Abstract
Background and Aims Eugenia sect. Phyllocalyx Nied. includes 14 species endemic to the Neotropics, mostly distributed in the Atlantic coastal forests of Brazil. Here the first comprehensive phylogenetic study of this group is presented, and this phylogeny is used as the basis to evaluate the recent infrageneric classification in Eugenia sensu lato (s.l.) to test the history of the evolution of traits in the group and test hypotheses associated with the history of this clade.
Methods A total of 42 taxa were sampled, of which 14 were Eugenia sect. Phyllocalyx for one nuclear (ribosomal internal transcribed spacer) and four plastid markers (psbA-trnH, rpl16, trnL-rpl32 and trnQ-rps16). The relationships were reconstructed based on Bayesian analysis and maximum likelihood. Additionally, ancestral area analysis and modelling methods were used to estimate species dispersal, comparing historically climatic stable (refuges) and unstable areas.
Key Results Maximum likelihood and Bayesian inferences indicate that Eugenia sect. Phyllocalyx is paraphyletic and the two clades recovered are characterized by combinations of morphological characters. Phylogenetic relationships support a link between Cerrado and south-eastern species and a difference in the composition of species from north-eastern and south-eastern Atlantic forest. Refugia and stable areas identified within unstable areas suggest that these areas were important to maintain diversity in the Atlantic forest biodiversity hotspot.
Conclusion This study provides a robust phylogenetic framework to address important historical questions for Eugenia s.l. within an evolutionary context, supporting the need for better taxonomic study of one of the largest genera in the Neotropics. Furthermore, valuable insight is offered into diversification and biome shifts of plant species in the highly environmentally impacted Atlantic forest of South America. Evidence is presented that climate stability in the south-eastern Atlantic forest during the Quaternary contributed to the highest levels of plant diversity in this region that acted as a refugium.
Keywords: Biome shifts, diversity patterns, Myrtaceae, Pleistocene
INTRODUCTION
Biogeography of the Neotropical flora, Myrtaceae, the Atlantic forest and refugia
The isolation of South America from Central America and Africa during the Mesozoic (250–65 Mya) left a strong imprint on the flora of the Neotropics, resulting in high levels of endemism in its humid forests (Burnam and Graham, 1999). Myrtaceae is one of the most important plant families in the Atlantic forest biome, considered one of five UNESCO priority biodiversity hotspots (Oliveira-Filho and Fontes, 2000). The evolutionary history of Myrteae, the neotropical Myrtaceae tribe, coincides with the evolution of the Atlantic forest and may be useful to understand other evolutionary aspects of the biome (Lucas and Bünger, 2015). A brief history of the evolution of Myrtaceae in the Neotropics follows. Palynostratigraphic evidence indicates that after its palaeotropical origin, the clades of contemporary Myrtaceae evolved and dispersed as far north as North America, reaching as far south as Antarctica by the Cretaceous–Paleogene boundary (65·5 Mya; Systma et al., 2004). Fossil pollen of the genus Myrtaceidites are found from the Late Eocene onwards in Australia, South America, New Zealand, Antarctica, Africa, North America, Europe and China (Thornhill and Macphail, 2012). The parent clade distribution then appears to contract into the Southern Hemisphere but is accompanied by an increase in morphological diversity and relative pollen abundance (Thornhill et al., 2012). Regarding the historical distribution of Myrteae, Lucas et al. (2007) used dispersal–vicariance analysis (DIVA 1·1; Ronquist, 1997) to hypothesize a Gondwanan origin for Myrteae followed by migration to South America. From an infrageneric point of view, these authors found southern and western South America to be ancestral distribution areas of one of the richest clades in Myrteae, the ‘Eugenia group’, before northwards movement and dispersal to South Asia and Africa. In south-eastern South America, the formation of the Atlantic forest was most influenced by the uplift of the Serra do Mar during the Oligocene, after the expansion of the Eugenia group. This geological event caused regional micro-regions and micro-climates to develop (Almeida and Carneiro, 1998) and has probably influenced the diversification of lineages.
During the Paleocene–Early Eocene, climatic conditions were probably warm enough to allow the expansion of megathermal angiosperms towards middle and high latitudes (Palazzesi and Barreda, 2007). After this interval, a progressive decline in temperature forced the migration of megathermal elements towards lower latitudes (Troncoso and Romero, 1998), e.g, south-eastern and north-eastern Brazil.
Based on the presence of presumed primitive species in the Atlantic forests, Gentry (1982) suggested that it could be a source area of Gondwanan taxa. The Atlantic forest flora includes early diverging lineages such as the Poaceae subfamily Anomochlooideae (Judziewicz and Clark, 2007), Goniorrhachis and Barnebydendron, the first diverging branches of the Detarieae clade of Leguminosae (Bruneau et al., 2008). Other Atlantic forest lineages appear to result from more recent colonization from other South American source areas such as birds (Cracraft and Prum, 1988), small mammals (Costa, 2003) and the genus Pagamea – Rubiaceae (Vicentini 2007). Based on these distributions, Pennington et al. (2006) conclude that the Atlantic forest flora comprises elements from both old and recently diverged lineages.
The southern portion of the Atlantic forest, an extensive centre of endemism that coincides with the Serra do Mar and Serra da Mantiqueira mountain ranges (Prance, 1982; da Silva et al., 2004), appears to be influenced more strongly by elements from extra-Amazonian biomes. As an example, some Andean-centred taxa can be found in the South Atlantic forest but are absent in the North Atlantic forest (from Rio Grande do Norte to northern Espírito Santo states), e.g. Myrceugenia (Landrum, 1981).
Within the Atlantic forest biodiversity hotspot, areas of higher angiosperm diversity and endemism are thought to correspond to areas of glacial refugia (Carnaval and Moritz, 2008; Staggemeier et al., 2015), where stable biota have contributed disproportionately to the evolutionary process, behaving as ‘museums’ and subsequently providing taxa to adjacent biomes (Keppel et al., 2012). An alternative explanation for high numbers of species found in so-called refugia is that soil conditions rather than climate change strongly influenced coastal ecosystems such as mangroves, sand banks and rain forests of south-east Brazil during the geological past (Sheel-Ybert, 2000). More recently, the refuge hypothesis is challenged by studies that predict historically unstable areas showing climatic stability during the Quaternary (Carnaval et al., 2014; Leite et al., 2016).
Eugenia as a case study for the refuge hypothesis
Angiosperms, the largest phylum of the Plant Kingdom, have approx. 250 000 species distributed in approx. 13 700 genera (Judd et al., 2008); only 57 of these genera contain >500 species (Frodin, 2004). The phenomenon of large genera and the fact that most large Angiosperm genera remain poorly known, particularly in terms of patterns of morphological and geographic diversification, represents a formidable challenge for botanists (Frodin, 2004). Reasons to overcome these challenges include the potential for large and geographically widespread genera to reveal more comprehensive geographical structure within phylogenies than has been possible to date, allowing examination of relationships to ecological processes (Schrire et al., 2005). Large genera also require better basic understanding to resolve long-standing taxonomic issues, allowing generic diversity to be the basis of future study in related fields such as ecology, evolution, physiology and conservation.
Eugenia sensu lato (s.l.) (sensu Mazine et al., 2014) comprises approx. 1100 species (Govaerts et al., 2015); it is both the largest Neotropical Myrtaceae genus and the most species-rich genus in Brazil (Sobral et al., 2014). So circumscribed, and as suggested by previous authors (Niedenzu, 1893; Kiaerskou, 1893; Proença, 1991; Sobral, 2003; Holst et al., 2003), Eugenia s.l. includes four previously accepted genera: Calycorectes O. Berg, Hexachlamys O. Berg, Phyllocalyx O. Berg and Stenocalyx O. Berg.
Taxonomic classification in Eugenia has traditionally emphasized inflorescence morphology (Berg, 1856). Recent molecular study (Mazine et al., 2014) shows that Eugenia comprises nine clades, not always supported by inflorescence architecture. The clades identified by Mazine et al. (2014) are to be treated as sections (F. F. Mazine et al., unpubl. res.). One of these corresponds to Eugenia sect. Phyllocalyx Nied. and comprises 14 species. This section is widely distributed through the Atlantic forest, from eastern Brazil to Paraguay with some species in the Cerrado and Amazon biomes. The section is characterized by peduncles with leaf-like bracts and showy sepals that are proportionally larger than the flowers (Berg, 1856).
The centre of Eugenia diversity is in Brazil from where approx. 388 taxa are recorded, of which 313 are endemic (Sobral et al., 2014), Eugenia occurs in all phytogeographic domains. The greatest species richness is found in the Atlantic forest and Amazonia, with 250 and 91 taxa, respectively, while 74 species are reported from the Cerrado (savanna-like vegetation). The importance of Myrtaceae and Eugenia s.l. as structural and ecological components of Atlantic rain forest of Brazil makes this group a good proxy for the diversity of other important angiosperm groups in the Atlantic rain forest (Oliveira-Filho and Fontes, 2000; Murray-Smith et al., 2009; Lucas and Bünger, 2015).
This study presents a molecular phylogeny of E. sect. Phyllocalyx including representatives of all other infrageneric groups (‘clades’ sensuMazine et al., 2014). The main goals of this study were to (1) test the monophyly of E. sect. Phyllocalyx (sensuBerg, 1856; Niedenzu, 1893); (2) provide a hypothetical chronology for major clade diversification within Eugenia; (3) test the Atlantic forest as the ancestral area for E. sect. Phyllocalyx; and (4) examine the evolutionary history of E. sect. Phyllocalyx in the context of stable Atlantic forest areas in the Quaternary.
MATERIALS AND METHODS
Taxonomic and molecular sampling
Forty-two Myrteae taxa were sampled including 28 Eugenia s.l., of which 12 (from 14 species) were Eugenia sect. Phyllocalyx, encompassing as much morphological and geographical variation as possible within the group (Supplementary Data Fig. S2). Approx. 85 % of E. sect. Phyllocalyx was represented. Thirteen additional genera from tribe Myrteae were additionally included. Myrtus communis was used as outgroup in all analyses following Lucas et al. (2011). Species names and nomenclature follow Sobral et al. (2014). The internal transcriber spacer (ITS) of the ribosomal nuclear region and four plastidial regions: psbA-trnH, rpl16, trnL-rpl32 and trnQ-rps16 were used (Table 1). The final analysis included 588 sequences, 57 % generated for this study. with the remaining 43 % obtained from published and unpublished works (Lucas et al. 2007; J. Q. Faria-Junior, UnB, Brasilia, Brazil, unpubl. res.; F. F. Mazine, Ufscar, Sorocaba, Brazil, unpubl. res.; M. F. Santos, Usp, Sao Paulo, Brazil, unpubl. res.).
Table 1.
PCR conditions
| Molecular marker | Primers | Conditions |
|---|---|---|
| ITS | AB101 | 5 min at 94 °C followed by 28 cycles of 1 min at 94 °C, 1 min at 48 °C, 1 min at 72 °C and a last stage of 7 min at 72 °C |
| AB102 | ||
| psbA-trnH | psbA (F) | 5 min at 94 °C followed by 28 cycles of 1 min at 94 °C, 1 min at 48 °C, 1 min at 72 °C and a last stage of 7 min at 72 °C |
| trnH (R) | ||
| rpl16 | rpL16F71 |
|
| rpL16R1516 | ||
| trnL-rpl32 | trnL(UAG) | 5 min at 94 °C followed by 28 cycles of 1 min at 94 °C, 1 min at 48 °C, 1 min at 72 °C and a last stage of 7 min at 72 °C |
| rpl32 | ||
| trnQ 5′-rps16 | trnQ 5′ MytrnQ Myrps16 rps16 | 5 min at 94 °C followed by 28 cycles of 1 min at 94 °C, 1 min at 48 °C, 1 min at 72 °C and a last stage of 7 min at 72 °C |
DNA sequencing
Total DNA was extracted from 0·3 g of silica gel-dried leaf material using a modified version of the 2× cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1987). Total DNA was further purified for long-term storage in RBG Kew’s DNA & Tissue Collections by equilibrium centrifugation in CsCl–ethidium bromide gradients (1·55 g mL–1) followed by butanol extraction of the ethidium bromide and dialysis to remove the caesium chloride. Amplification and purification of target DNA regions were executed according to the protocols outlined in Lucas et al. (2007, 2011) and Shaw et al. (2007) using internal primers for trnL-rpl32 and trnQ-rps16. The PCR conditions are listed in Table 1. Sequencing was performed according to Lucas et al. (2007). DNA sequences were assembled using MUSCLE (Edgar, 2004) and edited in Geneious 7·9 with subsequent manual adjustment when necessary. All sequences are deposited in GenBank; DNA samples are stored in RBG Kew’s DNA & Tissue Collections (see Appendix 1). Due to the inheritance of the plastid genome as a single linked unit, the four resulting plastid DNA matrices were combined into a single data set of 4190 bp. The nuclear and combined plastid regions were then analysed independently and simultaneously.
Phylogenetic analyses
Phylogenetic reconstruction using maximum likelihood (ML) and Bayesian inference was performed on all data sets. Models of nucleotide substitution were selected for each sequence partition with MrModeltest2, version 2·2 (Nylander, 2004). The model recommended under the Akkaike Information Criterion for the ITS, trnL-rpl32 and trnQ-rps16 region was GTR + G + I, and that for psbA-trnH and rpl16 was GTR + G. Data sets were analysed in combination with these model parameters fitted independently to each partition.
Maximum likelihood analyses were performed with RAxML v7.6.3 (Stamakis, 2006) using the rapid bootstrap algorithm with 1000 replicates, combined with a search of the best-scoring ML tree under default parameters. Bayesian analyses were performed using MrBayes version 3.2.1 (Ronquist et al., 2012). Two independent analyses each of four chains were conducted with 5 million generations of Monte Carlo Markov Chains (MCMCs) and a sampling frequency of 1000. Results were examined with Tracer v. 1·6 (Drummond and Rambaut, 2007), to ensure that the analyses reached convergence and that the effective sample size of each parameter was >200. A consensus tree with posterior probabilities (PPs) was generated using the ‘sumt’ option in MrBayes with the default burn-in of 10 % (500 trees in our case). The consensus tree and PPs were visualized with FigTree 1·4 (Rambaut, 2012).
Analyses conducted for each partition returned topologies inspected by eye and found to be congruent for major clades. The combined data set provided greater resolution and higher levels of support; our results and discussion therefore focus on the combined results. Phylogenetic reconstructions obtained with ML and Bayesian results are highly congruent.
To estimate temporal evolution of E. sect. Phyllocalyx. we used a Bayesian inference approach implemented in the package BEAST v.1.8.0 (Drummond et al., 2012) using the combined data set and applying the same partition and models as previously described for the Mr Bayes analyses (see above). An uncorrelated relaxed molecular clock assuming a lognormal distribution of rates and a Yule speciation model were applied. Four runs of 30 million generations were performed, sampling one tree every 1000 generations. Parameter convergence was confirmed by examining their posterior distribution in Tracer 1·6 (Drummond and Rambaut, 2007). MCMC sampling was considered sufficient when the effective sample size of each parameter was >200. All analyses were performed on the CIPRES portal (Miller et al., 2010). A maximum clade credibility tree with median branch lengths and 95 % highest posterior density interval on nodes was built using TreeAnnotator 1·8 (Drummond et al., 2012) based on the remaining set of trees after burn-in (for each run, a burn-in period of 3 million generations was applied). Calibration was performed using the fossil Paleomyrtinae princetonensis from the Paleocene (Crane et al., 1990; Pigg et al., 1993) to early Eocene (Manchester, 1999) of North America, comprising well-preserved fruits and seed probably related to Psidium and/or Mosiera (Pigg et al. 1993). The most recent common ancestor of Myrteae and the outgroup taxa (crown group of Myrteae) was constrained, following the recommendations of Forest (2009), using a lognormal distribution with a median of 55·8 Mya (corresponding to the lower bound of the Eocene), lower quartile (2·5 %) of 54·94 Mya and superior quartile of (97·5%) 61·9 Mya, achieved using an offset value of 54·8 Mya.
Ancestral area analysis
The ‘core Phyllocalyx’ clade identified by the phylogenetic analysis contained 12 species. A matrix was compiled indicating the presence or absence of each Eugenia species sampled for the phylogeny, in one of four geographic areas. The areas used were designed to indicate movement of species of the ‘core Phyllocalyx’ clade between the biomes in which they are found (Atlantic forest and Cerrado) but also included areas in which the other Eugenia species sampled occurred (Amazon and ‘not tropical America’). Dispersal Extinction Cladogenesis (DEC; Ree and Smith 2008) was implemented using the package RASP (Yu et al., 2015) run constraining range to two biogeographic units and dispersal between adjacent units (Atlantic/Amazon forest and Cerrado) set to 1·0, non-adjacent, proximal units (Atlantic and Amazon forest) set to 0·5 and non-adjacent distant units (Atlantic/Amazon forest/Cerrado and ‘not tropical America’) set to 0·1. The RASP output reports ancestral area probabilities as pie charts.
Ecological niche models (ENMs)
Data set
To access how the whole ‘core Phyllocalyx’ clade identified by the phylogenetic analysis could be a model group to identify a new refugium area during the Quaternary, the 14 species were modelled (the 12 from the phylogeny framework plus the two missing, i.e. Eugenia elongata Nied. and E. itacarensis Mattos). The floristic data set was extracted from NeoTropTree (Oliveira-Filho, 2015), a database of tree species (defined as free-standing woody plants >3 m in height) checklists for geo-referenced Neotropical sites compiled from the literature and herbarium specimen records. Data gathered for this study from multiple herbaria were also used (BHCB, BR, CEPEC, ESA, G, HB, HPL, HRCB, HUFU, K, LE, M, MBM, MBML, OUPR, OX, P, R, RB, SP, SPF, UEC and UPCB). In total, 609 records were included, encompassing all known occurrences (Supplementary Data Fig. S3) of E. sect. Phyllocalyx; the number of records per species is shown in Table 3.
Table 3.
Records used per species in the ecological model modelling (ENM)
| Species | Records |
|---|---|
| Eugenia elongata Nied. | 2 |
| Eugenia sp. nov. Bünger & Sobral | 2 |
| Eugenia expansa Spring. ex Mart. | 48 |
| Eugenia glandulosa Cambess. | 3 |
| Eugenia involucrata DC. | 451 |
| Eugenia itacarensis Mattos | 9 |
| Eugenia luschnathiana (O.Berg) Klotzsch ex B.D.Jacks. | 46 |
| Eugenia macrobracteolata Mattos | 6 |
| Eugenia magnibracteolata Mattos& D. Legrand | 7 |
| Eugenia membranifolia Nied. | 15 |
| Eugenia puberula Nied. | 5 |
| Eugenia regia Bünger & Sobral | 3 |
| Eugenia ruschiana Bünger & Mazine | 1 |
| Eugenia selloi B.D. Jacks. | 11 |
| Total records | 609 |
Bioclimatic variables
For all sites, we obtained the value at 1 km spatial resolution, of the 19 standard BIOCLIM variables, from the WorldClim database (Hijmans et al., 2005). We cropped the bioclimatic layers to span from 10°N to 40°S and from 70°W to 20°W (Fig. S3). This covers all known occurrences as well as the potentially accessible area for the species (Fig. S3); ‘core Phyllocalyx’ is actually restricted to South America, mostly to the coastal Atlantic forest in Brazil, with some species growing in rain forests from Argentina, Bolivia, Paraguay and Uruguay, and Cerrado from Brazil.
After assessing correlations between the bioclimatic variables, we retained ten of 19 variables, eliminating those with less biological relevance from groups of strongly inter-related variables (r > 0·9). This procedure was done to avoid over-parametrization of our modelling with redundant variables. The final selected variables were: annual mean temperature, mean diurnal range, isothermality, maximum temperature of warmest month, temperature annual range, mean temperature of driest quarter, annual precipitation, precipitation of wettest month, precipitation of wettest quarter and precipitation of warmest quarter.
Model construction
Ecological niche models (ENMs) usually involve the determination of associations between environmental conditions and information on the occurrence of species to identify areas critical to the maintenance of species populations (Peterson et al., 1999; Warren et al., 2010; Peterson and Soberóm, 2012), and this technique can be used to analyse the limiting factors or regulators that determine the spatial distribution of plants and animals (Guisan and Zimmermann, 2000).
We modelled the ecological niche of the 14 species (Table 3) using Maxent ver. 3.3 (Phillips et al., 2006). It has been demonstrated that Maxent is often critical to the maintenance of species populations (Elith et al., 2006, 2011; Pearson et al., 2006; Phillips and Dudík, 2008; Pena et al., 2014). In addition, an important reason for choosing Maxent was that it allowed us to use presence-only species data, which is of great utility because the vast majority of the biotic data available, including those used here, come in this form (Elith et al., 2006, Phillips and Dudík, 2008).
To calibrate and evaluate the quality of the models, we divided the data for each species into a training set (75 % of occurrences) and a test or validation set (25 % of occurrences). We constructed models five times and averaged the output to produce the final results used in downstream analyses. Next, for each species, we defined a threshold value above which grid cells were considered to have environmental characteristics suitable for the maintenance of viable populations of the species (Pearson et al., 2006). We used the ‘minimum training presence’ as the threshold selection method because it assumes that the species presence is restricted to sites at least as suitable as those at which the species has been observed so far (Pearson et al., 2006).
In order to produce models to infer the palaeodistribution of the ‘core Phyllocalyx’ species, we produced projections of the suitability of occurrence during the current (0 ka pre-industrial), mid-Holocene (6000 years ago – 6 kyr BP), Last Glacial Maximum (LGM; 21 000 years ago – 1 kyr BP) and Last Interglacial (LIG; 120 000 years ago – 120 kyr BP) time periods based on climatic simulations (Hijmans et al., 2005).
Palaeoclimatic data represent downscaled climate data from simulations with Global Climate Models (GCMs) based on the Coupled Model Intercomparison Project Phase 5 (CMIP5; Taylor et al., 2012). For the LIG model, we used the Otto-Bliesner et al. (2006) approach, while for LGM and Holocene we employed the Community Climate System Model – CCSM4 (Gent et al., 2011). All geographic information system (GIS) analyses were performed in ArcGIS v.10 (ESRI, 2011).
To indicate potential areas of stability for the species of E. sect. Phyllocalyx throughout the Quaternary, similar protocols to those used in recent studies for other Neotropical domains were adopted (Carnaval and Moritz, 2008; Werneck et al., 2011, 2012; Bueno et al., 2016) where converted models were produced from continuous outputs into presence/absence maps by applying the lowest presence threshold for each model. This approach maximizes agreement between observed and modelled distributions, balancing the cost arising from an incorrect prediction against the benefit gained from a correct prediction (Pearson et al., 2006). By summing the presence/absence maps obtained for current conditions, mid-Holocene (6000 years ago – 6 kyr BP), LGM (21 000 years ago – 21 kyr BP) and LIG (120 000 years ago – 120 kyr BP) projections, a map of areas showing historical stability was generated. This combined map depicted areas potentially occupied by E. sect. Phyllocalyx species during the climatic oscillations of the Quaternary. These historically stable areas, or refugia, were defined as those grid cells for which the presence of all species was inferred in all models and time projections.
We also used a threshold-independent method of model validation, the receiver operating characteristic (ROC) curve analysis. The ROC curve is obtained by plotting sensitivity values (the true-positive fraction) on the y-axis against their equivalent specificity values (1 – specificity, the false-positive fraction) on the x-axis for all possible thresholds (Fielding and Bell, 1997). The ROC analysis characterizes the predictive performance of a model at all possible thresholds by a single number, the area under the curve (AUC) (Fawcett, 2003; Phillips et al., 2006). A single AUC value was calculated for each species, representing the average across the five interations of model construction. The value of the AUC can fall between 0·5 and 1·0. If the value is 0·5, the model is no better than random, while models with values >0·75 are generally considered potentially useful, and models with a value near one are considered to be strongly supported (Fielding and Bell, 1997; Elith, 2002; Rushton et al., 2004; Phillips et al., 2006).
RESULTS
Phylogenetic relationships
Molecular marker statistics are summarized in Table 1; of the 3462 characters included in the plastid analysis, 466, 915, 807 and 1274 bp are from psbA-trnH, rpl16, trnL-rpl32 and trnQ-rps16, respectively. The ITS partition is more variable (16·7 % informative characters) than the four plastid regions (8·6 % informative characters for combined plastids; Table 2). Inspection of the topologies by eye and using the incongruence length difference (ILD) test (P > 0·001; partition-homogeneity test with heuristic search), performed using PAUP, indicates that the two partitions give congruent results. Differences between the resulting topologies were negligible; therefore. the combined ML tree is provided as Supplementary Data Fig. S1 and the Bayesian results (Fig. 1) form the basis of the discussion presented here.
Table 2.
Molecular data and parsimony-based tree characteristics
| ITS | psbA-trnH | rpl16 | trnL-rpl32 | trnQ-rps16 | Plastid | Combined molecular data sets | |
|---|---|---|---|---|---|---|---|
| Aligned characters (bp) | 738 | 466 | 915 | 807 | 1274 | 3462 | 4190 |
| No. of variable characters (%) | 256 (34·7) | 159 (34·1) | 117 (12·8) | 178 (22·0) | 271 (21·3) | 725 (21·0) | 560 (13·4) |
| No. of potentially parsimony-informative characters (%) | 123 (16·7) | 67 (14·4) | 31 (3·4) | 79 (9·8) | 87 (6·9) | 296 (8·6) | 421 (10·0) |
| Trees | 1315 | 2757 | 7626 | 3407 | 7626 | 156 | 24 |
| Length | 553 | 253 | 140 | 257 | 370 | 1066 | 1677 |
| Consistency index | 0·61 | 0·76 | 0·95 | 0·83 | 0·84 | 0·89 | 0·77 |
| Retention index | 0·57 | 0·83 | 0·95 | 0·80 | 0·78 | 0·86 | 0·71 |
ITS, internal transcribed spacer region.
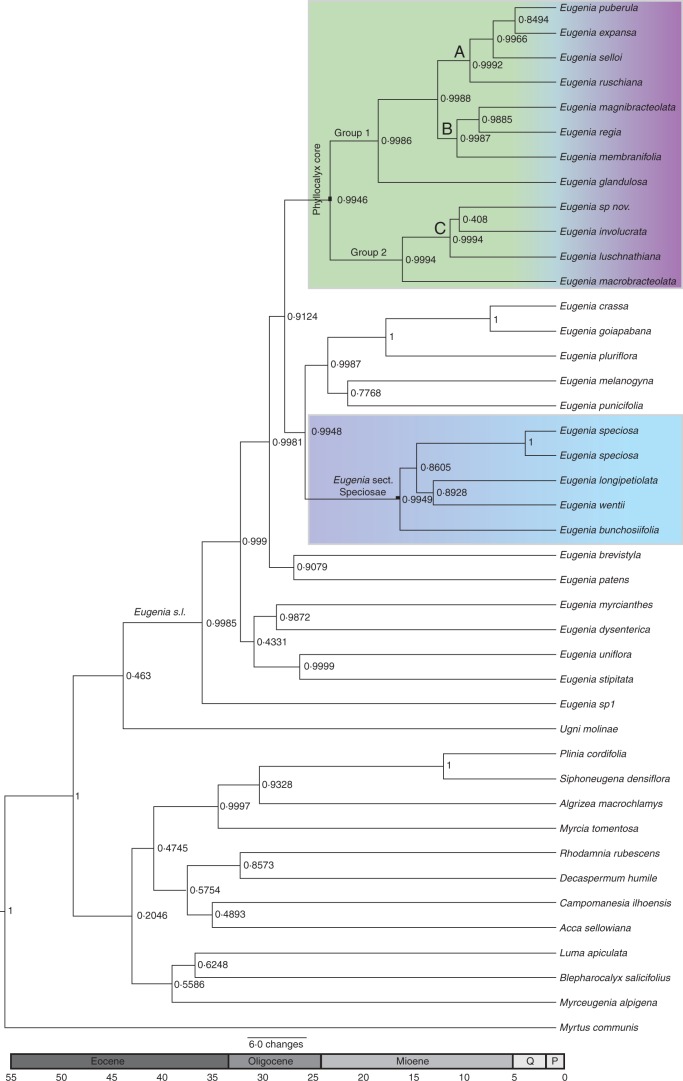
Fig. 1.
Maximum credibility clade tree with divergence time estimates and posterior probabilities obtained from BEAST analyses of Eugenia section Phyllocalyx based on combined analyses of nuclear (ITS) and plastid (psbA-trnH, rpl16, trnL-rpl32, trnQ-rpS16) regions and using Paleomyrtinaea princetonensis as a calibration point at the root of the tree (i.e. crown node of tribe Myrteae; 55·8 Mya). Posterior probability values are indicated on branches. P, Pliocene; Q, Quaternary.
Eugenia sect. Phyllocalyx sensu Berg emerges as a paraphyletic group (Fig. 1). The clade containing most species previously placed inside E. sect. Phyllocalyx, and the type-species of the section (Eugenia involucrata DC.) emerge as a well-supported monophyletic group (PPBAYES, 0·99; PPBEAST, 0·97; ML, 0·75). This clade corresponds to Eugenia sect. Phyllocalyx, and its taxonomic sense can be conserved; from hereon the name ‘core Eugenia sect. Phyllocalyx’ is used. A second well-supported clade (PPBAYES, 1; PPBEAST, 1; ML, 100) also included four species previously included in Eugenia sect. Phyllocalyx and was recently published as Eugenia sect. Speciosae Bünger & Mazine (Bünger et al., 2016). The new section emerges with high support (PPBAYES, 0·99; PPBEAST, 0·99; ML, 72) as sister to Eugenia sect. Umbellatae (F. F. Mazine et al., unpubl. res.). These clades correspond entirely, or with minor exception, to combinations of macro-morphological characters. Often such characters taken individually are not sufficiently informative to place a species in a particular group, but, when taken together, the sub-clades become morphologically diagnosable. Other clades identified but poorly supported in Mazine et al. (2014) are represented here with relationships between them generally receiving higher statistical support.
Within core Eugenia sect. Phyllocalyx, ‘Group 1’ receives good support (PPBAYES, 0·99; PPBEAST, 0·99; ML, 97) and includes sub-clades A, B and E. glandulosa Cambess. ‘Group 2’ also receives high support (PPBAYES, 1; PPBEAST, 0·99; ML, 100) and includes sub-clade C and E. macrobracteolata Mattos. Eugenia sect. Speciosae includes Eugenia wentii Amsh. (the only Amazonian species from what was E. sect. Phyllocalyx sensu Berg), E. bunchosiifolia Nied., E. longipetiolata Mattos and E. speciosa Cambess.; relationships within Eugenia sect. Speciosae are poorly supported.
In core Eugenia sect. Phyllocalyx, sub-clades A and B are composed of species exclusively from coastal Atlantic forest. Eugenia glandulosa appears sister to these clades and is exclusively found in Cerrado. Sub-clade C is composed of species from both Atlantic forest and dry biomes. Eugenia luschnathiana grows in the restingas of Northeast Brazil. Eugenia sp. nov. is from campos rupestres from Bahia and E. involucrata is widespread through Atlantic forest and other dry biomes (Cerrado, restinga and campos rupestres). Eugenia macrobracteolata from the Atlantic Forest is sister to sub-clade C.
Dating
Trees obtained from the BEAST analyses agree well with those from the Bayesian and ML phylogenetic hypotheses. The mean age for Eugenia s.l. is 35 million years (Mya) with 95 % confidence intervals (CIs) of 45–24 Mya (mid-Eocene to Early Miocene; Fig. 1). Core Eugenia sect. Phyllocalyx dates from the mid-Miocene (20·6 Mya; 95 % CI 27·5–14 Mya; mid-Oligocene to mid-Eocene; Fig. 1). Species endemic to the Atlantic forest (sub-clades A and B) are reported as younger (12·0 Mya; 95 % CI 16·0–8·0 Mya; mid-Miocene to Late Miocene) than species from Cerrado (E. glandulosa; 16·6 Mya; 95 % CI 23·0–10·0 Mya) and as older than others from dry biomes (sub-clade C; 10·6 Mya; 95 % CI 15·0–5·0 Mya). Eugenia sect. Speciosae dates from Miocene (16·9 Mya; 95 % CI 23–10 Mya).
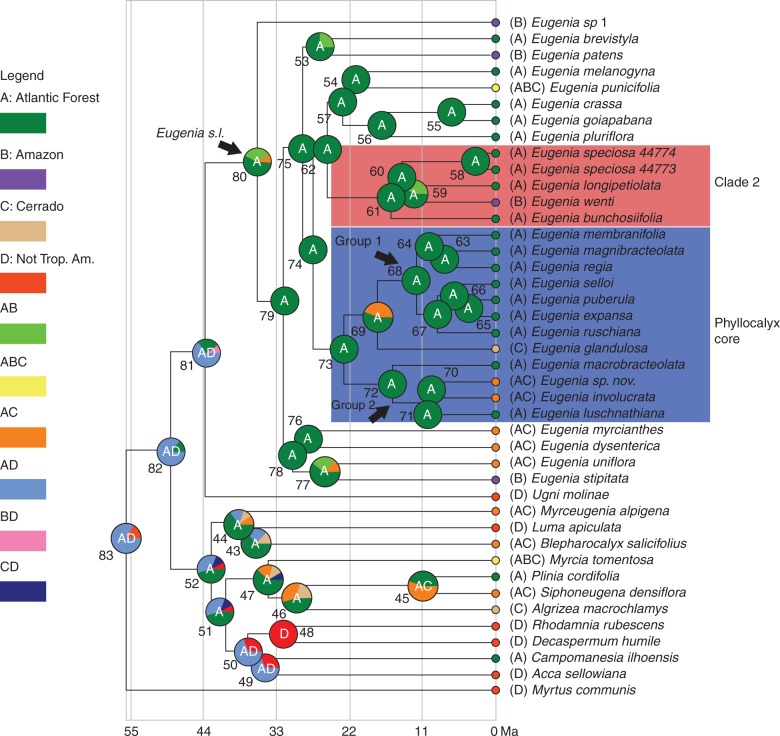
Ancestral area analysis
Most likely ancestral areas are indicated in Fig. 2.; ancestral area probabilities for key nodes are summarized in Supplementary Data Table S1. The most likely ancestral area of Eugenia s.l. (sensu Mazine et al., 2014) is seen to be the Atlantic forests (node 80: Atlantic forest, 58·4; Atlantic/Amazon forest, 32·37; Atlantic forest/Cerrado, 9·23). The majority of internal Eugenia nodes are reported to have originated exclusively (probability 100) in the Atlantic forest including Eugenia sect. Speciosae (node 61), the Phyllocalyx core (node 73), Groups 1 (node 68) and 2 (node 72). Within the Phyllocalyx core, only node 69, the ancestor of Group 1 plus Eugenia glandulosa, has the probability to have arisen in the Cerrado (Atlantic forest, 55·3; Atlantic forest/Cerrado, 44·7).
Fig. 2.
Ancestral area analysis (DEC) represented on the maximum credibility tree. Letters within the pie charts are the single or pair of most probable ancestral areas (A, Atlantic forest; B, Amazon forest; C, Cerrado; D, ‘non-tropical America’. Numbers adjacent to the pie chars identify each node. The range of each species is listed in parentheses next to the species name.
Modelling Eugenia sect. Phyllocalyx
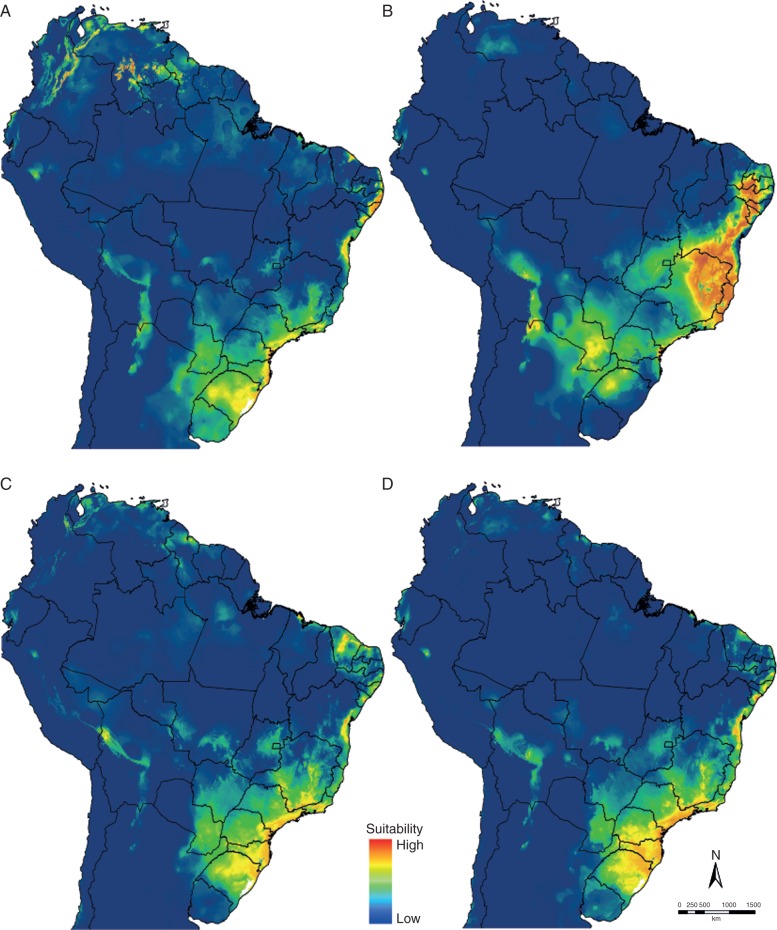
The quality of the models, according to the AUC values, were significant for all, confirmed by the correct assignment of testing data under the models, indicating that the models performed well in distinguishing species occurrence in relation to bioclimatic variables. The palaeomodels suggest changes in distribution ranges of core Phyllocalyx across the Quaternary (Fig. 3) but also show a constant area with high adequacy in south-eastern Brazil in contrast to the refugia hypothesis of Carnaval and Moritz (Fig. 4). However, the core Phyllocalyx clade appears to have experienced a remarkable change in its whole distribution during the LGM (Fig. 3B). Except during the LGM, the distribution ranges of core Phyllocalyx across the Quaternary appear very similar to current-day distributions. Areas that show a high probability of occurrence of core Phyllocalyx are in the north-eastern and south-eastern Atlantic coast (Fig. 3A, C, D) with species appearing in the sub-montane rainforests and restingas.
Fig. 3.
Predicted occupancy of the Phylocallyx clade across northern South America during: (A) the Last Interglacial (LIG; 130 ka BP); (B) the Last Glacial Maximum (LGM; 21 ka BP); (C) the mid-Holocene (6 ka BP); and (D) current climate (0 ka pre-industrial) scenarios. Warmer colours (red/yellow) of the logistic output correspond to regions with higher probability of occurrence.
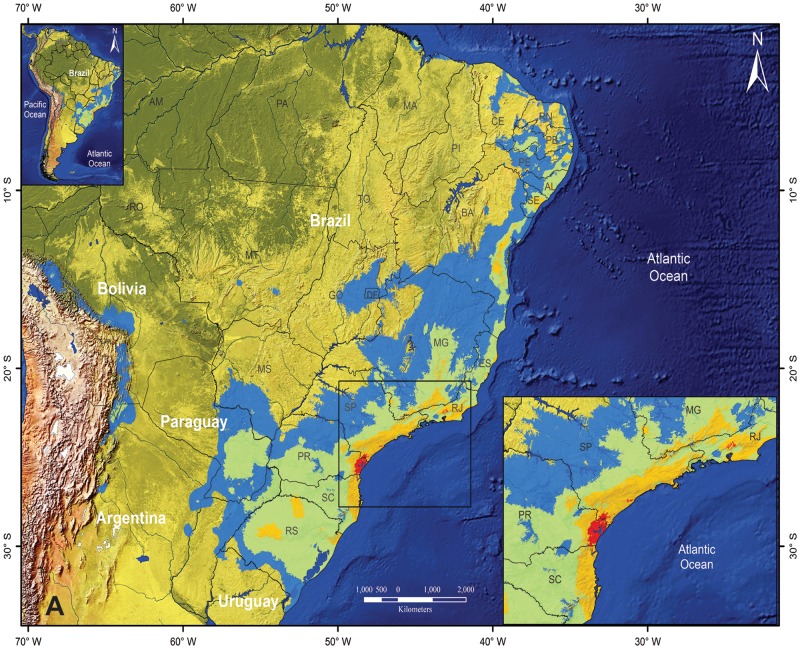
Fig. 4.
Predicted regions of historical stability in the Phylocallyx clade across the Quaternary, based on summed predicted occupancy of the clade across current (0 ka pre-industrial), mid-Holocene (6 ka BP), Last Glacial Maximum (LGM; 21 ka BP) and Last Interglacial (LIG; 130 ka BP) climatic scenarios. Areas in red are those where all species of the Phylocallyx clade are predicted to occur during all four time periods and represent postulated refugial areas for the Phylocallyx clade. Maps are for (a) South America, (b) central South America. Brazilian states are labelled as follows: Amazonas (AM), Bahia (BA), Ceará (CE), Distrito Federal (DF), Espírito Santo (ES), Goiás (GO), Maranhão (MA), Minas Gerais (MG), Mato Grosso (MT), Mato Grosso do Sul (MS), Pará (PA), Paraná (PR), Piauí (PI), Rio de Janeiro (RJ), Rio Grande do Norte (RN), Rio Grande do Sul (RS), Rondônia (RO), Santa Catariana (SC), São Paulo (SP), Tocantins (TO).
DISCUSSION
Although the molecular framework was based on five markers and does not represent a complete evolutionary history, results presented here provide a first comprehensive understanding of phylogenetic relationships within Eugenia sect. Phyllocalyx sensu Berg. The study uses the 14 species of Eugenia sect. Phyllocalyx set within a small but represtentive sample of the 1100 total global Eugenia species, to investigate evolutionary and ecological patterns in the genus. Myrtaceae sub-taxa have been used successfully as proxies for other groups (Murray-Smith et al., 2009; Lucas and Bünger, 2015), allowing the results presented here to be reasonably interpreted in this way. It is clear that the results presented here cannot be completely representative of the genus as a whole and that different Eugenia groups would have responded to different pressures or differently to the same pressures; however, this study represents the first biogeographical hypothesis of the genus and provides evolutionary and ecological models for consideration and testing by subsequent works, and data for inclusion in subsequent studies. Myrtaceae taxa have been used successfully as proxies for other groups (Murray-Smith et al., 2009; Lucas and Bünger, 2015), allowing the results presented here to be reasonably interpreted in this way.
The results are significant from a variety of perspectives, first suggesting that the classical taxonomy requires some rearrangement. According to Mazine et al. (2014) and F. F. Mazine et al. (unpubl. res.), Eugenia wentii (not treated as a group by Mazine et al., 2014) was more closely related to Eugenia clade 9 sensu Mazine et al. (2014) (= Eugenia sect. Umbellatae). Our results confirm this relationship and furthermore find the species to comprise part of a separate section inside Eugenia s.l. This section contains species from both the Atlantic and Amazon forests and supports suggested links between Amazonian and Atlantic forests (McVaugh, 1956). Although these two clades do not emerge in a monophyletic group, they share the floral characters of showy sepals and bracteoles that could be homoplastic characters in Eugenia s.l. Eugenia sect. Speciosae could be distinguished from the core Phyllocalyx by its linear sepals and bracteoles not persistent at anthesis. Further details and discussion are provided by Bünger et al. (2016).
The focus of the remainder of this study is the core Phyllocalyx group; the following discussion concerns the 14 species that comprise this group (Table 3), of which 12 are represented in the phylogenetic framework. The affinities of the missing species, based on morphological characters, are predicted as follows: Eugenia elongata and E. itacarensis both resemble E. regia (sub-clade B); shared characters include significantly larger flowers and sepals and larger bracteoles persistent at anthesis.
Dating and biogeography
Myrtaceae dates from the Cretaceous (86 Mya; Sytsma et al., 2004; Thornhill et al., 2012), apparently originating in paleotropical Africa (Thornhill and Macphail, 2012) or with a more general Gondwanan origin (Lucas et al., 2007). Results presented here suggest that Eugenia s.l. originated in the transition between the Eocene and Oligocene (35 Mya; Fig. 2). The fossil record of Myrteae is poor as fleshy fruits are rarely well preserved compared with dry fruits from other members of the family (e.g. tribe Eucalypteae). Only a single reliable fossil was available for the calibration procedure used here and, as a result, the biogeographical scenario outlined should be considered with some caution. Another putative Myrteae fossil, wood of Myrceugenelloxylon antarcticus from 72–66 Ma (Poole et al., 2003), was rejected due to doubts regarding its definite identification as Myrtaceae. The lack of an extensive, reliable fossil record might explain in part the discrepancies between results obtained here and those of Biffin et al. (2010) who found a mid-Miocene origin for Eugenia in a broader perspective.
Panti (2014) described Myrtaceae fossil leaves including those putatively, of Eugenia s.l., from the Río Turbio formation in Argentina; this formation is dated from the mid-Eocene. Despite the lower reliability associated with assigning leaf fossils to extant taxa, these finds correspond to results presented here that suggest a late Eocene origin for Eugenia s.l. An earlier origin for Eugenia s.l. and the distribution of the leaf fossils fit the dispersion hypothesis of Lucas et al. (2007) who suggest an origin of Eugenia after the Drake Passage opened isolating South America from Antarctica 42–17 Mya (Upchurch, 2008).
At the subgeneric level in Eugenia, and based on the sample presented here, older Atlantic forest lineages are represented by more geographically widespread species (Eugenia uniflora, approx. 26 Mya; E. brevistyla, approx. 24 Mya; and E. punicifolia, approx. 19·9 Mya). Lineages that appear more recently comprise species more likely to occur in centres of endemism such as the Serra do Mar in Paraná, São Paulo and Rio de Janeiro states (e.g. Eugenia expansa and E. puberula, approx. 4·3 Mya; E. selloi, approx. 6·5 Mya; and E. magnibracteolata, approx. 8 Mya). A large majority of core Eugenia sect. Phyllocalyx species are found only in the Atlantic forest, suggesting that the section originated in that biome (approx. 20 Mya). Within core Eugenia sect. Phyllocalyx (Fig. 1), two main groups emerge. Group 1 is comprised of almost all species from the Atlantic forest biome. Group 2 is comprised of species from drier biomes, restinga and Atlantic forest. Results from the dating analysis show Group 2 to be slightly younger (apprpx. 15·6 Mya) than Group 1 (approx. 16·7 Mya), suggesting that Group 2 emerged after and from ancestors in the Atlantic forest (Eugenia macrobracteolata is mostly found in the Serra do Mar, but also rarely occurs in Espírito Santo, with one specimen from Rio Grande do Norte; A. R. Araújo, pers. comm.). This scenario is supported by ancestral area analysis (Fig. 2) that shows most clades of Eugenia, including core Eugenia sect. Phyllocalyx and E. sect. Speciosae, to arise in the Atlantic forest biome. Because the specific sample is limited outside of core Eugenia sect. Phyllocalyx, caution is required when interpreting results; indeed, more detailed studies of Eugenia biogeography (e.g. F. F. Mazine et al., unpubl. res.) suggest a more pronounced role for the Amazon forest in early Eugenia diversification. However, for core Eugenia sect. Phyllocalyx and E. sect. Speciosae, sampling is almost complete (12 of 14 species and four of six species, respectively), and it is possible to be confident that these groups originated in the Atlantic forest biome with E. sp. nov. and E. involucrata subsequently expanding to drier environments. The pattern is supported by other studies that show Atlantic forest species colonizing the Cerrado at or around this time (Simon et al., 2009) and suggests a biome shift (Crisp et al., 2009; Donoghue and Edwards, 2014) between Atlantic forest and Cerrado. Eugenia involucrata occurs in multiple biomes; it is the most widespread species within core Phyllocalyx and emerges within Group 2. The most recent lineage in this group is composed of Eugenia sp. nov. and E. luschnathiana, species from the North Atlantic forest. The North Atlantic forest block (Thomas and Barbosa, 2008) is a relatively narrow strip of forest bounded to the west by the Caatinga domain. This result suggests that the two blocks (Thomas and Barbosa, 2008) of Atlantic forest form a ‘monophyletic’ group, unusual when previous biogeographic studies indicate the Atlantic forest to be a composite biogeographic area with significantly different species composition in the southern and northern blocks, suggesting relative evolutionarily independence of these areas (e.g. Cracraft and Prum, 1988; Costa, 2003; Perret et al., 2006; Nihei and Carvalho, 2007; Santos et al., 2007).
Eugenia glandulosa is the oldest lineage in Group 1 and is endemic to the Cerrado, suggesting the existence of the Cerrado biome approx. 16·7 Mya, or some precursor. This supports the findings of Ratter et al. (1997) who provide older dates for the origin of the Cerrado than the most recent hypotheses (e.g. Simon et al., 2009). The oldest estimates of the Cerrado suggest the biome to be a precursor to the adjacent Amazonian and Atlantic rain forests, during the Holocene (Ledru, 2002). Simon et al. (2009) found fire-adapted Cerrado lineages of Mimosa to have evolved independently at least 11 times and more recently than core Phyllocalyx (from 9·8 Mya to 0·4 Mya; late Miocene to Pliocene vs. 16·7 Mya to 9·2 Mya; mid-Miocene to late Miocene). Simon et al. (2009) also suggest that these woody legume lineages coincide with a hypothesized expansion of C4 grass-dominated savannah biomes that become ecologically dominant 4–8 Mya (Jaccobs et al., 1999). This is in contrast to Christin et al. (2008) who demonstrate that C4 grasses originated and began to diversify 32–25 Mya, coinciding more with the diversification time of core Phyllocalyx. If these hypotheses (an Atlantic forest origin for core Phyllocalyx and a biome shift between Atlantic forest and Cerrado) were correct, it appears that core Phyllocalyx arrived in the Cerrado via two routes. This has been reported in other lineages [e.g. Ruellia in Tripp (2007) and Stirax in Fritsch (2001)], but core Phyllocalyx is much older than these groups. The confidence intervals of the dating analysis are large, and, as a result, certainty in dates is reduced. Within the confidence intervals, difference in age of Groups 1 and 2 may be insignificant (Fig. 1).
The Carnaval–Moritz (2008) hypothesis of biological refugia delimits the north-eastern Brazilian area limited by the rivers Rio Doce and São Francisco during the LGM (Pleistocene). Areas of elevated species diversity of Myrcia sect. Aulomyrcia (O.Berg) Griseb. (Myrtaceae) in the Atlantic forests were shown (Staggemeier et al., 2015) to correspond to this hypothesis. However, the model here suggested indicates that south-east coastal regions of Brazil could comprise environmentally stable regions (Fig. 4) that acted as refuges for Eugenia sect. Phyllocalyx during the Quaternary period. The south-east coastal region from the southern state of São Paulo (SP) and northern state of Paraná (PR) comprise estuarine and lagoon complexes, with various micro-regions and micro-climates most probably influenced by the uplift of Serra do Mar during the Oligocene (approx. 70 Mya; Almeida and Carneiro, 1998). Palynological studies (Marchant et al., 2002; Behling and Negrelli, 2006) also indicate the stability of south-east coastal vegetation, with Myrtaceae pollen widely representative of the stratigraphy. Recent studies using vertebrates as model groups show historically unstable areas (southern state of SP and northern state of PR) with climatic stability during the Quaternary (Carnaval et al., 2014; Leite et al., 2016). These stable forest refuges would have been responsible for maintaining the pattern of diversity and endemism observed today. In fact, eight out of 14 species of core Phyllocalyx occur in the southern SP/northern PR region; this emphasizes that these species should provide appropriate proxy records of forest changes through time.
Concluding remarks
Results presented here advance understanding of species relationships in Eugenia s.l., but much progress must be made before the group is completely understood. Amazonian species remain poorly studied, as do relationships between Amazonian and Atlantic forest species.
Conclusions discussed here must be considered in light of high confidence intervals returned; however, the presented results confirm the paraphyletic nature of Eugenia sect. Phyllocayx sensu Berg and allowed a new infrageneric section to be proposed. Mazine et al. (2014) considered the emergence of Eugenia wentii close to E. sect. Umbellatae a likely error, but results presented here confirm this placement and demonstrate the importance of species sampling and molecular marker density in the phylogeny of large clades. Species niche modelling identified refuges within areas classically considered unstable during both the Quartenary and Pleistocene–Holocene–Present periods. As well as presenting further evidence for species diversity related to the north-eastern Brazilian refuge (Carnaval and Moritz, 2008; Staggemeier et al., 2015), this study also suggests that high species diversity in southern SP and northern PR may be linked to an area of environmental stability. This is supported by the analysis of Murray Smith et al. (2009) who reported centres of Myrtaceae species diversity in both the northern and southern blocks of the Atlantic. Evidence for refuges contributes to explanations of extreme plant species richness and endemism in the south and north-eastern coasts of Brazil and shows that plant diversification is not a simple random model with a constant rate of speciation and extinction (Donoghue and Sanderson, 2015). It is clear that multiple traits contribute to the success of both Eugenia and angiosperms in the Atlantic forest; these include morphological innovations, environmental changes and geographic movement, all traceable along the branches of a phylogenetic tree (Donughue and Edwards, 2014).
Biodiversity data typically contribute to conservation planning by allowing compilation of species lists and distributions that ultimately define an area as a diversity ‘hotspot’ (Myers et al., 2000), or otherwise labelled special (e.g. World Wildlife Fund ecoregions, 2015). Recently, evolutionary processes have also been implicated in conservation strategy, particularly in light of global climate change (Mace et al., 2003). Increasing understanding of Myrtaceae evolution in Neotropical forests, as well as biodiversity data produced (Lucas and Bünger, 2015), provides both streams of diversity data. The fact that Eugenia is the most species-rich genus of Brazilian angiosperms (Forzza, 2015) and one of the most ecologically important tree species taxa in the Atlantic forests (Oliveira-Filho and Fontes, 2000; Murray-Smith et al., 2009) compounds the significance of this information. For these reasons and in summary, this study provides a rich source of information for in situ conservation strategies under current and future scenarios of environmental change and anthropic pressure.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: maximum likelihood and Bayesian tree. Figure S2: plate of Eugenia sect. Phyllocalyx features and habitats. Figure S3: geographic distribution of Eugenia sect. Phyllocalyx. Table S1: ancestral area probabilities per numbered node of Fig. 2.
ACKNOWLEDGEMENTS
We are grateful to Laszlo Csiba, Dion Devey, Jim Clarkson, Laura Martinez, Rhina Duque, Jimmy, Ernesto and Becky for help in the lab, and Petra Broddle for help in the herbarium at Kew. Luis Palazzesi, Matheus Santos, Tânia Moura and Vanessa Staggemeier are thanked for discussion of and help with dating phylogenies; Marcos Sobral is gratefully acknowledged for all discussions throughout the period of the PhD of the first author. Herbarium curators that permitted examination of Eugenia specimens are thanked, as is Danilo Neves for discussion of modelling analyses, as well as all colleagues of the Laboratório de Sistemática from UFMG. This work was supported by CAPES and CNPq [014/2012] and a sandwich fellowship from MOB to develop part of this research at RBG Kew, London, UK [14045/2013-03]. M.L.B. thanks CNPq for a Post-doctoral scholarship (151002/2014-2).
Appendix 1. Vouchers of DNA samples used for phylogenetic analysis, with GenBank numbers
| Species | Collector | Voucher | ITS | psbA-trnH | rpl16 | trnL-rpl32 | trnQ-rps16* |
|---|---|---|---|---|---|---|---|
| Acca sellowiana (O. Berg) Burret | Chase 10349 | K | 10349 | 10349 | 10349 | 10349 | 10349 |
| Algrizea macrochlamys (DC.) Proença & Nic Lugh. | Giulietti 1648 | K | 16833 | 16833 | 16833 | 16833 | 16833 |
| Blepharocalyx salicifolius (Kunth) O. Berg | Lucas 78 | K | KX789287 | KX789292 | KX789317 | KX789346 | KX910667 |
| Campomanesia ilhoensis Mattos | Ibrahim 122 | K | 34650 | 34650 | 34650 | 34650 | 34650 |
| Decaspermum humile (Sweet ex G. Don) A.J. Scott | Belsham M82 | OTA | AM234128 | 19052 | AM489824 | 19052 | 19052 |
| Eugenia brevistyla D. Legrand | Mazine 993 | ESA, K | KJ187614 | 20683 | KJ469663 | 20683 | 20683 |
| Eugenia bunchosiifolia Nied. | Faria 2513 | BHCB, UB | KX789268 | KX789298 | KX789323 | KX789352 | KX910673 |
| Eugenia involucrata DC. | Bünger 551 | BHCB | KX789281 | KX789294 | KX789319 | KX789348 | KX910669 |
| Eugenia sp1 | Holst 9516 | K | 36243 | 36243 | 36243 | 36243 | 36243 |
| Eugenia crassa Sobral | Giacomin 1860 | BHCB | KX789269 | KX789296 | KX789321 | KX789350 | KX910671 |
| Eugenia expansa Spring ex Mart. | Bünger 634 | BHCB, K | KX789279 | KX789297 | KX789322 | KX789351 | KX910672 |
| Eugenia dysenterica DC. | Mazine 466 | ESA, K | KJ187620 | 20844 | KJ469669 | 20844 | 20844 |
| Eugenia sp. nov. | Moraes 600 | BHCB | KX789284 | KX789295 | KX789320 | KX789349 | KX910670 |
| Eugenia glandulosa Cambess. | Faria 37 | BHCB, UB | KX789277 | KX789299 | KX789324 | KX789353 | KX910674 |
| Eugenia goiapabana Sobral & Mazine | Bünger s/n | BHCB | KX789270 | KX789300 | KX789325 | KX789354 | KX910675 |
| Eugenia longipetiolata (O. Berg) Mattos | Bünger 626 | BHCB, K | KX789285 | KX789301 | KX789326 | KX789355 | KX910676 |
| Eugenia luschnathiana (O. Berg) Klotzsch ex B.D. Jacks. | Faria 3140 | BHCB, UB | KX789272 | KX789302 | KX789327 | KX789356 | KX910677 |
| Eugenia macrobracteolata Mattos | Faria 3050 | UB | KX789283 | KX789303 | KX789328 | KX789357 | KX910678 |
| Eugenia magnibracteolata Mattos & D. Legrand | Bünger 627 | BHCB, UB | KX789271 | KX789304 | KX789329 | KX789358 | KX910679 |
| Eugenia melanogyna (D. Legrand) Sobral | Mazine 969 | ESA, K | KJ187624 | KX789305 | KJ469673 | 20694 | 20694v |
| Eugenia membranifolia Nied. | Duarte 85677 | BHCB, ESA | 20942 | KX789306 | KX789330 | KX789360 | KX910680 |
| Eugenia myrcianthes Nied. | Faria 2850 | UB | 44019 | 44019 | 44019 | 44019 | 44019 |
| Eugenia patens Poir. | Lucas 104 | ESA, K | KJ187633 | 20947 | KJ469681 | KX789361 | KX910681 |
| Eugenia pluriflora DC. | Mazine 961 | ESA, K | KJ187636 | Mazine unpubl. | KJ469684 | Fiorella | Fiorella |
| Eugenia puberula Nied. | Bünger 629 | BHCB, K | KX789282 | KX789293 | KX789318 | KX789347 | KX910668 |
| Eugenia punicifolia (Kunth) DC. | Mazine 1065 | ESA, K | KJ187638 | – | KJ469686 | KX789361 | KX910682 |
| Eugenia regia Bünger & Sobral | Bünger 578 | BHCB | KX789276 | KX789307 | KX789333 | KX789362 | KX910683 |
| Eugenia selloi (O. Berg) B.D. Jacks. | Bünger 566 | BHCB, RB | KX789278 | KX789308 | KX789334 | KX789363 | KX910684 |
| Eugenia ruschiana Bünger & Mazine | Bünger 618 | BHCB, K | KX789280 | KX789309 | KX789335 | KX789364 | KX910685 |
| Eugenia speciosa Cambess. | Bünger 585 | BHCB | KX789274 | KX789310 | KX789336 | KX789365 | KX910686 |
| Eugenia speciosa Cambess. | Mota 2477 | BHCB | KX789275 | KX789311 | KX789337 | KX789366 | KX910687 |
| Eugenia stipitata McVaugh | Holst 8872 | SEL | KJ187645 | 35648 | KJ469694 | 35648 | 35648 |
| Eugenia uniflora L. | Lucas 207 | Cultivated K | AM234088 | – | AM489828 | KX789367 | KX910688 |
| Eugenia wentii Amshoff | Holst 9421 | K | KJ187651 | 35646 | KJ469701 | KX789368 | KX910689 |
| Luma apiculata (DC.) Burret | Chase 17313 | K | KX789288 | KX789312 | KX789340 | KX789369 | 17313 |
| Myrceugenia alpigena (DC.) L.R. Landrum | Lucas 167 | K | KX789289 | KX789313 | 19066 | KX789370 | 19066 |
| Myrcia tomentosa (Aubl.) DC. | Savassi s/n (ESA 85681) | K | 20697 | 20697 | 20697 | 20697 | 20697 |
| Myrtus communis L. | Chase 10347 | K | KX789286 | KX789314 | KX789342 | KX789371 | KX910690 |
| Plinia cordifolia (D. Legrand) Sobral | Mazine 957 | ESA, K | KX789291 | KX789315 | 20679 | KX789372 | 20679 |
| Rhodamnia rubescens (Benth.) Miq. | Belsham M83 | OTA | AM234127 | † | AM489879 | † | † |
| Siphoneugena densiflora O. Berg | Mazine 1050 | ESA, K | KX789290 | KX789316 | 20681 | KX789373 | 20681 |
| Ugni molinae Turcz. | J. Murillo 4213, Belsham M69 | CONC, OTA | JN660933 | JN660982 | AM489885 | JN661081 | JN661131 |
*Two internal primers for region trnQ-rps16 were used; see Table 1.
†Missing data.
Abbreviations: Institution/State: BHCB, Universidade Federal de Minas Gerais, MG, Brazil; CONC, Universidad de Concepción/Concepción, Chile; ESA, Instituto de Botânica/SP, Brazil; OTA, University of Otago/Dunedin, New Zealand; SEL, Marie Selby Botanical Gardens/Florida, USA; UB, Universidade de Brasília/DF, Brazil.
LITERATURE CITED
- Almeida FFM, Carneiro CDR. 1998. Origem e evolução da Serra do Mar. Revista Brasileira de Geociências 28: 135–150. [Google Scholar]
- Berg O. 1856. Revisio Myrtacearum Americae. Linnaea 27: 1–472. [Google Scholar]
- Biffin E, Lucas EJ, Craven LA, Costa IR, Harrington MG, Crisp MD. 2010. Evolution of exceptional species richness among lineages of fleshy-fruited Myrtaceae. Annals of Botany 106: 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behling H, Negrelli RRB. 2006. Vegetation and pollen rain relationship from the tropical Atlantic rain rorest in Southern Brazil. Brazilian Archives of Biology and Technology 49: 631–642. [Google Scholar]
- Bünger MO, Mazine FF, Lucas EV, Stehmann JR. 2016. Circumscription and synopsis of Eugenia section Speciosae Bünger & Mazine (Myrtaceae). Phytokeys 61: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham RJ, Graham A. 1999. The history of Neotropical vegetation: new development and status. Annals of the Missouri Botanical Garden 86: 546–549. [Google Scholar]
- Bruneau A, Mercure M, Lewis GP, Herendeen PS. 2008. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany 86: 697–718. [Google Scholar]
- Bueno ML, Pennington RT, Dexter KG, et al. 2016. Effects of Quaternary climatic fluctuations on the distribution of Neotropical savanna tree species. Ecography 39: 10.1111/ecog.01860. [Google Scholar]
- Carnaval AC, Moritz C. 2008. Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. Journal of Biogeography 35: 1187–1201. [Google Scholar]
- Carnaval AC, Waltari E, Rodrigues MT, et al. 2014. Prediction of phylogeographic endemism in an environmentally complex biome. Proceedings of the Royal Society B: Biological Sciences 281: 20141461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracraft J, Prum RO. 1988. Patterns and processes of diversification: speciation and historical congruence in some neotropical birds. Evolution 42: 603–620. [DOI] [PubMed] [Google Scholar]
- Crane PR, Manchester SR, Dilcher DL. 1990. A preliminary survey of fossil leaves and well-preserved reproductive structures from the Sentinel Butte Formation (Paleocene) near Almont, North Dakota. Fieldiana Geology 20: 1–64. [Google Scholar]
- Christin PA, Besnard G, Samaritani E, et al. 2008. Oligocene CO2 decline promoted C-4 photosynthesis in grasses. Current Biology 18: 37–43. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Arroyo MT, Cook LG, et al. 2009. Phylogenetic biome conservatism on a global scale. Nature 458: 754–756. [DOI] [PubMed] [Google Scholar]
- Costa LP. 2003. The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. Journal of Biogeography 30: 71–86. [Google Scholar]
- Donoghue MJ, Edwards EJ. 2014. Biome shifts and niche evolution in plants. Annual Review of Ecology, Evolution, and Systematic 45: 547–572. [Google Scholar]
- Donoghue MJ, Sanderson MJ. 2015. Confluence, synnovation, and depauperons in plant diversification. New Phytologist 207: 260–274. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Drummond A, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1·7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J. 2002. Quantitative methods for modeling species habitat: comparative performance and an application to Australian plants In: Ferson S, Burgman, M, eds. Quantitative methods for conservation biology. New York: Springer-Verlag, 39–58. [Google Scholar]
- Elith J, Graham CH, Anderson RP, et al. 2006. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29: 129–151. [Google Scholar]
- Elith J, Philips SJ, Hastie T, et al. 2011. A statistical explanation of MaxEnt for ecologists. Diversity and Distributions 17: 43–57. [Google Scholar]
- ESRI. 2011. ArcGIS Desktop: Release 10. Redlands, CA: Environmental Systems Research Institute. [Google Scholar]
- Fawcett T. 2003. ROC graphs: notes and practical considerations for data mining researchers. HP Laboratories Palo Alto, HPL-2003-4.
- Fielding AH, Bell JF. 1997. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environmental Conservation 24: 38–49. [Google Scholar]
- Forest F. 2009. Calibrating the Tree of Life: fossils, molecules and evolutionary timescales. Annals of Botany 104: 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzza RC. 2015. Lista de Espécies da Flora do Brasil. Jardim Botânico do Rio de Janeiro. http://floradobrasil.jbrj.gov.br/ (last accessed 21 August 2015)
- Fritsch PW. 2001. Phylogeny and biogeography of the flowering plant genus Styrax (Styracaceae) based on chloroplast DNA restriction sites and DNA sequences of the internal transcribed spacer region. Molecular Phylogenetic Evolution 19: 387–408. [DOI] [PubMed] [Google Scholar]
- Frodin DG. 2004. History and concepts of big plant genera. Taxon 53: 753–776. [Google Scholar]
- Gent PR, Danabasoglu G, Donner LJ, et al. 2011. The Community Climate System Model Version 4. Journal of Climate 24: 4973–4991. [Google Scholar]
- Gentry AH. 1982. Neotropical floristic diversity: phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Annals of the Missouri Botanical Garden 69: 557–593. [Google Scholar]
- Govaerts R, Sobral M, Ashton P, et al. 2015. World checklist of Myrtaceae. Royal Botanic Gardens, Kew. http://apps.kew.org/wcsp/ (last accessed 5 May 2015).
- Guisan A, Zimmermann NE. 2000. Predictive habitat distribution models in ecology. Ecological Modelling 135: 147–186. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Holst B K, Landrum L, Grifo F. 2003. Myrtaceae In: Berry PE, Yatskievych K, Holst B, eds. Flora of the Venezuelan Guayana, vol. 7 Missouri Botanical Garden Press, 1–99. [Google Scholar]
- Jacobs BF, Kingston JD, Jacobs LL. 1999. The origin of grass-dominated ecosystems. Annals of the Missouri Botanical Garden 86: 590–643. [Google Scholar]
- Judd WS, Campbell CS, Kellog E., Stevens PF, Donoghue MJ. 2008. Plant systematics: a phylogenetic approach, 3rd edn. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Judziewicz EJ, Clark LG. 2007. Classification and biogeography of new world grasses: Anomochlooideae, Pharoideae, Ehrhartoideae, and Bambusoideae. Aliso 23: 303–314. [Google Scholar]
- Keppel G, Van Niel KP, Wardell-Johnson GW, et al. 2012. Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecology and Biogeography 21: 393–404. [Google Scholar]
- Kiaerskou H. 1893. Enumeratio Myrtacearum brasiliensium In: Warming E. Symbolarum ad Floram Brasiliae Centralis Cognoscendam 39: 1–200. [Google Scholar]
- Landrum LR. 1981. A monograph of the genus Myrceugenia (Myrtaceae). Flora Neotropica Monographs 29: 1–137. [Google Scholar]
- Ledru MP. 2002. Late Quaternary history and evolution of the Cerrado as revealed by palynological records In: Oliveira PS, Marquis RJ, eds. The Cerrados of Brazil. New York: Columbia University Press, 33–50. [Google Scholar]
- Leite YLR, Costa PR, Loss AC, et al. 2016. Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proceedings of the National Academy of Sciences, USA 113: 1008–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo JM, Jiménez-Valverde A, Real R. 2008. AUC: a misleading measure of the performance of distribution models. Global Ecology of Biogeography 17: 145–151. [Google Scholar]
- Lucas EJ, Bünger MO. 2015. Myrtaceae in the Atlantic forest: their role as a ‘model’ group. Biodiversity and Conservation 24: 2165–2180. [Google Scholar]
- Lucas EJ, Harris SA, Mazine FF, et al. 2007. Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales). Taxon 56: 1105–1128. [Google Scholar]
- Lucas EJ, Matsumoto K, Harris SA, Nic Lughadha EM, Benardini B, Chase MW. 2011. Phylogenetics, morphology, and evolution of large genus Myrcia s.l (Myrtaceae). International Journal of Plant Sciences 172: 915–934. [Google Scholar]
- Mace GM, Gittleman JL, Purvis A. 2003. Preserving the tree of life. Science 300: 1707–1709. [DOI] [PubMed] [Google Scholar]
- Manchester S. 1999. Biogeographical relationships of North American Tertiary floras. Annals of the Missouri Botanical Garden 86: 472–522. [Google Scholar]
- Marchant R, Almeida L, Behling H, et al. 2002. Distribution and ecology of parent taxa of pollen lodged within the Latin American Pollen Database. Review of Palaeobotany and Palynology 121: 1–75. [Google Scholar]
- Mazine FF, Souza VC, Sobral M, Forest F, Lucas E. 2014. A preliminary phylogenetic analysis of Eugenia (Myrtaceae: Myrteae), with a focus on Neotropical species. Kew Bulletin 69: 9497. [Google Scholar]
- McVaugh R. 1956. Tropical American Myrtaceae: notes on generic concepts and descriptions of previously unrecognized species. Fieldiana Botany 29: 145–228. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees In: Proceedings of the Gateway Computing Environments Workshop (GCE) 14 November 2010. New Orleans, LA, 1–8. [Google Scholar]
- Murray-Smith C, Brummitt NA, Oliveira-Filho AT, et al. 2009. Plant diversity hotspots in the Atlantic coastal forests of Brazil. Conservation Biology 23: 151–163. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Fonseca GABd, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Niedenzu F. 1893. Myrtaceae In: Prantl K, Engler A. Die natürlichen Pflanzenfamilien 3: 57–105. [Google Scholar]
- Nihei SS, Carvalho CJB. 2007. Systematics and biogeography of Polietina Schnabl & Dziedzicki (Diptera, Muscidae): Neotropical area relationships and Amazonia as a composite area. Systematic Entomology 32: 477–501. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Oliveira-Filho AT. 2015. NoeoTropTree, flora arbórea da América do Sul cisandina tropical e subtropical: µm banco de dados envolvendo biogeografia diversidade e conservação. http://www.treeatlan.icb.ufmg.br/ (last accessed April 2015).
- Oliveira-Filho AT, Fontes MAL. 2000. Patterns of floristic differentiation among Atlantic forests in SE Brazil and the influence of climate. Biotropica 32: 793–810. [Google Scholar]
- Otto-Bliesner BL, Marsha SJ, Overpeck JT, Miller GH, Hu AX. 2006. Simulating arctic climate warmth and icefield retreat in the last interglaciation.Science 311: 1751–1753. [DOI] [PubMed] [Google Scholar]
- Palazzesi L, Barreda V. 2007. Major vegetation trends in the Tertiary of Patagonia (Argentina): a qualitative paleoclimatic approach based on palynological evidence. Flora 202: 328–337. [Google Scholar]
- Panti C. 2014. Myrtaceae fossil leaves from the Río Turbio Formation (Middle Eocene), Santa Cruz Province, Argentina.Historical Biology 28: doi: 10.1080/08912963.2014.976635. [Google Scholar]
- Pearson RG, Thuiller W, Araújo MB, et al. 2006. Model-based uncertainty in species range prediction. Journal of Biogeography 33: 1704–1711. [Google Scholar]
- Pena JCC, Kamiono LHY, Rodrigues M, Mariano-Neto E, de Sequiera MF. 2014. Assessing the conservation status of species with limited available data and disjunct distribution. Biological Conservation 170: 130–136. [Google Scholar]
- Pennington RT, Lewis GP, Ratter JA. 2006. An overview of the plant diversity, biogeography and conservation of Neotropical savannas and seasonally dry forests In: Pennington RT, Lewis GP, Ratter JA, eds. Neotropical savannas and seasonally dry forests: plant diversity, biogeography and conservation. The Systematics Association Special Volume, Series 69. Boca Rato, FL: CRC Press, 1–29. [Google Scholar]
- Perret M, Chautems A, Spichiger R. 2006. Dispersal–vicariance analysis in the tribe Sinningieae (Gesneriaceae): a clue to understanding biogeographical history of the Brazilian Atlantic Forest. Annals of the Missouri Botanical Garden 93: 340–358. [Google Scholar]
- Peterson AT, Soberón J, Sánchez-Cordero V. 1999. Conservatism of ecological niches in evolutionary time. Science 285: 1265–1267. [DOI] [PubMed] [Google Scholar]
- Peterson AT, Soberón J. 2012. Species distribution modeling and ecological niche modeling: getting the concepts right. Natureza & Conservação 10: 102–107. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modelling of species geographic distributions. Ecological Modelling 190:231–259. [Google Scholar]
- Phillips SJ, Dudik M. 2008. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31: 161–175. [Google Scholar]
- Pigg KB, Stockey RA, Maxwell SL. 1993. Paleomyrtinaea, a new genus of per-mineralized myrtaceous fruits and seeds from the Eocene of British Columbia and Paleocene of North Dakota. Canadian Journal of Botany 71: 1–9. [Google Scholar]
- Poole I, Mennega AMW, Cantrill DJ. 2003. Valdivian ecosystems in the Late Cretaceous and Early Tertiary of Antarctica: further evidence from myrtaceous and eucryphiaceous fossil wood. Review of Palaeobotany and Palynology 124: 9–27. [Google Scholar]
- Prance GT. 1982. A review of the phytogeographic evidences for Pleistocene climate changes in the Neotropics. Annals of the Missouri Botanical Garden 69: 594–624. [Google Scholar]
- Proença CEB. 1990. A revision of Siphoneugena Berg. Edinburgh Journal of Botany 47: 239–271. [Google Scholar]
- Rambaut A. 2012. FigTree. http://tree.bio.ed.ac.uk/software/figtree/ (last accessed October 2013).
- Ratter JA, Ribeiro JF, Bridgewater S. 1997. The Brazilian cerrado vegetation and threats to its biodiversity. Annals of Botany 80: 223–230. [Google Scholar]
- Ree RH, Smith SA. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57: 4–14. [DOI] [PubMed] [Google Scholar]
- Ronquist F. 1997. Dispersal–vicariance analysis: a new approach to the quantification of historical biogeography. Systematic Biology 46: 195–203. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. 2012. MrBayes 3·2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton SP, Ormerod SJ, Kerby G. 2004. New paradigms for modelling species distributions? Journal of Applied Ecology, 41:193–200. [Google Scholar]
- Santos AMM, Cavalcanti DR, da Silva JMC, Tabarelli M. 2007. Biogeographical relationships in north-eastern Brazil. Journal of Biogeography 34: 437–446. [Google Scholar]
- Scheel-Ybert R. 2000. Vegetation stability in the Southeastern Brazilian coastal area from 5500 to 1400 14C yr BP deduced from charcoal analysis. Review of Palaeobotany and Palynology 110: 111–138. [DOI] [PubMed] [Google Scholar]
- Schrire BD, Lavin M, Lewis GP. 2005. Global distribution patterns of the Leguminosae: insights from recent phylogenies. Biologiske Skrifter 55: 375–422. [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 94: 275–288. [DOI] [PubMed] [Google Scholar]
- da Silva JMC, de Sousa MC, Castelletti CHM. 2004. Areas of endemism for passerine birds in the Atlantic forest, South America. Global Ecology and Biogeography 13: 85–92. [Google Scholar]
- Simon MF, Grether R, de Queiroz LP, et al. 2009. Recent assembly of the Cerrado, a neo- tropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proceedings of the National Academy of Sciences, USA 106: 20359–20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral M. 2003. A família Myrtaceae no Rio Grande do Sul. UNISINOS. São Leopoldo, RS, Brasil. [Google Scholar]
- Sobral M, Proença C, Souza M, Mazine F, Lucas E. 2014. Myrtaceae In: Lista de Espécies da Flora do Brasil. Rio de Janeiro, Brazil: Jardim Botânico do Rio de Janeiro. [Google Scholar]
- Staggemeier V, Diniz-Filho J, Forest F, Lucas EJ. 2015. Phylogenetic analysis in Myrcia sect. Aulomyrcia (O.Berg) Griseb. and inferences on plant diversity in the Atlantic Rainforest. Annals of Botany 15: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Sytsma KJ, Litt A, Zjhra ML, et al. 2004. Clades, clocks, and continents: historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. International Journal of Plant Sciences 165: S85–S105 [Google Scholar]
- Taylor KE, Stouffer RJ, Meehl GA. 2012. An overview of CMIP5 and the experiment design. Bulletin of the American Meteorological Society 93: 485–498. [Google Scholar]
- Thomas WW, Barbosa MR de V. 2008. Natural vegetation types in the Atlantic forest of northeastern Brazil. Memoirs of the New York Botanical Garden 100: 6–20. [Google Scholar]
- Thornhill AH, Mcphail M. 2012. Fossil myrtaceous pollen as evidence for the evolutionary history of Myrtaceae: a review of fossil Myrtaceidites species. Review of Palaeobotany and Palynology 176–177: 1–23. [Google Scholar]
- Thornhill AH, Popple LW, Carter RJ, Ho SY, Crisp MD. 2012. Are pollen fossils useful for calibrating relaxed molecular clock dating of phylogenies? A comparative study using Myrtaceae. Molecular Phylogenetic Evolution 63: 15–27. [DOI] [PubMed] [Google Scholar]
- Tripp EA. 2007. Evolutionary relationships within the species-rich genus Ruellia (Acanthaceae). Systematic Botany 32: 628–649. [Google Scholar]
- Troncoso A, Romero EJ. 1998. Evolución de las comunidades florísticas en el extremo sur de Sudamé´rica durante el Cenofítico [Evolution of plant communities at southern South America during the Cenozoic]. Monographs in Systematic Botany from the Missouri Botanical Garden: 149–172. [Google Scholar]
- Upchurch P. 2008. Gondwanan break-up: legacies of a lost world? Trends in Ecology and Evolution 23: 229–236. [DOI] [PubMed] [Google Scholar]
- Vicentini A. 2007. Pagamea Aubl. (Rubiaceae), from species to processes, building the bridge. PhD Dissertation, University of Missouri Saint Louis, Missouri, USA. [Google Scholar]
- Warren DL, Glor RE, Turelli M. 2010. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33: 607–611. [Google Scholar]
- Werneck FP. 2011. The diversification of eastern South American open vegetation biomes: historical biogeography and perspectives. Quaternary Science Reviews 30: 1630–1648. [Google Scholar]
- Werneck FP, Nogueria C, Colli GR, Sites JW, Jr, Costa GC. 2012. Climatic stability in the Brazilian Cerrado: implications for biogeographical connections of South American savannas, species richness and conservation in a biodiversity hotspot. Journal of Biogeography 39: 1695–1706. [Google Scholar]
- World Wildlife Fund (WWF). 2015. http://www.worldwildlife.org/biomes (last accessed 7 November 2015).
- Yu Y, Harris AJ, Blair C, He X. 2015. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution 87: 46–49. [DOI] [PubMed] [Google Scholar]