Abstract
Background and Aims The centromere drive theory explains diversity of eukaryotic centromeres as a consequence of the recurrent conflict between centromeric repeats and centromeric histone H3 (CenH3), in which selfish centromeres exploit meiotic asymmetry and CenH3 evolves adaptively to counterbalance deleterious consequences of driving centromeres. Accordingly, adaptively evolving CenH3 has so far been observed only in eukaryotes with asymmetric meiosis. However, if such evolution is a consequence of centromere drive, it should depend not only on meiotic asymmetry but also on monocentric or holokinetic chromosomal structure. Selective pressures acting on CenH3 have never been investigated in organisms with holokinetic meiosis despite the fact that holokinetic chromosomes have been hypothesized to suppress centromere drive. Therefore, the present study evaluates selective pressures acting on the CenH3 gene in holokinetic organisms for the first time, specifically in the representatives of the plant genus Luzula (Juncaceae), in which the kinetochore formation is not co-localized with any type of centromeric repeat.
Methods PCR, cloning and sequencing, and database searches were used to obtain coding CenH3 sequences from Luzula species. Codon substitution models were employed to infer selective regimes acting on CenH3 in Luzula.
Key Results In addition to the two previously published CenH3 sequences from L. nivea, 16 new CenH3 sequences have been isolated from 12 Luzula species. Two CenH3 isoforms in Luzula that originated by a duplication event prior to the divergence of analysed species were found. No signs of positive selection acting on CenH3 in Luzula were detected. Instead, evidence was found that selection on CenH3 of Luzula might have been relaxed.
Conclusions The results indicate that holokinetism itself may suppress centromere drive and, therefore, holokinetic chromosomes might have evolved as a defence against centromere drive.
Keywords: CenH3, centromere drive, holokinetic chromosomes, positive selection, Luzula
INTRODUCTION
Fifteen years ago, Henikoff et al. (2001) proposed a model of centromere drive to explain why centromeric chromatin and fundamental kinetochore proteins – despite their highly conserved function in faithful chromosomal segregation – are rapidly evolving instead of being fixed at one optimal state. They suggested that kinetochore proteins are in evolutionary conflict with centromeric satellite DNA that exploits asymmetric female meiosis to secure its preferential transmission to the egg (which is the only surviving meiotic product) at the expense of its homologous counterpart. Expansions (or contractions) of centromeric repeats may lead to a larger (or smaller) kinetochore, which attracts more (or fewer) microtubules, resulting in their capture by the meiotic egg pole when it emanates more (or fewer) microtubules (Henikoff et al., 2001). A driving centromere then rapidly sweeps through a population along with possible negative effects and hitchhiking mutations (Henikoff et al., 2001). Therefore, mutations of kinetochore proteins that alter their affinity for binding to centromeric DNA are positively selected, if they restore the balance between homologous centromeres and suppress centromere drive along with the associated negative effects (Malik and Henikoff, 2009). The centromere drive model relying on recurrent genomic conflict thus provides an elegant explanation for the remarkable diversity of centromeres in eukaryotes with asymmetric meiosis.
Preferential inheritance of centromere-associated repetitive DNA via the female meiotic drive has been demonstrated in monkey flowers (Mimulus; Fishman and Saunders, 2008; Fishman, 2013). Size-dependent preferential transmission of centromere-like repeats has been documented on neo-centromeric knobs in maize (Kikudome, 1959). The ability of centromeric satellite repeats to affect the positioning of centromeric histone H3 (CenH3), which is the key kinetochore protein that serves as a scaffold for the formation of the entire kinetochore, has been recently shown in rice (Zhang et al., 2013). Aside from the evidence for mechanical aspects of centromere drive detectable more or less in real time, there is also evidence documenting predicted centromere drive impacts on an evolutionary time scale. The size divergence of satellite arrays is really larger in lineages with asymmetric meiosis, i.e. in those offering an opportunity for centromere drive (Melters et al., 2013). Similarly, adaptively evolving CenH3 has so far been detected only in lineages with asymmetric meiosis (Henikoff et al., 2001; Talbert et al., 2004, 2008; Hirsch et al., 2009; Schueler et al., 2010; Zedek and Bureš, 2012; Finseth et al., 2015) and not in lineages displaying symmetric meiosis (Talbert et al., 2004, 2008; Baker and Rogers, 2006), which is in accordance with the centromere drive model.
The evidence based on adaptive evolution of CenH3 actually suffers from incompleteness because the presence/absence of recurrent adaptive evolution of CenH3 depends not only on meiotic asymmetry/symmetry but also on the chromosomal structure in terms of kinetochore formation. Monocentric chromosomes form the kinetochore in a relatively small centromeric region (but see Neumann et al., 2012), while holokinetic chromosomes form the kinetochore along their entire length (Bureš et al., 2013; Heckmann et al., 2014). Although a specific DNA sequence is not required for kinetochore formation (Heslop-Harrison and Schwarzacher, 2011), centromeric satellites are able to affect CenH3 positioning (Zhang et al., 2013), thereby affecting kinetochore size in monocentrics and switching kinetochore formation from epigenetic to genetic control. In holokinetics, however, the dispersion of centromeric nucleosomes in holokinetic chromosomes may make it difficult for any particular sequence to gain an advantage and be amplified the way centromeric satellites can be in monocentrics because the holokinetic kinetochore is already everywhere. In accordance with that, kinetochore formation in monocentrics is mostly co-localized with arrays of centromeric sequences, while such co-localization is mostly absent in holokinetics (Heckman et al., 2013; but see Marques et al., 2015). In fact, all the studies that have addressed selection regimes acting on CenH3 have been carried out on monocentric organisms (e.g. Henikoff et al., 2001; Talbert et al., 2004; Hirsch et al., 2009; Schueler et al., 2010) or on Caenorhabditis (Zedek and Bureš, 2012) which is holokinetic in mitosis but forms a cup-like kinetochore in meiosis with ambiguous utilization of CenH3 (Chan et al., 2004; Monen et al., 2005). If the recurrent adaptive evolution of CenH3 is really a consequence of centromere drive, it should be missing (or at least less frequent) not only in lineages with symmetric meiosis but also in lineages with asymmetric meiosis that possess holokinetic chromosomes, such as the plant genus Luzula (Heckmann et al., 2013).
In the present study, we aimed to fill in the missing piece of the puzzle by evaluating selection regimes acting on CenH3 in the holokinetic plant genus Luzula (Juncaceae). To achieve that, we isolated the CenH3 gene from 13 Luzula species and employed branch-site models of codon substitution for detecting positive selection that work in a maximum likelihood framework (Anisimova and Yang, 2007; Kosakovsky Pond et al., 2011; Murrel et al., 2012). The results of this study may not only corroborate the centromere drive model, but may also shed more light on the adaptive value of holokinetic chromosomal structure that has evolved repeatedly and independently in various eukaryotic lineages.
MATERIALS AND METHODS
Plant material
Samples were collected from wild populations and botanical gardens and subsequently cultivated in pots in a greenhouse at the Department of Botany and Zoology, Masaryk University, Brno. A list of sample locations is included in Supplementary Data File S1.
Sequence preparation from plant material
Young leaves were ground to a fine powder in liquid nitrogen, and total genomic DNA or RNA was extracted using the DNeasy Plant Mini Kit (Qiagen) or the RNeasy Plant Mini Kit (Qiagen), respectively, following the manufacturer’s protocol. cDNA was prepared from isolated RNA using the QuantiTect Rev. Transcription Kit (Qiagen) according to the manufacturer’s instructions. We designed primers for PCR amplification of putative CenH3 sequences from analysed species based on CenH3 sequences from Luzula nivea, rice and maize as available in GenBank. The sequences of the designed primers are as follows: CENH3-F (5′-TGGCTCGGACGAAACACTTCYC-3′) and CENH3-deg-R (5′-CACCKATWCGCCTRGCAAGYTG-3′). The thermocycler was set to an initial denaturation step at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 57 °C for 30 s and 72 °C for 30 s, and a final elongation step at 72 °C for 5 min. PCR products were cloned into pCR®2.1 vectors and transformed into TOP10F' competent cells (Invitrogen) according to the manufacturer’s instructions. Selection of colonies for plasmid isolation was based on the blue–white test. Inserts of the desired length were evaluated using PCR with T7 and M13 primers. Sequencing was conducted by Macrogen, Inc.
Sequence alignment, phylogenetic and positive selection analyses
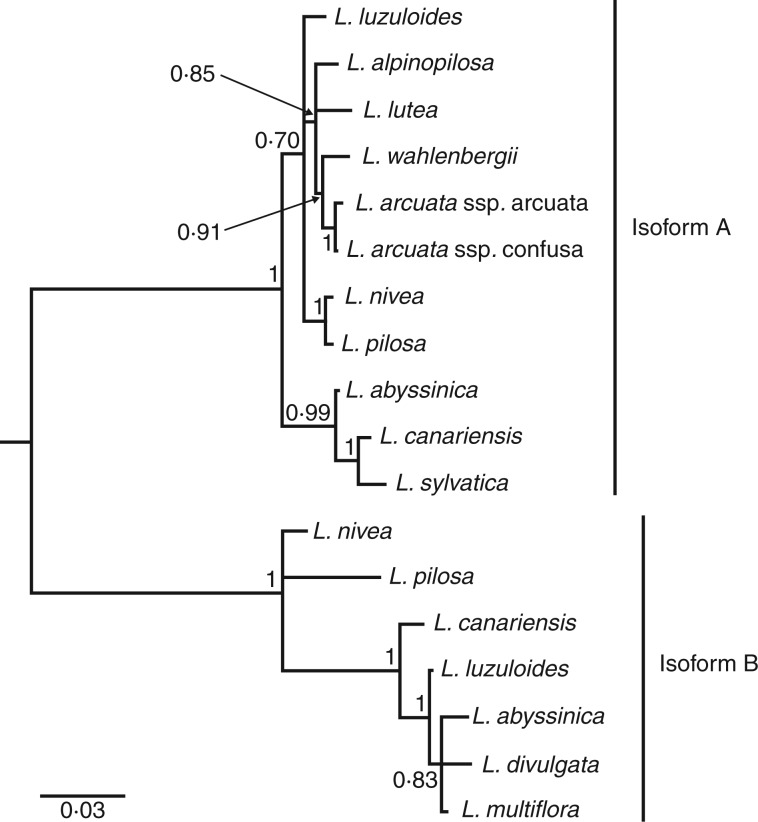
Intron–exon boundaries were identified manually based on the alignment of genomic sequences of CenH3 with available cDNA sequences. The codon alignments and phylogenetic trees were jointly inferred in BAli-Phy 2.3.5 (Redelings and Suchard, 2005; Redelings, 2014) using the M0 substitution model and RS07 model for indels; we ran ten independent chains until they converged and then pooled the results. We masked each codon with a reliability score <80 % as ‘NNN’ prior to selection analyses, and for selection analyses we used a 50 % consensus tree (Fig. 1). BAli-Phy alignments of CenH3 before and after masking the unreliable residues are supplied in Supplementary Data Files S2 and S3, respectively. The alignment and phylogenetic tree that we used for selection analyses are supplied in Supplementary Data File S4.
Fig. 1.
Consensus tree of CenH3 sequences from Luzula species inferred in BAli-Phy 2.3.5. Numbers indicate Bayesian posterior probabilities. Branches with posterior probabilities <50 % are collapsed. The scale bar indicates branch lengths.
We used codon substitution models implemented in software packages (see below) to infer the selective pressures acting on a protein/gene from the non-synonymous/synonymous substitution rate ratio (dN/dS = ω). Non-synonymous substitutions in a codon lead to amino acid changes, while synonymous substitutions do not. If there is no selective pressure (neutral evolution), non-synonymous and synonymous substitutions are expected to occur at the same rate, with ω = 1. Purifying selection, which keeps the protein as it is, is indicated by ω < 1, and positive selection favouring substitutions that change the amino acids in a protein is indicated by ω > 1. If purifying selection is relaxed, ω tends to be elevated towards 1. Likewise, if positive selection is relaxed, ω tends to decrease towards 1.
To determine positively selected branches in the CenH3 evolution in Luzula, we ran the data through the branch-site random effects likelihood (BS-REL) model of codon substitution (Kosakovsky Pond et al., 2011). BS-REL allows ω to vary across both codons and branches and infers selective regimes independently for each branch of a given phylogeny, pooling information across all codons (Kosakovsky Pond et al., 2011; Murrell et al., 2012). To assess the positive selection at the level of individual codons, we used the mixed effects model of evolution (MEME) model of codon substitution that allows ω to vary across both codons and branches, and infers selective regimes independently for each codon of a given alignment, pooling information over branches (Murrell et al., 2012). Finally, using the M0 model in PAML4.7, we have determined overall ω as an average across all codons and branches (Yang, 2007).
RESULTS
In total, we analysed 18 CenH3 sequences from 13 Luzula species. The two sequences from L. nivea were previously published (Nagaki et al., 2005; Moraes et al., 2011), and we obtained them from GenBank (accession nos AB201356.2 and HM988988.1), while the rest are reported here for the first time (Table 1). Eight sequences were obtained as cDNA, while we inferred the rest of the coding sequences from the PCR-amplified genomic DNA (Table 1). Phylogenetic analysis revealed two well-separated clades corresponding to previously described isoforms A and B from L. nivea (Nagaki et al., 2005; Moraes et al., 2011; Fig. 1). We found both isoforms in five species (L. abyssinica, L. canariensis, L. luzuloides, L. nivea and L. pilosa), while the rest of the analysed species were represented by either isoform A or isoform B (Table 1; Fig. 1).
Table 1.
GenBank accession numbers and sources of CenH3 sequences
| Species | Isoform A | Isoform B | ||
|---|---|---|---|---|
| L. abyssinica | KJ934237 | Genomic DNA | KJ934245 | cDNA |
| L. alpinopilosa | KJ934238 | Genomic DNA | – | – |
| L. arcuata ssp. arcuata | KJ934239 | Genomic DNA | – | – |
| L. arcuata ssp. confusa | KJ934240 | Genomic DNA | – | – |
| L. canariensis | KJ934235 | cDNA | KJ934246 | cDNA |
| L. divulgata | – | – | KJ934247 | cDNA |
| L. lutea | KJ934241 | Genomic DNA | – | – |
| L. luzuloides | KJ934242 | Genomic DNA | KJ934248 | cDNA |
| L. multiflora | – | – | KJ934249 | cDNA |
| L. nivea | AB201356.2 | cDNA | HM988988.1 | cDNA |
| L. pilosa | KJ934243 | Genomic DNA | KJ934250 | cDNA |
| L. sylvatica | KJ934236 | cDNA | – | – |
| L. wahlenbergii | KJ934244 | Genomic DNA | – | – |
BS-REL and MEME did not detect any positively selected branches or codons, respectively, because no branch or codon showed ω significantly higher than 1 (Supplementary Data File S5). It should be noted that both methods correct for multiple testing (Kosakovsky Pond et al., 2011; Murrell et al., 2012), which may be too conservative and lead to false-negative results. However, uncorrected P-values confirmed the absence of positively selected codons or branches, because none of them crossed the significance level at P = 0·05 (File S5). The overall ω, averaged across all codons and branches inferred using the M0 model in PAML4.7, was estimated to be ω = 0·485.
DISCUSSION
We revealed two isoforms of CenH3 in the genus Luzula that correspond to the previously reported isoforms A and B from L. nivea (Fig. 1; Nagaki et al., 2005, Moraes et al., 2011). It is likely that these isoforms originated from an ancient duplication event that had occurred prior to the divergence of analysed Luzula species. Clusters of L. nivea + L. pilosa and L. abyssinica + L. divulgata + L. multiflora in the isoform B sub-tree and clusters of L. nivea + L. pilosa and L. canariensis + L. sylvatica in the isoform A sub-tree (Fig. 1) correspond well with the species tree of the genus Luzula (Zaveska Drabkova and Vlček, 2010). However, it is difficult to assess whether the entire topologies of CenH3 isoform sub-trees agree with the species tree because the overall resolution of the published species tree is poor due to weak bootstrap support (Záveská Drábková and Vlček, 2010). The isoform A and B sub-trees differ from one another in the placement of L. luzuloides (Fig. 1), which might be due to incomplete lineage sorting, distinct selective pressures that have been acting on the two paralogues or a possible hybridization event in the evolutionary history of L. luzuloides.
We managed to isolate both isoform A and B from only five species, whereas for the remaining species we isolated only one of the two isoforms (Table 1; Fig. 1). It is unclear whether some species had lost one of the isoforms or if we failed to isolate it perhaps due to substitutions in primer-binding sites. We have isolated isoform B from cDNA (Table 1), implying that it is transcribed (at least in species where it was detected), and immunostaining with a specific antibody suggested that isoform B is also translated and incorporated into nucleosomes in L. nivea (Moraes et al., 2011). On the other hand, isoform A has been isolated as cDNA only from L. nivea (Nagaki et al., 2005), L. canariensis and L. sylvatica (this study), and the immunostaining experiments performed by Nagaki’s group could not confirm whether it was also translated because the authors did not use an isoform A-specific antibody (Moraes et al., 2011). The possession of two CenH3 isoforms appears to be common in plants, and either one or both of them can be functional (Lermontova and Schubert, 2013). Which of the two isoforms in Luzula species, whether just one or both of them, are involved in chromosome segregation and whether their functionality is species specific (or tissue specific or developmental stage specific) remains to be elucidated.
By applying branch-site models of codon substitution (Kosakovsky Pond et al., 2011; Murrell et al., 2012), we did not detect any signs of positive selection acting on CenH3 in Luzula (see the Results and File S5). This finding contrasts with CenH3 analyses in monocentric genera of roughly similar age, such as Oryza, Mimulus and Drosophila, whose authors found positively selected branches or codons in CenH3 (Hirsch et al., 2009; Beck and Llopart, 2015; Finseth et al., 2015). Moreover, in Oryza and Mimulus, positive selection has also been found after CenH3 duplication events (Hirsch et al., 2009; Finseth et al., 2015).
It is likely that CenH3 has not been in intragenomic conflict with centromeric repeats and that centromere drive has not been operating in the holokinetic genus Luzula. The observed absence of positive selection on CenH3 in Luzula is in accordance with a recent finding in Luzula elegans which lacks centromeric repeats that would co-localize with kinetochore formation along an entire chromosome (Heckmann et al., 2013). In contrast, Marques et al. (2015) have found a centromeric satellite repeat Tyba co-localized with kinetochore formation in holokinetic Rhynchospora pubera (Cyperaceae). The authors offer two explanations, either the preferential insertion of Tyba sequences into centromeric chromatin might have been facilitated by CenH3 itself or, alternatively, Tyba might define the preferential sites for CenH3 accumulation (Marques et al., 2015). Analysis of selection on CenH3 in Rhynchospora could help to resolve this dilemma; positively selected CenH3 could support Tyba-directed CenH3 accumulation in a centromere drive fashion, while absence of positive selection on CenH3 could suggest that CenH3 facilitated Tyba insertion.
The absence of positive selection on CenH3 in Luzula indicates that the holokinetic chromosomal structure per se might prevent centromere drive (Fig. 2) and, as previously speculated, that holokinetism itself might have evolved as a defence against centromere drive (Talbert et al., 2008; Malik and Henikoff, 2009; Bureš et al., 2013). Accordingly, although holokinetism has evolved independently in various lineages of plants and animals (Melters et al., 2012), it appears to have evolved only in lineages with asymmetric meiosis where the need for a defence against centromere drive can be assumed, not in lineages with symmetric meiosis, such as bryophytes, lycophytes, ferns, horsetails or fungi (Bureš et al., 2013), where such a defence has no sense. Nevertheless, even if centromere drive is suppressed in holokinetic lineages, holokinetism is likely to provide an opportunity for a different kind of meiotic drive propelling divergence in chromosomal size and number – holokinetic drive (Bureš and Zedek, 2014; Escudero et al., 2015). The potential of holokinetism to suppress centromere drive has a non-intuitive implication. Since the centromere drive is a form of meiotic drive, only meiotic (and not necessarily mitotic) holokinetism should be sufficient to suppress it, implying that there may be organisms that are functionally monocentric in mitosis but holokinetic in meiosis. The opposite situation, i.e. functional holokinetism in mitosis and monocentrism in meiosis, is known from true bugs (Hemiptera) or some nematodes (Heckmann et al., 2014).
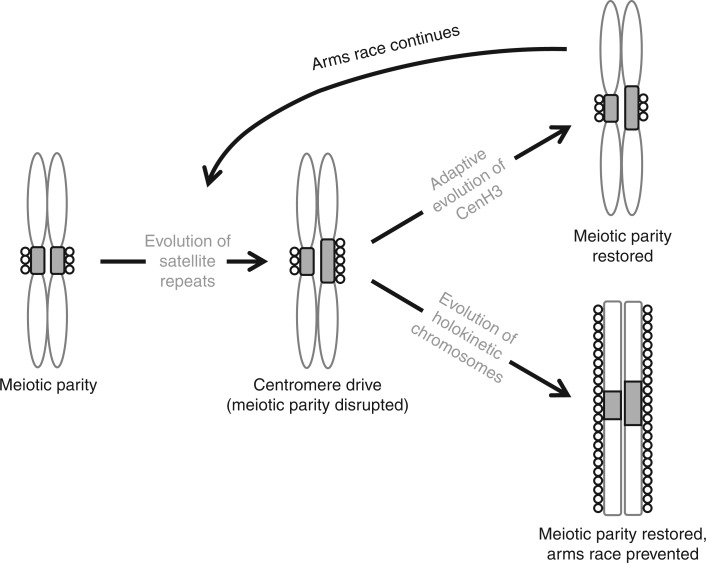
Fig. 2.
The model of centromere drive in monocentric chromosomes and its suppression by chromosomal holokinetism. In the first stage, an expansion of centromeric satellite repeats (grey rectangles) at one of the monocentric homologues leads to an expansion of CenH3 nucleosomes (circles) and thus to a larger kinetochore providing a transmission advantage in asymmetric meiosis. Then there are two possibilities: either (1) an adaptive CenH3 mutation changes its binding affinity for centromeric satellites thereby restoring meiotic parity and suppressing centromere drive until a new selfish satellite arises and the cycle begins again, or (2) the evolution of holokinetic chromosomes ends the arms race permanently by means of the extended kinetochore.
It is possible that CenH3 eventually becomes dispensable in holokinetic species, as has been shown in insects where the independent transitions from monocentric to holokinetic chromosomes are associated with the loss of CenH3 (Drinnenberg et al., 2014). To become lost, CenH3 must first escape from natural selection. The absence of positive selection in Luzula and a high average ω = 0·485 compared with other eukaryotic lineages (Zedek and Bureš, 2016) might indeed suggest a relaxed selection possibly indicating an initial phase of such a process in a clade much younger than holokinetic clades of insects; the holokinetic plant family Juncaceae originated roughly 90 million years ago (Escudero et al., 2012), while Phtiraphtera, the youngest holokinetic order of insects, originated about 110 million years ago (Misof et al., 2014). Of course, CenH3 from more holokinetic clades has to be analysed to confirm or disprove that this process is really associated with the evolution of holokinetism.
In summary, the absence of positive selection on CenH3 that we have detected in Luzula suggests that chromosomal holokinetism may prevent centromeric repeats from subverting the process of asymmetric meiosis to their own advantage. However, it still remains unclear whether (meiotic) holokinetism arises as an adaptation to selective pressures exerted by centromere drive or is an adaptation to something else and its ability to prevent centromere drive is merely a side effect. Since holokinetic organisms can easily cope with chromosomal fragmentations (Zedek et al., 2016), holokinetic chromosomes may be adaptive in clastogenic environments but, once they arise, their extended kinetochore makes centromere drive impossible (Fig. 2).
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. File S1: a list of sample locations where plant samples were collected. File S2: the alignment of analysed CenH3 sequences in fasta format inferred in BAli-Phy 2.3.5 using the M0 codon substitution model and RS07 model for indels. File S3: the masked alignment of analysed CenH3 sequences in fasta format inferred in BAli-Phy 2.3.5 using the M0 codon substitution model and RS07 model for indels. File S4: the nexus file containing the masked alignment and phylogenetic tree that we used for selection analyses. File S5: excel file containing raw outputs of BS-REL and MEME selection analyses.
ACKNOWLEDGEMENTS
We are grateful to Martin Dančák for providing specimens of Luzula abyssinica and L. canariensis. We also greatly appreciate the computational resources used for construction of alignment and phylogenetic tree in Bali-Phy that were provided by MetaCentrum (the program LM2010005) and CERIT-SC (the program Centre CERIT Scientific Cloud, part of the Operational Program Research and Development for Innovations, Reg. no. CZ.1.05/3.2.00/08.0144). This work was funded by the Czech Science Foundation, grant no. 13-29362S.
LITERATURE CITED
- Anisimova M, Yang Z. 2007. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Molecular Biology and Evolution 24: 1219–1228. [DOI] [PubMed] [Google Scholar]
- Baker RE, Rogers K. 2006. Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck EA, Llopart A. 2015. Widespread positive selection drives differentiation of centromeric proteins in the Drosophila melanogaster subgroup. Scientific Reports 5: 17197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P, Zedek F. 2014. Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution 68: 2412–2420. [DOI] [PubMed] [Google Scholar]
- Bureš P, Zedek F, Marková M. 2013. Holocentric chromosomes In: Leitch IJ, Greihuber J, Doležel J, Wendel J, eds. Plant genome diversity, Vol. 2. Heidelberg: Springer, 187–208. [Google Scholar]
- Chan RC, Severson AF, Meyer BJ. 2004. Condensin restructures chromosomes in preparation for meiotic divisions. Journal of Cell Biology 167: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, DeYoung D, Henikoff S, Malik HS. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLlife 3: e03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero M, Hipp AL, Waterway MJ, Valente LM. 2012. Diversification rates and chromosome evolution in the most diverse angiosperm genus of the temperate zone (Carex, Cyperaceae). Molecular Phylogenetics and Evolution 63: 650–655. [DOI] [PubMed] [Google Scholar]
- Escudero M, Maguilla E, Loureiro J, Castro M, Castro S, Luceño M. 2015. Genome size stability despite high chromosome number variation in Carex gr. laevigata. American Journal of Botany 102: 233–238. [DOI] [PubMed] [Google Scholar]
- Fishman L. 2013. Female meiotic drive in monkeyflowers: insight into the population genetics of selfish centromeres In: Jiang J, Birchler JA, eds. Plant centromere biology. Oxford: Wiley-Blackwell, 129–145. [Google Scholar]
- Fishman L, Saunders A. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562. [DOI] [PubMed] [Google Scholar]
- Finseth FR, Dong Y, Saunders A, Fishman L. 2015. Duplication and adaptive evolution of a key centromeric protein in Mimulus, a genus with female meiotic drive. Molecular Biology and Evolution 32: 2694–2706. [DOI] [PubMed] [Google Scholar]
- Heckmann S, Macas J, Kumke K, et al. 2013. The holocentric species Luzula elegans shows interplay between centromere and large-scale genome organization. The Plant Journal 73: 555–565. [DOI] [PubMed] [Google Scholar]
- Heckmann S, Jankowska M, Schubert V, Kumke K, Ma W, Houben A. 2014. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nature Communications 5: 4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. 2011. Organisation of the plant genome in chromosomes. The Plant Journal 66: 18–33. [DOI] [PubMed] [Google Scholar]
- Hirsch CD, Wu Y, Yan H, Jiang J. 2009. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Molecular Biology and Evolution 26: 2877–2885. [DOI] [PubMed] [Google Scholar]
- Kikudome GY. 1959. Studies on the phenomenon of preferential segregation in maize. Genetics 44: 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrel B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Molecular Biology and Evolution 28: 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Schubert I. 2013. CENH3 for establishing and maintaining centromeres In: Jiang J, Birchler JA. eds. Plant centromere biology. Oxford: Wiley-Blackwell, 67–82. [Google Scholar]
- Malik HS, Henikoff S. 2009. Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082. [DOI] [PubMed] [Google Scholar]
- Marques A, Ribeiro T, Neumann P, et al. 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proceedings of the National Academy of Sciences, USA 112: 13633–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SWL. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Research 20: 579–593. [DOI] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biology 14: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346: 763–767. [DOI] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nature Cell Biology 7: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Moraes IC, Lermontova I, Schubert I. 2011. Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Molecular Biology 75: 253–261. [DOI] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genetics 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Kashihara K, Murata M. 2005. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. The Plant Cell 17: 1886–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Navrátilová A, Schroeder-Reiter E, et al. 2012. Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genetics 8: e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings BD. 2014. Erasing errors due to alignment ambiguity when estimating positive selection. Molecular Biology and Evolution 31: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings BD, Suchard MA. 2005. Joint Bayesian estimation of alignment and phylogeny. Systematic Biology 54: 401–418. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED, Progra NCS. 2010. Adaptive evolution of foundation kinetochore proteins in primates. Molecular Biology and Evolution 27: 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. 2004. Adaptive evolution of centromere proteins in plants and animals. Journal of Biology 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bayes JJ, Henikoff S. 2008. Evolution of centromeres and kinetochores: a two-part fugue In: De Wulf P, Earnshaw WC, eds. The kinetochore: from molecular discoveries to cancer therapy. Berlin: Springer, 193–230. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Záveská Drábková L, Vlček C. 2010. Molecular phylogeny of the genus Luzula DC. (Juncaceae, Monocotyledones) based on plastome and nuclear ribosomal regions: a case of incongruence, incomplete lineage sorting and hybridisation. Molecular Phylogenetics and Evolution 57: 536–551. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bureš P. 2012. Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis. PLoS One 7: e30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Bureš P. 2016. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Scientific Reports 6: 33308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Veselý P, Horová L, Bureš P. 2016. Flow cytometry may allow microscope-independent detection of holocentric chromosomes in plants. Scientific Reports 6: 27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Talbert PB, Zhang W, et al. 2013. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proceedings of the National Academy of Sciences, USA 110: E4875–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.