Abstract
Background and Aims Limestone karst areas possess high floral diversity and endemism. The genus Primulina, which contributes to the unique calcicole flora, has high species richness and exhibit specific soil-based habitat associations that are mainly distributed on calcareous karst soils. The adaptive molecular evolutionary mechanism of the genus to karst calcium-rich environments is still not well understood. The Ca2+-permeable channel TPC1 was used in this study to test whether its gene is involved in the local adaptation of Primulina to karst high-calcium soil environments.
Methods Specific amplification and sequencing primers were designed and used to amplify the full-length coding sequences of TPC1 from cDNA of 76 Primulina species. The sequence alignment without recombination and the corresponding reconstructed phylogeny tree were used in molecular evolutionary analyses at the nucleic acid level and amino acid level, respectively. Finally, the identified sites under positive selection were labelled on the predicted secondary structure of TPC1.
Key Results Seventy-six full-length coding sequences of Primulina TPC1 were obtained. The length of the sequences varied between 2220 and 2286 bp and the insertion/deletion was located at the 5′ end of the sequences. No signal of substitution saturation was detected in the sequences, while significant recombination breakpoints were detected. The molecular evolutionary analyses showed that TPC1 was dominated by purifying selection and the selective pressures were not significantly different among species lineages. However, significant signals of positive selection were detected at both TPC1 codon level and amino acid level, and five sites under positive selective pressure were identified by at least three different methods.
Conclusions The Ca2+-permeable channel TPC1 may be involved in the local adaptation of Primulina to karst Ca2+-rich environments. Different species lineages suffered similar selective pressure associated with calcium in karst environments, and episodic diversifying selection at a few sites may play a major role in the molecular evolution of Primulina TPC1.
Keywords: Primulina, calcium, karst, Ca2+-permeable channel, TPC1, molecular evolution, positive selection
INTRODUCTION
Elucidating the selective and neutral forces underlying molecular evolution is fundamental to understanding the genetic basis of adaptation. As plants are sessile organisms during most of their life cycles, edaphic factors, such as physical and chemical characteristics, are thought to confer strong selective pressures on their local adaptation to different environments, but relatively little is known about which genes are involved in such adaptation (Turner et al., 2010). Limestone karst topography is mainly composed of calcium carbonate sedimentary rock and has unique geological conditions and natural environments. Karst soils, which were formed from carbonate bedrock, are usually characterized by higher concentrations of calcium (Ca) and magnesium (Mg), higher pH and lower water storage capacities than the surrounding non-karst soils (Hao et al., 2015). Thus, the unique physical and chemical profiles of karst soils generate a range of microclimatic and edaphic habitats that are centres of high diversity and endemism. Plants on these limestone rock outcrops are usually adapted to severe summer drought, extreme temperature fluctuations and unusual soil chemistry (because of proximity to bedrock). The underlying genetic basis of edaphic adaptation for karst endemics, however, remains poorly understood.
Calcium is an essential macronutrient element in plants; it plays a crucial role in regulating nearly all aspects of plant growth and development (Hepler, 2005). For example, Ca2+ plays an important role in determining the structure and function of the plant cell wall and regulating polarized plant cell growth and elongation (Hepler, 2005; Konrad et al., 2011). Furthermore, as the most ubiquitous secondary messenger, Ca2+ senses and responds to a range of abiotic and biotic environmental stimuli (Kudla et al., 2010). Calcium is absorbed by roots from soil solution and then transported to other organs through xylem in the form of Ca2+ or an organic acid–calcium complex. Calcium may traverse the roots either through the symplast pathway or through the apoplast pathway. In plant cells, calcium is mainly stored in the cell wall and vacuoles, and the endoplasmic reticulum is also necessary in regulating the balance of the intracellular calcium ion concentration.
The influx and efflux of cellular Ca2+ are mainly mediated by membrane transport proteins: Ca2+-permeable channels, Ca2+-ATPase and H+/Ca2+ antiporters (Furuichi et al., 2008; Spalding and Harper, 2011). Calcium-permeable channels in plant cells modulate the cytosolic Ca2+ concentration by regulating Ca2+ influx through the plasma membrane and Ca2+ release from intracellular Ca2+ pools; thus, Ca2+-permeable channels would be responsible for the elevation of cytosolic Ca2+ (Swarbreck et al., 2013). Plants possess several calcium channels, including two-pore channels (TPCs), cyclic nucleotide-gated channels (CNGCs), glutamate receptor homologues (GLRs) and mechanosensitive Ca2+-permeable channels (MSCCs). These channels are predominantly involved in signal transduction and play important roles in various crucial cellular responses, such as hormone activity, flowering, pathogen defence and salt stress (Jammes et al., 2011; Swarbreck et al., 2013).
According to their gating mechanisms, Ca2+-permeable channels can be further classified into three subtypes: voltage-dependent, ligand-activated and stretch-activated types (Furuichi et al., 2008). The slow-activating vacuolar (SV) channel is a voltage-dependent calcium channel with slow activation kinetics. The SV channel mainly localizes in vacuolar membranes and seems to be widespread in terrestrial plants, including ferns and liverworts (Pottosin and Schonknecht, 2007). Plant SV channels were first cloned from Arabidopsis thaliana and are encoded by AtTPC1; the SV channel was therefore also named the two-pore channel 1 (TPC1) (Furuichi et al., 2001). The Saccharomyces cerevisiae calcium-channel protein CCH1 shares high homology with the α-subunit of the animal voltage-dependent Ca2+ channel, and the growth rate of yeast cch1 mutant, which is defective in Ca2+ uptake, can be suppressed in medium with lower Ca2+ concentration (Fischer et al., 1997). AtTPC1 can rescue Ca2+ uptake activity after heterologous expression in yeast mutant cch1, suggesting that AtTPC1 encodes a Ca2+-permeable channel that has the ability to transport Ca2+ across the plasma membrane of the cch1 mutant (Furuichi et al., 2001). Using a similar heterologous expression system, homologues of AtTPC1 were identified successively from rice (OsTPC1) (Hashimoto et al., 2004; Kurusu et al., 2004), tobacco (NtTPC1s) (Kadota et al., 2004) and wheat (TaTPC1) (Wang et al., 2005). Calcium-permeable channels of CNGCs, GLRs and MSCCs all belong to large multigene families in most plant species. For example, the CNGC, GLR and MSCC families consist of 20 members (subdivided into five subfamily groups), 20 homologues (subdivided into three clades) and two members (MCA1 and MCA2) in the Arabidopsis genome, respectively (Lacombe et al., 2001; Mäser et al., 2001). However, except in tobacco (Nicotiana tabacum) and mosses (Physcomitrella patens), where there are two and nine copies of TPCs, respectively (Verret et al., 2010), only one member of the TPCs was identified in Arabidopsis and most of the other plants (Furuichi et al., 2001; Schönknecht, 2013).
All known plant TPCs have similar transmembrane topology, comprising two fused domains, each with six transmembrane helices and one highly conserved pore loop (P-loop). Between the two homologous transmembrane domains is the cytosolic loop, containing two Ca2+-binding EF hands (Hedrich and Marten, 2011). Previous studies mainly focused on characterizing the functions of TPC1 in model species (Kadota et al., 2004; Kurusu et al., 2004; Peiter, 2011; Wang et al., 2005); however, the molecular evolution of TPC1 in plants has not been studied.
The genus Primulina is a typical limestone karst or karst cave plant. This genus is a monophyletic group comprising more than 150 species widely distributed throughout the limestone karst areas in southern China. However, almost all Primulina species exhibit specific soil-based habitat associations, with the majority of species occurring in calcareous soils that originated from limestone bedrock (Hao et al., 2015). Previous biological stoichiometry analysis revealed that the leaf concentration of Ca2+ exhibited great variation among species, ranging from 1·4 to 20·5 % (dry mass) and the average concentration of leaf Ca2+ in the Primulina genus was ∼3-fold higher than that found in other plants in China (Hao et al., 2015). The leaf Ca2+ concentration in Primulina may reflect its availability in the soil (Hao et al., 2015). We hypothesized that the extremely high Ca2+ environment in karst soil is a strong selective pressure for Primulina and has consequently led to species divergence based on selection on species-specific physiological traits.
In this study, we chose the Ca2+-permeable channel TPC1 from the Primulina genus as the target to explore whether the gene involved in local adaptation to high-calcium soil environments. For this purpose, we obtained full-length coding sequences from 76 Primulina species, covering the main distribution areas and soil habitat types in China, and then used molecular evolutionary methods to test whether positive selection or selective constraints have arisen in the gene. As far as we know, this is the first study of the molecular evolution of plant TPC1 and the first attempt to explore the adaptive molecular evolutionary mechanism of Primulina to a karst high-calcium soil habitat.
MATERIALS AND METHODS
Plant materials
A total of 76 Primulina species were collected in the field investigation and used in molecular analysis. The closely related species Hemiboea henryi was used as the outgroup in this study. These 76 Primulina species represent nearly the whole of the distribution range of this genus and the typical soil environments in China (Supplementary Data Fig. S1). Specific information on the plant materials used in this study is given in Supplementary Data Table S1.
RNA extraction and reverse transcription
Fresh leaves were harvested and immediately frozen in liquid nitrogen, and then stored at −70 °C until use. Total RNA was extracted using RNAiso Plus (Takara) according to the protocol recommended by the manufacturer. First-strand cDNA was synthesized using PrimeScript Reverse Transcriptase (Takara) according to the manufacturer’s instructions.
TPC1 amplification and sequencing
Transcriptome resources of 11 Primulina species were constructed in previous work (Ai et al., 2015). The full-length coding sequence of AtTPC1 (AT4G03560) was used as the query in a BLAST transcriptome search. Seven full-length sequences and four partial-length sequences were obtained from Primulina species. The full-length sequences were then aligned using ClustalW implemented in MEGA v5.2 (Tamura et al., 2011), and specific amplification primers were designed using Primer Premier v5.0 on conserved regions of the alignment of full-length Primulina TPC1 gene sequences: TPC-S0 (5′-ATCAGTTGGCACCACATTCACT-3′) and TPC-A0 (5′- GACAAAAACATGCTCACATTACACA-3′) (Supplementary Data Fig. S2). The length of the seven full-length sequences that were obtained varied from 2220 to 2247 bp according to the alignment, and we therefore designed another two pairs of internal sequencing primers to ensure the convenience of sequencing: TPC-F2 (5′-GCTGGTTGGCGTTCGTTATCT-3′) andTPC-R2 (5′-AATGTAAGGCCACGGGGAGA-3′), and TPC-F3 (5′-TTCTGCTCAAGGGTTTTGGC-3′) and TPC-R3 (5′-CCAACGCGATTCCTTGTATGC-3′) (Supplementary Data Fig. S2).
To obtain the full-length coding sequences of TPC1 from leaf cDNA of Primulina, polymerase chain reactions (PCRs) were carried out in a total volume of 50 μL, consisting of ∼5 ng of cDNA, 1 μL of 1·25 U μL−1 PrimerSTAR GXL DNA Polymerase (TaKaRa), 4 μL of 2·5 mm dNTP mixture (TaKaRa), 10 μL of 5× PrimeSTAR GXL buffer (TaKaRa) and 1·5 μL of each primer (TPC-S0 and TPC-A0), made up to 50 μL with sterile distilled water. The PCR reactions were carried out on a LabCycler Standard Plus (SensoQuest) with the following PCR reaction conditions: initial denaturation step of 1 min at 98 °C, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 15 s, extension at 68 °C for 2·4 min, and final extension at 68 °C for 10 min.
The amplified products were assessed by gel electrophoresis on 1 % agarose gels containing Goldview. The PCR products were then sequenced in both directions after purification using the amplification primers (TPC-S0, TPC-A0) and two additional pairs of internal sequencing primers (TPC-F2 and TPC-R2, TPC-F3 and TPC-R3). The sequencing results were assembled using SeqMan v7.1.0 implemented in the Lasergene software package (DNASTAR) and proof-read manually.
Sequence alignment
The assembled sequences were firstly translated into amino acid sequences using MEGA v5.2, and then multiple sequence alignment of the amino acid sequences was performed using the software MAFFT v7.158 (Katoh and Standley, 2013). After checked carefully by eye, the amino acid alignment was converted into the corresponding codon-based nucleotide multiple sequence alignment using the program PAL2NAL (http://www.bork.embl.de/pal2nal/) (Suyama et al., 2006).
Test for substitution saturation
Substitution saturation will decrease phylogenetic information contained in sequences, and phylogeny constructed by sequences that have experienced full substitution saturation will not reflect phylogenetic relationships (Xia et al., 2003). Furthermore, saturation of synonymous sites will lead to the underestimation of the synonymous substitution rate (dS), which will elevate the nonsynonymous/synonymous substitution rate ratio (dN/dS), and thus may result in misleading positive selection analysis (Lynn et al., 2004). Therefore, we first examined substitution saturation in the aligned sequences using DAMBE v5.3.48 (Xia, 2013). The entropy-based index of substitution saturation (Iss) was used together with plotting transition and transversion rates against a corrected genetic distance to evaluate whether the sequences showed signs of saturation.
Test for recombination
It is well known that recombination can affect phylogenetic reconstruction (Posada and Crandall, 2002) and positive selection analysis (Shriner et al., 2003; Anisimova et al., 2003). To overcome the noise of recombination, we used the Genetic Algorithms for Recombination Detection (GARD) (Kosakovsky Pond et al., 2006a, b) and the Single Breakpoint (SBP) (Kosakovsky Pond et al., 2006a) methods implemented in the Datamonkey web server (http://www.datamonkey.org/) (Delport et al., 2010) to test the occurrence of recombination breakpoints in the alignments of Primulina TPC1 sequences and also determine the precise breakpoint locations.
Phylogenetic reconstruction
MrModeltest v2.3 (Nylander, 2008) was used to test the most appropriate molecular evolutionary model for our data set using the Akaike information criterion (AIC). MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003) was used to reconstruct phylogenetic relationships. The Markov chain Monte Carlo (MCMC) search was run for 10 000 000 generations and sampled every 100 generations. The remaining trees were concatenated to construct the majority rule consensus tree and calculate the posterior probability after the burn-in samples (25 %) had been discarded. The final phylogenetic tree was displayed with TreeView v1.6.6 (Page, 1996).
Selection analysis at nucleic acid level
Codon-based multiple sequence alignment of PrimulinaTPC1 and the Bayes phylogenetic tree were used in subsequent molecular evolution analysis. The ratio of non-synonymous to synonymous substitution rates, ω (ω = dN/dS), is an effective indicator of selective pressure in molecular level, where values of 1, <1 and >1 indicate neutral selection, purifying selection and positive selection, respectively (Yang and Bielawski, 2000). A series of codon-based maximum likelihood models implemented in the codeml program of PAML software v4.7 (Yang, 2007) were employed to evaluate the influence of natural selection acting on Primulina TPC1. Different initial ω values were used in each model to avoid local optima (Suzuki and Nei, 2001).
Firstly, the branch models, which allow the ω ratio to vary among branches in the phylogeny (Yang, 1998; Yang and Nielsen, 1998), were used to test whether the selective pressure is variable among different branches within the gene tree, and the one-ratio model (M0) and the free-ratio model (Mf) were compared in this analysis. The M0 model assumes all branches and sites of the tree have the same ω ratio, while the Mf model assumes each branch has an independent ω ratio. A likelihood ratio test (LRT) was used to compare the two models (degrees of freedom = total number of branches − 1) and the model with higher likelihood value was assumed to fit the data better (Yang and Nielsen, 1998; Bielawski and Yang, 2003).
Secondly, the site-specific models, which allow the ω ratio to vary among codons or amino acid sites, were employed to test the variance of selective pressure among sites and also to identify the particular sites that are under positive selection (Nielsen and Yang, 1998; Yang et al., 2000). Four pairs of nested models implemented in site models were compared using LRTs: M0 (one-ratio model) versus M3 (discrete model), M1a (neutral model) versus M2a (selection model), M7 (β) versus M8 (β and ω) and M8a (β and ω = 1) versus M8 (β and ω). With appropriate degrees of freedom (the difference in the number of parameters between the two models), the LRT was conducted by comparing twice the log likelihood difference between nested models with the χ2 distribution (Nielsen and Yang, 1998; Yang et al., 2000; Yang, 1998). The significant LRT suggests that the alternative model fits the data significantly better than the corresponding null model and the ω ratio is significantly different from 1. In this case, the Bayes empirical Bayes (BEB) method (Yang et al., 2005) was used to identify sites under positive selection. Sites from the class with ω > 1 and having a higher posterior probability (>95 %) were inferred to be under positive selection.
As predefined biological hypotheses about which Primulina TPC1 genes may be subject to natural selection were unavailable, the Branch-site REL (BS-REL) model (Kosakovsky Pond et al., 2011) implemented in HyPhy v2.2.1 software (Kosakovsky Pond et al., 2005a) was employed to test episodic diversifying selection. BS-REL does not require the identification of foreground branches (branches under positive selection) and background branches (branches that lack positive selection) a priori.
In addition, four other methods implemented in the Datamonkey web server (Delport et al., 2010) were also applied to detect sites under positive selection, including the single likelihood ancestor counting (SLAC) method, the fixed effect likelihood (FEL) method (Kosakovsky Pond and Frost, 2005b), the mixed effects model of evolution (MEME) (Murrell et al., 2012) method and the fast unconstrained Bayesian approximation (FUBAR) method (Murrell et al., 2013). For the SLAC, FEL and MEME methods, sites with P-values <0·1 were considered as candidates under positive selection, while for the FUBAR method a posterior probability of >0·9 was used as the cut-off value.
Selection analysis at amino acid level
Although the ω ratio is a very useful measurement to detect selective pressure at the protein level, this method does not consider the impact of non-synonymous substitution on the physicochemical properties of amino acids and cannot be used to determine how the overall structure and function of the protein was influenced by the identified selective pressure. In addition, when the sequences are highly conserved, the signal of positive selection may not be detected using the ω ratio method (McClellan et al., 2005). In this study, Primulina TPC1 was further analysed using the program TreeSAAP v3.2 (Woolley et al., 2003) to evaluate positive selection at the amino acid level, and also to provide further support of the results we obtained with PAML and Datamonkey.
Multiple sequence alignment of TPC1 and the Bayes phylogenetic tree were used in TreeSAAP analysis. Ancestral states were reconstructed using the Baseml program implemented in PAML with the general-time reversible (GTR) model. The default 31 physicochemical amino acid properties were used and the magnitudes of non-synonymous substitution changes were divided into eight magnitude categories according to the change in a specific amino acid property. Categories 1–3 indicate conservative substitutions while categories 6–8 indicate radical substitutions. A Z-score was then calculated for each category; the value indicated the direction of selection, a positive Z-score meaning positive selection and a negative Z-score meaning negative selection. Only amino acid properties that fell into categories 6–8 and with significant positive Z-scores were considered to be under positive destabilizing selection. Subsequently, sliding window analyses with window size 20 were performed to identify regions that differed significantly from a nearly neutral model in the protein, and then the particular amino acid sites within the regions that were under positive selection for each property were identified.
TPC1 secondary structure prediction
According to the phylogenetic relationships of Primulina species (Kang et al., 2014), Primulina heterotricha is located in the basal position in the phylogeny. Therefore, the P. heterotricha TPC1 amino acid sequence, named PhTPC1, was used as a template to predict the secondary structure of Primulina TPC1 in this study. Kyte–Doolittle hydropathy plots (Kyte and Doolittle, 1982) were used to display the hydropathic character and simultaneously indicate potential transmembrane regions of PhTPC1. The THMHH program (Krogh et al., 2001) was also used to predict the transmembrane regions, and the conserved transmembrane domains of PhTPC1 were determined by comparing with the structures of AtTPC1 (Furuichi et al., 2001; Peiter, 2011) and NtTPC1s (Kadota et al., 2004). Finally, TOPO2 (Johns, 2010) was used to display the predicted secondary structure of PhTPC1 and the identified sites under positive selection were mapped on the structure.
RESULTS
Amplification and sequencing of Primulina TPC1
Only single bands were obtained from cDNA of Primulina species after PCR amplification, and the PCR results were sent to direct sequence in both directions after recycling. According to the amplification results and the transcriptome resources of Primulina (Ai et al., 2015), we can conclude that Primulina TPC1 should be a single-copy gene. After assembly, the untranslated regions (UTRs) were removed and only the open reading frame (ORF) regions remained. The results showed that the length of coding sequences varied between 2220 and 2286 bp, indicating that Primulina TPC1 coding regions may contain insertion/deletion mutations. We obtained full-length coding sequences of TPC1 from 76 Primulina species and one Hemiboea species; all sequences have been deposited in NCBI and accession numbers are listed in Table S1. The obtained sequences were used in subsequent phylogenetic reconstruction and selection analysis after stop codons had been deleted.
Multiple sequence alignment and phylogenetic reconstruction
As insertion/deletion may exist in Primulina TPC1 sequences, all sequence alignments were firstly performed at amino acid level, and then the amino acid alignments were converted to the corresponding alignment of codons. According to the codon-based sequence alignments, the insertion/deletion regions were mainly located at the 5′-end of the sequences and resulted in the unequal length of coding regions among species.
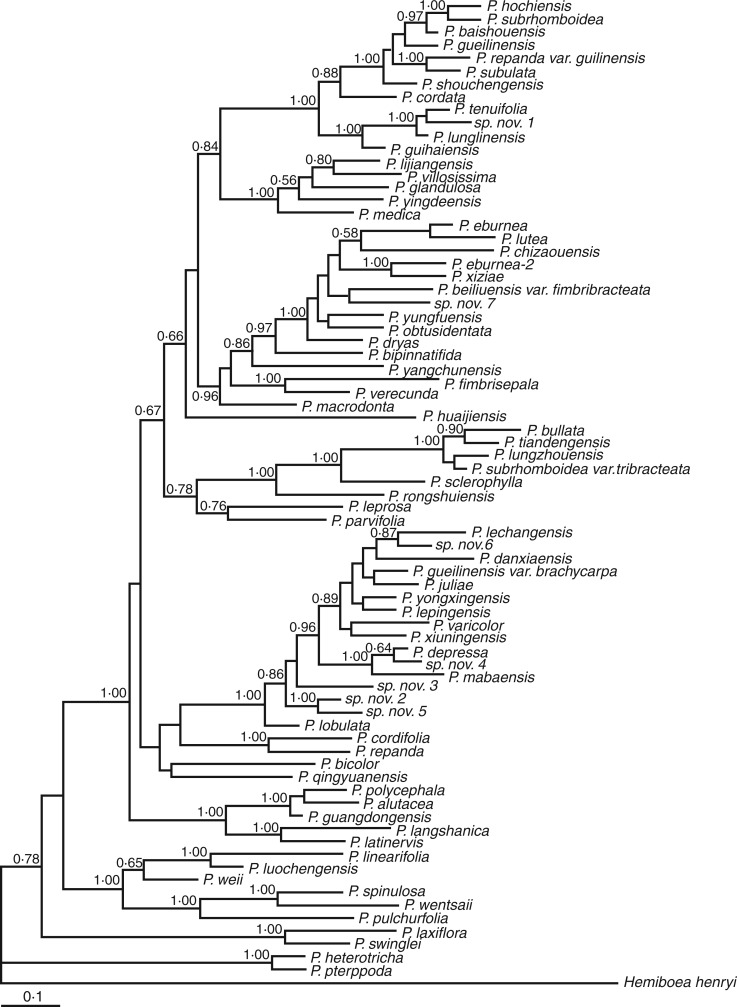
Before carrying out tests for phylogeny and positive selection, the substitution saturation and recombination of TPC1 data sets were examined to test whether the sequences were suitable for phylogeny and selection analyses. Our results showed that the index of substitution saturation (Iss) was significantly lower than the critical Iss (Iss.c) (Supplementary Data Table S2). Furthermore, transition and transversion rates of TPC1 sequences plotted against GTR genetic distance showed that both transitions and transversions increased linearly with increasing genetic distance (Supplementary Data Fig. S3). These results indicated that there is no significant substitution saturation in Primulina TPC1 alignments. For recombination analysis, SBP and GARD analyses identified potential recombination sites at positions 147 and 369 bp, respectively, within TPC1 sequences (Supplementary Data Fig. S4). Considering that the insertion/deletion mutation existed at the 5′ end as well as the recombination signals identified in the sequences, the first 1–369 bp regions were deleted and only the largest block of non-recombination after 370 bp was used in the subsequent Bayesian phylogenetic reconstruction (Fig. 1). The best-fit nucleotide substitution model identified by the AIC, implemented in MrModeltest, was GTR + I + G.
Fig. 1.
Bayesian phylogenetic tree of TPC1 from Primulina species. Bayesian posterior probabilities above 0·5 are labelled at the nodes.
Selection at nucleic acid level
The ω ratio of Primulina TPC1 was 0·233 under the one-ratio model M0, indicating that Primulina TPC1 sequences were mainly under purifying selection. The LRT test showed that different branches may have undergone similar selective pressure, as the free-ratio model (Mf) did not fit the data significantly better than M0 (−2ΔlnL = 170·552, P = 0·120) (Table 1). The BS REL method did not identify any branches under positive selection at the P ≤ 0·05 level.
Table 1.
Likelihood ratio test statistics for models of variable selective pressure among branches
| Model | Np | lnL | d.f. | −2ΔlnL | P-value |
|---|---|---|---|---|---|
| M0 | 153 | −7026·870 | |||
| Mf | 303 | −6941·594 | 150 | 170·552 | 0·120 |
Np, number of estimated parameters; lnL, log likelihood score.
To test the possibility of diversifying selection on some specific sites or regions of the sequences, four pairs of LRT tests (M0 versus M3, M1a versus M2a, M7 versus M8, and M8a versus M8) were performed. The comparison between M0 and M3 revealed that M3 was significantly better than M0 (−2ΔlnL = 354·088, P < 0·001) (Table 2), indicating that the ω ratio was not homogeneous among sites along TPC1 sequences. The positive selection models M2a and M8 were significantly better than the null models M1a and M7 (Table 2), respectively, suggesting that Primulina TPC1 has undergone selective pressure and may have sites under positive selection. The comparison between M8 (allowing ω > 1) and M8a (restricting ω to 1) could exclude the possibility of ω = 1 and avoid false-positive results. The LRT results showed that the positive model M8 was a significantly better fit than the null hypothesis M8a (−2ΔlnL = 64·208, P < 0·001). With the BEB posterior probability criterion at 95 % cut-off, the M2a model identified seven sites (80, 291, 324, 347, 553, 601 and 618) under positive selection, while the M8 model identified nine positive selection sites (31, 80, 291, 305, 324, 347, 553, 601 and 618) (Table 2).
Table 2.
Phylogenetic tests of positive selection for TPC1 in Primulina using site models
| Model | Np | lnL | Parameter | Models compared | d.f. | −2ΔlnL | P-value | Positively selected sites (posterior probability) |
|---|---|---|---|---|---|---|---|---|
| M0 | 153 | −7026·87 | ω = 0·233 | None | ||||
| M3 | 157 | −6849·826 | p0 = 0·906, ω0 = 0·072, p1 = 0·077, ω1 =1·266, p2 = 0·018, ω2 = 4·875 | M0, M3 | 4 | 354·088*** | 0·0000 | 31E (1·000); 44T (1·000); 60W (0·997); 80V (1·000); 147Y (0·999); 222S (0·999); 234K (0·99); 244D (0·989); 291H (1·000); 305Q (1·000); 322S (0·98); 324A (1·000); 347V (1·000); 360V (0·969); 368E (1·000); 374G (0·993); 541M (0·962); 553D (1·000); 562V (1·000); 565V (1·000); 600E (0·989); 601D (1·000); 606D (1·000); 611L (0·992); 618I (1·000) |
| M1a | 154 | −6884·202 | p0 = 0·88, ω0 = 0·052, p1 = 0·12, ω1 = 1 | Not allowed | ||||
| M2a | 156 | −6850·154 | p0 = 0·888, ω0 = 0·065, p1 = 0·092, ω1 = 1, p2 = 0·021, ω2 = 4·527 | M1a, M2a | 2 | 68·096*** | 0·0000 | 80V (0·997); 291H (1·000); 324A (1·000); 347V (0·999); 553D (0·998); 601D (0·953); 618I (0·999); |
| M7 | 154 | −6893·876 | p = 0·081, q = 0·374 | Not allowed | ||||
| M8 | 156 | −6852·224 | p0 = 0·968, p = 0·250, q = 1·624, p1 = 0·032, ω = 3·757 | M7, M8 | 2 | 83·304*** | 0·0000 | 31E (0·982); 80V (1·000); 291H (1·000); 305Q (0·972); 324A (1·000); 347V (1·000); 553D (1·000); 601D (0·997); 618I (1·000); |
| M8a | 155 | −6884·328 | p0 = 0·880, p = 5·528, q = 99·000, p1 = 0·200, ω = 1·000 | M8a, M8 | 1 | 64·208*** | 0·0000 | Not allowed |
Amino acids refer to the sequence of P. baishouensis.
Np, number of estimated parameters; lnL, log likelihood score; pi is the proportion of sites in each category.
***Significant at P < 0·0001.
In addition, another four methods implemented in Datamonkey were used to test the selective signal and identify sites under positive selection. Similar to the results obtained with the one-ratio model (ω = 0·233), the mean value of dN/dS was 0·263 (Table 3), this again suggesting an overall pattern of purifying selection on Primulina TPC1. Nevertheless, SLAC, FEL and MEME identified one site (347), six sites (31, 44, 305, 347, 360 and 565) and seven sites (31, 44, 96, 194, 347, 553 and 565) under positive selection with P < 0·1, respectively. In addition, FUBAR identified four positive selective sites (31, 44, 305 and 347) with posterior probability >0·9 (Table 3).
Table 3.
Positive selection analysis using SLAC, FEL, MEME and FUBAR methods
| Mean dN/dS | Positive selection sites | |||
|---|---|---|---|---|
| SLAC (P-value) | FEL (P-value) | MEME (P-value) | FUBAR (posterior probability) | |
| 0·263 | 347 (0·054) | 31 (0·046); 44 (0·077); 305 (0·085); 347 (0·011); 360 (0·081); 565 (0·066) | 31 (0·056); 44 (0·079); 96 (0·002); 194 (0·074); 347 (0·017); 553 (0·019); 565 (0·075) | 31 (0·967); 44 (0·917); 305 (0·905); 347 (0·993) |
SLAC, single likelihood ancestor counting; codons with P-values < 0·1; FEL, fixed-effect likelihood; codons with P-values < 0·1; MEME, mixed effects model of evolution; codons with P-values < 0·1; FUBAR: fast unconstrained Bayesian approximation; codons with posterior probability > 0·9.
Selective analysis at amino acid level
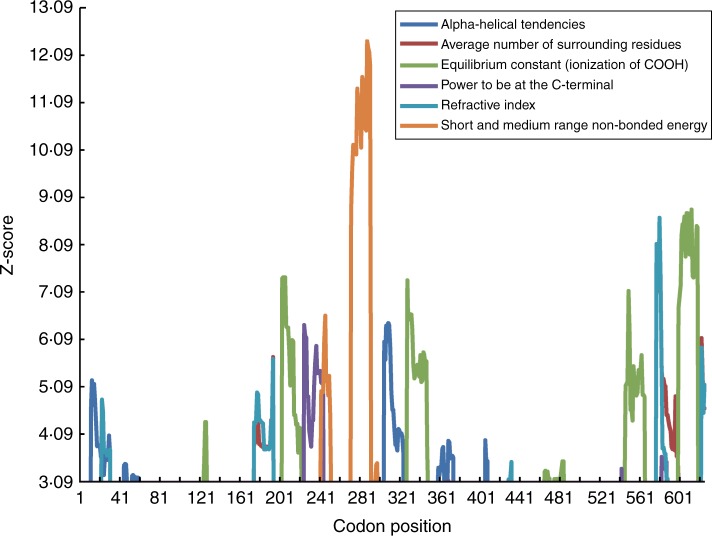
A complementary amino acid level approach implemented in TreeSAAP was further used to test positive selection at the amino acid level. Under the radical changes categories of 6–8, TreeSAAP detected that six amino acid properties had significantly positive Z-scores from the analysed 31 physiochemical properties available in the software (Table 4): equilibrium constant (ionization of COOH) (pK'); average number of surrounding residues (Ns); power to be at the C-terminal (αc); α-helical tendencies (Pα); short- and medium-range non-bonded energy (Esm); and refractive index (μ). Among the six properties, three were structure-related (Ns, αc and Pα) and the other three were biochemical-related (pK', Esm and μ). It is worth noting that pK' belongs to category 8 with significant support (P < 0·001) (Table 3). The sliding window analysis (sliding window size = 20) of the six identified physiochemical properties indicated that positive destabilizing selection was mainly located in the intermediate region and the C-terminus region of TPC1 (Fig. 2). A total of 34 amino acid sites were identified to be under positive destabilizing selection under at least one amino acid property (Supplementary Data Table S3). However, under an empirical threshold of three properties, only two sites (31 and 44) were identified as being under positive selection (Table S3).
Table 4.
Properties under positive selection determined in TreeSAAP
| Property | Category | Z-score |
|---|---|---|
| Equilibrium constant (ionization of COOH) (pK') | 8 | 8·07*** |
| Average number of surrounding residues (Ns) | 7 | 2·933** |
| Power to be at the C-terminal (αc) | 6 | 2·583** |
| α-Helical tendencies (Pα) | 6 | 2·016* |
| Short- and medium-range non-bonded energy (Esm) | 6 | 2·012* |
| Refractive index (μ) | 6 | 1·835* |
*P < 0·05; **P < 0·01; ***P < 0·001.
Fig. 2.
Sliding window plots of the Z-scores of radically changed amino acid properties showing regions under positive destabilizing selection (Z-score values >3·09) in Primulina TPC1.
Secondary structure of TPC1
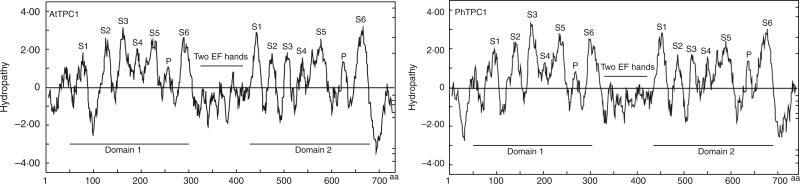
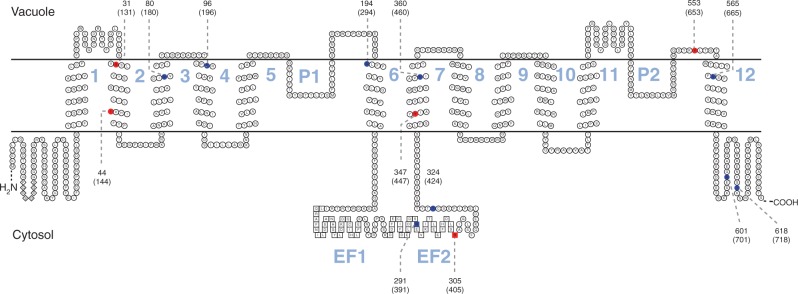
The Kyte–Doolittle hydropathy plots indicated that PhTPC1 has similar structure to AtTPC1, as shown in Fig. 3. Both of the proteins have two conserved homologous domains (I and II), and each domain contains six transmembrane segments (S1–S6) and a pore loop (P) between S5 and S6. The two homologous domains are connected by two EF-hand motifs, which have the ability to bind Ca2+ and may regulate the activity of the channel. The identified positive selective sites were labelled on the predicted secondary structure of PhTPC1, and the sites were found to be mainly located in the transmembrane domains and the EF-hand 2 region (Fig. 4).
Fig. 3.
Kyte–Doolittle hydropathy plots for AtTPC1 and PhTPC1. aa, amino acid.
Fig. 4.
Topology model of P. heterotricha TPC1 (PhTPC1) secondary structure and the location of the potential positive selection sites. Red and blue sites represent amino acids that evolved under positive selection, and the sites identified by at least three methods are shown in red. EF hand domains are shown as squares. Insertion or deletion regions in the N-terminal region are shown as diamonds. The codon number of each positively selected site is marked near the dotted line. Numbers without brackets correspond to the positions of positively selected sites in the sequence alignment used in selection analyses, and numbers within brackets correspond to the positions of positively selected sites in full-length sequences of PhTPC1.
DISCUSSION
Limestone karst areas have unique geological features and variable climatic conditions, which provide multitude ecological niches for organisms and support high floral species richness and endemism. Soils in karst areas were developed from weathered carbonate rock containing high levels of calcium. Calcium is not normally deficient in natural conditions and may be available in most soils, whereas excessive Ca in calcium-rich soils may not only prevent seed germination and reduce plant growth rate but also restrict plant communities (White and Broadley, 2003). Karst edaphic specialists, i.e. calcicoles, have evolved a suite of traits to respond to calcium-rich environments. For example, many plants can accumulate calcium in the form of calcium oxalate in vacuoles to sequester excess Ca (Franceschi and Nakata, 2005), while calcium is mainly stored in the cell wall in the form of calcium pectate in Primulina (Qi et al., 2013). Moreover, for Lonicera confusa, which inhabits karst areas, excess Ca can not only be deposited within plants via storage in leaf glands and trichomes, but can also be excreted via stomata (Wu et al., 2010). In this study, significant positive selection signals were detected in the Primulina Ca2+-permeable channel TPC1, and a few sites were identified to be under positive selection. These results indicate that TPC1 may play a role in local adaptation of Primulina to karst calcium-rich environments.
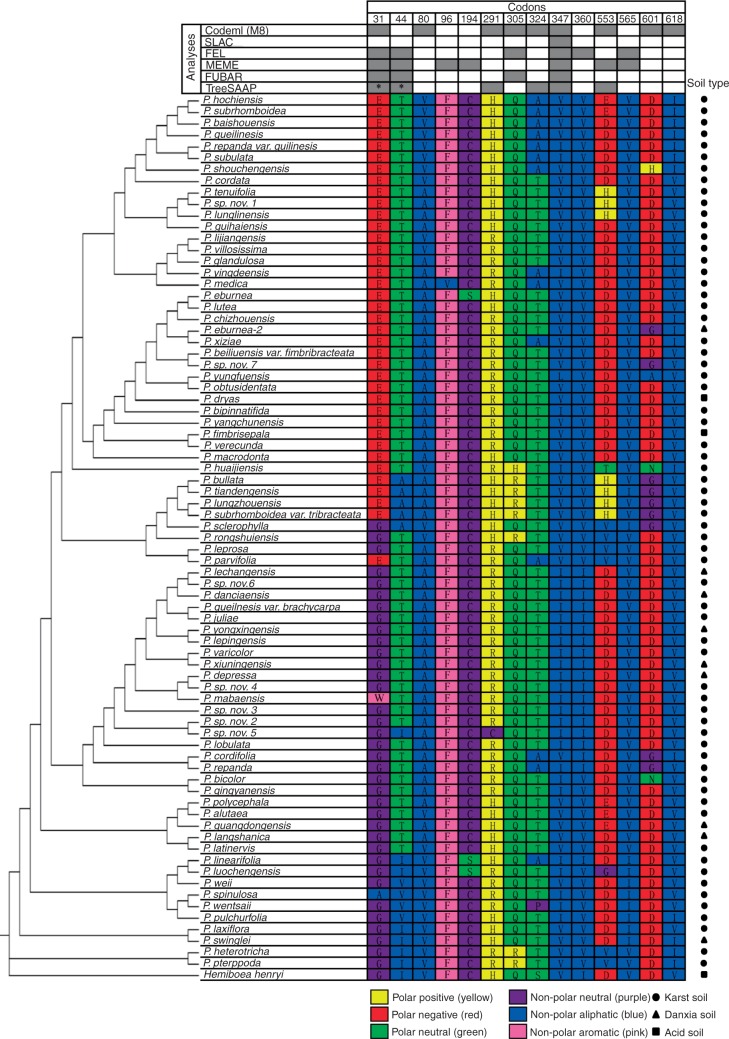
Several sites under positive selection were identified in Primulina TPC1 using different methods (Fig. 5). At nucleic acid level, the positive selective models M8 and M2a in PAML were significantly better than the corresponding null models M7 and M1a, respectively, and all identified positive selective sites at a posterior probability of 95 % (Table 1). The other four methods implemented in Datamonkey also identified signals of positive selection and identified sites under selection (Table 2). At the amino acid level, six physicochemical properties were found to be under positive-destabilizing selection and a total of 34 positive selection amino acid residues were identified under these properties, and it was noted that there are two sites (31 and 44) were identified by at least three different properties (Table S3). Among the identified amino acid sites, six sites (31, 44, 291, 324, 347 and 553) were also identified at the nucleic acid level (Fig. 5). Combining the results from nucleic acid and amino acid levels, a total of five sites (31, 44, 305, 347 and 553) were identified by at least three different methods, and site 347 was identified by all the methods used in this study (Fig. 5). These results indicate that the significant sites under positive selection may play a role in local adaptation of Primulina to karst calcium-rich environments. No branches were identified to be under positive selection using the BS-REL method, and branch-site model A did not find any branches under positive selection (data not shown), suggesting that different branches in the phylogeny tree may be subject to similar selective pressures. Primulina species are calciphilous plants and exhibit a high degree of soil specialization, and most of the species (∼84 %) have been found to occur only in karst soil (Hao et al., 2015). Therefore, Primulina TPC1 may have been subject to similar selective pressures related to Ca, and a few sites under episodic diversify selection may play a role in local adaptation of Primulina to calcium-rich habitats.
Fig. 5.
Amino acids in the same evolutionary positions showing strong selection signatures at the nucleic acid and amino acid levels. Results obtained with PAML (M8), SLAC, FEL, MEME, FUBAR and TreeSAAP (at least one property under selection) are marked with a black box at sites showing positive selection. Sites with at least three properties under selection in TreeSAAP are marked with an asterisk. Background colours represent amino acid properties: polar positive (yellow), polar negative (red), polar neutral (green), non-polar neutral (purple), non-polar aliphatic (blue) and non-polar aromatic (pink).The three typical soil types – karst, Danxia and acid soils – where the species grow are marked with circles, triangles and squares, respectively.
In terrestrial plants TPC1 has a highly conserved structure and consists of 12 transmembrane domains and two P-loops, as well as two EF hands. The P-loop domains are highly conserved as they control Ca2+ permeation (White et al., 2002). No sites under selection were found to be located in the P-loop domains; this result reflects the structural conservativeness and functional importance of the P-loop domains. For the Ca2+-binding EF-hand motifs (EF-hand 1 and EF-hand 2), two sites under selection were located in EF-hand 2 and no site was located in EF-hand 1. The two EF-hands have different Ca2+ affinities, as previous studies showed that only EF-hand 2 can bind Ca2+ directly and is responsible for the Ca2+-dependent activity of plant TPC1, whereas EF-hand 1 plays a modulation role (Schulze et al., 2011). Whether and how the identified positive selection sites in EF-hand 2 affect the Ca2+ affinities and the Ca2+-receptor activity of the TPC1 protein needs further research. The other sites under selection are mainly distributed in transmembrane domains and the C-terminus regions of the channel. The two terminal domains are essential for TPC1 function, and the N-terminal region is crucial for vacuolar targeting of the protein while the C-terminus is not required for targeting and dimerization (Larisch et al., 2012). The impact of the identified sites on the structure and function of TPC1 needs to be tested in future work.
We should keep in mind that statistical tests cannot provide direct evidence that a gene has experienced adaptive evolution, and experimental verification and functional tests should be used to validate statistical results (Yang, 2006). Positively selected sites provide ideal candidates for further mutagenesis and structural analysis to determine the specific roles of the sites and genes in local adaptation, as these sites are usually located near functionally or structurally relevant regions (Fonseca et al., 2008; Casasolia et al., 2009; Morgan et al., 2010; Moury and Simon, 2011). Site-directed mutagenesis has usually been used to verify the results of selection analysis (Ivarsson et al., 2003; Barkman et al., 2007; Lan et al., 2013). The Ca2+ transport activities of AtTPC1 (Furuichi et al., 2001), NtTPC1s (Kadota et al., 2004), OsTPC1 (Kurusu et al., 2004) and TaTPC1 (Wang et al., 2005) were verified by heterogeneously expressing them in the yeast cch1 mutant. Therefore, site-directed mutagenesis and the heterogeneous expression system of the yeast mutant could be used in future work to test the sites identified as being under positive selection in this study.
In conclusion, our results show that the Ca2+-permeable channel TPC1 may participate in local adaptation of Primulina to karst Ca2+-rich environments. Selection analysis of Primulina TPC1 showed that selective pressures show no significant difference among branches, suggesting that different branches may have undergone similar selective pressures related to calcium in karst habits. However, a significant signature of positive selection was detected at both nucleic acid and amino acid level, and several sites under positive selection were identified with multiple approaches. These results indicate that episodic diversifying selection at a few sites may play a major role in the adaptive evolution of Primulina TPC1 in karst Ca2+-rich environments.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: species names, coordinates and GenBank accession numbers used in this study. Table S2: detection of saturation substitution with the program DAMBE. Table S3: sites showing positive selection in TreeSAAP. Figure S1: locations of sampled Primulina species. Figure S2: schematic diagram of P. heterotricha TPC1 (2636 bp). Boxes and lines indicate coding sequences and UTRs, respectively. Locations of primers used in this study are shown in the diagram. Figure S3: transitions (S) and transversion (V) of TPC1 sequences plotted against GTR genetic distance. Figure S4: GARD and SBP analysis for detection of recombination in Primulina TPC1. GARD and SBP found evidence of recombination breakpoints at 369 and 147 bp, respectively.
ACKNOWLEDGEMENTS
This work was supported by grants from the NSFC- Guangdong Natural Science Foundation Joint Project (U1501211).
LITERATURE CITED
- Ai B, Gao Y, Zhang XL, Tao JJ, Kang M, Huang HW. 2015. Comparative transcriptome resources of eleven Primulina species, a group of ‘stone plants' from a biodiversity hot spot. Molecular Ecology Resources 15: 619–632. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang ZH. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164: 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkman TJ, Martins TR, Sutton E, Stout JT. 2007. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Molecular Biology and Evolution 24: 1320–1329. [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang ZH. 2003. Maximum likelihood methods for detecting adaptive evolution after gene duplication. Journal of Structural and Functional Genomics 3: 201–212. [PubMed] [Google Scholar]
- Casasolia M, Federicib L, Spinellia F, et al. 2009. Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proceedings of the National Academy of Sciences of the USA 106: 7666–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Schnell N, Chattaway J, Davies P, Dixon G, Sanders D. 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Letters 419: 259–262. [DOI] [PubMed] [Google Scholar]
- Fonseca RR, Johnson WE, O'Brien SJ, Ramos MJ, Antunes A. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Nakata PA. 2005. Calcium oxalate in plants: formation and function. Annual Review of Plant Biology 56: 47–71. [DOI] [PubMed] [Google Scholar]
- Furuichi T, Cunningham KW, Muto S. 2001. A putative two pore channel AtTPC1 mediates Ca2+ flux in Arabidopsis leaf cells. Plant & Cell Physiology 42: 900–905. [DOI] [PubMed] [Google Scholar]
- Furuichi T, Kawano T, Tatsumi H, Sokabe M. 2008. Roles of ion channels in the environmental responses of plants. In: B Martinac, ed. Sensing with ion channels. Heidelberg: Springer, 47–67. [Google Scholar]
- Hao Z, Kuang Y, Kang M, Niu S. 2015. Untangling the influence of phylogeny, soil and climate on leaf element concentrations in a biodiversity hotspot. Functional Ecology 29: 165–176. [Google Scholar]
- Hashimoto K, Saito M, Matsuoka H, Iida K, Iida H. 2004. Functional analysis of a rice putative voltage-dependent Ca2+ channel, OsTPC1, expressed in yeast cells lacking its homologous gene CCH1 . Plant & Cell Physiology 45: 496–500. [DOI] [PubMed] [Google Scholar]
- Hedrich R, Marten I. 2011. TPC1-SV channels gain shape. Molecular Plant 4: 428–441. [DOI] [PubMed] [Google Scholar]
- Hepler PK. 2005. Calcium: acentral regulator of plant growth and development. The Plant Cell 17: 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson Y, Mackey AJ, Edalat M, Pearson WR, Mannervik B. 2003. Identification of residues in glutathione transferase capable of driving functional diversification in evolution. A novel approach to protein redesign. Journal of Biological Chemistry 278: 8733–8738. [DOI] [PubMed] [Google Scholar]
- Jammes F, Hu HC, Villiers F, Bouten R, Kwak JM. 2011. Calcium-permeable channels in plant cells. FEBS Journal 278: 4262–4276. [DOI] [PubMed] [Google Scholar]
- Johns SJ. 2010. TOPO2, transmembrane protein display software. http://www.sacs.ucsf.edu/TOPO2/. [Google Scholar]
- Kadota Y, Furuichi T, Ogasawara Y, et al. 2004. Identification of putative voltage-dependent Ca2+-permeable channels involved in cryptogein-induced Ca2+ transients and defense responses in tobacco BY-2 cells. Biochemical and Biophysical Research Communications 317: 823–830. [DOI] [PubMed] [Google Scholar]
- Kang M, Tao JJ, Wang J, et al. 2014. Adaptive and nonadaptive genome size evolution in Karst endemic flora of China. New Phytologist 202: 1371–1381. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad KR, Wudick MM, Feijo JA. 2011. Calcium regulation of tip growth: new genes for old mechanisms. Current Opinion in Plant Biology 14: 721–730. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD, Muse SV. 2005a. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. 2005b. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular Biology and Evolution 22: 1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006a. Automated phylogenetic detection of recombination using a genetic algorithm. Molecular Biology and Evolution 23: 1891–1901. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006b. GARD: a genetic algorithm for recombination detection. Bioinformatics 22: 3096–3098. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Molecular Biology and Evolution 28: 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijine G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. 2010. Calcium signals: the lead currency of plant information processing. The Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu T, Sakurai Y, Miyao A, Hirochika H, Kuchitsu K. 2004. Identification of a putative voltage-gated Ca2+-permeable channel (OsTPC1) involved in Ca2+ influx and regulation of growth and development in rice. Plant & Cell Physiology 45: 693–702. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Lacombe B, Becker D, Hedrich R, et al. 2001. The identity of plant glutamate receptors. Science 292: 1486–1487. [DOI] [PubMed] [Google Scholar]
- Lan T, Wang XR, Zeng QY. 2013. Structural and functional evolution of positively selected sites in pine glutathione S-transferase enzyme family. Journal of Biological Chemistry 288: 24441–24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larisch N, Schulze C, Galione A, Dietrich P. 2012. An N-terminal dileucine motif directs two-pore channels to the tonoplast of plant cells. Traffic 13: 1012–1022. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Lloyd AT, Fares MA, O'Farrelly C. 2004. Evidence of positively selected sites in mammalian α-defensins. Molecular Biology and Evolution 21: 819–827. [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder J, et al. 2001. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology 126: 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan DA, Palfreyman EJ, Smith MJ, Moss JL, Christensen RG, Sailsbery JK. 2005. Physicochemical evolution and molecular adaptation of the cetacean and artiodactyl cytochrome b proteins. Molecular Biology and Evolution 22: 437–455. [DOI] [PubMed] [Google Scholar]
- Morgan CC, Loughran NB, Walsh TA, Harrison AJ, O'Connell MJ. 2010. Positive selection neighboring functionally essential sites and disease-implicated regions of mammalian reproductive proteins. BMC Evolutionary Biology 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Simon V. 2011. dN/dS-based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of potato virus Y. Molecular Biology and Evolution 28: 2707–2717. [DOI] [PubMed] [Google Scholar]
- Murrell B, Wertheim J, Moola S, et al. 2012. Detecting individual sites subject to episodic diversifying selection. PloS Genetics 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, et al. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Molecular Biology and Evolution 30: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang ZH. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. 2008. MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Page R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Peiter E. 2011. The plant vacuole: emitter and receiver of calcium signals. Cell Calcium 50: 120–128. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 2002. The effect of recombination on the accuracy of phylogeny estimation. Journal of Molecular Evolution 54: 396–402. [DOI] [PubMed] [Google Scholar]
- Pottosin II, Schonknecht G. 2007. Vacuolar calcium channels. Journal of Experimental Botany 58: 1559–1569. [DOI] [PubMed] [Google Scholar]
- Qi QW, Hao Z, Tao JJ, Kang M. 2013. Diversity of calcium speciation in leaves of Primulina species (Gesneriaceae). Biodiversity Science 21: 715–722. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Schönknecht G. 2013. Calcium signals from the vacuole. Plants 2: 589–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze C, Sticht H, Meyerhoff P, Dietrich P. 2011. Differential contribution of EF-hands to the Ca2+-dependent activation in the plant two-pore channel TPC1. The Plant Journal 68: 424–432. [DOI] [PubMed] [Google Scholar]
- Shriner D, Nickle DC, Jensen MA, Mullins JI. 2003. Potential impact of recombination on sitewise approaches for detecting positive natural selection. Genetical Research 81: 115–121. [DOI] [PubMed] [Google Scholar]
- Spalding EP, Harper JF. 2011. The ins and outs of cellular Ca2+ transport. Current Opinion in Plant Biology 14: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nei M. 2001. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Molecular Biology and Evolution 18: 2179–2185. [DOI] [PubMed] [Google Scholar]
- Swarbreck SM, Colaco R, Davies JM. 2013. Plant calcium-permeable channels. Plant Physiology 163: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics 42: 260–263. [DOI] [PubMed] [Google Scholar]
- Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C. 2010. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytologist 187: 23–43. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Yu JN, Chen T, et al. 2005. Functional analysis of a putative Ca2+ channel gene TaTPC1 from wheat. Journal of Experimental Botany 56: 3051–3060. [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR. 2003. Calcium in plants. Annals of Botany 92: 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Demidchik V, Nichols C, Davies JM. 2002. Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochimica et Biophysica Acta 1564: 299–309. [DOI] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19: 671–672. [DOI] [PubMed] [Google Scholar]
- Wu G, Li MT, Zhong FX, Fu CH, Sun J, Yu LJ. 2010. Lonicera confusa has an anatomical mechanism to respond to calcium-rich environment. Plant and Soil 338: 343–353. [Google Scholar]
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Molecular Biology and Evolution 30: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution 26: 1–7. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 2006. Computational molecular evolution. Oxford: Oxford University Press. [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends in Ecology & Evolution 15: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. Journal of Molecular Evolution 46: 409–418. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen A-MK. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Wong WS, Nielsen R. 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution 22: 1107–1118. [DOI] [PubMed] [Google Scholar]