Abstract
Currently, drug discovery approaches commonly assume a monotonic dose-response relationship. However, the assumption of monotonicity is increasingly being challenged. Here we show that for two simple interacting linear signaling pathways that carry two different signals with different physiological responses, a non-monotonic input-output relation can arise with simple network topologies including coherent and incoherent feed-forward loops. We show that non-monotonicity of the response functions has severe implications for pharmacological treatment. Fundamental constraints are imposed on the effectiveness and toxicity of any drug independent of its chemical nature and selectivity due to the specific network structure.
In pharmacology and toxicology the effect of a drug is typically described by a monotonic (linear or non-linear) dose-response curve1. For monotonic dose-response relations, the effectiveness reaches its maximum at a certain dose, and increasing the dose beyond this point does not (significantly) increase the effectiveness further. At this dose-saturation point, the maximum effectiveness depends on the chemical nature of the drug with respect to the target choice. Assuming the most appropriate target for drug treatment is chosen, improvements of the effectiveness of the treatment can be explored by focusing on the selectivity of the drug for the target. Indeed, a long-standing dogma in pharmacology is that selectivity implies effectiveness and safety. Therefore, in order to maximize the effectiveness (while minimizing side effects), rational drug design traditionally has been focused on the discovery of maximally selective and potent drugs2. Significant investments are made by pharmaceutical industry in the synthesis and screening of a large number of potential drugs, aiming for higher effectiveness and lower toxicity.
Structural analysis of biochemical networks helps in target selection thereby improving effectiveness and toxicity. The choice of the target is an important determinant of the resulting effectiveness and toxicity. A significant step forward in rationalizing the choice of drug targets was realized following advances in molecular biology and network theory3,4,5,6. In recent years, many components of signaling pathways and their interactions are discovered and represented using detailed knowledge of the molecular connections within the signaling network7,8. These developments allow for selecting targets on a stronger rational basis. For example, bridging nodes, that connect modular subregions of the signaling network, are promising drug targets from the standpoints of effectiveness and side effects, since their disruption would specifically prevent information flow between network modules of interest (high effectiveness) while it does not lead to any global change in the network (low toxicity)9. In the case of antibiotics and anti-cancer drugs, (network) hub nodes are considered interesting targets, and indeed for commercialized antibiotics and anti cancer drugs, the average number of interaction partners for protein targets are 4 and 8 respectively10.
A key question is whether network dynamics and topology reveal constraints on the form of the pharmacological response and provide insights into toxicity and effectiveness11,12,13,14. If the network analysis could provide additional insight into the network dynamics, on the form of the dose-response relation, on fundamental limits of achievable effectiveness and possible toxicity, and on how to administer the designed drugs, it would significantly reduce the cost of the drug development. How the form of the dose-response relation depends on the network topology and dynamics has remained to be systematically explained. One fundamental characteristics of the dose-response relation is its (non)monotonicity. While a monotonic form has been typically assumed in pharmacology, there is growing evidence that many bio-molecular pathways have non-monotonic dose-response relationships15,16,17,18,19 with potential functional significance. An example of its use is transmitting different biochemical signals through one and the same pathway, and yet respond to them specifically—a phenomenon that is called multiplexing in telecommunication and computer networks20.
To address this fundamental question, it is instructive to consider small signaling networks with a specific topology, investigate the pharmacological implications of such a network topology, and evaluate the advantages and limitations of the selective (and non-selective) pharmacology approaches. A simple, yet general and dynamically rich example would entail two linear signaling pathways with inter-pathway interactions. Canonical signaling pathways in cells are known to interact with each other via direct regulation or via their shared components (cross-talk)21. Large-scale surveys of inter-pathway interactions, revealed extensive crosstalk between pathways22,23. Cross-talk between signaling pathways results in non-linearity and dynamical monotonicity in response to stimuli due to mechanisms that are not well understood23.
These, and other topological interactions, though important24, are not the only crucial ingredient in the functioning of signaling networks. Indeed, the topological structure by itself not necessarily dictates the dynamics of the network25; the dynamical behavior is determined by the kinetic parameters of the network as well. Moreover, cells are able to multiplex signals along pathways. For example, the highly conserved MAPK (Mitogen Activated Protein Kinase) pathway is predicted to be one such pathway with multiplexing capabilities26. The pathway includes an incoherent feed-forward loop involving Ras-Raf-Akt. The design of drugs that target components of a MAPK pathway is an area of intense research20,27,28,29.
In this article we take two linear signaling pathways, allow for inter-pathway interactions (both shared components and biochemical interactions) and determine the input-output relations for many different topologies. Then, for each topology we look for interactions and dynamic parameters that generate a non-monotonic input-output relation, providing mechanistic insight into the origin of the non-monotonicity. Next, to study the effect of non-monotonicity on effectiveness and toxicity of pharmacological approaches we study two simple examples of two linear pathways, with either direct interaction and/or a shared component. We study a network with two inputs and two outputs where one pathway directly regulate the other pathway and study the occurrence of non-monotonicity and its effect on drug treatment in detail. In the remainder we describe the fundamental constraints, due to the non-monotonicity of the dose-response relationship, on the effectiveness and toxicity depending on the target choice of each network. We find that therapeutic windows for certain therapies are narrow and the preferred concentration range might occur after a broad toxic regime.
Non-monotonicity in simple networks
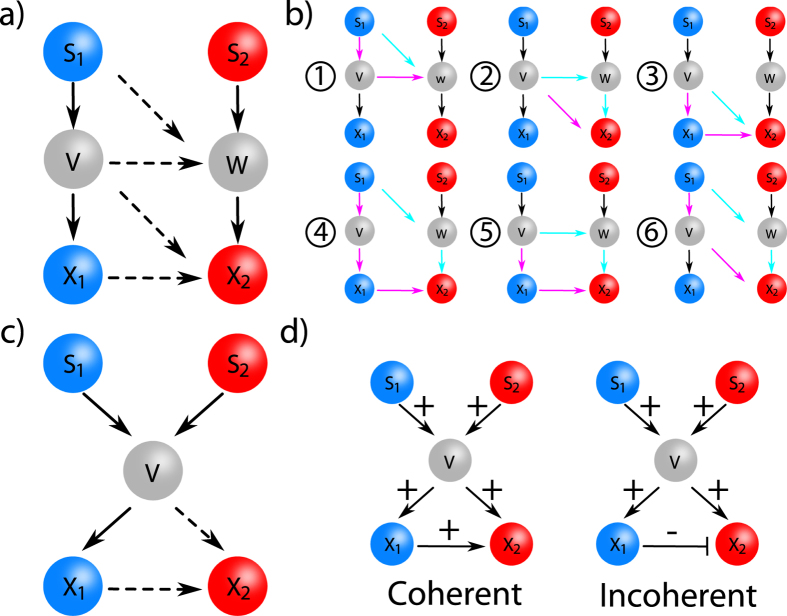
We start by studying two simple networks to show the theoretical abundance of non-monotonic behavior in networks consisting of two signaling pathways which share components and have biochemical interactions. For this we take two 3-node signaling pathways. Both signaling pathways have one input signal (S1,S2, respectively) and one response (X1,X2, respectively) each. Here the simplest interaction is a direct interaction between a node from one path and a node from another path. This is represented in the first network consisting of 6 nodes (Fig. 1a). In this network S1 affects both X1 and X2, but S2 only affects X2. A less trivial interaction arises when both signaling pathways share the same node. This is investigated in the other network, consisting of 5 nodes (Fig. 1c), where both signals influence both responses. In both networks we exclude direct regulation of any signal to either response. This corresponds to common processes, e.g. extracellular signals are transmitted via a receptor to the intracellular domain, or intracellular signals are transmitted to the DNA in the nucleus via messenger proteins.
Figure 1. Overview of networks. Each network consists of two pathways S1 → X1 (Left) and S2 → X2 (Right).
The pathways may interact via regulatory interactions and a shared component. Solid arrows are either activating or repressive, while dashed arrows can be activating, repressive or absent. a) 6 node topology. b) 6 different possible incoherent feed-forward loops marked by colored arrows (magenta activating, light blue repressing) c) 5 node topology. d) example of an incoherent and coherent feed-forward loop for a 5 node topology (+ activating, − repressing).
Each response is either in an active form 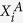 or inactive form Xi. We model the interactions between the different components using the well-known Monod-Wyman-Changeux (MWC) model (see Methods), which has shown to agree with experiments on proteins30,31,32. The MWC model is a statistical-physical model for the probability of a protein to be in its active state, which depends both on the intrinsic energies and on the binding of other molecules. Before we proceed, we formally have to define a non-monotonic response. We imagine each response to be in one of three states, ON, OFF, NONE, with the following definition
or inactive form Xi. We model the interactions between the different components using the well-known Monod-Wyman-Changeux (MWC) model (see Methods), which has shown to agree with experiments on proteins30,31,32. The MWC model is a statistical-physical model for the probability of a protein to be in its active state, which depends both on the intrinsic energies and on the binding of other molecules. Before we proceed, we formally have to define a non-monotonic response. We imagine each response to be in one of three states, ON, OFF, NONE, with the following definition
 |
When the level of the active form of components, 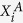 , is higher than a fraction α > 0.5 of the total concentration of a specific component i, Xi,T, i.e. if
, is higher than a fraction α > 0.5 of the total concentration of a specific component i, Xi,T, i.e. if 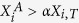 , then the response is considered ON. Similarly if
, then the response is considered ON. Similarly if 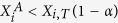 it is considered OFF. In the intermediate ramp,
it is considered OFF. In the intermediate ramp, 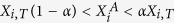 , the response is neither OFF nor ON; this is the NONE state. This state is included to take into account that in the presence of noise, an intermediate response may not lead to a significant phenotypic response at the cell population level. We note however, that for α = 0.5, there is no NONE state and that none of the conclusions presented below depend on the precise choice of alpha.
, the response is neither OFF nor ON; this is the NONE state. This state is included to take into account that in the presence of noise, an intermediate response may not lead to a significant phenotypic response at the cell population level. We note however, that for α = 0.5, there is no NONE state and that none of the conclusions presented below depend on the precise choice of alpha.
As an example we describe a non-monotonic response of X1 as function of S2. This is defined as a double change in the state of X1, ON-OFF-ON or OFF-ON-OFF due to an increase or decrease in S2, while S1 is kept at a constant level.
It is often recognized that non-monotonicity is the result of incoherent feed-forward loops; and this can be explained qualitatively, both for dynamical and steady-state responses33,34. For this reason in the 5 and 6 node networks we focus on the influence of (in)coherent feed-forward loops on possible steady-state non-monotonic behavior. A feed-forward topology consists of three components (A,B,C, where A regulates B directly and C via B. In a coherent feed-forward loop the regulation of B and C by A are equal (e.g. both activating), while in an incoherent loop the regulations are opposite (activating and repressing).
6-node topology
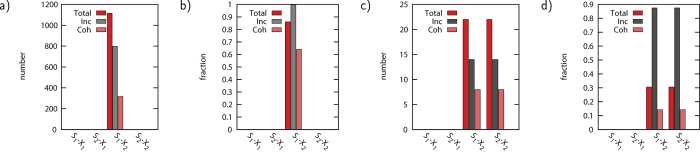
The 6-node network (Fig. 1a) consists of two linear pathways S1 → X1 (Left) and S2 → X2 (Right). Further, from the left cascade (S1 → X1) at various positions regulatory interactions to the right (S2 → X2) cascade can be present. We exclude interactions from the right to the left pathway to prevent bistability in the results. We sample parameters over all possible topologies (N = 24 × 34 = 1296) (see Supporting Material). For this 6-node network, 6 different incoherent feed-forward loops could be present, depending on the sign of each interaction arrow (Fig. 1b). In total, 800 out of 1296 topologies have at least one incoherent feed-forward loop, while each incoherent feed-forward loop is present in 288 topologies.
In Fig. 2a,b the number of topologies that have a non-monotonic response are shown, and we split the results into topologies that have an incoherent feed-forward loop and topologies that do not have this. The first observation is that non-monotonic interactions only occur for changes in S1 that lead to changes in X2. The absence of the other three possible non-monotonic responses can be easily explained. First, S2 has no regulatory effect on X1 and therefore a non-monotonic S2-X1 response is not possible. Second, S1 to X1 is a linear cascade with only one species acting onto another, excluding non-monotonicity between S1 and X1. An equivalent reasoning holds for the cascade S2-X2. Although there are more difficult interactions present, for any given value of the components S1, V and X1, a linear cascade between S2 and X2 remains. Almost all of the 800 (798) topologies with at least one incoherent feed-forward loop have at least one parameter set that leads to non-monotonic behavior (see SI).
Figure 2.
Histogram with the appearance of non-monotonicity for the 5 and 6 node network specified for each combination Si − Xj. a,b) 6 node network, α = 0.55, n = 2, minimum number of samples >25000. c,d) 5 node network, α = 0.55, n = 2, minimum number of samples 47430. a,c) Number of topologies that show non-monotonicity. b,d) Fraction of topologies that show non-monotonicity with respect to total number of all/incoherent/coherent topologies (see Fig. 1).
Unexpectedly, we see that not only topologies with incoherent feed-forward loops show non-monotonicity, but also topologies that have only coherent interactions. This effect depends on the specific parameters within the topology, and requires an in-depth explanation. As an example we take a coherent feed-forward loop with a specific set of parameters that lead to non-monotonic behavior. X down-regulates Z, but up-regulates Y, while Y down-regulates Z. Further Z is active, in the absence of both X and Y. Importantly, the binding and activation strength of X and Y for Z are not equal; Y has both a much smaller binding affinity to Z and a much larger down-regulating affect on Z compared to X. Let us discuss the dose-response of Z following an increase in X. Without X present (and thus no presence of Y) Z is active. An increase in X leads to an increase in Y and a decrease in Z. The upregulation of Y by X saturates at a given concentration of X. If Y saturates while X still increases, then X becomes the dominant negative regulator of Z. Since the negative regulation of X is much smaller than that of Y, Z increases again, hence giving rise to a non-monotonic response (see SI for an illustration).
Although coherent feed-forward loops can lead to non-monotonic behavior, non-monotonic behavior is more likely to occur for incoherent feed-forward loops. This can be observed from the fraction of different topologies that show non-monotonic behavior, as well as from the number of parameter sets for each topology that show non-monotonic behavior (see SI).
One can analytically prove that for non-monotonicity it is required that a component is simultaneously regulated by two other components, of which the steady-state value of one of these components depends non-linearly on the other component (see SI).
5-node topology
In the 5 node network (Fig. 1c) the component V is shared between the pathways S1 → X1 and S2 → X2. In total 72 topologies can be constructed. X1 is only regulated by V, and non-monotonic behavior for X1 can therefore be excluded. There exists only a single possibility for an incoherent feed-forward loop, which is present if the regulatory effect of the pathway V → X1 → X2 is opposite to that of the pathway V → X2, leading to 16 topologies that have an incoherent feed-forward loop.
In Fig. 2c,d we show histograms of the number (and fraction) of topologies that leads to non-monotonic behavior. We first observe that, as expected, non-monotonic behavior is only observed for X2. Out of the possible 72 topologies, 22 show non-monotonicity in the response X2 as function of S1, and 22 are non-monotonic as function of S2.
Not every topology with an incoherent feed-forward loop shows non-monotonicity. This does not imply that non-monotonic behavior for these topologies is impossible. It does show, however, that for these topologies the fraction of parameters within the total sampling space that allow for non-monotonic behavior is so small that brute force sampling did not reveal these (see SI). We again note that there are coherent topologies that show non-monotonicity.
The networks studied here can be seen as networks of interactions between species, but can also be viewed as coarse-grained representations of more complex networks. Specifically, a link between two nodes can be interpreted as an interaction between two species, but it may also represent a linear pathway. The input-output relation of a linear pathway—in the absence of interactions with other pathways—is monotonic and can thus be coarse-grained. Indeed, this coarse-graining does not affect the (non)-monotonicity of a multi-pathway network. Based upon our simple networks, with coherent feed-forward loops, incoherent feed-forward loops and/or shared nodes, we show that these networks can show non-monotonic dose-response relations. We next study the implications of non-monotonicity for pharmacodynamics of two example topologies.
Pharmacological implications of non-monotonicity
Before proceeding, we first introduce two important quantities: effectiveness and toxicity. Effectiveness is quantified as the ability to change the state of a selected response by applying a drug, while toxicity is quantified as unwanted change in the state of any other non-selected response. Throughout the paper we assume that the responses X1 and X2 are non-targettable and therefore a network can only be targeted at the signal level (S1, S2) or the intermediate components (V,W). Then, by definition, an effective agonist (antagonist) for S1 or V increases (decreases) the concentration of S1 or V to such a level, that a desired change of the response X1 is observed.
We are interested in the effect of drugs that specifically change the state of one and only one signal output (effectiveness) without affecting the state of the other output (toxicity).
6-node network
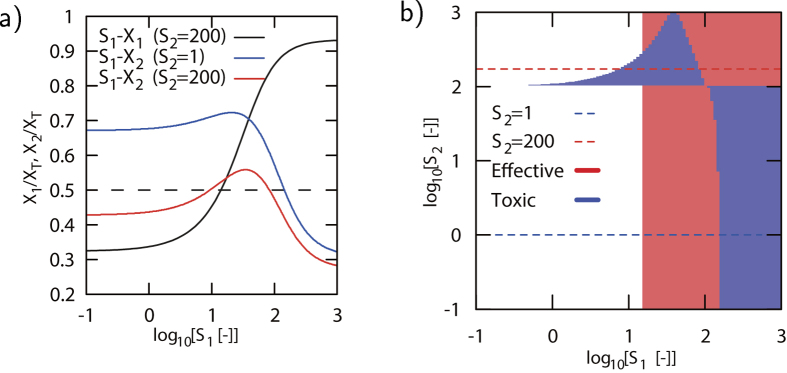
We first select one 6 node network that has a single incoherent feed-forward loop (Fig. 1b. 3); the activating connections S1 − V, S2 − W, V − X1 and X1 − X2, while V and W both repress X2.
Figure 3. Analysis of non-monotonicity in the 6 node network from Fig. 1b.
3 and its pharmacological implications. α = 0.5, n = 2.a) Dose-response curve for X1 as function of S1 for S2 = 200 (black solid), and X2 as function of S1 for S2 = 200 (red solid) and of S1 for S2 = 1 (blue solid), showing the non-monotonic response for X2 as function of S1. b) Contour plot showing the toxicity and effectivity of the state of X2 as function of an agonist for S1 for various concentrations of S2. The two horizontal dashed lines at S2 = 200 and S2 = 1 compare to the curves in a).
This network, for specific parameters, displays a non-monotonic relation between S1 and X2. To illustrate this behavior, Fig. 3a shows the dose-response curves of S1 − X1 for S2 = 200 [a.u.] (black solid), S1 − X2 for S2 = 200 (red solid) and S1 − X2 for S2 = 1 (blue solid). The non-monotonic behavior of X2 as a function of S1 is clearly observed if concentration S2 = 200, in contrast to the case where S2 = 1. The consequences of this non-monotonic behavior for pharmacology are shown in Fig. 3b. In this contour plot the effective concentration regime for an agonist for S1 is shown (red shading). As expected, a minimum concentration is required to activate X1, but due to the network structure there is no dependence on S2. The toxic region (blue shading) has a completely different shading. Since the effect of an agonist for S1 is studied, toxicity is defined as a change in the state of X2 compared to the state of X2 in the absence of S1, but for equal concentration of S2. For the two cases S2 = 1,200 in Fig. 3a this means a toxic response occurs when crossing the black dashed line α = 0.5, changing the state of X2 with respect to its initial state. This is further illustrated by drawing the corresponding red and blue dashed lines in Fig. 3b for S2 = 1,200. Fig. 3b shows that at large concentrations of S2 (>100) the non-monotonic response leads to a toxic concentration range at intermediate concentrations. A small window exist for which the drug becomes toxic, before it is effective. Interestingly, due to the non-monotonic response, the drug becomes non-toxic and effective at even larger concentrations. At low concentrations of S2 the toxic regime occurs at large agonist concentrations, and the drug is effective before a toxic effect occurs.
Effect of intrinsic differences between individuals
In the previous paragraph we showed that specific drugs are toxic for this 6-node network. From a pharmaceutical perspective, not only the toxicity as function of total dose, but also as a function of intrinsic fluctuations in network kinetics is relevant. In this section we study this in more detail.
It is well known that inter- and intra-individual variations exist for kinetic parameters due to variations in physicochemical properties of the internal environment, like the temperature, pH and concentration of components35. For example, despite the fact that human body temperature is tightly controlled, human body temperature varies between healthy individuals for nearly a degree centigrade36. This variation has significant implications for kinetics, as the rate constants depend exponentially dependent on temperature. Indeed, one can estimate based upon temperature changes that differences up to ≈10% are possible in the rate constants. Similarly enzymatic reaction rates depend on pH37. In the following, we study the effect of variations in the kinetic parameters in general and investigate their effect on toxicity and effectiveness.
Since our analysis depends on specific dissociation constants (e.g. between S1 and V), we here set out to study the influence of these variations on the effectiveness and toxicity of an antagonist. As in the previous paragraph, we study the effect of an agonist for S1, assuming (the red dashed line in Fig. 3b). Additionally, we study the effect of an agonist for V, assuming S1 = 0.1 and S2 = 200. To mimic the intrinsic variations in the dissociation constants, we vary each parameter (P) (see SI) in the system following
 |
where η is a random number sampled from a uniform distribution between [−1:1] and λ is the maximal percentage change in each parameter. As a result of this procedure, each parameter Pnew is bounded between Pnew[Porig(1 − λ):Porig(1 + λ)].
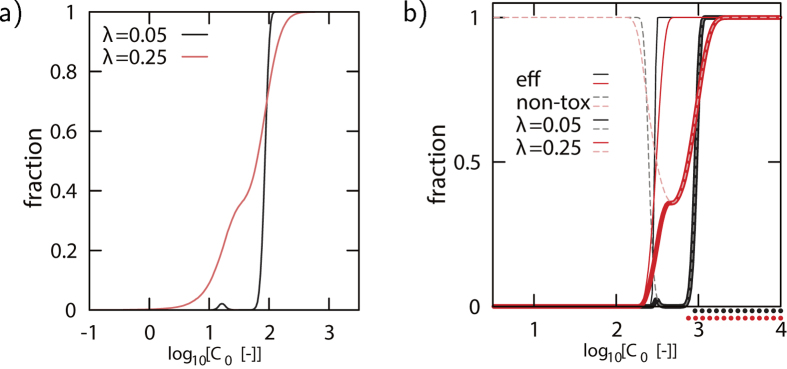
The current analysis provides a way to determine the therapeutic window as a function of the variation variable λ. The therapeutic window of a drug is the range of drug concentrations that elicits a therapeutic response without undesired toxic effects. It is quantified as the drug range between the ED50 and the TD50 point, where TD50 denotes the dose at which toxic response is observed in 50% of the population and ED50 denotes the dose at which a therapeutic response is observed in 50% of the population. The results are shown in Fig. 4.
Figure 4. The effect of intrinsic variability in the 6 node network from Fig. 1b.
3. α = 0.5, n = 2. A relative change λ in all the intrinsic parameters (see Eq. (2)) leads to a reduction of the effectiveness and/or an increase in the toxicity. Shown is the fraction of parameter combinations for which a drug with concentration C0 is effective and non-toxic. a) Lines show for an agonist for S1 (with S2 = 200) the fraction of individuals for which—at the given dose C0—an effective and non-toxic treatment is obtained. b) Similar results for an agonist for V, however the results for effectiveness (thin lines) and non-toxicity (thick lines) are shown separately. The solid thick lines indicates the dose for which the drug is non-toxic and effective. The dotted thick lines denotes the therapeutic window (S1 = 0.1,S2 = 200).
An agonist for S1 (Fig. 4a) requires for increasing λ (increasing the variability) an increase in minimum drug concentration for a 100% effective and non-toxic dose. In other words, a higher variability leads to a larger concentration that is effective for all individuals. Interestingly, the concentration for which only a small fraction of individuals has an effective and non-toxic treatment increases. This is due to the fact that larger differences between individuals exist, and the probability that a relative low concentration works for a single individual increases. As a result, the therapeutic window is larger for larger variability. In Fig. 4b a similar dependence on the variability for an agonist for V is shown. Here for two values of λ both the effectiveness (solid thin) and non-toxic (dashed thin) fractions as function of the agonist concentration are shown, together with the fraction of both effective and non-toxic simultaneously (solid thick). The non-monotonic response is clearly observed by following the non-toxicity line. The fraction of simultaneous effective and non-toxic (solid thick) for low concentration rises because of the increase in the effectiveness, but is then limited by the increase of toxicity. Only when the toxicity decreases again, can the drug become both effective and non-toxic. The therapeutic window (below the graph,thick dotted)), shows that for larger variability the window is larger. Note that the steepness of the curves means that a small change in the curve can imply a large difference in the fraction of non-toxic and effective cells (see SI).
A 5-node network: the multiplexing topology
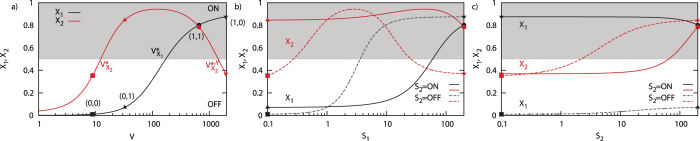
The multiplexing topology is the incoherent 5-node network as shown in Fig. 1d. Downstream of V an incoherent feed-forward loop exist, where V activates X1 and X2, while X1 represses X2. This network has a shared node (V) and an incoherent feed-forward loop. A closer look at this topology reveals a non-monotonic relation between X2 and S1 and between X2 and V (see Fig. 5). Here we study the effects of these non-monotonic relations on the effectiveness and toxicity of drugs.
Figure 5. Dose-response curves for X1 (black), X2 (red) as a function of.
a) V, b) S1, and c) S2 for the incoherent topology in Fig. 1d. Shading indicates the regime for which the response X is ON, assuming α = 0.5. a) The dose-response curve for X1 (black) and X2 (red) as function of V. Further the critical concentrations 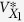 ,
, 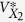 ,
, 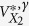 are indicated where the state switches from ON to OFF, or vice versa. The symbols indicate the state of the input signals, ■ = (S1 = OFF, S2 = OFF), ▲ = (S1 = OFF, S2 = ON) ● = (S1 = ON, S2 = OFF) ▼ = (S1 = ON, S2 = ON) (see Eq. 3). The non-monotonic relation between X2 and V is clearly observed. b) The dose-response curve for for X1 (black) and X2 (red) as function of S1. Solid lines indicate S2 = ON, dashed lines S2 = OFF. The non-monotonic relation between X2 and S1 is clearly observed. c) Similar to b), but with the roles of S1 and S2 reversed.
are indicated where the state switches from ON to OFF, or vice versa. The symbols indicate the state of the input signals, ■ = (S1 = OFF, S2 = OFF), ▲ = (S1 = OFF, S2 = ON) ● = (S1 = ON, S2 = OFF) ▼ = (S1 = ON, S2 = ON) (see Eq. 3). The non-monotonic relation between X2 and V is clearly observed. b) The dose-response curve for for X1 (black) and X2 (red) as function of S1. Solid lines indicate S2 = ON, dashed lines S2 = OFF. The non-monotonic relation between X2 and S1 is clearly observed. c) Similar to b), but with the roles of S1 and S2 reversed.
The most important result is that a specific drug could effectively lead to a change in the response of Xi, but as a result also change the state of the other response Xj≠i; leading to a drug that is effective but toxic. Importantly, normally drugs are applied in a gradual manner, and, upon the desired change in a specific response the treatment is stopped. At this drug concentration however, the treatment could be toxic. More intriguing, a toxic response could occur before the effective response is observed, or the toxicity could disappear for higher concentrations.
For a specific parameter set, this network is capable of multiplexing signals20. This indicates that the response X1 reflects the state of S1 independent of the state of S2, and similarly for X2. Since we have two binary input signals, we can define four specific input patterns,
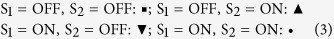 |
Assume S1 and S2 are both in the OFF state, and we target S2 with an agonist. As a result, X2 switches from OFF to ON, and at the moment of switch, X1 has not changed (see Fig. 5c, starting from the ■). This agonist therefore is effective and non-toxic. Similarly, an agonist for S1, while X2 is OFF, switches X1 from OFF to ON (Fig. 5b). But now, at the moment of the transition X2 has changed state as well. The agonist therefore is effective, but toxic. A further increase of agonist would eventually remove the toxicity, while remaining effective (see Fig. 5b, starting from ■). This is a direct consequence of the non-monotonic relation between S1 and X2.
Lastly, we discuss drugs for the intermediate component V, showing an interesting discrepancy between an agonist and antagonist. Assume we start in the state where S1 and X1 are OFF, S2 is ON and X2 is ON (see Fig. 5a, ▲) and we would like to use a drug that switches X2 from ON to OFF. Applying an antagonist for V, decreasing V drives the network to state ■. During this process, X2 switched from ON to OFF, while X1 has not changed state. The antagonist is thus effective and non-toxic. On the contrary, an agonist for V, driving the network to ▼-state, leads to an effective, but toxic response, since then both X1 and X2 change state. The direction of driving in this example determines the toxicity of the drug, but does not change the effectiveness. Again, this is an immediate consequence of the non-monotonic relation between V and X2, combined with the initial position of the system. We summarize the results in Table 1 (see SI for complete table).
Table 1. Summarized overview of the effects of agonists for the signals S 1,S2, and agonists/antagonist for V.
| S1 | S2 | Transition | Goal | Result | |
|---|---|---|---|---|---|
| OFF | OFF → ON | ■ → ▲ | X2: OFF → ON | Effective, non-toxic | |
| ON | OFF → ON | ▼ → ● | X2: OFF → ON | Effective, non-toxic | |
| OFF → OFF | ON | ■ →▼ | X1: OFF → ON | Effective, toxic | |
| OFF → ON | ON | ▲ → ● | X1: OFF → ON | Effective, non-toxic | |
| S1 | S2 | State | Goal | Transition of V | Result |
| OFF | OFF | ■ | X1: OFF → ON | ■ → ● | Effective, toxic |
| OFF | OFF | ■ | X2: OFF → ON | ■ → ▲ | Effective, non-toxic |
| OFF | ON | ▲ | X1: OFF → ON | ▲ → ● | Effective, non-toxic |
| OFF | ON | ▲ | X2: ON → OFF | ▲ → ■ | Effective, non-toxic |
| OFF | ON | ▲ | X2: ON → OFF | ▲ → ▼ | Effective, toxic |
So far we have assumed that we can change a single component (e.g. 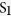 ) by applying a drug, while the other component(s) remain(s) constant. This assumption of course is not necessarily valid. If the other signal is non-constant, effective drug-doses can become ineffective. Imagine that starting in a state with both
) by applying a drug, while the other component(s) remain(s) constant. This assumption of course is not necessarily valid. If the other signal is non-constant, effective drug-doses can become ineffective. Imagine that starting in a state with both 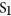 and
and 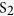 in the ON state (● in Fig. 5a), we add antagonist for
in the ON state (● in Fig. 5a), we add antagonist for 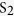 which drives
which drives 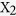 to the OFF-state (▼). If
to the OFF-state (▼). If 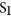 is not constant in the ON-state because, for example, it has a circadian rhythm, then there can be periods during the circadian cycle where the antagonist is non-effective, since during the gradual oscillations of
is not constant in the ON-state because, for example, it has a circadian rhythm, then there can be periods during the circadian cycle where the antagonist is non-effective, since during the gradual oscillations of 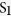 from ON to OFF (state ▼ to ■), the response
from ON to OFF (state ▼ to ■), the response 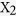 switches from OFF to ON to OFF.
switches from OFF to ON to OFF.
Influence of agonist dose and administration interval
In the previous paragraph we showed that specific drugs are toxic for this 5-node network with multiplexing capacity. From a pharmaceutical perspective, not only the toxicity as function of total dose, but also as a function of dose-interval is interesting. In this paragraph we study this in more detail.
Consider the case with initial state ■ and final state ▲, where we provide an agonist for S2 (see Fig. 5c). In the previous section we have shown that this treatment is effective and non-toxic, but this observation changes if we consider the dynamics of drug administration. For a typical clinical treatment, we imagine that the average agonist concentration equals the required effective concentration C0. However, for a non-continuous uptake and some type of degradation/dilution of agonist over time, this implies that the actual agonist concentration varies as a function of time (see Methods).
Importantly, it is not the average dose C0, but the actual dose C(t) (see Eq. (13)) that determines the toxicity and effectiveness of the drug. Indeed, for such a delivery process we need to specify toxicity and effectiveness in more detail. A drug is effective, if both for the minimal and maximal dose of the drug (thus for the full dose-range of the protocol) the required effectiveness is observed (e.g. X2 from OFF to ON). A drug is considered toxic if within the dose-interval (at least at the minimal or at the maximal drug dose) a toxic effect is observed.
In the remaining part of this section we discuss the dependence of toxicity and effectiveness on the required dose C0, the time between administrations τ and the dilution rate k.
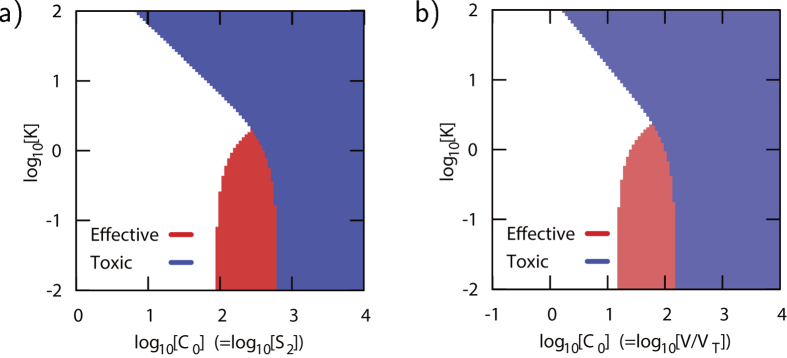
In Fig. 6a we show the effectiveness and toxicity of an agonist for S2 as a function of C0 and K = kτ for a network that initially is in state ▼ (S1 is ON and S2 is OFF). Applying an agonist for S2 to the system from state ▼ leads to a decrease in the concentration V affecting both X1 and X2. To switch X2 from OFF to ON a minimal drug concentration is required. Indeed, in Fig. 6a, we observe that for too low average concentrations C0 no change in X2 is observed (absence of red shading), leading to a lower bound on the minimum concentration (see Eq. (15)) for an effective treatment. Further, for too large average concentrations C0 at K or relatively small average concentrations at large K the drug is toxic (blue shading). The red shaded regime shows the “total dose-interval”-space for which the agonist is always effective (X2 OFF to ON), and never toxic (X1 remains ON).
Figure 6.
a,b) Contour plots for the effectiveness (red shading) an toxicity (blue shading) as function of average dose C0 and administration parameter K = kτ (see Fig. 7) for a system that a) is initially in state ▼ (see Fig. 5) with an agonist for S2, and b) is initially in state ■ with an agonist for V. Parameters: α = 0.55, n = 2.
Instead of targeting S2, one could also directly target V, in order to change X2. An agonist for V, trying to bring the system from state ■ to state ▲, has a very narrow window of effectiveness (Fig. 6b). This narrow window originates from the different interaction between V and X2, and X1 and X2. The increase in the concentration X2 by increasing concentration V is suppressed by a an increase in the concentration X1, which in turn represses X2. Since for a large agonist dose C0, X1 is pushed from the OFF into the ON state, this concentration is toxic.
These results show that it is possible that a drug for which the required dose C0 is non-toxic, if administered at constant levels , can become toxic, depending on the administration procedure.
Polypharmacology strategies
In the preceding discussions, the drug was assumed to be selective for a single-target, resulting in the exploration of single-target therapeutic strategies. We discussed effective, non-toxic pharmacological protocols for different scenarios. In some cases, we observed that single-target therapies are inherently toxic (see Table 1). For these scenarios, we study the effect of a multi-target approach, following the concept of polypharmacology. Recently evidence has emerged indicating that the effectiveness of a significant number of approved drugs is rooted in their promiscuity38,39,40,41,42. These observations led to the development of polypharmacology. In practice however, the paradigm of polypharmacological drug design has intrinsic difficulties: developing a drug that influences several targets in a desired way is a tedious and expensive task 43. Thus any prior knowledge about the achievable effectiveness and toxicity would be highly valuable.
Here we ask if polypharmacological approaches can provide a non toxic therapeutic strategy when selective therapy is toxic. We focus on the case where we would like to use an agonist for S1 to drive X1 from OFF to ON; we thus would like to bring the system from state ■ to state ▲, a situation we have shown to be toxic if an agonist for S1 is administered. From a polypharmacological perspective, besides targeting S1 the drug can target both V and S2, either in a antagonistic or agonistic fashion. This creates four different scenarios.
Unfortunately, in all scenarios the non-selective therapy remains toxic. The origin of the toxicity lies in the ordering of the so called critical concentrations, e.g. the critical concentration 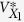 is the concentration V for which X1 switches state. Since, by construction of the multiplexing motif,
is the concentration V for which X1 switches state. Since, by construction of the multiplexing motif, 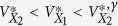 (see Fig. 5a), at the first transition to the OFF-state for X1, X2 is in the ON-state, while it initially was in the OFF-state. For a gradual increase in the drug, this toxicity can not be overcome, even if polypharmacologic drugs are considered.
(see Fig. 5a), at the first transition to the OFF-state for X1, X2 is in the ON-state, while it initially was in the OFF-state. For a gradual increase in the drug, this toxicity can not be overcome, even if polypharmacologic drugs are considered.
Here we explain the response of the current topology to polypharmacologic drugs. If the agonist for S1 would also act as an agonist for V, the result would be that for a smaller concentration of agonist the toxic response is observed. If the drug would act as agonist for S1 but an antagonistic for V, either no effective response is observed (if the antagonistic effect is too strong), or an effective response is observed, yet the drug is also toxic. Even worse, this polypharmacologic drug can be toxic, but not effective, if the concentration V is decreased below 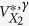 , but not below
, but not below 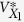 . Similar arguments can be formulated for an agonistic or antagonistic effect of the drug with respect to S2. In any case, due to the ordering of the critical values V*, the drug is toxic at the crossing of
. Similar arguments can be formulated for an agonistic or antagonistic effect of the drug with respect to S2. In any case, due to the ordering of the critical values V*, the drug is toxic at the crossing of 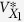 . Clearly, for this topology, polypharmacological approaches would not overcome the toxicity.
. Clearly, for this topology, polypharmacological approaches would not overcome the toxicity.
Conclusion
In this study we have discussed the occurrence of non-monotonicity in interacting signaling pathways in the context of drug targeting. We have shown that non-monotonic input-output relations can arise not only in incoherent feed-forward loops, but also in coherent feed-forward loops. We note that network topologies that contain coherent and incoherent feed-forward loops, do not necessarily show non-monotonicity. Binding and activation of molecular components are found to critically influence the form of the response function as well.
The described non-monotonicity is often left unnoticed experimentally. Our study considered a network topology, which incorporates features (incoherent feed-forward, multiple ligand activation) that are commonly observed in many cellular processes, which has a non-monotonic dose-response relation. The state of the art high-throughput methods for generating dose-response curves, namely robotic microplate-based and microfluidics-based approaches, construct dose-response relation based on small datasets44,45,46. In these analyses therefore, delicate non-monotonic features in dose-response curves are probably left undetected. With the emergence of high-resolution methods we expect to increasingly discover complex response functions47,48.
Non-monotonicity imposes fundamental constraints on the pharmacodynamics of a drug as well as the choice of administration routes and protocols, irrespective of the chemical nature of the designed drug. The models discussed in this article are very simple in nature, and by construction we know all its parameters. Even though our models are simple (they constitute either 5 species with 5 interactions or 6 species and 6 interactions), the resulting dynamics are rich. While the effectiveness and toxicity of drug in systems with monotonic input-output relations can typically be predicted intuitively, the behavior of non-monotonic response systems are much more difficult to interpret and in such systems, the response has to be studied over a wide range of conditions (e.g., drug concentration range), and that our intuition can be misleading: For example, and observed decrease in effectiveness and/or increase in toxicity upon increasing drug dose may not imply that a higher drug dose is even worse, as it would be for systems with monotonic input-output relations. In fact, the qualitative behavior of incoherent feed-forward and multiplexing motifs often does not follow from the network architecture alone, but also depends on the quantitative details of the interactions. We find that certain pharmacological manipulations are inherently toxic, and neither single-target nor polypharmacology strategies can be taken to avoid the toxicity. Our study reveals that the optimal drug dose for a certain pharmacological manipulation may be far beyond the onset of toxicity. We have shown that the therapeutic windows for certain therapies are narrow and the preferred concentration range might occur after a broad toxic regime, suggesting that for a newly designed drug, one has to explore administration parameter space carefully to detect the effectiveness of the drug. This is important because apparent dose limitations imposed by toxicity likely discourage clinical trials exploring higher dosage levels and therefore the effectiveness of the drug will remain unnoticed due to inadequate exposure. Clearly, the precise size of a therapeutic window depends on the details of the system of interest, and must be determined by experiment. Our results imply that to determine its size, a much larger parameter scan (e.g., drug concentration range) may be warranted than typically employed hitherto.
Methods
Modeling the networks
In Fig. 1 we introduce the 5-node and 6-node networks taken into consideration. We distinguish between two types of interactions, up-regulatory (activation) and down-regulatory (repression). To model these interactions we use the MWC model. Using this statistical physics model we determine the probability of a component to be in the active form. A key concept of the MWC model is that each component can always be either in the active or in the inactive form, independent of the interaction with other components. However, the interaction (e.g. through binding) of another component can shift the balance to either the active or the inactive form.
The probability pA for a component to be in the active form is computed from the partition functions, which enumerate all possible ways a receptor can be in either the active (ZA) or inactive (ZI) form:
 |
The explicit forms of the partition functions under the MWC model are presented in the Supplementary Material. For intuition, we provide an example here: the partition functions for a component X that is under the regulation of V are
 |
 |
where the parameter 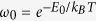 is the Boltzmann factor corresponding to the energy difference E0 between the active and inactive form, and the parameters
is the Boltzmann factor corresponding to the energy difference E0 between the active and inactive form, and the parameters 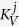 are the dissociation constants of V in form j ∈ {A,I}. In Eq. (5) , the two terms correspond to the component being active when X is unbound and when X is bound. The factor n reflects the cooperativity of n V components interacting. The same holds for Eq. (6) with the receptor being inactive. Importantly, if
are the dissociation constants of V in form j ∈ {A,I}. In Eq. (5) , the two terms correspond to the component being active when X is unbound and when X is bound. The factor n reflects the cooperativity of n V components interacting. The same holds for Eq. (6) with the receptor being inactive. Importantly, if 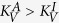 , the component V represses X, since V is more likely to bind in the inactive form.
, the component V represses X, since V is more likely to bind in the inactive form.
Combining Eq. (4) and Eqs. (5,6), the total concentration of active components XA is
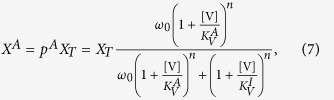 |
where XT is the total concentration of X. In the SI we connect the MWC-model to the more common Hill-function formalism.
Time dependence of drug-administration
The drug concentration, assuming exponential dilution over time, is
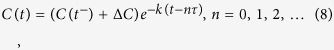 |
where C(t−) is the concentration at t− = nτ, just before drug administration, ΔC is the administered dose at t = nτ and k is the dilution rate. We assume periodicity on the interval τ, meaning that the concentration just before adding ΔC, C(t−) equals the concentration after dilution of the dose ΔC over a time window τ. We thus obtain the concentration just before drug administration (t−) and directly after drug administration (t+)
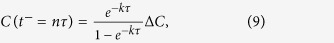 |
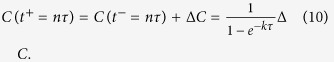 |
Combining Eqs. (8, 9, 10), we obtain an expression for the dynamics of the drug
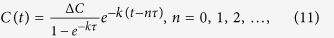 |
Further we require that the average drug concentration in the time interval [nτ : (n + 1)τ] is C0 (note that the total administered dose is CT = τC0),
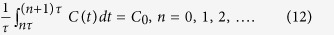 |
Combining Eq. (11) and Eq. (12) , we write for the drug concentration as a function of time
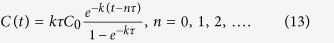 |
As a result, the maximum and minimum concentration in the time interval [nτ : (n + 1)τ] are
 |
 |
We define a new parameter K = kτ, which combines the effect of the dilution rate k and dose-interval τ, reflecting that fast dilution (large k) has a similar effect on the results as a long time-interval (large τ). In Fig. 7 we show the agonist concentration as a function of time. The dependence on the time-dilution parameter K is clearly visible, where for longer intervals/faster dilution an increase in both the maximum and the minimum drug concentration is observed.
Figure 7. Time dependence of the agonist concentration for different average dose levels C0.
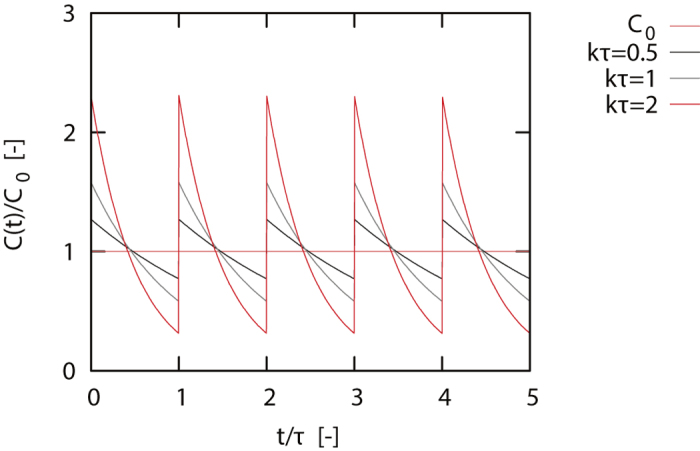
Longer interval times between administration or faster dilution lead to much larger fluctuations in the dose-level.
Additional Information
How to cite this article: van Wijk, R. et al. Non-monotonic dynamics and crosstalk in signaling pathways and their implications for pharmacology. Sci. Rep. 5, 11376; doi: 10.1038/srep11376 (2015).
Supplementary Material
Acknowledgments
The authors thank Wiet de Ronde for his involvement at the early stage of this project and Jeroen van Zon for his helpful comments on the manuscript. RvW, SJT and PRtW are supported by the research program of the Foundation for Fundamental Research on Matter (FOM), which is part of The Netherlands Organisation for Scientific Research (NWO).
Footnotes
Author Contributions A.M. conceived and designed the project. A.M. and R.v.W. performed the research and analyzed the data. All authors discussed the results and wrote the manuscript.
References
- Porter R. S. et al. The Merck Manual of Diagnosis and Therapy 19th edn (ed. Porter R. S. ) (Merck Publishing, 2011). [Google Scholar]
- Hopkins A. L., Mason J. S. & Overington J. P. Can we rationally design promiscuous drugs ? Curr. Opin. Struc. Biol. 16, 127–136 (2006). [DOI] [PubMed] [Google Scholar]
- Csermely P., Korcsmáros T., Kiss H. J. M., London G. & Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol. Therapeut. 138, 333–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin J. A., Hunter T., Palsson B. O. & Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Bio. 6, 99–111 (2005). [DOI] [PubMed] [Google Scholar]
- Mashaghi A. R., Ramezanpour A. & Karimipour V. Investigation of a protein complex network. Eur. Phys. J. B. 41, 113–121 (2004). [Google Scholar]
- Ramezanpour A., Karimipour V. & Mashaghi A. Generating correlated networks from uncorrelated ones. Phys. Rev. E. 67, 046107 (2003). [DOI] [PubMed] [Google Scholar]
- Hyduke D. R. & Palsson B. Ø. Towards genome-scale signalling-network reconstructions. Nat. Rev. Gen. 11, 297–307 (2010). [DOI] [PubMed] [Google Scholar]
- Lokody I. Integrating GWASs and protein interactions. Nat. Rev. Gen. 15, 5 (2014). [Google Scholar]
- Hwang W. C., Zhang A. & Ramanathan M. Identification of information flow-modulating drug targets: a novel bridging paradigm for drug discovery. Clin. Pharmacol. Ther. 84, 563–572 (2008). [DOI] [PubMed] [Google Scholar]
- Fliri A. F., Loging W. T. & Volkmann R. A. Cause-effect relationships in medicine: a protein network perspective. Trends Pharmacol. Sci. 31, 547–555 (2010). [DOI] [PubMed] [Google Scholar]
- Yin N. et al. Synergistic and antagonistic drug combinations depend on network topology. Plos Comput. Biol. 9, e93960 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ouyang Q. & Wang H. Dose-response aligned circuits in signaling systems. Plos One , 7, e34727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- Kaplan S., Bren A., Dekel E. & Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol. Syst. Biol. 4, 203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley C. What doesn’t kill makes you stronger. EMBO Rep. 4, 921–924 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Andersen M. E., Heber S. & Zhang Q. Non-monotonic dose-response relationship in steroid hormone receptor-mediated gene expression. J. Mol. Endocrinol. 38, 569–585 (2007). [DOI] [PubMed] [Google Scholar]
- vom Saal F. S. et al. Prostate enlargement in mice due to fetal exposure to low doses of estradiol or diethylstilbestrol and opposite effects at high doses. P. Natl. Acad. Sci. USA. 94, 2056–2061 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill Y. B., Petre C. E., Monk K. R., Puga A. & Knudsen K. E. The xenoestrogen bisphenol A induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Mol. Cancer Ther. 1, 515–524 (2002). [PubMed] [Google Scholar]
- Calabrese E. J. Getting the dose-response wrong: why hormesis became marginalized and the threshold model accepted. Arch. Toxicol. 83, 227–247 (2009). [DOI] [PubMed] [Google Scholar]
- de Ronde W., Tostevin F. & ten Wolde P. R. Multiplexing biochemical signals. Phys. Rev. Lett. 107, 1–4 (2011). [DOI] [PubMed] [Google Scholar]
- Schwartz M. A. & Madhani H. D. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38, 725–748 (2004). [DOI] [PubMed] [Google Scholar]
- Fraser T. D. C. & Germain R. N. Navigating the network: signaling cross-talk in hematopoietic cells. Nat. Immunol. 10, 327–331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M., Lin K. M., Hsueh R. C., Sternweis P. C. & Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat. Cell Bio. 8, 571–580 (2006). [DOI] [PubMed] [Google Scholar]
- Kollmann M., Lø vdok, , Bartholomé K., Timmer J. & Sourjik V. Design principles of a bacterial signalling network. Nature 438, 504–507 (2005). [DOI] [PubMed] [Google Scholar]
- de Ronde W., Tostevin F. & ten Wolde P. R. Feed-forward loops and diamond motifs lead to tunable transmission of information in the frequency domain. Phys. Rev. E. 86, 21913 (2012). [DOI] [PubMed] [Google Scholar]
- Marshall C. J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185 (1995). [DOI] [PubMed] [Google Scholar]
- Ostrem J. M., Peters U., Sos M. L., Wells J. A. & Shokat K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel J., Cromm P. M., Zimmermann G., Grossmann T. N. & Waldmann H. Small-molecule modulation of Ras signaling. Nat. Chem. Bio. 10, 613–622 (2014). [DOI] [PubMed] [Google Scholar]
- Shima F. et al. Small-molecule modulation of Ras signaling. P. Natl. Acad. Sci. USA 110, 8182–8187 (2014). [Google Scholar]
- Tu Y. The nonequilibrium mechanism for ultrasensitivity in a biological switch: sensing by Maxwell’s demons. P. Natl. Acad. Sci. USA 105, 11737–11741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. H., Sourjik V. & Wingreen N. S. A dynamic-signaling-team model for chemotaxis receptors in Escherichia coli. P. Natl. Acad. Sci. USA 107, 17170–17175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoge M. L., Endres R. G. & Wingreen N. S. Receptor-receptor coupling in bacterial chemotaxis: evidence for strongly coupled clusters. Biophys. J. 90, 4317–4326 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuit (Chapman & Hall/CRC, 2007). [Google Scholar]
- Ishihara S., Fujimoto K. & Shibata T. Cross talking of network motifs in gene regulation that generates temporal pulses and spatial stripes. Genes Cells 10, 1025–1038 (2005). [DOI] [PubMed] [Google Scholar]
- Marangoni A. G. Enzyme Kinetics: A Modern Approach (John Wiley & Sons, Inc, 2003). [Google Scholar]
- Kasper D. et al. Harrison’s Principles of Internal Medicine 19th edn (McGraw-Hill Education, 2015). [Google Scholar]
- Nelson D. L. & Cox M. M. Lehninger Principles of Biochemistry 6th edn (W.H.Freeman & Co. Ltd, 2013). [Google Scholar]
- Roth B. L., Sheffler D. J. & Kroeze W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004). [DOI] [PubMed] [Google Scholar]
- Keith C. T., Borisy A. A. & Stockwell B. R. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 4, 71–78 (2005). [DOI] [PubMed] [Google Scholar]
- Mencher S. K. & Wang L. G. Promiscuous drugs compared to selective drugs (promiscuity can be a virtue). BMC Clin. Pharmacol. 5, 3 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton T. “Promiscuous” anticancer drugs that hit multiple targets may thwart resistance. JAMA 292, 419–422 (2004). [DOI] [PubMed] [Google Scholar]
- Frantz S. Drug discovery: playing dirty. Nature 437, 942–943 (2005). [DOI] [PubMed] [Google Scholar]
- Margineanu D. G. Systems biology impact on antiepileptic drug discovery. Epilepsy Res. 98, 104–115 (2012). [DOI] [PubMed] [Google Scholar]
- Malo N., Hanley J. A., Cerquozzi S., Pelletier J. & Nadon R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175 (2006). [DOI] [PubMed] [Google Scholar]
- Inglese J. et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. P. Natl. Acad. Sci. USA 103, 11473–11478 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P., Hill C. & Janc J. W. Practical robust fit of enzyme inhibition data. Methods Enzymol. 383, 366–381 (2004). [DOI] [PubMed] [Google Scholar]
- Miller O. J. et al. High-resolution dose-response screening using droplet-based microfluidics. P. Natl. Acad. Sci. USA 109, 378–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton R. C. R. & deMello A. J. Microfluidics: analog-to-digital drug screening. Nature 483, 43–44 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.