Abstract
Cytochrome P450 2A6 (CYP2A6) catalyzes nicotine metabolism and contributes to the metabolism of the tobacco-specific lung carcinogen, NNK. Genetic variation in CYP2A6 may affect smoking behavior and contribute to lung cancer risk. A nested case-control study of 325 lung cancer cases and 356 controls was conducted within a prospective cohort of 18,244 Chinese men in Shanghai, China. Quantified were 4 allelic variants of CYP2A6 [*1(+51A), *4, *7, and *9] and urinary total nicotine, total cotinine, total trans-3'-hydroxycotinine (3HC), and total NNAL (an NNK metabolite). Calculated were total nicotine equivalents (TNE), the sum of total nicotine, total cotinine and total 3HC; and the total 3HC:total cotinine ratio as a measure of CYP2A6 activity. The nicotine metabolizer status (normal, intermediate, slow and poor) was determined by CYP2A6 genotypes. The smoking-adjusted odds ratios (95% confidence intervals) of lung cancer for the highest vs. lowest quartile of total nicotine, total cotinine, total 3HC, TNE, and total NNAL were 3.03 (1.80-5.10), 4.70 (2.61-8.46), 4.26 (2.37-7.68), 4.71 (2.61-8.52), and 3.15 (1.86-5.33) (all Ptrend < 0.001), respectively. Among controls CYP2A6 poor metabolizers had a 78% lower total 3HC:total cotinine ratio and 72% higher total nicotine (Ptrend ≤0.002). Poor metabolizers had an odds ratio of 0.64 (95% confidence interval=0.43-0.97) for lung cancer, which was statistically non-significant (odds ratio=0.74, 95% confidence interval=0.48-1.15) after adjustment for urinary TNE and smoking intensity and duration. The lower lung cancer risk observed in CYP2A6 poor metabolizers is partially explained by the strong influence of CYP2A6 genetic polymorphisms on nicotine uptake and metabolism.
INTRODUCTION
Tobacco smoking is the most important causal factor for lung cancer. It is estimated that 90% of all lung cancer deaths are attributable to cigarette smoking.1 However, there is considerable variation in susceptibility to lung cancer among smokers. For current smokers it is estimated that the cumulative risk of lung cancer by age 75 years is about 15%.2 The large inter-individual variation in smoking-related lung cancer risk may be determined in part by variability in the uptake and metabolism of tobacco carcinogens. There are 72 established carcinogens in cigarette smoke.3 Among these, polycyclic aromatic hydrocarbons (PAH) and the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are widely considered to be among the most important causative agents for lung cancer.4,5 We previously reported that the urinary level of total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), a metabolite of NNK, and the urinary level of phenanthrene tetraol, a biomarker of PAH, were associated with increased risk of lung cancer in two prospective cohorts of Chinese smokers.6, 7
Cytochrome P450 (CYP) enzymes catalyze the α-hydroxylation of both NNK and NNAL,8 which generates reactive metabolites that covalently modify DNA and lead to the initiation of lung carcinogenesis. These enzymes include CYP2A6 and CYP2B6 in the liver and CYP2A6, CYP2A13 and CYP2B6 in the lung.9, 10 In human liver microsomes, up to 70% of NNK α-hydroxylation is inhibited by CYP2A6 antibodies.9 CYP2B6 is similar in catalytic efficiency to CYP2A6, but is typically less abundant in the liver.9, 11 CYP2A13 is a more efficient catalyst of NNK α-hydroxylation than is CYP2A6, however CYP2A6 protein is more abundant in the human lung.11 These data suggest that CYP2A6 may play an important role in the catalysis of NNK α-hydroxylation, and the development of lung cancer in smokers.
Nicotine, although non-carcinogenic, is a psychoactive substance that is responsible for tobacco addiction and influences tobacco use patterns. Nicotine is primarily metabolized by CYP2A6-catalyzed 5′-oxidation.12 The product of this reaction is then oxidized, either by CYP2A6 or aldehyde oxidase, to cotinine.12, 13 In most smokers, the conversion of nicotine-tocotinine accounts for 70-80% of inhaled nicotine.12 CYP2A6 is also the primary enzyme that converts cotinine to trans-3'-hydroxycotinine (3HC), which is the most abundant urinary metabolite of nicotine.12 Nicotine, cotinine, and 3HC all are glucuronidated.12 The sum of urinary total nicotine (nicotine plus nicotine-glucuronide), total cotinine (cotinine plus cotinine-glucuronide), and total 3HC (3HC plus its glucuronide) is called “total nicotine equivalents” (TNE). TNE of smokers represents approximately 80% of the excreted nicotine dose,12 and is the best marker for daily nicotine intake and total tobacco smoking exposure.14
Genetic polymorphisms in CYP2A6 gene may contribute to inter-individual variation in risk of lung cancer among smokers by altering cigarette consumption and smoking patterns resulting from altered nicotine metabolism and excretion and/or affecting the activation/detoxification pathway of NNK. Several epidemiological studies have examined the relationship between individual polymorphisms of CYP2A6 and lung cancer risk. The results are inconsistent. Some studies found a reduced risk of lung cancer in individuals carrying the low-activity CYP2A6 alleles,15-17 while others reported an increased or null risk association with lung cancer.18-20 The inconsistent findings among these studies could be due, in part, to the difficulties and measurement error in genotyping due to the altered gene structure resulting from the deletion polymorphism in the CYP2A6 gene as well as different genetic variants across different race/ethnic populations.
At present, there are more than 80 known single nucleotide polymorphisms (SNPs) in the CYP2A6 gene located at 19q13.2 (http://www.cypalleles.ki.se/cyp2a6.htm). This highly polymorphic gene results in large inter-individual and interethnic variations in the protein expression and activities of CYP2A6. Genetic polymorphisms of the CYP2A6 gene that result in no or reduced CYP2A6 activity alter nicotine metabolism and tobacco consumption in smokers.16, 21, 22 The ratio of 3HC to cotinine in plasma or in urine has been used to assess the relative activity of CYP2A6.12, 21 In urine a somewhat better measure of CYP2A6 is the ratio of total 3HC to cotinine, since a portion of the 3HC is excreted as its glucuronide conjugate.12, 21 Alternatively the urinary ratio of total 3HC to total cotinine has been used to phenotype CYP2A6 activity.23
The overarching goal of this research project is to evaluate the relationship between biomarkers of tobacco smoking exposure and genetic variants of CYP2A6 on the risk of lung cancer development. The present report focused on the analyses that examined: 1) the association between urinary TNE and other metabolites of tobacco constituents and risk of lung cancer; 2) the CYP2A6 genotype-phenotype (urinary total 3HC:total cotinine ratio) association to verify the accuracy of CYP2A6 genotype; 3) the association between CYP2A6 genotype-determined metabolizer status and risk of lung cancer.
MATERIAL AND METHODS
Study Population
Subjects were drawn from the Shanghai Cohort Study.24 Briefly, the Shanghai Cohort consisted of 18,244 men enrolled from 1 January 1986 through 30 September 1989, who were 45-64 years of age and resided in the city of Shanghai, China. In addition to in-person interviews eliciting information on use of tobacco and alcohol, a 10-ml blood sample and a single-void urine specimen were collected from each participant at baseline. The Shanghai Cohort Study has been approved by the Institutional Review Boards at the Shanghai Cancer Institute and the University of Pittsburgh. The present study has been approved by the Institutional Review Boards at the Shanghai Cancer Institute, the University of Minnesota and the University of Pittsburgh.
Nested Case-Control Study
Identification of incident lung cancer cases was accomplished through annual in-person re-interviews of all surviving cohort members and routine review of reports from the population-based Shanghai Cancer Registry. To date, losses to follow-up totaled 839 individuals (4.6%) of the entire cohort after 25 years of study. The nested case-control design was previously described.7 As of December 31, 2006, 706 cohort participants developed lung cancer. Among them, 574 were current smokers, 43 were former smokers, and 89 were never smokers at baseline. Only current smokers at baseline were eligible for the present study because urinary metabolites of nicotine and NNK were undetectable in never smokers and long-term quitters. For each of 574 cases of current smokers, we randomly selected one control subject from all cohort members who were current smokers at biospecimen collection, free of cancer and alive at the time of cancer diagnosis of the index case. Controls were matched to the index case by age at enrollment (±2 years), date of biospecimen collection (±1 month) and neighborhood of residence at recruitment.
Laboratory Measurements
Urinary metabolites of cigarette smoke constituents
Urine samples of all study subjects were retrieved from the biospecimen bank. Specimens from matched control subjects and their index cases were always assayed in the same batch. All urine aliquots were identified only by unique codes, and laboratory personnel had no knowledge of the case/control status of the test samples. The assay for quantifying total NNAL in urine was identical to the one previously described.6 Quantification of total nicotine, total cotinine, and total 3HC in urine that had been treated with β-glucuronidase was carried out by gas chromatography-mass spectrometry or liquid chromatography tandem mass spectrometry as previously described.7, 25 Urinary creatinine (Cr) was assayed by Fairview-University Medical Center Diagnostic Laboratories (Minneapolis) with a Kodak Ektachem 500 chemistry analyzer.
CYP2A6 Genotyping
Genomic DNA was extracted from buccal cells or serum samples using QIAmp DNA mini kit (Qiagen Inc, Valencia, CA, US). Quality and quantity of purified DNA were evaluated using a Nanodrop UV-spectrometer (Thermo Fisher Scientific Inc., Wilmington, DE, US). DNA samples were stored at −20°C until analysis.
Genotyping of CYP2A6 was accomplished with several different methods. There are two structural variants of CYP2A6: *4 (gene deletion) and *12 (a hybrid allele with CYP2A7). These structural variants were measured using quantitative PCR assays designed by ABI (Applied Biosystems); all qPCR assays were repeated 4 times. An initial assay using a probe specific to intron 1 of CYP2A6 (hs0754274_cn) was used to determine the number of CYP2A6 gene copies present relative to the housekeeping gene RNaseP. Those with no amplification of hs0754274_cn would be designated as having two structural variants (i.e. *4/*4); this was not observed in the present study. Those with a single copy of CYP2A6 were carried forward to a second qPCR assay using ABI probe hs07545275_cn which is specific for intron 7 of the CYP2A6 gene (i.e., does not cross-react with CYP2A7). Those subjects with 1 copy of intron 7 in the second assay were designated *1/*4, and those with 2 copies of intron 7 were designated *1/*12 (the allele *12 was not observed in the present study). We then assessed additional variants that do not alter the CYP2A6 gene structure. To maximize assay specificity and minimize DNA input, the *7 (rs5031016) and *9 (rs28399433) alleles were genotyped using nested PCR followed by pyrosequencing (Pyromark Q96MD instrument, Qiagen). The *1(+51A) variant was captured with an allelic discrimination (TaqMan) assay for rs1137115.
We assessed the performance of genotyping using two approaches. For quality control (QC) purpose, DNA samples of Han Chinese ethnicity from the Coriell Institute (Coriell.org) were inserted in all test samples in one-tenth by frequency. In addition, 55 paired buccal-derived and serum-derived DNA samples from 55 unique study subjects were evenly dispersed among all test samples to assess the concordance of genotypes from the two different DNA sources. Of the 55 paired DNA samples, discordant genotypes were observed for the structural variant (CYP2A6*4) in two paired DNA samples, and the *7 and *9 each in three paired samples whereas no discordant genotypes of *1(+51A) were detected. All six subjects with discordant genotypes were excluded from the present analysis.
CYP2A6*9 was considered a ‘decrease of function (D)’ allele whereas *4 and *7 were considered ‘loss of function (L)’.21, 26 A more recently identified SNP rs1137115, a synonymous variant in exon 1 of CYP2A6 gene that defines CYP2A6*1A(51A), is associated with lower mRNA expression and slower in vivo nicotine metabolism,27 and was considered as a D allele in the present study. Thus the CYP2A6 genotype grouping was based on these predicted pharmacokinetic impacts: (1) ‘normal metabolizers’ were defined as having neither a D nor L allele (i.e., *1/*1); (2) ‘intermediate metabolizers” had only one D allele (i.e., *1A(51A) or *9); (3) ‘slow metabolizers’ had either one L allele or two D alleles (e.g., *1/*4 or *9/*9); and (4) ‘poor metabolizers’ had either one L plus one D allele or two L alleles (e.g., *9/*7 or *7/*4).
Of the 574 case-control pairs, 40 cases and 19 controls did not have urine samples for total cotinine measurement. Among those with cotinine measurement, 16 urine samples (5 from cases and 11 from controls) had urinary total cotinine levels below 35 ng/mL, indicating that they were from nonsmokers (or very light/infrequent smokers) at the time of urine collection. In addition, 136 subjects (71 cases and 65 controls) did not have urinary total 3HC due to the depletion of their urine samples after measurement of other urinary biomarkers in previous studies.6, 7, 28 Among the remaining 937 subjects, we did not obtain DNA from 67 subjects (48 cases and 19 controls). In addition, 84 cases (19.8%) and 99 controls (21.6%) were excluded where CYP2A6 genotype could not be determined. The most frequent missing genotype was CYP2A6*4 (42 cases, 53 controls), followed by *7 (with additional 22 cases and 24 controls), *9 (16 cases, 8 controls), and *1(+51A) (11 cases, 10 controls). The main reason for these undeterminable genotype calls was the limited amount of DNA extracted from serum samples that were still available after previous genotyping assays.28 Among the genotypes, one case and five controls had discordant genotypes from the repeated tests of duplicate samples. After excluding those subjects, the present study included 325 cases and 356 control subjects.
Statistical analysis
Urinary metabolites of cigarette smoke constituents including total nicotine, total cotinine, total 3HC and their sum, TNE, were expressed in nmol/mg Cr, and total NNAL in pmol/mg Cr to correct for varying water contents of individual spot urine samples. The metabolic total 3HC:total cotinine ratio in urine was calculated to evaluate the CYP2A6 activity. We also calculated the ratio of total NNAL over TNE as a detoxification index for NNK through urinary excretion of NNAL after controlling for nicotine uptake. Given the markedly skewed distributions of these urinary biomarkers, formal statistical testing was performed on logarithmically transformed values, and geometric means are presented.
The χ2 test and the t-test were used to compare the distributions of selected variables between lung cancer cases and controls. The analysis of covariance (ANCOVA) method was used to examine the difference in the concentrations of urinary biomarkers or their ratios across different CYP2A6 genotype predicted metabolizers among control subjects.
We used unconditional logistic regression method, for maximizing the number of subjects included in the analysis, to calculate odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) and P values to assess the relationship between urinary metabolites of cigarette smoke constituents and lung cancer risk with adjustment for matching factors including age, year of sample collection, and neighborhood of residence at enrollment. For each urinary biomarker, study subjects were grouped into quartiles according to its distribution among control subjects. The linear trend test for the association between urinary biomarker and lung cancer risk was based on the ordinal value of quartiles. Similarly we examined the association between CYP2A6 genotype-predicted metabolizers and lung cancer risk because the phenotypic measure of CYP2A6, the total 3HC:total cotinine ratio, is somewhat influenced by genetic variants of UDP glucuronosyltransferase (UGT) 2B10, the enzyme that catalyzes cotinine glucuronidation. However, the variants are relatively rare in Asian populations.25 The logistic regression models that assessed the association between the CYP2A6 genotype-determined metabolizer status and lung cancer risk included different sets of covariates as follows: 1) demographic variables and/or self-reported history of smoking intensity and duration that provided the risk estimates similar as those reported in previous studies; 2) urinary TNE, the best biomarker for daily nicotine intake and total tobacco smoking exposure that reduced the measurement error in tobacco smoking based on self-report only; and 3) further adjustment for total NNAL that would show the independent effect of CYP2A6 on the risk of lung cancer.
Statistical analyses were carried out using SAS software version 9.3 (SAS Institute, Cary, NC). All P-values reported are two-sided, and those that were less than 0.05 were considered to be statistically significant.
RESULTS
Of the 325 cases, 214 (66%) were histopathologically confirmed while the remaining 111 (34%) were diagnosed based on radiography or computer-assisted tomography evidence. Among the histopathologically confirmed cases, 102 (47.7%) were squamous cell cancers, 80 (37.4%) adenocarcinomas, 15 (7.0%) small cell cancers, and 17 (7.9%) other cell types. The mean age (standard deviation) at cancer diagnosis of all case patients was 67.7 (6.6) years. The average time interval between baseline biospecimen collection and cancer diagnosis was 10.5 (5.5) years, ranging from 1 month to 20.5 years.
Compared with controls, cases had greater numbers of cigarettes smoked per day, years of smoking, and pack-years of smoking at baseline. The percentage of regular drinkers of alcohol was comparable between cases and controls. Among drinkers of alcohol, cases consumed more drinks per day than their controls (Table 1).
Table 1.
Baseline demographic and lifestyle characteristics and urinary biomarkers of current smokers who developed lung cancer (Cases) and those who remained cancer-free (Controls), The Shanghai Cohort Study 1986-2014
| Characteristics or biomarkers* | Cases | Controls | P † |
|---|---|---|---|
| Number of subjects | 325 | 356 | ... |
| Mean age (SD), years | 56.7 (4.9) | 56.7 (4.9) | 0.919 |
| Sex, % | |||
| Men | 100 | 100 | --- |
| Women | --- | --- | |
| Mean body mass index (SD), kg/m2 | 21.3 (2.7) | 21.8 (3.0) | 0.020 |
| Level of education, % | |||
| No formal education | 10.2 | 6.9 | 0.108 |
| Primary (1-6 years) | 34.7 | 31.3 | |
| Secondary and above | 55.1 | 61.8 | |
| Mean no. of cigarettes/day (SD) | 19.8 (8.7) | 15.4 (7.1) | <0.001 |
| Mean no. of years of smoking (SD) | 35.2 (8.3) | 32.2 (10.0) | <0.001 |
| Mean no. of pack-years of cigarettes (SD) | 35.3 (18.4) | 25.2 (14.6) | <0.001 |
| Alcohol drinking, % | |||
| Nondrinkers | 41.9 | 43.3 | 0.710 |
| Regular drinkers | 58.2 | 56.7 | |
| Mean no. of drinks/day (SD)‡ | 3.3 (2.9) | 2.5 (2.3) | 0.004 |
| Urinary biomarkers |
Geometric mean (95% CI) |
||
| Total nicotine (nmol/mg Cr) | 12.44 (11.06-14.00) | 7.56 (6.76-8.46) | <0.001 |
| Total cotinine (nmol/mg Cr) | 13.08 (11.88-14.40) | 7.76 (7.08-8.52) | <0.001 |
| Total 3HC (nmol/mg Cr) | 16.60 (14.80-18.60) | 9.34 (8.38-10.42) | <0.001 |
| TNE (nmol/mg Cr)§ | 46.80 (42.80-51.16) | 28.52 (26.20-31.04) | <0.001 |
| Total NNAL (pmol/mg Cr)¶ | 0.20 (0.20-0.22) | 0.14 (0.14-0.16) | <0.001 |
Abbreviations: SD, standard deviation; NNAL, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol; 3HC, trans-3'-hydroxycotinine; TNE, total nicotine equivalents.
Two-sided Ps were based on t test for continuous variables or chi-square test for categorical variables.
Among regular alcohol drinkers only.
The sum of total nicotine, total cotinine and total 3HC.
Sixteen subjects (8 cases and 8 controls) were excluded due to missing NNAL.
The geometric means of total nicotine, total cotinine, total 3HC, TNE, and total NNAL in urine of cases were statistically significantly higher than those in controls (Table 1). Lung cancer risk increased with increasing level of urinary biomarkers (Table 2). The smoking-adjusted ORs (95% CIs) for lung cancer for the highest relative to the lowest quartile of total nicotine, total cotinine, total 3HC, TNE, and total NNAL were 3.03 (1.80-5.10), 4.70 (2.61-8.46), 4.26 (2.37-7.68), 4.71 (2.61-8.52), and 3.15 (1.86-5.33), respectively (all Ptrend < 0.001). The positive association between total NNAL and lung cancer risk remained statistically significant after adjustment for TNE; the TNE-adjusted OR for the highest quartile of total NNAL was 2.06 (95% CI = 1.17-3.63; Ptrend = 0.025).
Table 2.
Urinary levels of nicotine metabolites and total NNAL in relation to the risk of developing lung cancer, The Shanghai Cohort Study 1986-2014
| Biomarkers in quartile (cut-off values)* | Cases | Controls | OR† (95% CI) |
|---|---|---|---|
| Total nicotine (nmol/mg creatinine) | |||
| 1st (<3.85) | 30 | 86 | 1.00 |
| 2nd (3.85-9.56) | 77 | 91 | 2.10 (1.22-3.61) |
| 3rd (9.57-18.47) | 91 | 90 | 2.35 (1.38-4.00) |
| 4th (>18.47) | 127 | 89 | 3.03 (1.80-5.10) |
| P for trend | <0.001 | ||
| Total cotinine (nmol/mg creatinine) | |||
| 1st (<3.89) | 20 | 80 | 1.00 |
| 2nd (3.89-9.07) | 49 | 93 | 2.00 (1.08-3.71) |
| 3rd (9.08-15.81) | 110 | 94 | 3.35 (1.86-6.04) |
| 4th (>15.81) | 146 | 89 | 4.70 (2.61-8.46) |
| P for trend | <0.001 | ||
| Total 3HC (nmol/mg creatinine) | |||
| 1st (<4.17) | 19 | 79 | 1.00 |
| 2nd (4.17-10.79) | 62 | 89 | 2.31 (1.25-4.28) |
| 3rd (10.80-20.13) | 89 | 89 | 3.02 (1.65-5.51) |
| 4th (>20.13) | 155 | 99 | 4.26 (2.37-7.68) |
| P for trend | <0.001 | ||
| TNE (nmol/mg creatinine)‡ | |||
| 1st (<15.64) | 19 | 78 | 1.00 |
| 2nd (15.64-33.27) | 51 | 93 | 1.91 (1.02-3.58) |
| 3rd (33.28-54.28) | 96 | 90 | 3.26 (1.79-5.93) |
| 4th (>54.28) | 159 | 95 | 4.71 (2.61-8.52) |
| P for trend | <0.001 | ||
| Total NNAL (pmol/mg creatinine)§ | |||
| 1st (<0.09) | 33 | 86 | 1.00 |
| 2nd(0.10-0.15) | 69 | 88 | 1.87 (1.10-3.18) |
| 3rd (0.16-0.25) | 96 | 92 | 2.09 (1.24-3.51) |
| 4th (>0.25) | 119 | 82 | 3.15 (1.86-5.33) |
| P for trend | <0.001 |
Abbreviations: NNAL, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol; 3HC, trans-3′-hydroxycotinine; TNE, total nicotine equivalents; OR, odds ratio; CI, confidence interval.
Adjusted for number of cigarettes per day, number of years of smoking, and matching factors.
The sum of total nicotine, total cotinine and total 3HC.
Sixteen subjects (8 cases and 8 controls) were excluded from this analysis due to missing total NNAL.
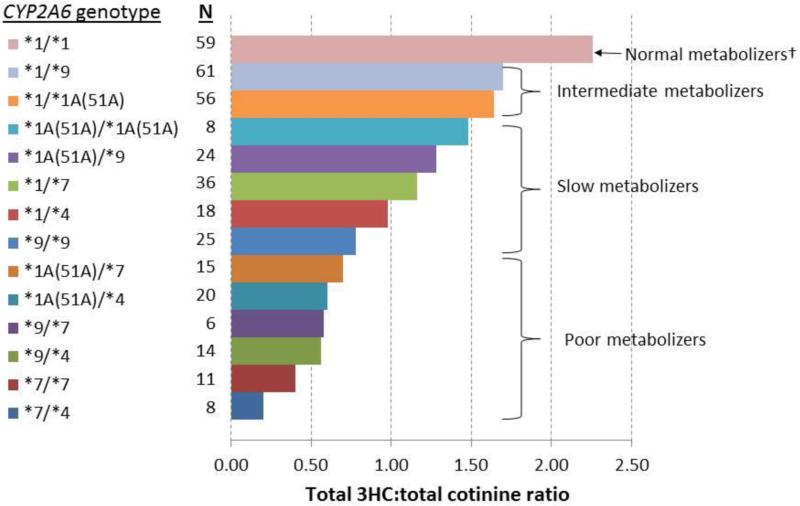
The allele frequencies of CYP2A6*1A(51A), *4, *7, and *9 among controls and cases are shown in Supplemental Table S1. In both controls and cases, *9 was the most common variant allele, followed by *1A(51A) and *7; *4 was the least frequent allele (6.2% in controls). The distributions of CYP2A6 genotypes in controls and cases, respectively, were in Hardy-Weinberg equilibrium (P >0.15) (Supplemental Table S2). The difference in the total 3HC:total cotinine ratio, a quantitative measure of CYP2A6 enzyme activity, was more than 10 fold between the CYP2A6 *1/*1 (normal function) and *7/*4 (loss of function) genotypes in both controls and cases (Figure 1 and Supplemental Table S2). Compared with the normal metabolizers (i.e., CYP2A6*1/*1), the CYP2A6 genotype predicted intermediate metabolizers had a 26% reduced total 3HC:total cotinine ratio, slow metabolizers had a 53% reduced total 3HC:total cotinine ratio, and poor metabolizers had an 78% reduced total 3HC:total cotinine ratio among controls (Figure 1 and Table 3).
Figure 1.
Geometric means of total 3HC:total cotinine ratio by CYP2A6 genotype among control subjects, the Shanghai Cohort Study
Abbreviations: 3HC, total trans-3'-hydroxycotinine.
† See the genotype grouping described in Methods section.
Table 3.
Average levels of smoking intensity and duration and urinary metabolites of cigarette smoke constituents by CYP2A6 genotypes-predicted metabolizers status among control subjects, The Shanghai Cohort Study 1986-2014
| Smoking related variables* |
CYP2A6 genotype predicted metabolizer status† |
P for trend | |||
|---|---|---|---|---|---|
| Normal | Intermediate | Slow | Poor | ||
| Number of subjects (%) | 58 (16.3) | 117 (32.9) | 109 (30.6) | 72 (20.2) | |
| Number of cigarettes/day, mean (SD) | 14.9 (1.0) | 15.5 (0.7) | 15.6 (0.7) | 15.1 (0.7) | 0.869 |
| Age start smoking (years), mean (SD) | 25.3 (1.1) | 24.3 (0.7) | 25.0 (0.8) | 24.6 (0.9) | 0.880 |
| Number of years of smoking, mean (SD) | 31.2 (1.4) | 32.8 (0.9) | 31.7 (1.0) | 32.9 (1.2) | 0.614 |
| Pack-years of smoking, mean (SD) | 24.3 (2.2) | 25.9 (1.4) | 25.3 (1.4) | 24.7 (1.5) | 0.955 |
| Urinary biomarkers (nmol/mg creatinine)‡ | |||||
| Total nicotine | 6.1 (4.5-8.3) | 6.3 (5.1-7.9) | 8.3 (6.6-10.4) | 10.5 (7.9-13.8) | 0.002 |
| Total cotinine | 7.5 (5.8-9.6) | 7.3 (6.1-8.7) | 8.7 (7.3-10.5) | 7.4 (5.9-9.3) | 0.660 |
| Total 3HC | 16.8 (12.8-22.1) | 12.2 (10.1-14.8) | 9.3 (7.7-11.4) | 3.8 (2.9-4.8) | <0.001 |
| TNE§ | 33.1 (26.1-41.8) | 28.2 (23.9-33.4) | 29.1 (24.5-34.5) | 24.9 (20.2-30.8) | 0.132 |
| Total NNAL (pmol/mg creatinine)¶ | 0.16 (0.14-0.20) | 0.14 (0.12-0.16) | 0.16 (0.14-0.18) | 0.14 (0.12-0.16) | 0.283 |
| Total NNAL:TNE ratio (pmol/nmol × 1000)¶ | 4.94 (3.86-6.34) | 5.28 (4.42-6.30) | 5.12 (4.26-6.14) | 5.50 (4.40-6.86) | 0.633 |
| Total 3HC:total cotinine ratio | 2.26 (1.88-2.70) | 1.68 (1.48-1.90) | 1.06 (0.94-1.22) | 0.50 (0.44-0.60) | <0.001 |
Abbreviation: 3HC, trans-3′-hydroxycotinine; NNAL, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol; TNE, total nicotine equivalents.
See the Methods and Figure 1 for the CYP2A6 grouping.
Geometric mean (95% confidence interval)
The sum of total nicotine, total cotinine and total 3HC.
Eight subjects were excluded from this analysis due to missing NNAL.
Urinary biomarkers of nicotine metabolism as well as smoking history across the 4 different CYP2A6-predicted metabolizer stata are shown in Table 3. There was no difference in smoking history (age at starting to smoke, number of cigarettes per day, years of smoking, and pack-years of smoking) between metabolizer status groups. Compared with normal metabolizers, poor metabolizers had a statistically significant 72% higher level of urinary total nicotine, a significant 77% lower total 3HC, and a non-significant 25% lower TNE (Ptrend = 0.132). There was no significant difference in urinary total cotinine, total NNAL and the ratio of total NNAL to TNE among the CYP2A6 predicted metabolizer groups.
The associations between CYP2A6-determined metabolizer status and risk of lung cancer are shown in Table 4. The risk of lung cancer in intermediate and slow metabolizers was comparable with the normal metabolizers. Compared with these three genotype groups combined, poor metabolizers had a statistically significant lower odds ratio of lung cancer (OR = 0.64, 95% CI = 0.43-0.97). Adjustment for number of cigarettes per day and number of years of smoking slightly weakened the association, but the OR remained statistically borderline significant (OR=0.68, 95% CI = 0.44-1.04, P=0.074). Further adjustment for urinary total cotinine did not change the risk estimate (cotinine-adjusted OR = 0.68, 95% CI = 0.44-1.05, P=0.082). Urinary TNE explained some additional effect of CYP2A6 poor metabolizer status on lung cancer risk, suggesting TNE may be a better biomarker than total cotinine for tobacco smoking. However, there was still a statistically non-significant 26% reduced risk of lung cancer for the CYP2A6-determined poor metabolizers (TNE-adjusted OR = 0.74, 95% CI = 0.48-1.15, P=0.18). Additional adjustment for total NNAL did not change the CYP2A6-lung cancer risk (Table 4).
Table 4.
CYP2A6 genotypes-predieted metabolizer status in relation to the risk of developing lung cancer, The Shanghai Cohort Study 1986-2014
| CYP2A6 predicted metabolizer status* | Median of 3HC:total cotinine ratio | Ca/Co† | OR (95% CI)‡ | OR (95% CI)§ | OR (95% CI)¶ |
|---|---|---|---|---|---|
| Normal | 2.19 | 52/58 | 1.00 | 1.00 | 1.00 |
| Intermediate | 1.64 | 120/117 | 1.17 (0.74-1.85) | 1.15 (0.70-1.89) | 1.12 (0.68-1.85) |
| Slow | 1.15 | 108/109 | 1.12 (0.70-1.78) | 1.12 (0.68-1.85) | 1.13 (0.68-1.88) |
| Poor | 0.49 | 45/72 | 0.71 (0.42-1.22) | 0.82 (0.47-1.46) | 0.84 (0.47-1.49) |
| P for trend | 0.204 | 0.488 | 0.587 | ||
| Normal/intermediate/slow | 1.50 | 280/284 | 1.00 | 1.00 | 1.00 |
| Poor | 0.49 | 45/72 | 0.64 (0.43-0.97) | 0.74 (0.48-1.15) | 0.76 (0.49-1.19) |
| P | 0.034 | 0.184 | 0.232 |
See the Methods and Figure 1 for the CYP2A6 grouping.
Ca/Co, number of cases/number of controls; OR, odds ratio; CI, confidence interval.
Adjusted for matching factors only including age, year of enrollment, and neighborhood of residence at recruitment.
In addition to matching factors, ORs were adjusted for number of cigarettes per day, number of years of smoking, and urinary total nicotine equivalent.
In addition to all variables described above, ORs were adjusted for urinary total NNAL (8 cases and 8 controls with missing total NNAL were classified as a separate group in the adjustment)
The association between the CYP2A6 predicted metabolizer status and risk of lung cancer stratified by levels of exposure to cigarette smoking is shown in Table 5. There was no statistically significant association between the CYP2A6 metabolizer status and lung cancer risk in low or high cigarette smoking exposure groups defined by number of cigarettes per day, number of years of smoking, pack-years of smoking, urinary total TNE, or urinary total NNAL (all Ps > 0.05). None of the interaction terms between CYP2A6 genotype-determined metabolizer status and levels of exposure to cigarette smoking was statistically significant (all P for interaction > 0.15).
Table 5.
CYP2A6 genotypes-predieted metabolizer status and risk of lung cancer stratified by levels of exposure to smoking in the Shanghai Cohort Study
| Stratifying variable | Odds ratio* (95% confidence interval) by CYP2A6 metabolizer status† |
P for interaction | ||||
|---|---|---|---|---|---|---|
| Normal | Intermediate | Slow | Poor | P for trend | ||
| <20 cigarettes/day | 0.173 | |||||
| Cases/Controls | 18/33 | 30/55 | 44/54 | 18/38 | ||
| OR (95% CI) | 1.00 | 1.14 (0.54-2.41) | 1.71 (0.83-3.53) | 0.94 (0.41-2.13) | 0.721 | |
| ≥20 cigarettes/day | ||||||
| Cases/Controls | 34/25 | 90/62 | 64/55 | 27/34 | ||
| OR (95% CI) | 1.00 | 1.07 (0.58-1.98) | 0.84 (0.45-1.59) | 0.59 (0.29-1.22) | 0.084 | |
| <30 years of smoking | 0.949 | |||||
| Cases/Controls | 10/23 | 23/40 | 30/45 | 5/23 | ||
| OR (95% CI) | 1.00 | 1.47 (0.58-3.72) | 1.62 (0.66-3.96) | 0.52 (0.15-1.78) | 0.596 | |
| ≥30 years of smoking | ||||||
| Cases/Controls | 42/35 | 97/77 | 78/64 | 40/49 | ||
| OR (95% CI) | 1.00 | 1.10 (0.63-1.91) | 1.06 (0.60-1.88) | 0.71 (0.38-1.32) | 0.249 | |
| <30 pack-years of smoking | 0.479 | |||||
| Cases/Controls | 20/39 | 37/69 | 51/69 | 17/48 | ||
| OR (95% CI) | 1.00 | 1.18 (0.59-2.35) | 1.60 (0.82-3.13) | 0.75 (0.34-1.64) | 0.831 | |
| ≥30 pack-years of smoking | ||||||
| Cases/Controls | 32/19 | 83/48 | 57/40 | 28/24 | ||
| OR (95% CI) | 1.00 | 1.05 (0.53-2.08) | 0.83 (0.41-1.68) | 0.68 (0.31-1.50) | 0.210 | |
| Low TNE (<median) | ||||||
| Cases/Controls | 12/26 | 26/57 | 24/47 | 8/41 | 0.331 | |
| OR (95% CI) | 1.00 | 1.05 (0.44-2.51) | 1.22 (0.50-2.93) | 0.42 (0.15-1.20) | 0.159 | |
| High TNE (≥median) | ||||||
| Cases/Controls | 40/32 | 94/60 | 84/62 | 37/31 | ||
| OR (95% CI) | 1.00 | 1.27 (0.71-2.26) | 1.07 (0.60-1.89) | 0.97 (0.50-1.90) | 0.708 | |
| Low total NNAL (<median)‡ | ||||||
| Cases/Controls | 19/27 | 37/54 | 34/53 | 12/40 | 0.217 | |
| OR (95% CI) | 1.00 | 1.08 (0.50-2.30) | 0.98 (0.46-2.06) | 0.49 (0.20-1.21) | 0.122 | |
| High total NNAL (≥median)‡ | ||||||
| Cases/Controls | 32/30 | 79/58 | 72/54 | 32/32 | ||
| OR (95% CI) | 1.00 | 1.22 (0.66-2.26) | 1.22 (0.66-2.26) | 0.93 (0.46-1.88) | 0.841 | |
Odds ratios (OR) were adjusted for matching factors including age, year of enrollment, and neighborhood of residence at recruitment; CI, confidence interval; TNE, total nicotine equivalents.
See the Methods section for the CYP2A6 functional classifications.
Sixteen subjects (8 cases and 8 controls) were excluded from these analysis due to missing total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL).
We also examined and found no difference in the association between the CYP2A6 genotype-determined metabolizer status and risk of lung cancer by histological type. The CYP2A6-risk association was similar for lung adenocarcinoma (28% reduced risk) and squamous cell carcinoma (23% reduced risk). Neither reaches significance given reduced sample sizes.
DISCUSSION
Utilizing the resources of the Shanghai Cohort Study, we demonstrated that CYP2A6 genotype-determined poor metabolizer status was associated with a statistically significant, 36% reduced risk of lung cancer. Adjustment for number of cigarettes per day and number of years of smoking slightly weakened the association, but there was a statistically borderline significant 32% reduced risk of lung cancer. Additional adjustment for urinary TNE, the best biomarker of daily nicotine intake and total tobacco smoking exposure, further diminished the effect of the CYP2A6 poor metabolizer status on lung cancer risk, but there was still a statistically non-significant 26% reduced risk of lung cancer. Further adjustment for urinary total NNAL did not alter the risk estimate for lung cancer. Although statistically non-significant, a 26% risk reduction of lung cancer associated with the CYP2A6-determined poor metabolizer status suggests a potential important role of CYP2A6 in the development of lung cancer. Future studies with a prospective study design and a larger sample size are warranted to confirm the role of CYP2A6 in the development of lung cancer in smokers.
CYP2A6 catalyzes the α-hydroxylation-mediated bioactivation of the tobacco specific lung carcinogen NNK and NNAL.8 It's not possible to directly measure either NNK or NNAL α-hydroxylation pathway in smokers. If CYP2A6 is an important catalyst of NNK α-hydroxylation, one would predict that poor CYP2A6 metabolizers would have a reduced amount of NNK metabolized by the α-hydroxylation pathway and increased conversion to NNAL.29 In addition, the further metabolism of NNAL by α-hydroxylation would also be decreased in these individuals. Therefore, we investigated the association of CYP2A6 predicted metabolizer status and the urinary ratio of NNAL to TNE (TNE is serving as a proxy for NNK dose). We did not detect any association between this indirect measure of NNK bioactivation and CYP2A6 activity. Future studies are warranted to develop biomarkers that directly measure the NNK activation pathway, such as DNA adducts, that can clarify the role of CYP2A6 in lung carcinogenesis through the NNK α-hydroxylation pathway.
Consistent with the plasma ratio of 3HC to cotinine,12, 21 the present study demonstrated urinary total 3HC:total cotinine ratio as an excellent measure for CYP2A6 enzyme activity. There was a more than 10-fold difference in the total 3HC:total cotinine ratio between the CYP2A6 *1/*1 and *7/*4 genotypes, and a strong dose-dependent relation between CYP2A6 genotypes and phenotypic measures (Figure 1). These data suggest that the urinary total 3HC:total cotinine ratio is a robust phenotypic measure for CYP2A6 activity for ad-libitum smoking.
Nicotine is the main addictive compound in tobacco.30 Approximately 80% of nicotine is inactivated to cotinine in a reaction principally mediated by the enzyme CYP2A6,12 and an association between genetic variation in CYP2A6 activity and smoking behavior has been reported in several, but not all study populations.15-17, 31 An early meta-analysis found no association between individuals with CYP2A6 variant alleles and smoking status or amount smoked.32 In a more recent study of African Americans, cigarette consumption, age of initiation, and nicotine-dependence scores did not differ among CYP2A6 genotype or phenotype groups.33 Whereas, a recent study in a Chinese population found that the CYP2A6 genotype-predicted poor metabolizers consumed fewer cigarettes per day, initiated smoking at a later age, had a shorter duration of smoking, and were more likely to quit smoking than normal metabolizers (these findings were specific to men).34 In the present study, we did not find statistically significant differences in the number of cigarettes smoked per day, age at starting to smoke, number of years of smoking, and pack-years of smoking across different CYP2A6 genotypes in control subjects. However, TNE were 25% lower in poor metabolizers compared to normal metabolizers, although the difference did not reach statistical significance level. The lack of an association for TNE and other smoking measures could be due to the late age of smoking initiation of our study population (23.4 years), and limited access to cigarettes due to rationing and the relatively high cost of cigarettes in Shanghai, China before the 1980s.
The frequencies of CYP2A6 alleles vary widely among different racial/ethnic populations. We focused on the CYP2A6 *4, *7, and *9 alleles because they are common in Asian populations.34 We also genotyped rs1137115, a SNP that captures the CYP2A6*1A(+51A) allele, which results in a significant reduction in nicotine metabolism in European American smokers, as we previously reported.27, 35 Consistent with the data in European Americans, homozygous variant individuals had approximately one-third the total 3HC:total cotinine ratio of individuals with the CYP2A6 *1/*1 genotype (Supplemental Table S2).
A number of studies have assessed the association between CYP2A6 genetic polymorphisms and risk of lung cancer in different populations; however, the results are inconsistent and inconclusive. For example, the earliest report on the CYP2A6 deletion polymorphism and lung cancer risk was a hospital-based case-control study in a Japanese population and reported a 50% reduced risk of lung cancer associated with CYP2A6 deletion (*4).36 A similar study in a French population found no association between CYP2A6 deletion and lung cancer risk.18 In contrast, a hospital-based case-control study in China reported that Han Chinese with the CYP2A6*4 deletion had a two-fold increased risk of lung cancer.19 Four reports of meta-analysis for the association between CYP2A6 genetic polymorphisms and risk of lung cancer were recently published, although these analyses primarily comprised the same individual studies.37-40 All four meta-analyses reported a statistically significant approximately 50% lower crude odds ratio of lung cancer for poor metabolizers (e.g., carriers of two loss-of-function alleles or one loss-of-function allele plus one decreased function allele) in Asians, especially in smokers.38, 39 These smoking-unadjusted results were similar to our findings. Comprehensive adjustment for exposure to cigarette smoking measured by urinary TNE, number of cigarettes per day and years of smoking explained a10% reduced risk of lung cancer associated with the CYP2A6-determined poor metabolizer status. Although statistically non-significant, there is still a 26% reduced risk of lung cancer for smokers with the CYP2A6-determined poor metabolizers after adjustment for the best biomarker of daily nicotine intake and total tobacco smoking exposure, suggesting that CYP2A6 may play a role in the risk of lung cancer independent of its impact on cigarette consumption and smoking patterns.
Several genome-wide association (GWA) studies of lung cancer provided strong evidence of a susceptibility region in 15q25.1 that includes three cholinergic nicotine-receptor genes (CHRNA3, CHRNA4, and CHRNA5) encoding nicotinic acetylcholine receptors,41-43 and in 5p15.33 that contains two genes (TERT and CLPTM1L).44-46 A pooled analysis confirmed these loci in whites but not in Asian populations.47 While CYP2A6 has not been associated with lung cancer in GWA studies, CYP2A6 SNPs have been identified in GWA of cigarette consumption48 and nicotine metabolism.49 The most potent CYP2A6 alleles for CYP2A6 function are structural alterations, which may obscure detection using single nucleotide polymorphisms (SNP)-GWA approaches. There was one GWA study involving more than 17,000 smokers and 997 lung cancer cases in a Japanese population that investigated both copy number polymorphism (CNP) and SNP in CYP2A6 locus in relation to smoking quantity and lung cancer risk.50 That study identified two susceptibility alleles including one CNP (rs8102683) and one SNP (rs11878604). The deletion CNP of rs8102683 was linked to CYP2A6*4 and the variant allele of rs11878604 was linked to deleterious alleles including CYP2A6*7 and *9 and other SNPs. Compared with the wild-type haplotype [having at least one copy of rs8102683 and wild-type allele (T) of rs11878604], individuals carrying either rs8102683 deletion or rs11878604 variant allele (C) (or both) smoked 3-4 fewer cigarettes per day, and had approximately 50% reduced risk of lung cancer. The estimated reduced risk of lung cancer was not adjusted for number of cigarettes smoked per day, thus an altered smoking pattern could be confounding the observed effect of CYP2A6 genetic variants on lung cancer risk. That study also suggested that the CYP2A6*4 was not well tagged by nearby SNPs in the 1000 Genomes Project.50
The inconsistent results of CYP2A6 and lung cancer risk among previous studies could be attributed to several factors: 1) possible measurement error in genotyping of CYP2A6 due to the altered gene structure resulting from the deletion polymorphism; 2) different distributions of CYP2A6 variants and allele frequencies across different racial/ethnic groups. For example, the CYP2A6*7 allele is absent in both Caucasian and African populations but is common in Asian populations (5.7-12.5%), whereas CYP2A6*2 and *12 are present in Caucasians (3-5%) but absent or extremely rare in Asians;22, 34 3) inclusion of smokers and nonsmokers in prior studies; 4) the lack of adjustment for total tobacco smoking exposure such as urinary TNE; 5) potential selection bias due to hospital-based retrospective study design; and 6) small sample size and low statistical power. The present study circumvented most of these limitations by using a prospective study design, including only current smokers whose smoking status was biochemically verified, enrolling only Han Chinese, and genotyped for CYP2A6 variants that are relatively common in Asian populations. We used a novel genotyping approach that more accurately determined genetic variants of CYP2A6. In addition, the present study included a new SNP, CYP2A6*1(+52A) that has not been studied with lung cancer risk. More importantly, the CYP2A6 genotype-determined metabolizer status was confirmed by the urinary 3HC:total cotinine ratio, a functional measure of CYP2A6 activity, indicating the high quality of genotyping and phenotyping data in the present study. However, a limitation of the present study was the relatively small sample size.
The present study was the first to show that urinary TNE was a strong predictor of lung cancer risk. TNE is a comprehensive measure of nicotine uptake and a better measure of tobacco smoke exposure than reported cigarettes per day or total cotinine, which was demonstrated in the adjustment for TNE versus total cotinine on the association between CYP2A6 poor metabolizers and lung cancer risk. Future studies should consider using urinary TNE as a biomarker of total tobacco smoking exposure for disease risk assessment.
In summary, using prospectively collected urine and DNA samples from participants of the Shanghai Cohort Study, we demonstrated that common genetic polymorphisms of CYP2A6 significantly reduced the metabolic conversion of cotinine to 3HC, and resulted in increased excretion of total nicotine and decreased excretion of TNE. The comprehensive adjustment for total tobacco smoking exposure using the validated urinary biomarker TNE plus self-reported smoking intensity and duration partially explained the significantly reduced risk of lung cancer in smokers with the CYP2A6 genotype-determined poor metabolizer status. A statistically non-significant 26% reduced risk of lung cancer for the poor metabolizers requires future studies to confirm the role of CYP2A6 in the development of lung cancer.
Supplementary Material
NOVELTY AND IMPACT.
Smoking dramatically increases lung cancer risk, yet only about one-fifth of long-term heavy smokers develop the disease. Cytochrome P450 2A6 (CYP2A6) catalyzes nicotine metabolism. Genetic variation in CYP2A6 may affect smoking behavior and contribute to lung cancer risk. The authors examined the association between CYP2A6 genotype determined metabolizer status, verified by phenotypic measures, and the risk of developing lung cancer in a prospective cohort of Chinese men in Shanghai, China. There was a statistically significant reduced risk of lung cancer in CYP2A6 poor metabolizers that is partially explained by smoking intensity and duration, and urinary total nicotine equivalents, the best biomarker of daily nicotine intake and total tobacco smoking exposure. These data clarify the role of CYP2A6 genetic polymorphisms in the metabolism of tobacco smoke constituents and the development of lung cancer in Chinese smokers.
ACKNOWLEDGEMENTS
Grant support: USPHS grants R01CA 043092, R01 CA129534, R01 CA144034, R01 CA81301, and UM1 CA182876.
We thank Katherine Wickham for carrying out all nicotine and metabolite analyses, and Xue-Li Wang of the Shanghai Cancer Institute for supervising the field work of the Shanghai Cohort Study. We also thank the Shanghai Cancer Registry for assistance with the identification of lung cancer cases via database linkage.
REFERENCES
- 1.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risk to Humansed. Vol. 83. IARC Scientific Publications; Lyon, France: 2004. Tobacco smoke and involuntary smoking. pp. 36–40. [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12:399–408. doi: 10.1016/S1470-2045(10)70126-1. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Research opportunities related to establishing standards for tobacco products under the Family Smoking Prevention and Tobacco Control Act. Nicotine Tob Res. 2012;14:18–28. doi: 10.1093/ntr/ntq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–44. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 5.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humansed. Vol. 89. IARC Scientific Publications; Lyon, France: 2007. Smokeless tobacco and tobacco-specific nitrosamines. pp. 421–583. [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J-M, Koh WP, Murphy SE, Fan Y, Wang R, Carmella SG, Han S, Wickham K, Gao YT, Yu MC, Hecht SS. Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;69:2990–5. doi: 10.1158/0008-5472.CAN-08-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J-M, Gao YT, Murphy SE, Carmella SG, Wang R, Zhong Y, Moy KA, Davis AB, Tao L, Chen M, Han S, Nelson HH, et al. Urinary levels of cigarette smoke constituent metabolites are prospectively associated with lung cancer development in smokers. Cancer Res. 2011;71:6749–57. doi: 10.1158/0008-5472.CAN-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jalas JR, Hecht SS, Murphy SE. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem Res Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 9.Dicke KE, Skrlin SM, Murphy SE. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-butanone metabolism by cytochrome P450 2B6. Drug Metab Dispos. 2005;33:1760–4. doi: 10.1124/dmd.105.006718. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, D'Agostino J, Wu H, Zhang QY, von Weymarn L, Murphy SE, Ding X. CYP2A13: variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J Pharmacol Exp Ther. 2007;323:570–8. doi: 10.1124/jpet.107.127068. [DOI] [PubMed] [Google Scholar]
- 11.Gervot L, Rochat B, Gautier JC, Bohnenstengel F, Kroemer H, de Berardinis V, Martin H, Beaune P, de Waziers I. Human CYP2B6: expression, inducibility and catalytic activities. Pharmacogenetics. 1999;9:295–306. [PubMed] [Google Scholar]
- 12.Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 13.von Weymarn LB, Brown KM, Murphy SE. Inactivation of CYP2A6 and CYP2A13 during nicotine metabolism. J Pharmacol Exp Ther. 2006;316:295–303. doi: 10.1124/jpet.105.091306. [DOI] [PubMed] [Google Scholar]
- 14.Scherer G, Engl J, Urban M, Gilch G, Janket D, Riedel K. Relationship between machine-derived smoke yields and biomarkers in cigarette smokers in Germany. Regul Toxicol Pharmacol. 2007;47:171–83. doi: 10.1016/j.yrtph.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Fujieda M, Yamazaki H, Saito T, Kiyotani K, Gyamfi MA, Sakurai M, Dosaka-Akita H, Sawamura Y, Yokota J, Kunitoh H, Kamataki T. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 2004;25:2451–8. doi: 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- 16.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–6. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wassenaar CA, Ye Y, Cai Q, Aldrich MC, Knight J, Spitz MR, Wu X, Blot WJ, Tyndale RF. CYP2A6 reduced activity gene variants confer reduction in lung cancer risk in African American smokers--findings from two independent populations. Carcinogenesis. 2015;36:99–103. doi: 10.1093/carcin/bgu235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loriot MA, Rebuissou S, Oscarson M, Cenee S, Miyamoto M, Ariyoshi N, Kamataki T, Hemon D, Beaune P, Stucker I. Genetic polymorphisms of cytochrome P450 2A6 in a case-control study on lung cancer in a French population. Pharmacogenetics. 2001;11:39–44. doi: 10.1097/00008571-200102000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Tan W, Chen GF, Xing DY, Song CY, Kadlubar FF, Lin DX. Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. Int J Cancer. 2001;95:96–101. doi: 10.1002/1097-0215(20010320)95:2<96::aid-ijc1017>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, Lin D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–61. [PubMed] [Google Scholar]
- 21.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80:457–67. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8:1385–402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 23.Derby KS, Cuthrell K, Caberto C, Carmella SG, Franke AA, Hecht SS, Murphy SE, Le Marchand L. Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:3526–35. doi: 10.1158/1055-9965.EPI-08-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J-M, Ross RK, Wang XL, Gao YT, Henderson BE, Yu MC. Morbidity and mortality in relation to cigarette smoking in Shanghai, China. A prospective male cohort study. JAMA. 1996;275:1646–50. [PubMed] [Google Scholar]
- 25.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, Le Marchand L. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35:2526–33. doi: 10.1093/carcin/bgu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, Kwon JT, McLeod HL, Yokoi T. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80:282–97. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Bloom AJ, Harari O, Martinez M, Zhang X, McDonald SA, Murphy SE, Goate A. A compensatory effect upon splicing results in normal function of the CYP2A6*14 allele. Pharmacogenet Genomics. 2013;23:107–16. doi: 10.1097/FPC.0b013e32835caf7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.London SJ, Yuan J-M, Chung FL, Gao YT, Coetzee GA, Ross RK, Yu MC. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: a prospective study of men in Shanghai, China. Lancet. 2000;356:724–9. doi: 10.1016/S0140-6736(00)02631-3. [DOI] [PubMed] [Google Scholar]
- 29.Hecht SS, Trushin N, Rigotty J, Carmella SG, Borukhova A, Akerkar S, Rivenson A. Complete inhibition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced rat lung tumorigenesis and favorable modification of biomarkers by phenethyl isothiocyanate. Cancer Epidemiol Biomarkers Prev. 1996;5:645–52. [PubMed] [Google Scholar]
- 30.Benowitz NL. Pharmacology of nicotine: addiction, smoking-induced disease, and therapeutics. Annu Rev Pharmacol Toxicol. 2009;49:57–71. doi: 10.1146/annurev.pharmtox.48.113006.094742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11:400–9. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 32.Carter B, Long T, Cinciripini P. A meta-analytic review of the CYP2A6 genotype and smoking behavior. Nicotine Tob Res. 2004;6:221–7. doi: 10.1080/14622200410001676387. [DOI] [PubMed] [Google Scholar]
- 33.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, Tyndale RF. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85:635–43. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, David SP, Tyndale RF, Wang H, Zhou Q, Ding P, He YH, Yu XQ, Chen W, Crump C, Wen XZ, Chen WQ. Associations of CYP2A6 genotype with smoking behaviors in southern China. Addiction. 2011;106:985–94. doi: 10.1111/j.1360-0443.2010.03353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloom J, Hinrichs AL, Wang JC, von Weymarn LB, Kharasch ED, Bierut LJ, Goate A, Murphy SE. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet Genomics. 2011;21:403–16. doi: 10.1097/FPC.0b013e328346e8c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto M, Umetsu Y, Dosaka-Akita H, Sawamura Y, Yokota J, Kunitoh H, Nemoto N, Sato K, Ariyoshi N, Kamataki T. CYP2A6 gene deletion reduces susceptibility to lung cancer. Biochem Biophys Res Commun. 1999;261:658–60. doi: 10.1006/bbrc.1999.1089. [DOI] [PubMed] [Google Scholar]
- 37.Liu ZB, Shu J, Wang LP, Jin C, Lou ZX. Cytochrome P450 2A6 deletion polymorphism and risk of lung cancer: a meta-analysis. Mol Biol Rep. 2013;40:5255–9. doi: 10.1007/s11033-013-2625-0. [DOI] [PubMed] [Google Scholar]
- 38.Liu T, Xie CB, Ma WJ, Chen WQ. Association between CYP2A6 genetic polymorphisms and lung cancer: a meta-analysis of case-control studies. Environ Mol Mutagen. 2013;54:133–40. doi: 10.1002/em.21751. [DOI] [PubMed] [Google Scholar]
- 39.Liu YL, Xu Y, Li F, Chen H, Guo SL. CYP2A6 deletion polymorphism is associated with decreased susceptibility of lung cancer in Asian smokers: a meta-analysis. Tumour Biol. 2013;34:2651–7. doi: 10.1007/s13277-013-0815-y. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Zang W, Liu J, Xie D, Ji W, Pan Y, Li Z, Shen J, Shi Y. Association of CYP2A6*4 with susceptibility of lung cancer: a meta-analysis. PLoS ONE. 2013;8:e59556. doi: 10.1371/journal.pone.0059556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 43.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truong T, Hung RJ, Amos CI, Wu X, Bickeboller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, Dienemann H, Kaaks R, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–71. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loukola A, Buchwald J, Gupta R, Palviainen T, Hallfors J, Tikkanen E, Korhonen T, Ollikainen M, Sarin AP, Ripatti S, Lehtimaki T, Raitakari O, et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. 2015;11:e1005498. doi: 10.1371/journal.pgen.1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumasaka N, Aoki M, Okada Y, Takahashi A, Ozaki K, Mushiroda T, Hirota T, Tamari M, Tanaka T, Nakamura Y, Kamatani N, Kubo M. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS ONE. 2012;7:e44507. doi: 10.1371/journal.pone.0044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.