’ (Grossi and Terruzzi 2014). In addition to behavioural traits, GI abnormalities such as diarrhoea, constipation, bloating and abdominal pain are common in autism and they seem to contribute to, and exacerbate, overall behaviour of children (irritability, sleeplessness, posturing) (Van De Sande, Van Buul and Brouns 2014). A crosstalk exists between the gut microbiota and central nervous system (CNS) mediated via a range of different chemical, immunological and signalling interactions that form part of the gut–brain axis. Several studies have demonstrated the role of the gut microbiota in neurodevelopment and mental health (Foster and McVey Neufeld 2013), and there is increasing evidence associating gut microbial dysbiosis with GI problems that might affect autistic children.
Bacteria such as Clostridium spp., Desulfovibrio spp. and Streptococcus spp. are dominant in the guts of children with ASD. Finegold et al. (2002) found nine unique species of clostridia in autistic children compared with controls. Song, Liu and Finegold (2004), using qPCR analysis, found higher levels of Clostridium bolteae and Clostridium clusters I and XI. Furthermore, Parracho et al. (2005), using FISH analysis, found greater number of species derived from the C. histolyticum group (Clostridium clusters I and II). Desulfovibrio group was found to be 10 times higher in the gut microbiota of autistic children compared with controls (Finegold et al. 2010; Finegold 2011).
High-throughput sequencing has been used in more recent studies to determine bacterial composition of faecal samples from autistic children. The genera Prevotella, Coprococcus and unclassified Veillonellaceae have been found in lower abundance in autistic individuals (Kang et al. 2013) with high genus Sutterella spp. (Williams et al. 2012; Wang et al.2013). In addition, Bifidobacterium species decreased in ASD, comparing with the non-autistic control (De Angelis et al. 2013).
Metabolic associations have also been identified with ASD and may be attributed to gut dysbiosis in autistic individuals. Abnormalities have been reported in tryptophan metabolism where higher amount of indole derivatives in the blood and higher levels of IAG (indolyl-acryloyl-glycine) in the urine of autistic children have been identified. Increased abundance of Clostridium spp. in the ASD-associated microbiota may contribute to these metabolic alterations as these organisms can metabolise tryptophan (Bingham 2003). Metabolomic studies also identified alterations in nicotinic acid metabolism (Yap et al. 2010) and amino acid deficiencies in autism with restricted diets, modified gut microbial population and GI symptoms being suggested as potential contributors (Ming et al. 2012).
Modulation of gut microbiota is an interesting potential strategy to reduce presence of harmful microorganisms and their metabolites that might be involved in negative stimulation of CNS and affect behaviour (Shaw, Kassen and Chaves 1995; Sandler et al. 2000). Treating GI disorders in ASD with antibiotics or pro/prebiotics has been postulated to regulate microbiota and improve gut symptoms, but the evidence is scarce, especially for prebiotics.
The bifidogenic properties of B-GOS (Bimuno®, Clasado Biosciences Ltd, Buckinghamshire, UK) have been investigated in vitro and in human intervention studies involving healthy volunteers, and conditions that have a purported microbial input such as IBS, travellers’ diarrhoea and obesity (Tzortzis et al. 2005; Depeint et al. 2008; Vulevic et al. 2008; Drakoularakou et al. 2009; Silk et al. 2009; Vulevic et al. 2013). Recently, B-GOS was also shown to reduce cortisol secretion and anxiety in healthy volunteers (Schmidt et al. 2015). Cortisol is a reliable marker of stress and hypothalamic pituitary adrenal axis activity. B-GOS supplementation lowered cortisol reactivity and modulated attention to emotional stimuli compared with a placebo group, supporting the hypothesis that gut microbiota might have a role in behavioural traits (Schmidt et al. 2015).
Our study aimed to assess the effects of B-GOS (65% GOS content) on gut microbial ecology and metabolic end products of microbial fermentation. We used in vitro, three-stage, continuous gut model systems, inoculated with faecal samples of autistic and non-autistic children, which simulated different physicochemical characteristics of the proximal, transverse and distal colons.
MATERIALS AND METHODS
Substrate
The B-GOS product was supplied by Clasado Biosciences Ltd. The mixture was in syrup format consisting of 65% (w/v) GOS, 10.1% (w/v) lactose, 22% (w/v) glucose and 1.8% (w/v) galactose.
Faecal inoculation
Faecal samples were obtained from three non-autistic children and three autistic child donors (male, aged 5–10 years old) who were free of any metabolic and gastrointestinal diseases, were not taking probiotic or prebiotic supplements and had not taken antibiotics 6 months before faecal sample donation. Autistic children had formal diagnosis of mild autism. None of the children followed any specific or restricted diet.
All parents were then provided written informed consent for use of their children's faeces in the study. This study was approved by The University of Reading research Ethics Committee (UREC 15/20). Faecal samples were placed in an anaerobic jar (AnaeroJarTM 2.5 L, Oxoid Ltd) including a gas-generating kit (AnaeroGenTM, Oxoid). An aliquot of 20 g of samples was diluted in 100 mL anaerobic PBS (0.1 mol/L phosphate buffer solution, pH 7.4, w/w) and homogenised (Stomacher 400, Seward, West Sussex, UK) for 2 min at 240 paddle beats per minute. Samples were added to anaerobic fermenters within 15 minutes of voiding.
Three-stage continuous culture gut model system
Physicochemical conditions in the colon were replicated in a continuous culture system, comprised of a cascade of three glass fermenters of increasing working volume connected in series. A small-scale version of the validated system described by Macfarlane, Macfarlane and Gibson (1998) was used in this study, with vessels (V) representing the proximal (V1, 80 mL, pH = 5.5), transverse (V2, 100 mL, pH = 6.2) and distal colon (V3, 120 mL, pH = 6.8). The systems were inoculated with 20% (wt:v) faecal homogenate from either non-autistic and autistic children volunteers in a growth medium (Macfarlane, Macfarlane and Gibson 1998). Following inoculation, the colonic model was run as a batch culture for 24 h in order to stabilise bacterial populations prior to the initiation of medium flow. After 24 h (T0), the medium flow was initiated and the system ran for at least 8 full volume turnovers to allow for steady state to be achieved (SS1). Short-chain fatty acid (SCFA) profiles (±5%) were assessed before starting B-GOS administration. Taking into account the operating volume (300 mL) and the retention time (48 h, flow rate 6.25 mL/h) of the colonic model system, a syrup containing GOS (2 g/daily, equivalent to 1 g of GOS) was added daily into V1. The syrup was added to the system for at least a further 8 volume turnovers upon which steady state 2 (SS2) was achieved. Aliquots of 4.5 mL were removed at SS1 and SS2.
SCFAs analysis by HPLC
The production of SCFAs in the fermentations was determined by HPLC (Merck, NJ) as previously described by Rodriguez-Colinas et al. (2013). Twenty microlitres of each sample was injected with a run time of 45 min. Peaks were integrated using Atlas Lab managing software (Thermo Lab Systems, Mainz, Germany). Quantification of the samples was obtained through calibration curves of lactic, acetic, propionic, butyric and formic acids in concentrations 12.5, 25, 50, 75 and 100 mM, respectively.
In vitro enumeration of bacterial population by FISH-FCM
Bacterial composition in the gut models was analysed for using fluorescence in situ hybridization combined with flow cytometry (FISH-FCM). Seven hundred and fifty microlitres of samples were centrifuged at 1136 × g for 5 min. Pellets were resuspended in 375 μL of filtered PBS (using a 0.22-μm PVDF membrane) and fixed in 1125 μL of 4% (v/v) paraformaldehyde. After 4 h of incubation at 4°C, samples were washed twice using 1 mL of PBS, resuspended in 600 μL PBS-ethanol (1:1, v/v) and stored at –20°C. Permeabilisation steps were performed using 30 μL of the fixed samples added to 500 μL PBS and centrifuged at 1136 × g for 3 min. Pellets were resuspended using 100 μL of filtered TE-FISH (Tris/HCl 1 M pH 8, EDTA 0.5 M pH 8, distilled H2O, 0.22 μm PVDF membrane) containing lysozyme (1 mg/mL of 50 000 U/mg protein) and incubated for 10 min at room temperature. Solutions containing the samples were then vortexed and centrifuged at 1136 × g for 3 min. Pellets were washed with 500 μL PBS and centrifuged (1136 × g, 3 min). Hybridisations were performed by resuspending the pellets in 150 μL of hybridisation buffer (5 M NaCl, 1 M Tris/HCl pH 8, 30% formamide, ddH2O, 10% SDS), vortexed and centrifuged (1136 × g, 3 min). Pellets were then resuspended in 1 mL of hybridisation buffer and 50 μL aliquoted into Eppendorf tubes. The probes used (Sigma Aldrich Ltd, Poole, Dorset, UK) are reported in Table 1 (Devereux et al. 1992; Wallner, Amann and Beisker 1993; Langendijk et al. 1995; Poulsen et al. 1995; Manz et al. 1996; Franks et al. 1998; Stoffel et al. 1998; Daims et al.1999; Harmsen et al. 1999, 2000, 2002; Hold et al.2003; Lay et al. 2005; Walker et al. 2005; Kong et al. 2012). NON EUB338 and EUB338 I-II-III linked at their 5΄ end either to Alexa488 or Alexa647. Group-specific probes were linked with Alexa647 at their 5΄ end. Four microlitres of each probe and 4 μL of Eub338 I-II-III (linked to Alexa488) were added to the working solution and incubated overnight at 35°C in a heating block. After 12 h of incubation, an aliquot of 150 μL hybridisation buffer was added to the working solution, vortexed and centrifuged (1136 × g, 3 min). One hundred and fifty microlitres of supernatant were removed from each sample and the remaining volume was centrifuged (1136 × g, 3 min). The pellets were washed with 200 μL of washing buffer (5 M NaCl, 1 M Tris/HCl pH 8, 0.5 M EDTA pH 8, ddH2O, 10% SDS), homogenised by vortexing and incubated for 20 min at 37°C in a heating block. Afterwards the samples were centrifuged (1136 × g, 3 min) and supernatants were removed. Negative control samples (no probes added) were screened by FCM to detect background before the probe samples were resuspended in an appropriate amount of PBS. Samples were stored at 4°C until determinations. Numbers of specific and total bacteria were determined taking into account dilution factor, calculated from different volumes used in samples preparation steps, and events/μL obtained from NON EUB338 and EUB338 I-II-III probes analysed by FCM.
Table 1.
Oligonucleotide probes used in this study for FISH-FCM analysis of bacterial populations. +: These probes are used together in equimolar concentration of 50 ng/μL.
| Probe name | Sequence (5΄ TO 3΄) | Target group | Reference |
|---|---|---|---|
| Non Eub | ACTCCTACGGGAGGCAGC | Wallner, Amann and Beisker (1993) | |
| Eub338 I + | GCT GCC TCC CGT AGG AGT | Most bacteria | Daims et al. (1999) |
| Eub338 II + | GCA GCC ACC CGT AGG TGT | Planctomycetales | Daims et al. (1999) |
| Eub338 III + | GCT GCC ACC CGT AGG TGT | Verrucomicrobiales | Daims et al. (1999) |
| Bif164 | CAT CCG GCA TTA CCA CCC | Most Bifidobacterium spp. and Parascardovia denticolens | Langendijk et al. (1995) |
| Lab158 | GGTATTAGCAYCTGTTTCCA | Most Lactobacillus, Leuconostoc and Weissella spp.; Lactococcus lactis; all Vagococcus, Enterococcus, Melisococcus, Tetragenococcus, Catellicoccus, Pediococcus and Paralactobacillus spp, | Harmsen et al. (1999) |
| Bac303 | CCA ATG TGG GGG ACC TT | Most Bacteroidaceae and Prevotellaceae, some Porphyromonadaceae | Manz et al. (1996) |
| Clit135 | GTTATCCGTGTGTACAGGG | Some of the Clostridium lituseburense group (Clostridium cluster XI) | Manz et al. (1996) |
| Erec482 | GCT TCT TAG TCA RGT ACCG | Most of the Clostridium coccoides-Eubacterium rectale group (Clostridium clusters XIVa and XIVb) |
Manz et al. (1996) |
| Chis150 | TTATGCGGTATTAATCTYCCTTT | Most of the Clostridium histolyticum group (Clostridium clusters I and II) | Franks et al. (1998) |
| Rrec584 | TCA GAC TTG CCG YAC CGC | Roseburia subcluster | Franks et al. (1998) |
| Prop853 | ATT GCG TTA ACT CCG GCAC | Clostridial cluster IX | Walker et al. (2005) |
| Ato291 | GGT CGG TCT CTC AAC CC | Atopobium, Colinsella, Olsenella and Eggerthella spp.; Cryptobacterium curtum; Mycoplasma equigenitalium and Mycoplasma elephantis | Harmsen et al. (2000) |
| Fprau655 | CGCCTACCTCTGCACTAC | Faecalibacterium prausnitzii and related sequences | Hold et al. (2003) |
| DSV687 | TAC GGA TTT CAC TCC T | Most Desulfovibrionales (excluding Lawsonia) and many Desulfuromonales | Devereux et al. (1992) |
| EC1531 | CACCGTAGTGCCTCGTCATCA | Escherichia coli BJ4 | Poulsen et al. (1995) |
| Rbro730 + | TAAAGCCCAGYAGGCCGC | Clostridium sporosphaeroides, Ruminococcus bromii, Clostridium leptum | Harmsen et al. (2002); Lay et al. (2005) |
| Rfla729 + | AAA GCC CAG TAA GCC GCC | Ruminococcus albus, R. flavefaciens | Harmsen et al. (2002); Lay et al. (2005) |
| SUBU1237 | CCC TCT GTT CCG ACC ATT | Burkholderia spp., Sutterella spp. | Stoffels et al. (1998) |
| Vei723 | ACA CAG TCC AGA AAG GCG | Veillonellaceae | Kong et al. (2012) |
Metabolic analysis by 1H-NMR
Three consecutive days of the three biological replicates for each group (autistic and non-autistic) of all time points (before and after treatment) were analysed by 1H-NMR (n = 27, each group). Fermentation supernatants were defrosted, vortexed and centrifuged at 599 × g for 5 min. The supernatants were filtered using 0.22 μm low protein binding Durapore polyvianylidene fluoride (PVDF) membranes (Millex; EMD Millipore, Billerica, MA, USA) and 400 μL transferred into fresh Eppendorf tubes. Filtered samples were combined with 200 μL of phosphate buffer (0.2 M (pH 7.4) in D2O plus 0.001% TSP), mixed by vortexing, centrifuged at 1136 × g for 10 min and then 550 μL was transferred into 5 mm NMR tubes for analysis. All NMR spectra were acquired on a Bruker Avance DRX 500 MHz NMR spectrometer (Bruker Biopsin, Rheinstetten, Germany) operating at 500 MHz. They were acquired using a standard 1D pulse sequence [recycle delay (RD)-90°-t1-90°-tm-90°-acquire free induction decay (FID)] with water suppression applied during RD of 2 s, a mixing time Tm of 100 ms and a 90 pulse set at 7.70 μs. For each spectrum, a total of 128 scans were accumulated into 64 k data points with a spectral width of 12 001 ppm. The FIDs were multiplied by an exponential function corresponding to 0.3 Hz line broadening.
Data preprocessing and analysis
All spectra were manually phased, baseline corrected and calibrated to the chemical shift of TSP (3-(trimethylsilyl)-[2,2,3,3,−2H4]-propionic acid, δ 0.00). Spectra were digitised using an in-house MATLAB (version R2014a, The Mathworks, Inc.; Natwick, MA) script. The spectral region containing the water resonance was removed to minimise distortions in the baseline arising from imperfect water saturation. Median fold normalization was performed for both groups: non-autistic and autistic children. Before and after administration of B-GOS, principal components analysis (PCA) using mean-centred data was applied. Orthogonal projection to latent structure discriminant analysis (OPLS-DA) models was constructed using unit variance scaling for pairwise comparisons of the different experimental groups and time points. Correlation coefficients plots were generated from the model outputs by back scaling transformation to display the contribution of each variable (metabolites) to sample classification (e.g. before and after treatment). Colour represents the significance of correlation (R2) for each metabolite to class membership. Predictive strength (Q2Y) of the models was obtained using a 7-fold cross-validation method and these were validated using permutation testing (number of permutations = 10 000).
Statistical analysis
Data from HPLC and FMC-FISH analyses were analysed using paired t-test in order to assess significance of results, comparing the two time points SS1 and SS2, before and after treatment, respectively. Statistical significance was at P < 0.05 for all analyses. Analyses were performed using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
RESULTS
Bacterial enumeration
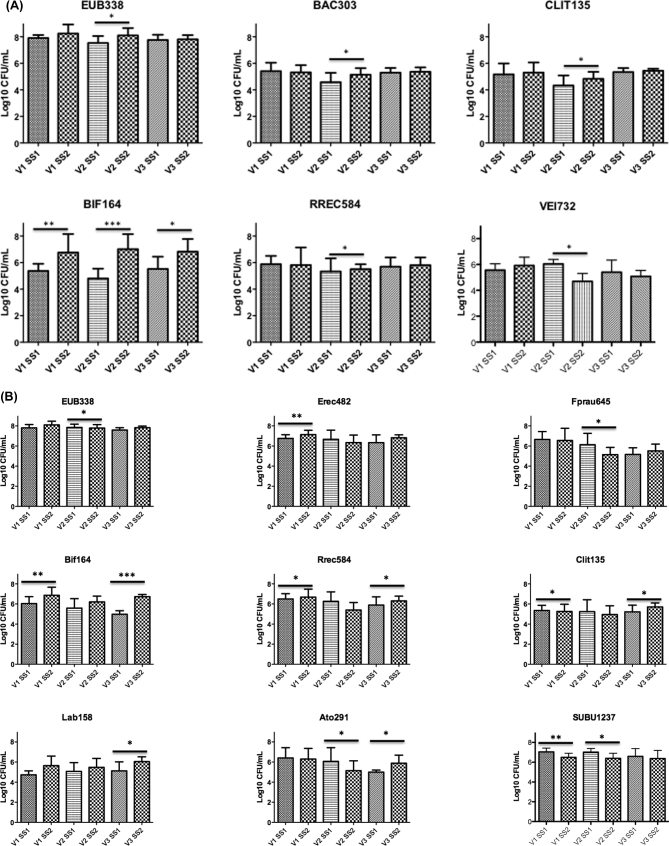
Changes in bacterial compositions in gut model systems are reported in Fig. 1. The data showed lower numbers of bifidobacteria in ASD models compared with non-autistic ones. Significant increases in the Bifidobacterium spp., following the addition of B-GOS to models containing both autistic and non-autistic samples, were seen. In autistic models, a significant increase of bifidobacteria occurred from 5.32 to 7.27 log10 cells/mL (P < 0.01), from 4.81 to 6.79 log10 CFU/mL (P < 0.001) and from 5.57 to 6.83 log log10 cells/mL (P < 0.05), in V1, V2 and V3, respectively. A slight but significant increase in Clostridium cluster XI in V2 for autistic children was also found, as well as significant decrease in V2 in Veillonellaceae group from 6.06 to 5 log10 CFU/mL (P < 0.05). In non-autistic models, there was a significant increase in numbers of bifidobacteria in V1, from 5.83 to 7.16 log log10 cells/mL (P < 0.01), and in V3, from 4.97 to 6.73 log10 cells/mL (P < 0.001) and in lactic acid bacteria (Lab158) in V3 from 5.13 to 6.01 log10 cells/mL (P < 0.05). Additionally, B-GOS slightly increased Roseburia spp. in V1 and V3 (P < 0.05) and reduced Atopobium spp. from 6.06 to 5.28 log10 cells/mL and Faecalibacterium prausnitzii from 6.78 to 5.27 log10 cells/mL (P < 0.05 for both) in the second vessel, while increasing Atopobium spp. from 5 to 5.92 log10 cells/mL (P < 0.05) in the third vessel of non-autistic models. In these models, numbers of Clostridium coccoides–Eubacterium rectale were also increased from 6.76 to 7.08 log10 cells/mL (P < 0.01) in V1 and Sutterella spp. significantly decreased in V1 from 7.05 to 6.49 (P < 0.01) and V2 from 7.02 to 6.37 log10 CFU/mL (P < 0.05) after B-GOS administration. There was a general trend to increase all other bacterial groups analysed in all vessels but this was not significant. Exceptions were seen for Bacteroides (V1), Clostridial cluster IX (V1), F. prausnitzii (V1), Escherichia coli (V3), Ruminococcus spp., Clostridium leptum (V2), Sutterella spp. and Veillonellaceae (all vessels) in autistic models, and for Clostridium coccoides–Eubacterium rectale (V2), Atopobium spp. (V1), Clostridial cluster IX (V2), Clostridium cluster XI (V1, V2), E. coli (V2), Sutterella spp. and Veillonellaceae (all vessels) in non-autistic models that slightly decreased.
Figure 1.
Bacterial groups detected by FISH-FCM (Log10 CFU/mL) in culture broth recovered from each vessel (V1, V2 and V3) of a colonic model before (SS1) and after (SS2) the daily administration of B-GOS (2 g/d, equivalent to 1 g GOS). Significant difference after the treatment: * P < 0.05; **P < 0.01; ***P < 0.001. Probes: total bacteria (Eub338I-II-III), Bifidobacterium spp. (Bif164), Lactobacillus spp. (Lab158), most Bacteroidaceae and Prevotellaceae (Bac303), Clostridium coccoides–Eubacterium rectale group (Erec482), Roseburia subcluster (Rrec584), F. prausnitzii (Fprau655), Clostridium cluster XI (Clit135), Sutterella spp. (SUBU1237), Veillonellaceae (VEI732), Atopobium spp. (Ato291). (A) Autistic children; (B) non-autistic children.
SCFAs production
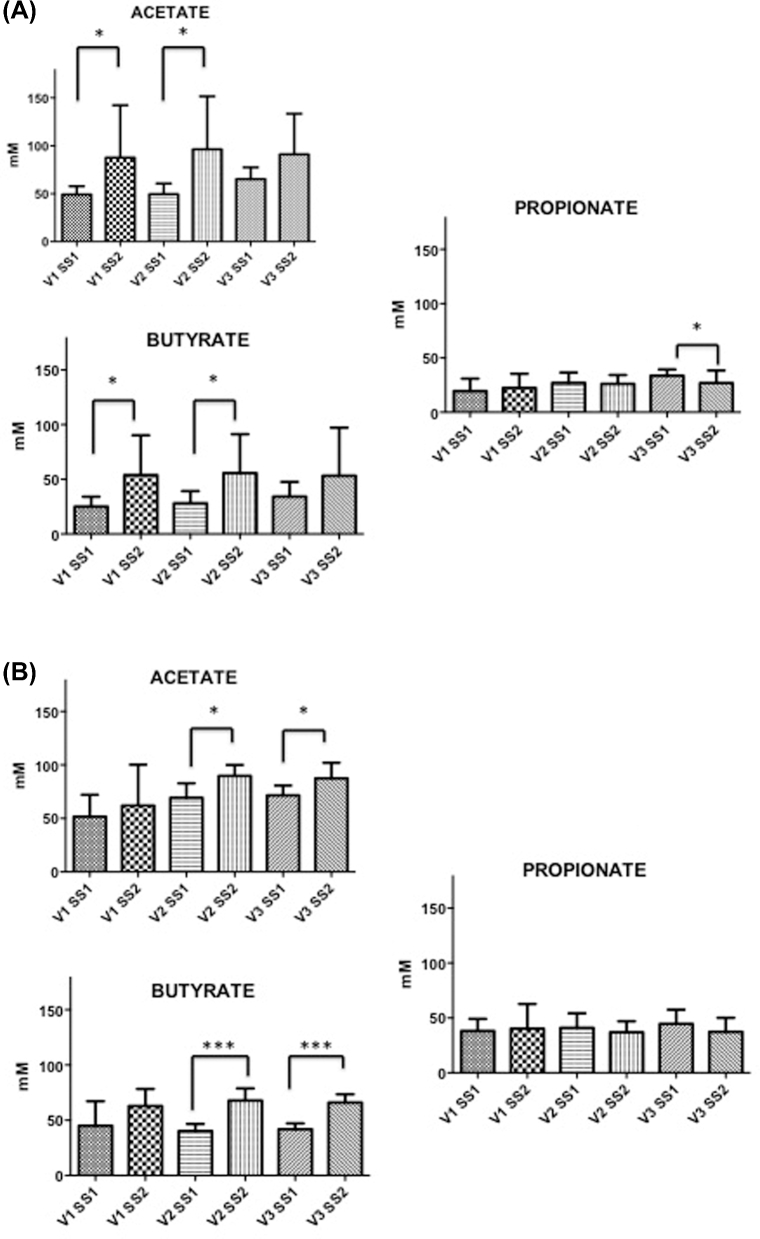
SCFA concentrations are shown in Fig. 2. Our data show a lower concentration of butyrate and propionate in autistic models, compared with non-autistic models, but no differences in acetate before adding B-GOS into the system. After the administration of B-GOS, acetate and butyrate were the main end products of microbial fermentation. Supplementation of B-GOS to gut models inoculated with faecal samples from autistic children led to a significant increase of acetate and butyrate in V1 and V2, simulating the proximal and transverse colons (P < 0.05), respectively, while concentration of propionate was decreased (P < 0.05) in V3 mimicking distal colon. In models simulating the colon of non-autistic children, the fermentation of B-GOS mediated significant production of acetate (P < 0.05) and butyrate (P < 0.001) in V2 and V3, simulating the transverse and distal colon, respectively. There was no effect on propionate.
Figure 2.
HPLC analysis. Acetate, propionate and butyrate concentrations in culture broths recovered from vessels (V1, V2 and V3) of in vitro gut model systems before (SS1) and after (SS2) administration of B-GOS (1 g/daily GOS). Results are reported as means (mM) of the data (n = 3): (A) autistic children and (B) non-autistic children. Significant difference after the treatment: * P < 0.05; ***P < 0.001.
1H-NMR spectroscopic profiles
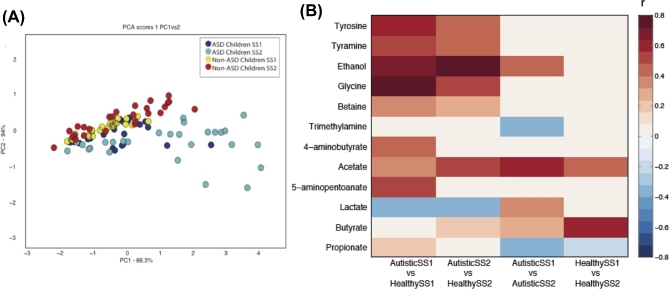
PCA was performed on mean-centred data to summarise variance with the dataset. The scores plot (PC1 versus PC2) shown in Fig. 3A showed separation between autistic and non-autistic models after treatment, indicating that B-GOS supplementation contributed to the largest source of variance in the metabolic data. Comparison of the spectra profiles from gut models before and after treatment identified that a number of metabolites changed following B-GOS supplementation to characterise the metabolic variation associated with ASD, B-GOS supplementation and differences in microbial response to B-GOS between the ASD and non-ASD microbiota. The results of these analyses are summarised in Fig. 3B. A significant OPLS-DA model was obtained comparing the metabolic profiles of the autistic and non-autistic models at baseline (Q2Y = 0.07; P < 0.05; Fig. SC-I, Supporting Information). Supernatants from the autistic models contained greater amounts of ethanol, glycine, tyrosine, tyramine, 5-aminopentoanate, acetate, 4-aminobutyrate and betaine, and lower amounts of butyrate compared with the non-autistic models. B-GOS supplementation was found to modulate the metabolic profile of the autistic models (Q2Y = 0.08; P < 0.05) increasing ethanol, lactate, acetate and butyrate and decreasing propionate and trimethylamine (Fig. SB-I, Supporting Information). Increased butyrate and acetate production was also observed in the non-autistic models following the addition of BGOS (Q2Y = 0.12; P < 0.01; Fig. SB-II, Supporting Information). Comparing the metabolic profiles of the autistic and non-autistic models after B-GOS feeding (Q2Y = 0.17; P < 0.01) revealed that some of the metabolic variation was reduced (Fig. SC-II, Supporting Information). There was no longer variation in 4-aminobutyrate between the models; however, the difference in ethanol and acetate between autistic and non-autistic models was increased being higher in the autistic models.
Figure 3.
1H-NMR data analysis. (A) PCA score plot shows a separation between models inoculated with stool samples of non-ASD and ASD children after administration of B-GOS. Dark and light blue dots represent replicates of samples from gut models inoculated with faecal samples of autistic children, before (SS1) and after (SS2) treatment, respectively. Yellow and red dots represent replicates of samples from gut models inoculated with faecal samples of non-autistic children, before (SS1) and after (SS2) treatment, respectively. (B) Correlation coefficients indicating the associations of identified metabolites with autism and their alteration upon B-GOS administration. SS1: before treatment; SS2: after treatment. White cells represent no significant correlations.
DISCUSSION
Recent studies have focused on the effect of pre/probiotics on the gut–brain axis (Liu, Cao and Zhang 2015). This study investigated the influence of B-GOS on a small scale, in vitro, gut model system inoculated with faeces from autistic and non-autistic children. The results showed a positive modulation of bacterial populations, using an automated FISH method combined with FCM. We also assessed metabolic profiles and key metabolites in both test groups.
Lower concentrations of SCFAs have previously been found in children with ASD by Adams et al. suggesting a reduced fermentation capacity by the ASD microbiota. It was hypothesised that this was due to a compromised microbiota characterised by a lower number of bifidobacteria, consistent with microbial signatures observed here (Adams et al. 2011). Concomitant with these population changes, functional alterations were also observed in both autistic and non-autistic models with acetate and butyrate, the main end products of microbial fermentation, being increased.
Recent studies have focussed on SCFAs and their effect on the CNS. These fermentation products can cross the blood–brain barrier and might influence early brain development. The synthesis of neuroactive compounds such as dopamine and serotonin can be modulated by SCFA and they are able to produce reversible psychological and physiological changes in rats similar to those found in ASDs (Wang et al. 2011). Experimental evidence using intraventricular infusion in rats indicates that propionic acid can produce brain and behavioural changes similar to ASD (MacFabe et al. 2008).
Recent ASD studies have shown increase in numbers of Sutterella spp. and decrease in Veillonellaceae group. In this study, the results did not show any significant differences between ASD and non-ASD group. However, a general decrease in those bacterial groups after treatment was highlighted, suggesting that B-GOS administration might have an impact on the growth of these ASD-associated bacteria.
Following B-GOS feeding, the microbiota of autistic children produced greater amount of ethanol and lactate while the amount of amino acids and the SCFA propionate, present in the model, was reduced. These metabolic alterations were not observed when the faecal microbiota of non-autistic individuals were fed B-GOS. In a healthy colon, lactate production is generally low due to its conversion to other organic acids by many bacteria and because lactate can be used as a substrate for dissimilation of sulphate by some bacteria (e.g. Desulfovibrio spp.) (Fite et al. 2004; Marquet et al. 2009; Flint et al. 2014). In children with ASD, the presence of lactate is interesting because its accumulation has been associated with neurological problems, in particular studies show the effect of lactate infusions on anxiety and panic disorders (Cowley et al. 1987; Dillon et al.1987). Cowley and colleagues in their findings showed that lactate infusion in patients suffering from panic disorder provokes higher panic symptoms reaction compared with controls (Dillon et al. 1987). Dillon et al. have showed similar results in in vivo, where panic and anxiety reaction has been measured using Acute Panic Inventory scores. After lactate infusions, the scores were much higher in patients with panic and anxiety disorders compared with normal controls (Cowley et al. 1987).
The lysine degradation product, 5-aminopentanoic acid, was also higher in the autistic compared with the non-autistic models. This metabolite can be produced both endogenously and through the bacterial catabolism of lysine. It is believed to act as a methylene homologue of γ-aminobutyric acid (GABA) and functions as a weak GABA agonist (Callery and Geelhaar 1985). Interestingly, GABA was also higher in the autistic models compared with the non-autistic models pre-treatment but these differences were not present following B-GOS treatment. Certain bacteria, such as lactobacilli, are able to produce molecules that act as neurotransmitters and directly affect the brain (Wall et al. 2014). In our results, its reduction might be due to changes in gut microbiota composition.
Ethanol was found in higher amount in ASD children comparing with non-ASD children. The vast majority of bacteria form ethanol from acetyl-CoA and the glycolytic pathway (Macfarlane and Macfarlane 2003). Microorganisms are able to oxidase ethanol and the impact of bacterial overgrowth on ethanol production has previously been studied (Baraona et al. 1986). Metabolism of ethanol can lead to the production of toxic end products such as acetaldehyde, which may affect the gastrointestinal mucosa. The role of acetaldehyde in ASD has been recently evaluated in particular for its role in oxidative stress and DNA damage. Under healthy conditions, ethanol is converted into acetic acid in the liver by a two-step process involving alcohol dehydrogenase and aldehyde dehydrogenase (ALDH). Mutation of the ALDH gene has been shown to increase the accumulation of acetaldehyde and result in cancers within different regions of the gastrointestinal tract and Alzheimer's disease (Jurnak 2015). The potential role of this toxic compound in neurological disorders, including autism, warrants further exploration.
CONCLUSIONS
This in vitro study showed promising and positive results in that supplementing the microbiota of children with ASD with 65%B-GOS may manipulate the gut bacterial population and alter metabolic activity towards a configuration that might represent a health benefit to the host. However, further work will be required to assess such changes in an in vivo human intervention study.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
AUTHORS' CONTRIBUTIONS
RG carried out the experiments and drafted the manuscript. DC helped in experimental work. JRS assisted with NMR analyses. GRG and AC were involved in designing and coordination of the study and revising the manuscript critically for important intellectual content. JV and GT are employed by Clasado Biosciences Ltd, who provided the B-GOS product, marketed as Bimuno(R), used within this research. There are no patents, products in development or other marketed products to declare. This does not alter the authors adherence to all the FEMS policies on sharing data and materials. All the authors reviewed the final version of the manuscript. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest. None declared.
REFERENCES
- Adams JB, Johansen LJ, Powell LD et al. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol 2011;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraona E, Julkunen R, Tannenbaum L et al. Role of intestinal bacterial overgrowth in ethanol production and metabolism in rats. Gastroenterology 1986;90:103–10 [DOI] [PubMed] [Google Scholar]
- Bingham M. Functional food: dietary intervention strategies in autistic spectrum disorders. Gibson RG. Food Science & Technology Bulletin - Funcional Food. Reading:IFIS, 2003, 1–11 [Google Scholar]
- Callery PS, Geelhaar LA. 1-Piperideine as an in vivo precursor of the gamma aminobutyric acid homologue 5-aminopentanoic acid. J Neurochemistry 1985;45:946–8 [DOI] [PubMed] [Google Scholar]
- Cowley DS, Hyde TS, Dager SR et al. Lactate infusions: the role of baseline anxiety. Psychiat Res 1987;21:169–79 [DOI] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R et al. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 1999;22:434–44 [DOI] [PubMed] [Google Scholar]
- De Angelis M, Piccolo M, Vannini L et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One 2013;8:e76993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depeint F, Tzortzis G, Vulevic J et al. Prebiotic evaluation of a novel galactooligosaccharide mixture produced by the enzymatic activity of Bifidobacterium bifidum NCIMB 41171, in healthy humans: a randomized, double-blind, crossover, placebo-controlled intervention study. Am J Clin Nutr 2008;87:785–91 [DOI] [PubMed] [Google Scholar]
- Devereux R, Kane MD, Winfrey J et al. Genus- and group-specific hybridisation probes for determinative and environmental studies of sulphate-reducing bacteria. Syst Appl Microbiol 1992;15:601–9 [Google Scholar]
- Dillon D, Gorman D, Liebowitz M et al. Measurement of lactate-induced panic and anxiety. Psychiat Res 1987;20:97–105 [DOI] [PubMed] [Google Scholar]
- Drakoularakou A, Tzortzis G, Rastall RA et al. A double-blind, placebo-controlled, randomized human study assessing the capacity of a novel galacto-oligosaccharide mixture in reducing travellers’ diarrhoea. Eur J Clin Nutr 2009;64:146–52 [DOI] [PubMed] [Google Scholar]
- Finegold SM. Desulfovibrio species are potentially important in regressive autism. Med Hypotheses 2011;77:270–4 [DOI] [PubMed] [Google Scholar]
- Finegold SM, Dowd SE, Gontcharova V et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010;16:444–53 [DOI] [PubMed] [Google Scholar]
- Finegold SM, Molitoris D, Song Y et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis 2002;35:S6–S16 [DOI] [PubMed] [Google Scholar]
- Fite A, Macfarlane GT, Cummings JH et al. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 2004;53:523–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP et al. Links between diet, gut microbiota composition and gut metabolism. P Nutr Soc 2014;74:13–22 [DOI] [PubMed] [Google Scholar]
- Foster JA, McVey Neufeld KA. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci 2013;36:305–12 [DOI] [PubMed] [Google Scholar]
- Franks AH, Harmsen HJM, Gerwin C et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microb 1998;64:3336–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi E, Terruzzi V. The role of intestinal dysbiosis in the pathogenesis of autism: Minireview. Int J Microbiol Adv Immunol 2014;2:41–44 [Google Scholar]
- Harmsen HJM, Elfferich P, Schut F et al. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb Ecol Health D 1999;11:3–12 [Google Scholar]
- Harmsen HJM, Raangs GC, He T et al. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microb 2002;68:2982–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJM, Wildeboer-Veloo AM, Grijpstra J et al. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microb 2000;66:4523–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold GL, Schwiertz A, Aminov RI et al. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microb 2003;69:4320–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurnak F. The pivotal role of aldehyde toxicity in autism spectrum disorder: the therapeutic potential of micronutrient supplementation. Nutr Metab Insights 2015;8:57–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Park JG, Ilhan ZE et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One 2013;8:e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Xia Y, Seviour R et al. In situ identification of carboxymethyl cellulose-digesting bacteria in the rumen of cattle fed alfalfa or triticale. FEMS Microbiol Ecol 2012;80:159–67 [DOI] [PubMed] [Google Scholar]
- Langendijk PS, Schut F, Jansen GJ et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microb 1995;61:3069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay C, Sutren M, Rochet V et al. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol 2005;7:933–46 [DOI] [PubMed] [Google Scholar]
- Liu X, Cao S, Zhang X. Modulation of gut microbiota–brain axis by probiotics, prebiotics, and diet. J Agr Food Chem 2015;63:7885–95 [DOI] [PubMed] [Google Scholar]
- MacFabe DF, Rodríguez-Capote K, Hoffman JE et al. A novel rodent model of autism: Intraventricular infusions of propionic acid increase locomotor activity and induce neuroinflammation and oxidative stress in discrete regions of adult rat brain. Am J Biochem Biotechnol 2008;4:146–66 [Google Scholar]
- Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 1998;35:180–7 [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc 2003;62:67–72 [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W et al. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 1996;142:1097–106 [DOI] [PubMed] [Google Scholar]
- Marquet P, Duncan SH, Chassard C et al. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett 2009;299:128–34 [DOI] [PubMed] [Google Scholar]
- Ming X, Stein TP, Barnes V et al. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res 2012;11:5856–62 [DOI] [PubMed] [Google Scholar]
- Parracho HMRT, Bingham MO, Gibson GR et al. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol 2005;54:987–91 [DOI] [PubMed] [Google Scholar]
- Poulsen LK, Licht TR, Rang C et al. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol 1995;177:5840–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Colinas B, Kolida S, Baran M et al. Analysis of fermentation selectivity of purified galacto-oligosaccharides by in vitro human faecal fermentation. Appl Microbiol Biot 2013;97:5743–52 [DOI] [PubMed] [Google Scholar]
- Sandler RH, Finegold SM, Bolte ER et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol 2000;15:429–35 [DOI] [PubMed] [Google Scholar]
- Schmidt K, Cowen PJ, Harmer CJ et al. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology 2015;232:1793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W, Kassen E, Chaves E. Increased urinary excretion of analogs of Krebs cycle metabolites and arabinose in two brothers with autistic features. Clin Chem 1995;41:1094–104 [PubMed] [Google Scholar]
- Silk DBA, Davist A, Vulevic J et al. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharm Ther 2009;29:508–18 [DOI] [PubMed] [Google Scholar]
- Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microb 2004;70:6459–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffels M, Amann R, Ludwig W et al. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl Environ Microb 1998;64:930–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzortzis G, Goulas AK, Gee JM et al. A novel galactooligosaccharide mixture increases the bifidobacterial population numbers in a continuous in vitro fermentation system and in the proximal colonic contents of pigs in vivo. J Nutr 2005;135:1726–31 [DOI] [PubMed] [Google Scholar]
- Van De Sande MMH, Van Buul VJ, Brouns FJPH. Autism and nutrition: the role of the gut-brain axis. Nutr Res Rev 2014;27:199–214 [DOI] [PubMed] [Google Scholar]
- Vulevic J, Drakoularakou A, Yaqoob P et al. Modulation of the fecal microflora profile and immune function by a novel trans-galactooligosaccharide mixture ( B-GOS ) in healthy elderly volunteers. Am J Clin Nutr 2008;88:1438–46 [DOI] [PubMed] [Google Scholar]
- Vulevic J, Juric A, Tzortzis G et al. A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr 2013;143:324–31 [DOI] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, Mcwilliam EC et al. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microb 2005;71:3692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall R, Cryan JF, Ross RP et al. Bacterial neuroactive compounds produced by psychobiotics. Mark Lyte. Microbial Endocrynology: The Microbiota-Gut-Brain Axis in Health and Disease. New York:Springer, 2014, 221–39 [DOI] [PubMed] [Google Scholar]
- Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993;14:136–43 [DOI] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ et al. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microb 2011;77:6718–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ et al. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism 2013;4:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Parekh T et al. Application of novel PCR-based methods for detection, quantitation and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012;3:e00261–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap IKS, Angley M, Veselkov KA et al. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J Proteome Res 2010;9:2996–3004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.