Abstract
Friedreich's ataxia (FRDA) is the most common autosomal recessive ataxia. This severe neurodegenerative disease is caused by an expansion of guanine-adenine-adenine (GAA) repeats located in the first intron of the frataxin (FXN) gene, which represses its transcription. Although transcriptional silencing is associated with heterochromatin-like changes in the vicinity of the expanded GAAs, the exact mechanism and pathways involved in transcriptional inhibition are largely unknown. As major remodeling of the epigenome is associated with somatic cell reprogramming, modulating chromatin modification pathways during the cellular transition from a somatic to a pluripotent state is likely to generate permanent changes to the epigenetic landscape. We hypothesize that the epigenetic modifications in the vicinity of the GAA repeats can be reversed by pharmacological modulation during somatic cell reprogramming. We reprogrammed FRDA fibroblasts into induced pluripotent stem cells (iPSCs) in the presence of various small molecules that target DNA methylation and histone acetylation and methylation. Treatment of FRDA iPSCs with two compounds, sodium butyrate (NaB) and Parnate, led to an increase in FXN expression and correction of repressive marks at the FXN locus, which persisted for several passages. However, prolonged culture of the epigenetically modified FRDA iPSCs led to progressive expansions of the GAA repeats and a corresponding decrease in FXN expression. Furthermore, we uncovered that differentiation of these iPSCs into neurons also results in resilencing of the FXN gene. Taken together, these results demonstrate that transcriptional repression caused by long GAA repeat tracts can be partially or transiently reversed by altering particular epigenetic modifications, thus revealing possibilities for detailed analyses of silencing mechanism and development of new therapeutic approaches for FRDA.
Keywords: : Friedreich's ataxia, expanded GAA repeats, FXN silencing, somatic cell reprogramming, induced pluripotent stem cells
Introduction
Friedreich's ataxia (FRDA) is the most common autosomal recessive ataxia with a prevalence of 1–2 in 50,000 people [1]. This severe, multisystem, progressive disease is caused by an expansion of guanine-adenine-adenine (GAA) repeats located in the first intron of the frataxin (FXN) gene [2,3]. Although the FXN coding sequence in FRDA patients remains unchanged, transcription of the FXN gene is significantly repressed as a consequence of this large GAA repeat expansion, typically ranging from 70 to >1,500 triplets [1,4–6]. Chromatin changes, resembling the epigenetic silencing landscape at repetitive elements, are a hallmark of the molecular pathogenesis of FRDA [7–11]. Current models postulate that expanded GAAs block initiation and progression of transcription by formation of noncanonical DNA or DNA-RNA hybrid structures, leading consequently to the recruitment of silencing machineries and establishing heterochromatin-like landscape at the FXN locus [12–15].
We and others have demonstrated that expanded GAAs induce epigenetic changes in the vicinity of the repeat tract [8,9,16–20]. Posttranslational histone modifications typical for heterochromatin (H3K9me3 and H3K27me3) are enriched in the sequences flanking the repeats, while active chromatin marks (acetylation of histones H3 and H4) in this region are underrepresented in FRDA samples. Hypermethylation of cytosine residues within CpG dinucleotides located upstream of the expanded GAAs has also been detected in FRDA cells [21,22]. It has been demonstrated that heterochromatinization of DNA sequences flanking the repeats can be induced by inserting long GAA tracts, even outside of their natural sequence context, into a reporter gene [8,23]. Epigenetic changes detected in these model systems are similar to those observed in the expanded GAA repeat tract of the FXN gene in FRDA cells. Reactivation of FXN expression by alleviating epigenetic silencing or by removing the intronic GAAs represents the ultimate therapeutic goal for FRDA. Although chromatin changes are a hallmark of FRDA molecular pathogenesis, data related to the deposition of silencing and erasing of activating histone marks at the FXN locus are sparse. Thus far, only histone deacetylases 1 and 3 (HDAC1 and 3), histone macroH2A, and polycomb group ring finger 2 have been implicated in FXN silencing [24,25]. Potentially, inhibition of the histone H3K9 methyltransferase G9a when accompanied by targeting DNA-RNA hybrids can also stimulate FXN expression [12].
Somatic cell reprogramming is associated with major remodeling of the epigenome [26]. Thus, modulating chromatin modification pathways during the cellular transition from a somatic to a pluripotent state is likely to generate long-term or even permanent changes to the epigenetic landscape. It has been recently demonstrated that induced pluripotent stem cells (iPSCs) generated from Fragile X syndrome (FXS) fibroblasts maintained epigenetic silencing of the FMR1 gene [27]. Similar to FRDA, FXS is a trinucleotide repeat disease caused by large (>200 repeats) expansions of cytosine guanine guanine (CGG) repeats in the 5′ UTR of the FMR1 gene [27]. The CGG expansion leads to epigenetic silencing as shown by increased CpG methylation, enrichment of histone H3K9me3, and decreased histone H3 acetylation [28,29].
Unlike the iPSCs, several lines of human FXS embryonic stem cells (ESCs) containing expanded CGGs showed transcriptionally active FMR1 accompanied by a lack of repressive chromatin marks at this locus [27,30,31]. Apart from underscoring differences between human iPSCs and ESCs, this result indicates that epigenetic changes that occur during development of the ESCs can erase silencing marks present in somatic cells and reactivate FMR1 expression despite the presence of expanded CGG repeats. Furthermore, differentiation of the FXS ESCs into the neuronal lineage led to silencing of FMR1, typical for somatic cells of FXS patients [27,31].
To circumvent the unavailability of FRDA ES cells in the attempt to reactivate FXN expression in the presence of expanded GAAs, we reprogrammed FRDA patient fibroblasts into iPSCs in the presence of various small molecule inhibitors that affect DNA methylation, histone acetylation, and histone methylation. When supplemented during the reprogramming protocol, sodium butyrate (NaB) and tranylcypromine (brand name, Parnate) treatment resulted in a significant increase of FXN expression, associated with the correction of repressive histone modifications at the FXN locus exclusively in FRDA iPSCs. Furthermore, we discovered that differentiation of the treated iPSCs into neurons resulted in resilencing of the FXN gene. Similarly, extended culturing of the FRDA iPSCs led to progressive expansions of the GAA repeat and resilencing of the FXN locus.
Taken together, these results demonstrate that transcriptional repression caused by long GAA repeat tracts can be, at least partially, reversed during somatic cell reprogramming, revealing a window for therapeutic intervention. However, secondary silencing of FXN due to somatic expansion of GAAs or differentiation is inevitable. Importantly, reactivating the expanded FXN gene using selective epigenetic modifiers will help elucidate pathways responsible for silencing of this locus and identify new targets for possible therapeutic intervention.
Materials and Methods
Tissue culture cell lines and conditions
FRDA GM04078 and control GM08399 fibroblasts were obtained from the Coriell Institute for Medical Research. The FRDA cells are homozygous for the GAA expansion in the FXN gene with alleles containing ∼340 (GAA1) and 430 (GAA2) repeats. Fibroblasts were cultured in Dulbecco's Modified Eagle's Medium (Life Technologies), supplemented with l-glutamine, 10% FBS (Hyclone), 1× of nonessential amino acids (Life Technologies), and 1× penicillin and streptomycin (Hyclone).
Reprogramming of human fibroblasts to iPSCs
Human iPSCs were attained from GM04078 fibroblasts by retroviral transduction of Oct3/4, Sox2, Klf-4, and c-Myc transcription factors as previously described [32,33], with minor modifications. Cells were treated with various compounds potentially affecting epigenetic modifications: sodium butyrate (cat. B5887; Sigma-Aldrich); histone deacetylase inhibitor IV (cat. 382170; Calbiochem); Tubastatin A trifluoroacetate salt (cat. 10559; Cayman chemical); tranylcypromine hydrochloride (Parnate) (cat. 040033; Stemgent); RG108 (cat. 040001; Stemgent); BIX01294 (cat. 040002; Stemgent); and Epigenetic Multiple Ligand (cat. 324888; Calbiochem), and dimethyl sulfoxide (DMSO) as vehicle control. The chemicals were individually added into the reprogramming media to expose GM04078 cells for 7 consecutive days, between days 7 and 14 of the reprogramming protocol. Fresh compounds were supplemented with each daily media change. Cells were cultured for ∼3 weeks until iPSC colonies were clearly visible. Twenty iPSC colonies per condition were manually picked and transferred to matrigel (hESC-qualified Matrix, cat. 354277; BD)-coated 24-well plates containing the mTeSR1 medium (cat. 05850; Stem Cell Technologies). In parallel, the remaining population of the iPSCs was used for analyses of FXN expression. Culturing of iPSCs was conducted without the use of feeder cells in mTeSR1 media according to manufacturer's recommendations. Typically two to four individual clonal lines per reprogramming condition were established. The entire reprogramming experiment with all compounds and controls was conducted twice. Data represent the average of two independent reprogramming experiments and two to four iPSC clones per each reprogramming condition.
Human iPSC characterization
The expression of pluripotency markers in the iPSCs was conducted using a previously described immunostaining protocol [32,33]. Briefly, exponentially growing iPSCs were washed with phosphate-buffered saline (PBS) and fixed for 10 min at 4°C in 4% formaldehyde (cat. BP531; Fisher Scientific). Fixation solution was removed and ice-cold methanol was added for 10 min and cells were kept at −20°C. After washing thrice with PBS at room temperature, the cells were incubated for 30 min with a blocking buffer containing 5% goat or donkey serum and 0.5% Triton X-100. Standard analyses included Oct3/4 and Sox2 and Nanog staining. In addition, live immunostaining with alkaline phosphatase (Alkaline Phosphatase Live Stain, cat. A14353; Life Technologies) and Tra-1-60 (StainAlive Tra-1-60 antibody, cat. 09-0068; Stemgent) was performed on each clone. Pluripotency analyses of all iPSC lines generated in this study were conducted using the PluriPCR™ kit (MTI-GlobalStem) to quantify expression of five genes strongly indicative of pluripotency: Oct-3/4, Nanog, DNMT3b, Dppa4, and Rex1. Determinations were conducted using 7500 Fast or StepOne Plus Real-Time PCR Systems (Applied Biosystems) according to manufacturer's recommendations. Results were evaluated using the MTI-GlobalStem-provided Excel template (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). Retroviral transgene expression in the established iPSC lines was analyzed exactly as described in Ref. [34]. Briefly, the expression levels of the OSKM transcription factors were determined using quantitative reverse transcription–polymerase chain reaction (qRT-PCR). Transgene-specific expression of OSKM mRNA in fibroblasts 7 days posttransduction was used as a reference. The qRT-PCRs were performed using primers described in Ref. [35] and are listed in Supplementary Table S1.
RNA isolation and qRT-PCR analyses
Total RNA was isolated using the RNeasy Mini kit (cat. 74104; Qiagen) and subsequently processed with DNase (cat. AM1907; Ambion). All qRT-PCR analyses were performed using Power SYBR® Green RNA-to-CT™ 1-Step Kit, according to the manufacturer's protocol (cat. 4389986; Applied Biosystems). In general, a single reaction mixture included 50 ng RNA in a 10 μL total volume. All reactions were conducted in triplicate alongside “No RT” control reactions. A typical qRT-PCR protocol included a reverse transcription step at 48°C for 30 min followed by a 10-min incubation at 95°C and 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 20 s, and extension at 60°C for 1 min. Reactions were run on a 7500 Fast or StepOnePlus Real-Time PCR System (Applied Biosystems). The mRNA expression of each target was normalized to the levels of GAPDH, β-actin, or L19 transcripts. Relative expression levels were calculated by 2−ΔΔCt method. The PCR primers for FXN mRNA as well as retroviral transgene expression analyses are indicated in Supplementary Table S1.
Amplification of the GAA repeat region
To determine the number of GAA repeats in FRDA fibroblasts and iPSCs, genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (cat. K182001; Life Technologies). PCR analyses were performed using primers: FXN_short, amplifying 498 bp of the GAA repeat flanking sequences. Reactions were performed as described in Refs. [34,36] using the FailSafe PCR System and mix D (cat. FS99100; Epicentre). The amplification products were resolved on 1% agarose gels. The size of PCR product was determined using GelAnalyzer (GelAnalyzer.com) and the number of GAAs was calculated using the following formula: (base pairs of PCR product −498)/3.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the EZ ChIP™ kit protocol (Upstate Biotechnology, Inc./EMD Millipore) as we described in Ref. [9]. Typically 3–5 × 106 cells were used per ChIP reaction with the exception of iPSC-derived neurons, in which 1.5–2 × 106 cells were used. Briefly, proteins and DNA were cross-linked with 1% formaldehyde for 10 min at room temperature, followed by a 5-min incubation with 125 mM glycine at room temperature. Whole lysates were prepared using a cell lysis buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, and 1% SDS) and sonicated to obtain 100–300 bp DNA fragments using a Bioruptor Sonicator (Diagenode). The fragmented chromatin was diluted 10 times with a dilution buffer (16.7 mM Tris-HCl at pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, and 0.01% SDS) and immunoprecipitated overnight with 5 μg of antibodies recognizing histones and specific histone posttranslational modifications. The immunoprecipitates were immobilized using protein A agarose beads, washed with buffers containing low salt, high salt, and LiCl, and chromatin was eluted from the beads with an elution buffer (100 mM NaHCO3 and 1% SDS). The formaldehyde cross-links were reversed by adding NaCl to a final concentration of 200 mM and incubating the reactions at 65°C for at least 5 h. The reactions were then treated with 1 μL RNase A (10 mg/mL) at 37°C for 30 min followed by incubation at 42°C for 1 h in a Tris-EDTA buffer supplemented with 1 μL Proteinase K (10 mg/mL). Finally, the DNA was purified by phenol/chloroform extraction followed by ethanol precipitation. The quantitative polymerase chain reactions (qPCRs) were conducted using the Power SYBR Green-CT Kit using previously tested primer pairs amplifying three regions of the FXN gene: Promoter, Upstream, and Downstream of the GAA repeats [9] (Supplementary Table S1). The abundance of each histone modification was calculated by normalizing the quantity of the immunoprecipitated sample to the quantity of total histone H3 and then expressed as relative to the Input sample. The antibodies used in ChIP experiments were as follows: anti-rabbit IgG as a negative control (Cell Signaling), anti-total H3 (Cell Signaling), anti-H3K9ac (Cell Signaling), anti-H3K14ac (Active Motif), anti-H3K4me2 (Active Motif), anti-H3K4me3 (Active Motif), and anti-H3K9me3 (Active Motif).
Statistical analyses
Statistical analyses were conducted using GraphPad Prism 6. Statistical significance was determined by performing Student's t-test and P < 0.05 was considered significant.
Results
Expansion of GAA repeats silences expression of the FXN gene in iPSCs
It has been previously demonstrated that long GAA repeats (∼200 triplets or more) present in intron 1 of the human FXN gene, or artificially inserted into intron 1 of mouse Fxn or into a reporter construct, silenced their expression at the transcriptional level [8,23,37]. Reduced expression is associated with specific chromatin changes in the vicinity of the repeat sequence, predominantly enrichment in heterochromatin-specific histone marks (e.g., H3K9me3) and reduced representation of histones bearing marks of active chromatin (e.g., H3K9ac or H3K14ac) (Fig. 1A) [8,10,16,32]. This phenomenon has been observed in all cell lines and somatic tissues derived from FRDA patients [18]. In addition, we and others demonstrated that reprogramming FRDA primary fibroblasts into iPSCs using standard conditions of retrovirus-mediated delivery of the reprogramming factors Oct4, Sox2, Klf4, and c-Myc (OSKM) [38,39] does not affect the epigenetic status or transcriptional repression of the FXN locus containing expanded GAA repeats [32,40].
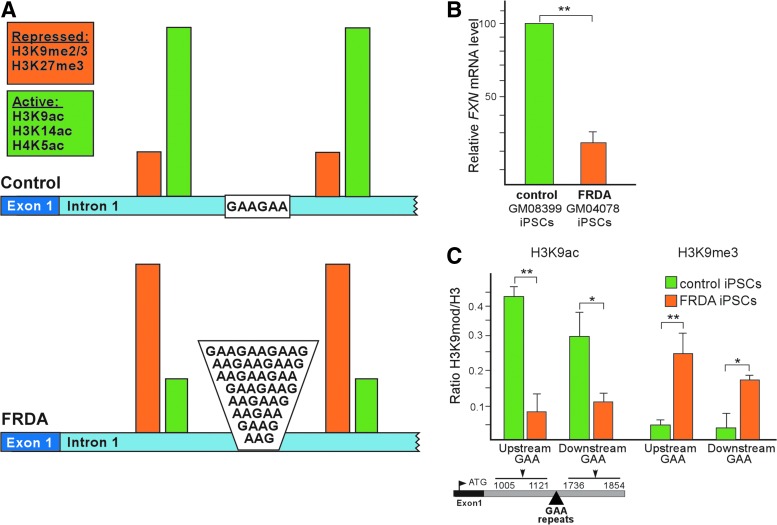
FIG. 1.
Transcriptional repression of the FXN gene in FRDA iPSCs. (A) Schematic representation of the epigenetic changes observed in FRDA cells containing expanded GAA repeats in intron 1 of the FXN gene compared to controls with short GAAs. (B) Quantitative RT-PCR results comparing FXN mRNA expression in control iPSCs derived from GM08399 fibroblasts and FRDA iPSCs derived from GM04078 fibroblasts. (C) ChIP analyses of histone H3K9ac and H3K9me3 modifications in the control and FRDA iPSCs. Data are expressed as the mean ± SD. For all histone modification analyses shown in Figs. 3–5, ChIP data are presented relative to input DNA and normalized to the total histone H3 in each region. A diagram of intron 1 of the FXN gene is presented below along with the exact locations of primers used for ChIP qPCR analyses. Due to differences in the reported transcription start sites, the positions indicated on the diagram are relative to the adenine residue of the translational start codon (position 1). Asterisks (*) designate P < 0.05, **P < 0.01. ChIP, chromatin immunoprecipitation; FRDA, Friedreich's ataxia; FXN, frataxin; GAA, guanine-adenine-adenine; iPSCs, induced pluripotent stem cells; RT-PCR, reverse transcription–polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
To verify these results and set a foundation for our subsequent studies, we reprogrammed the FRDA fibroblast line GM04078 along with control GM08399 fibroblasts (both cell lines obtained from Coriell Cell Repositories) and determined expression of FXN. The FRDA fibroblast cell line contains two expanded GAA tracts of ∼340 and ∼430 repeats and were derived from a 30-year-old male donor. As expected, a significant reduction (∼75%) in the level of FXN mRNA in FRDA iPSCs was detected by qRT-PCR when compared to the iPSCs derived from control cells lacking expanded GAAs (Fig. 1B). In addition, ChIP using antibodies specific for histone H3K9me3 and histone H3K9ac demonstrated overrepresentation of heterochromatic histone modifications and underrepresentation of histones, signifying active chromatin both upstream and downstream of the expanded GAAs in FRDA iPSCs relative to control cells (Fig. 1C). These results confirm that reprogramming of FRDA fibroblasts to a pluripotent state under standard conditions does not reactivate the silenced, mutated FXN locus.
Modulation of the epigenetic environment during reprogramming of FRDA fibroblasts
Reprogramming to pluripotency is associated with massive changes of the epigenetic environment from a somatic to an embryonic-like state [26]. We demonstrated above that the mutated FXN gene, harboring large GAA expansions, escapes reactivation and remains silenced in the pluripotent state. On the other hand, specific compounds affecting predominantly acetylation of histones have been recurrently shown to partially restore expression of the mutated FXN in patient cells [10,16,41,42]. In fact, the promising effects of several HDAC inhibitors (HDACi) warranted initial clinical trials of the most potent compounds [43]. Therefore, we hypothesized that epigenetic changes associated with the GAA repeats can be reversed by modulating the chromatin environment during somatic cell reprogramming. We conducted reprogramming of FRDA fibroblasts using retrovirus transduction to deliver the OSKM factors in the presence of several small molecule inhibitors that selectively target histone acetylation, histone methylation, as well as DNA cytosine methylation. We initially selected 12 compounds known to interfere with specific epigenetic gene silencing pathways. Cells undergoing reprogramming were exposed to the compounds between day 7 and 14 of the reprogramming scheme (Fig. 2A, full details in Materials and Methods section), and we were able to obtain and establish individual iPSC clones in the case of 8 different reprogramming conditions, including DMSO-treated control (Table 1). High expression of pluripotency markers was detected for each iPSC line generated in the presence of the different compounds (Supplementary Fig. S1). Silencing of the retroviral transgenes was also confirmed in resulting iPSCs as described in Ref. [34].
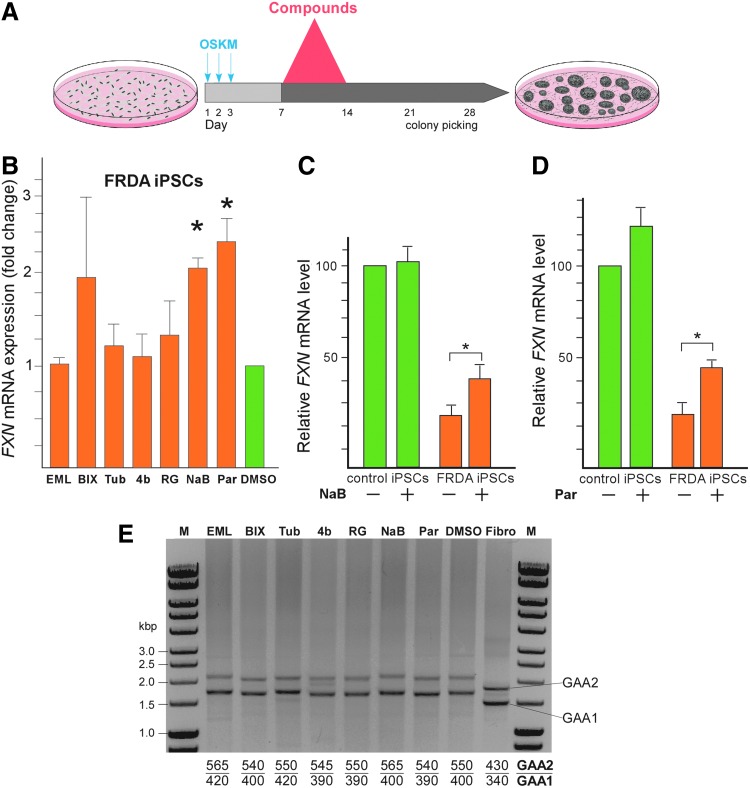
FIG. 2.
Reprogramming of FRDA fibroblasts in the presence of epigenetic inhibitors partially reactivates expression of the FXN gene. (A) Schematic of the reprogramming regimen. Fibroblasts were transduced in triplicate using retroviruses encoding the Yamanaka reprogramming (OSKM) factors [38,39]. Treatment with different compounds that inhibit epigenetic modifying enzymes (listed in Table 1) was conducted between days 7 and 14 of the reprogramming protocol. The daily media change during this period included a fresh dose of the respective compound. Colonies were picked and transferred into 24-well plates ∼28 days after transduction. (B) Analysis of FXN mRNA expression in FRDA iPSCs reprogrammed in the presence of different compounds (abbreviations listed in Table 1). The data represent the average of two independent reprogramming experiments with two to four individual iPSC clones per experiment. Asterisks (*) indicate significant difference relative to DMSO controls, with P < 0.05. (C) Real-time qRT-PCR demonstrates an increase in FXN mRNA levels in FRDA cells following reprogramming of FRDA fibroblasts in the presence of NaB or (D) Parnate. (E) Determination of the GAA repeat size by PCR amplification after reprogramming of FRDA fibroblasts in the presence of various small molecule inhibitors of epigenetic regulators. GAA1 indicates the shorter, while GAA2 indicates the longer of the two FXN alleles. The number of GAAs per allele was calculated using GelAnalyzer and is indicated below the gel. Note the large GAA expansions that occur during the somatic reprogramming process when compared to the initial repeat sizes in the parental fibroblast cells (Fibro lane). DMSO, dimethyl sulfoxide; NaB, sodium butyrate; OSKM, Oct3/4, Sox2, Klf4, c-myc transcription factors.
Table 1.
Miniscreen for Epigenetic Regulators of FXN Silencing During Reprogramming
| Compound (abbreviation) | Target | Process | Concentration |
|---|---|---|---|
| HDAC inhibitor 4b (4b)a | HDAC3 | Histone acetylation | 5 μM |
| Sodium butyrate (NaB) | HDAC class I | Histone acetylation | 0.5 mM |
| Tranylcypromine, Parnate (Par) | LSD1 | Histone H3K4 methylation | 4 μM |
| Tubastatin A (TubA) | HDAC6 | Histone acetylation | 5 μM |
| RG108 (RG) | DNMT | CpG methylation | 5 μM |
| BIX01294 (BIX) | G9a | Histone H3K9 methylation | 4 μM |
| Epi-ML (EML)b | Multiple | Multiple targets | 5 μM |
| DMSO | Control | — | — |
Only compounds that resulted in successful establishment of iPSC clones are shown.
HDACi 4b has been demonstrated to increase expression of the FXN gene in FRDA lymphoid cells and neurons, but not in FRDA fibroblasts or iPSCs.
Epigenetic Multiple Ligand (Epi-ML; Calbiochem) is a cell-permeable bis-arylidene compound that inhibits several mammalian histone-modifying enzymes, including SET7, PRMT1, p300/CBP, and SIRT1/2.
DMSO, dimethyl sulfoxide; EML, Epigenetic Multiple Ligand; FRDA, Friedreich's ataxia; FXN, frataxin; HDACi, histone deacetylase inhibitors; iPSCs, induced pluripotent stem cells.
Frataxin expression was determined at the earliest possible time point defined as passage 0 (p0). The p0 time point is defined when the cultured iPSCs reach confluence in 3 wells of a 12-well plate, allowing for DNA and RNA isolation from 2 wells with continuation of the culture in the remaining 3rd well. A comparative analysis of FXN expression demonstrated that two compounds, NaB and Parnate (Table 1), confer significant upregulation (∼2–3-fold) of FXN expression in FRDA iPSCs using GAPDH, β-actin, and L19 transcript levels as normalizers (Fig. 2B and Supplementary Fig. S2). The entire reprogramming experiment was independently conducted twice with two to four iPSC clones established per each reprogramming protocol. In addition, none of these compounds was able to increase FXN mRNA levels in FRDA fibroblasts before reprogramming (data not shown). It is important to note that combined treatment of somatic cells with NaB and Parnate did not result in the formation of viable iPSCs. Treatment with NaB or Parnate during reprogramming of control fibroblasts had no significant effect on FXN expression in control iPSCs (Fig. 2C, D). In conclusion, we were able to partially alleviate the transcriptional deficit in FRDA iPSCs by modulating the epigenetic environment during the reprogramming process.
GAA repeats expand during somatic reprogramming
We demonstrated previously that reprogramming of the FRDA fibroblasts induces large expansions of the GAA repeats [32]. Therefore, it was essential to evaluate the length of the GAA repeats in FRDA iPSCs reprogrammed under different conditions to determine whether the increase in FXN expression resulted from significant instability of long GAA repeats between the clones. The lengths of the expansions were determined by long GAA repeat PCR and high-resolution gel electrophoresis analyses, and the results confirmed a significant increase in the size of the GAA repeat tract during reprogramming under all conditions (Fig. 2E). The FRDA fibroblasts carry 340 (GAA1 allele) and 430 (GAA2 allele) repeats. Indeed, reprogramming stimulated large GAA expansions. The average number of repeats detected in GAA1 was 400 GAAs (∼18% increase) and in GAA2 was 550 repeats (∼13% increase) (Fig. 2E). Importantly, no contractions or smaller expansions were found in FRDA iPSCs reprogrammed in the presence of the NaB or Parnate compared to DMSO control or other compounds used in experiments (Fig. 2E). All GAA size analyses were conducted at p0, immediately after reprogramming. Taken together, these results imply that activating effects of NaB and Parnate treatment on FXN transcription is likely to be direct by changes in the chromatin environment of the FXN gene and independent of changes in the repeat length.
NaB and Parnate treatment influences chromatin modifications at the FXN locus in iPSCs
Both of the small molecules that demonstrated efficacy in elevating FXN transcription are inhibitors of enzymes that modify lysine residues on histone proteins. The short chain fatty acid NaB inhibits the HDAC activity, and treatment with this molecule leads to global increases in histone H3 and H4 acetylation [44]. Parnate is a nonselective monoamine oxidase inhibitor that irreversibly inhibits the demethylase activity of lysine-specific demethylase 1 (LSD1/KDM1A) [45,46]. LSD1/KDM1A demethylates mono- and dimethylated K4 and K9 of histone H3 [47,48].
To determine whether reprogramming in the presence of NaB or Parnate directly affected histone modifications at the FXN gene in FRDA iPSCs, we conducted ChIP analyses using histone H3K9ac-, H3K14ac-, H3K4me2-, H3K4me3-, and H3K9me3-specific antibodies in these cells alongside DMSO-treated cells as controls (Figs. 3 and 4). We determined levels of the selected histone modifications at three regions of the FXN locus frequently used to represent the epigenetic status of this gene: promoter (positions −358 to −232 relative to the A +1 of the ATG start codon), upstream of the GAAs (region 1,005 to 1,121 relative to the start codon), and downstream of the GAA repeats (positions 1,736 to 1,854 relative to the start codon). Positions downstream of the repeats are defined for an allele harboring six GAAs in the FXN locus.
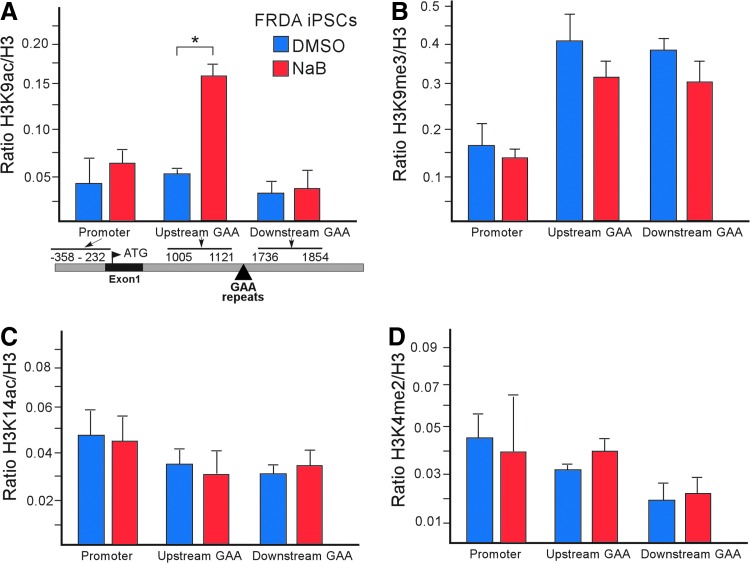
FIG. 3.
Chromatin status at the FXN gene is affected by sodium butyrate treatment during reprogramming of FRDA fibroblasts. (A) ChIP analysis reveals overrepresentation of the histone H3K9ac mark at the FXN locus upstream of the GAA repeats in FRDA iPSCs (p0) following a reprogramming regimen that included NaB treatment (red bars). The primer locations are shown in the diagram below the plot (see legend to Fig. 1). (B–D) No significant changes in histone H3K9me3, H3K14ac, or H3K4me2 are detected at the FXN locus, as determined by ChIP, in FRDA iPSCs reprogrammed in the presence of NaB (red bars) when compared to cells treated with DMSO (blue bars). A trend toward lower abundance of H3K9me3 in FRDA iPSCs reprogrammed in the presence of NaB is noted. *Indicates P < 0.05. DMSO, dimethyl sulfoxide.
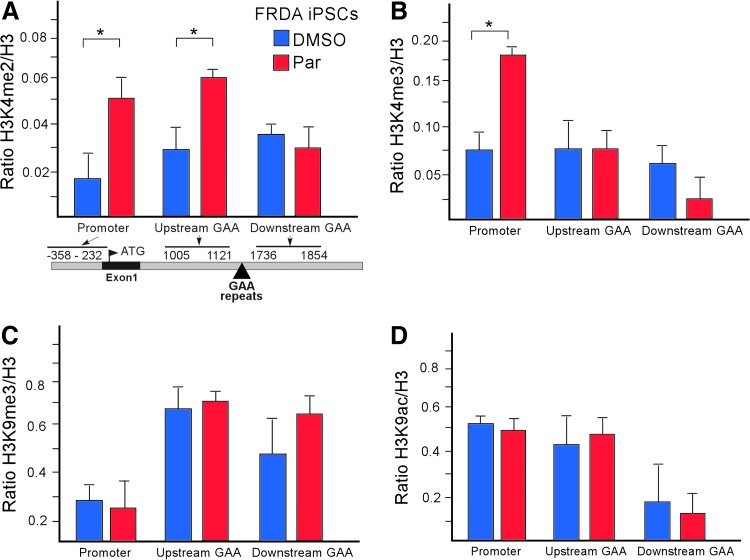
FIG. 4.
Chromatin status at the FXN gene is affected by Parnate treatment during reprogramming of FRDA fibroblasts. (A, B) ChIP analyses of FRDA iPSCs (p0) treated with Parnate during the reprogramming protocol (red bars; compare to blue bars of DMSO controls) reveals overrepresentation of the histone H3K4me2 mark in the promoter region and upstream of the GAA repeats, and enrichment in histone H3K4me3 upstream of the GAAs in the FXN locus. No changes in (C) histone H3K9me3 or (D) histone H3K9ac were detected by ChIP at the FXN locus following Parnate treatment. Locations of the primers used in ChIP qPCR are shown in the diagram below the plot (see legend to Fig. 1). *Indicates P < 0.05.
In the case of FRDA iPSCs reprogrammed in the presence of NaB, a significant increase in histone H3K9ac upstream of the GAA repeats with a simultaneous trend of decreased H3K9me3 was observed without accompanied changes in histone H3K14ac or H3K4me2 (Fig. 3A–D). As expected, in the case of the LSD1/KDM1A inhibitor, we observed enrichment of histone H3K4me2 and H3K4me3 in FRDA iPSCs treated with Parnate when compared to the DMSO controls, especially in the promoter and upstream GAA regions of FXN (Fig. 4A, B). This result is as expected as the region upstream of the GAA repeats is in immediate proximity of the regulatory sequences in the FXN gene typically enriched in H3K4me2,3 histone marks [49]. However, no changes in histone H3K9me3 were detected at the FXN locus upon Parnate treatment (Fig. 4C). As the LSD1/KDM1A demethylase is a component of several multiprotein complexes that include HDACs1/2 [50–52], the status of histone H3K9ac was determined at the FXN promoter and within the GAA repeat flanking sequences. No changes in histone H3K9ac were observed following Parnate treatment, in agreement with previous global analyses performed in P19 human embryonic carcinoma cells (Fig. 4D) [45]. Hence, treatment of the FRDA cells during somatic reprogramming with selected compounds affecting histone lysine acetylation or methylation, partially reactivates expression of the FXN gene by changing particular modifications within the chromatin landscape in the vicinity of the GAA repeats.
Differentiation to neuronal lineage restores silencing of the expanded GAA locus
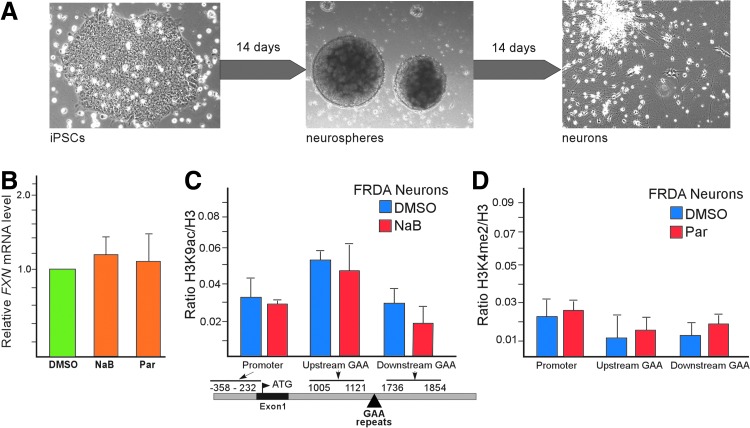
Frataxin deficiency affects predominantly terminally differentiated, nondividing cells of the nervous system, heart, and pancreas [2]. Therefore, we considered the possibility that differentiation of FRDA iPSCs may affect expression of the FXN gene. To address this issue, we conducted differentiation of FRDA iPSCs into neuronal cells as described in Ref. [53] (Fig. 5A). We differentiated FRDA iPSCs that were obtained in the presence of DMSO, NaB, or Parnate. The neuronal differentiation capacity of the resulting iPSCs was similar irrespective of the inhibitor treatment delivered during reprogramming. Quantitative RT-PCR analyses demonstrated that the initial ∼2-fold difference in FXN expression observed in NaB- and Parnate-treated iPSCs relative to DMSO controls diminished upon neuronal differentiation (Fig. 5B). Moreover, ChIP analyses using antibodies specific for histone H3K9ac (NaB and DMSO treatments) and histone H3K4me2 (Parnate and DMSO treatments) showed no significant differences in these histone marks between neuronal cells differentiated from FRDA iPSCs that were exposed to either inhibitor or to DMSO (Fig. 5C, D). These data indicate that while proliferating, pluripotent cells were capable of maintaining the chromatin state acquired at the FXN locus during somatic cell reprogramming, but differentiated and nonproliferating neuronal cells rapidly erased the differences established in the pluripotent state.
FIG. 5.
Differentiation of NaB- and Parnate-treated FRDA iPSCs induces resilencing of the FXN gene. (A) Brightfield microscopy images depicting the morphological changes that occur during the two-step differentiation protocol by intermediate neurosphere neuronal progenitor cultures to neuronal cells as described in Ref. [53]. (B) Real-time qRT-PCR analysis of FXN mRNA expression in neuronal cells derived from FRDA iPSCs obtained in the presence of DMSO, NaB, and Parnate. (C) ChIP analysis of histone H3K9ac in neurons derived from FRDA iPSCs obtained in the presence of DMSO (blue bars) and NaB (red bars). Regions of the FXN gene are defined in Fig 3. (D) ChIP analysis of histone H3K4me2 in neurons derived from FRDA iPSCs obtained in the presence of DMSO (blue bars) and Parnate (red bars). Regions of the FXN gene are defined in Fig. 3.
Progressive GAA repeat expansions reduce expression of the FXN gene in FRDA iPSCs
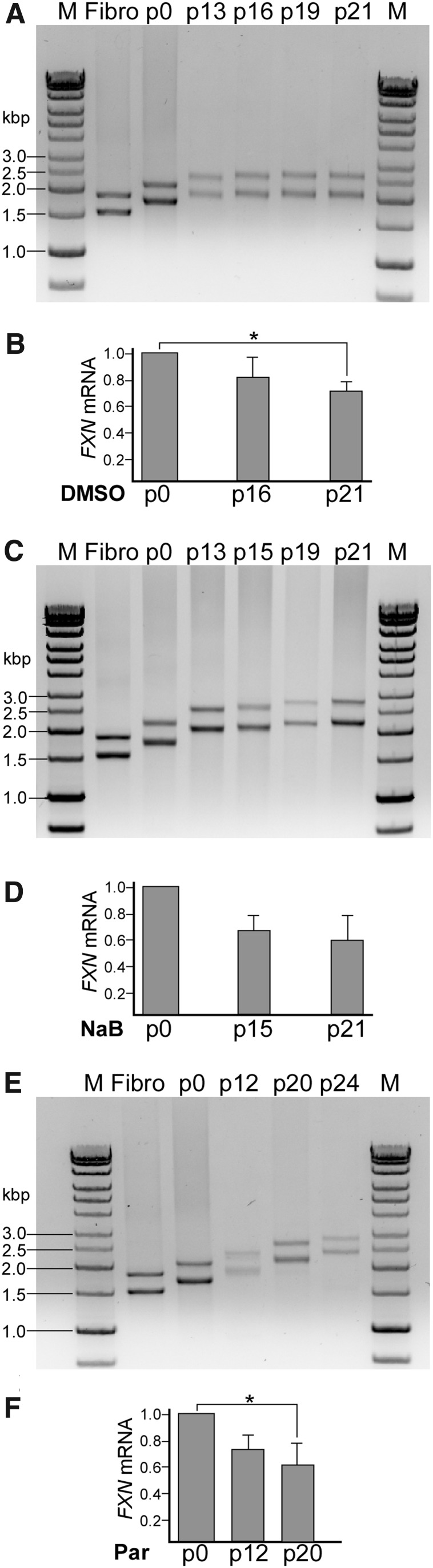
Although the extent of reactivation of FXN expression in FRDA iPSCs reprogrammed in the presence of NaB or Parnate is slightly lower than the improvement of FXN expression in lymphocytes or neurons treated with specific HDACi, the effect in FRDA iPSCs lasts significantly longer [41,54]. Increased FXN expression in FRDA iPSCs could be detected even after several passages (between iPSC colony picking and establishment of p0 cells; Fig. 2A), as opposed to a few days in the case of HDACi-treated somatic cells [54]. However, a systematic decrease in FXN expression was noticed with increasing passage number in both NaB- and Parnate-treated FRDA iPSCs (Fig. 6B, D, F). In contrast to FRDA iPSCs, in control iPSCs harboring short GAAs, no changes in FXN mRNA were observed in the span of 26 passages (Supplementary Fig. S3).
FIG. 6.
Progressive expansion of the GAA repeats in FRDA iPSCs correlates with decreased expression of FXN mRNA. (A, C, E) Analyses of the GAA repeat size in FRDA iPSCs subcultured for up to 24 passages (p). Genomic DNA from the iPSC cultures was isolated at the indicated passage numbers and the repeat length was determined using long GAA PCR as described in Materials and Methods. (B, D, E) In parallel, at the indicated passages, total RNA was extracted to measure FXN mRNA levels. Determination of GAA repeat expansion and FXN mRNA expression in FRDA iPSCs obtained in the presence of DMSO (A and B, respectively), NaB (C, D, respectively), and Parnate (E, F, respectively). *Indicates P < 0.05.
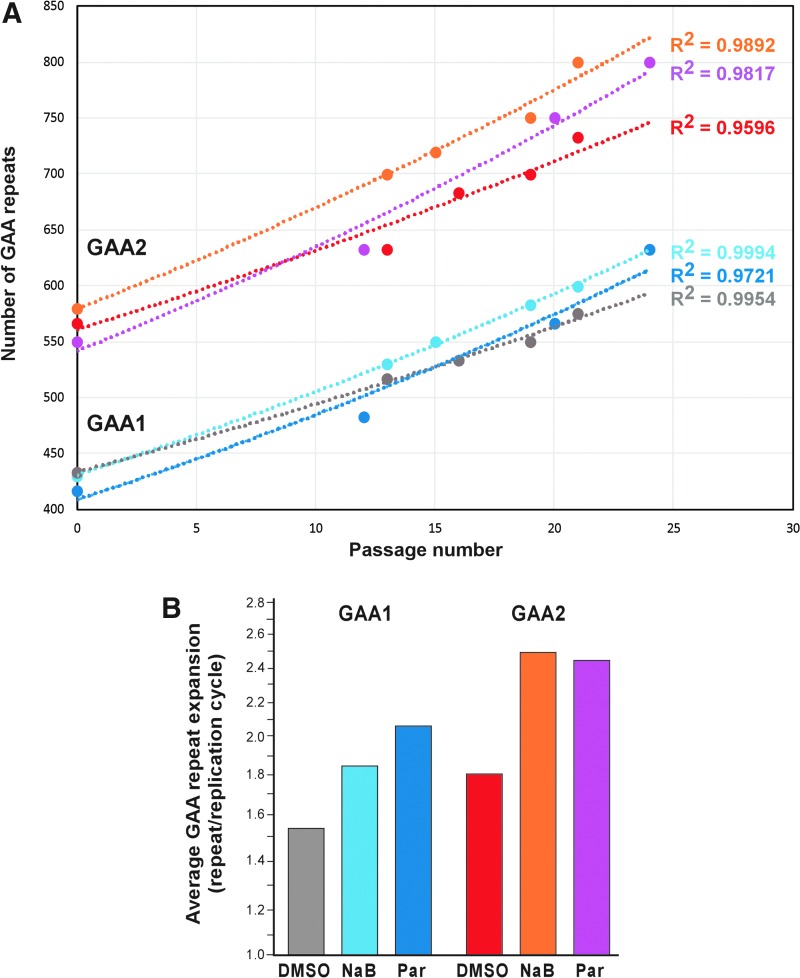
As we have observed progressive GAA expansions in FRDA iPSCs [32], we conducted analyses of GAA repeat length in FRDA iPSCs generated under different conditions (DMSO, NaB, and Parnate; Fig. 6A, C, E). The PCR analyses demonstrated a significant increase in the number of repeats with the shorter tract expanding by up to ∼170 triplets (510 bp) over 20 passages, and the longer GAA2 allele expanding by up to ∼250 repeats (750 bp) (Fig. 7A). On average, an extension by 8 to 10 GAA repeats (for GAA1 and GAA2, respectively) was detected per single passage of the iPSCs. Considering that human iPSCs divide every ∼29 h [55] and the cells were passaged in 5-day intervals, we calculated the average GAA repeat expansion rate per replication cycle (Fig. 7B). The expansion rates varied from 1.6 to 2.5 GAA repeats per replication cycle, which is very similar to the rate of 1.7–2.3 repeats/replication cycle reported earlier [56].
FIG. 7.
Progressive expansion of the GAA repeat tract during FRDA iPSC culture. (A) The number of GAA trinucleotide repeats in GAA1 and GAA2 alleles was determined using GelAnalyzer (Fig. 6) and plotted as a function of the passage number. The R2 value indicates correlation between these parameters. The gray and red plots represent progression of expansion in FRDA iPSCs obtained in the presence of DMSO (GAA1 and GAA2, respectively); light blue and orange plots designate FRDA iPSCs obtained in the presence of NaB; and dark blue and purple plots represent FRDA iPSCs obtained in the presence of Parnate. (B) Average GAA repeat expansion size per replication cycle. Designation of colors as indicated above.
Analogous to prior studies, we observed that the allele harboring longer GAAs (GAA2) exhibited higher expansion rates than GAA1 (Fig. 7B). Also, consistent with our previous report, the short GAA repeat tracts in control iPSCs remain stable during extended culture ([32] and data not shown). Thus, our data suggest that a dramatic increase in the number of GAA repeats is likely to be responsible for the downregulation of FXN expression observed in FRDA iPSCs, especially that a strong correlation exists between the size of GAA1 and FXN expression in patient cells, tissues, and all model systems studied [5,36].
Discussion
Although chromatin changes and transcriptional silencing are hallmarks of the molecular pathogenesis of FRDA, little is known about the mechanism and pathways involved in heterochromatinization of the FXN locus. We demonstrated herein that, using selected compounds that inhibit epigenetic modifiers, we were able to partially reactivate expression of the mutated FXN gene during somatic cell reprogramming. Increased FXN mRNA levels were associated with increased histone acetylation and increased histone H3K4 methylation at the FXN locus, both of which are linked to transcriptional activation. The increase in FXN transcript levels persisted for several passages of the pluripotent cells without subsequent inhibitor treatment. However, over time, progressive expansions of the GAAs occurred, which nearly doubled the initial number of the repeats observed in the parental fibroblast line, and concomitantly, FXN transcript levels declined (Fig. 6).
The increase in GAA repeat length during FRDA iPSC culture is very consistent regardless of the reprogramming conditions (Fig. 6). Moreover, an average expansion of 1.6–2.5 GAAs per replication cycle together with bias toward greater expansion rate in the longer of the two GAA tracts (GAA2), matches very well with results of previous studies [32,56]. Remarkably, the increase of the repeat size correlates extremely well with passage number (R2 = 0.95–0.99; Fig. 7A). Taken together, these results implicate GAA repeat expansion as a primary driver of reduced FXN transcription and suggest that recurrent dosing of epigenetic inhibitors is likely necessary to maintain chromatin in a permissive transcriptional state at the FXN locus in FRDA cells. Progressive expansions of the GAA repeats, if persistent throughout life in some tissues or organs, may have a deleterious and clinically relevant role by aggravating FRDA symptoms.
Interestingly, we observed an increase in the expansion rate in the iPSCs reprogrammed in the presence of NaB and Parnate, when compared to DMSO controls. One hypothesis to be tested is a potential effect of collision between transcription and replication machineries in the vicinity or within the GAA tract. Our recent studies [57] demonstrated that the replication program in FRDA iPSCs differs from control iPSCs. The majority of replication forks encountering the expanded GAA region progress in the direction opposite to the movement of transcription machinery. It is tempting to speculate that the increase of transcription observed in iPSCs reprogrammed in the presence of NaB and Parnate leads to more collisions with replication forks resulting in a higher rate of repeat expansions. However, further studies designed to precisely manipulate replication and transcription interplay at the FXN locus will be necessary to determine whether head-on collisions between RNA and DNA synthesis machineries indeed occur and, if so, whether they influence the GAA tract expansion rate. Comprehensive analyses in FRDA animal models will also be necessary to assess whether long-term reactivation of the FXN gene affects stability of the expanded GAA repeats.
Specific HDACi predominantly targeting class I and class III have been developed over the past decade as lead compounds to reverse transcriptional silencing of the FXN gene [16,25,43,58]. However, some of these compounds demonstrate selectivity by increasing FXN expression in certain cell types without affecting FXN transcription in other lineages [15]. Similarly, in our study, compounds capable of reactivating FXN expression during somatic cell reprogramming of FRDA fibroblasts did not have an effect on FXN expression in differentiated FRDA fibroblasts. Moreover, HDACi 4b was discovered as an activator of FXN expression in primary FRDA lymphocyte cells [16], but did not increase FXN expression when administered during reprogramming or in fibroblast cells (Fig. 2; [16]).
Given that chromatin modifications at an individual gene locus vary in a cell- and/or tissue-specific manner, and are perhaps influenced by cell cycle progression, certain compounds might be more or less effective in correcting deficient FXN transcription in FRDA cell models of different origins. Therefore, detailed cell-specific analyses of potential drug candidates need to be conducted in multiple cell types and animal models to avoid dismissal of potentially efficacious candidates based on limited data. More importantly, it is likely that a specific cocktail of FXN activators, rather than single compound, will be necessary to ensure appropriate stimulation of FXN expression across different tissues and organs.
Results of studies on CGG expansions in FXS ESCs and iPSCs show that expanded repeats are not inherently poised for transcriptional silencing and heterochromatin formation [27,31]. Epigenetic changes during the development of ESCs can erase silencing marks present in somatic cells and reactivate FMR1 expression despite the presence of the expanded CGG repeats, indicating that mechanisms that prevent a transcriptional block, such as R-loop formation, must exist in these cells [27,31]. Furthermore, differentiation of FXS ESCs into the embryoid bodies leads to silencing of FMR1, typical for somatic cells of FXS patients [31]. Similarly, in the case of FRDA iPSCs reprogrammed in the presence of NaB and Parnate, the FXN locus is resilenced after differentiation to neuronal cells (Fig. 5).
Human FRDA ESCs have not been isolated to date. Thus, the status of the FXN locus at the earliest stage of the development cannot be determined in human cells. The fundamental difference, from the perspective of ESC physiology, between the FMR1 and FXN genes is the location of the former on the X chromosome. Inactivation and reactivation of this chromosome during early stages of development may be an important step allowing for reactivation of the FMR1 gene, despite the presence of expanded CGG repeats. Hence, the FXN gene located on chromosome 9 may not be subjected to the same reactivation mechanisms and processes as the FMR1 gene. More recently, reactivation of the mutated FMR1 gene was demonstrated during conversion of primed to naive human iPSCs [59]. Again, one of the hallmarks of this process is the upregulation of XIST and inactivation of X-linked gene expression [60]. Therefore, even in the naive state, the expanded GAA-induced silencing of the FXN gene might not be reversed. Future studies using naive FRDA iPSCs will help to establish potential similarities between silencing and reactivation of the FMR1 and FXN genes harboring expanded repeat sequences. Obtaining FRDA cells expressing FXN, despite the presence of expanded GAAs, would help to elucidate the molecular pathways involved in pathological silencing of this locus and lead to the identification of possible novel targets for therapeutic intervention.
Overall, the ∼2-fold level of reactivation of FXN expression, we and others have achieved using inhibitors of chromatin modifiers, is moderate. Although from a therapeutic perspective, any increase in the FXN level is beneficial to the patients. Recent work on GAA-specific oligonucleotides reported a higher, three- to four-fold increase in FXN mRNA and protein levels [15]. These results indicate that sequence-specific secondary structures (R-loops formed at the GAA repeats) are the primary instigators of transcriptional silencing, and targeting the initial steps not only corrects pathological chromatin changes in FRDA cells but also improves efficacy [15]. Perhaps combining both approaches, targeting the genomic structure and enhancing epigenetic remodeling of the FXN locus, would result in a synergistic effect and increase the level of FXN reactivation, as well as prolong the therapeutic window between administration of the drugs, thus reducing potential side effects.
Supplementary Material
Acknowledgments
We would like to thank Dr. Sharon Dent for her support. These studies were supported by NIH 7R01NS081366 from NINDS (to M.N.), Friedreich's Ataxia Research Alliance and FARA Ireland (to M.N. and J.S.B.), and grant from the Polish National Science Centre 2015/19/B/NZ1/02804 (to M.N.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. (1996). Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 5254:1423–1427 [DOI] [PubMed] [Google Scholar]
- 2.Marmolino D. (2011). Friedreich's ataxia: past, present and future. Brain Res Rev 1–2:311–330 [DOI] [PubMed] [Google Scholar]
- 3.Pandolfo M. (2009). Friedreich ataxia: the clinical picture. J Neurol 256:3–8 [DOI] [PubMed] [Google Scholar]
- 4.Durr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A. and Koenig M. (1996). Clinical and genetic abnormalities in patients with Friedreich's ataxia. N Engl J Med 16:1169–1175 [DOI] [PubMed] [Google Scholar]
- 5.Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, Campanella G. and Cocozza S. (1996). The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet 3:554–560 [PMC free article] [PubMed] [Google Scholar]
- 6.Epplen C, Epplen JT, Frank G, Miterski B, Santos EJ. and Schols L. (1997). Differential stability of the (GAA)n tract in the Friedreich ataxia (STM7) gene. Hum Genet 6:834–836 [DOI] [PubMed] [Google Scholar]
- 7.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB. and Gottesfeld JM. (2006). DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich's ataxia. Proc Natl Acad Sci U S A 31:11497–11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soragni E, Herman D, Dent SY, Gottesfeld JM, Wells RD. and Napierala M. (2008). Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res 19:6056–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim E, Napierala M. and Dent SY. (2011). Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res 19:8366–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandi C, Pinto RM, Al-Mahdawi S, Ezzatizadeh V, Barnes G, Jones S, Rusche JR, Gottesfeld JM. and Pook MA. (2011). Prolonged treatment with pimelic o-aminobenzamide HDAC inhibitors ameliorates the disease phenotype of a Friedreich ataxia mouse model. Neurobiol Dis 3:496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saveliev A, Everett C, Sharpe T, Webster Z. and Festenstein R. (2003). DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 6934:909–913 [DOI] [PubMed] [Google Scholar]
- 12.Groh M, Lufino MM, Wade-Martins R. and Gromak N. (2014). R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 5:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler JS. and Napierala M. (2015). Friedreich's ataxia—a case of aberrant transcription termination? Transcription 2:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chutake YK, Lam C, Costello WN, Anderson M. and Bidichandani SI. (2014). Epigenetic promoter silencing in Friedreich ataxia is dependent on repeat length. Ann Neurol 4:522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Matsui M. and Corey DR. (2016). Activating frataxin expression by repeat-targeted nucleic acids. Nat Commun 7:10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL. and Gottesfeld JM. (2006). Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol 10:551–558 [DOI] [PubMed] [Google Scholar]
- 17.De Biase I, Chutake YK, Rindler PM. and Bidichandani SI. (2009). Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One 11:e7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D. and Pook M. (2008). The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet 5:735–746 [DOI] [PubMed] [Google Scholar]
- 19.Kumari D, Biacsi RE. and Usdin K. (2010). Repeat expansion affects both transcription initiation and elongation in Friedreich ataxia cells. J Biol Chem 6:4209–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punga T. and Buhler M. (2010). Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol Med 4:120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene E, Mahishi L, Entezam A, Kumari D. and Usdin K. (2007). Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 10:3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, Filla A, Sacchetti S, Keller S, Avvedimento VE, Chiariotti L. and Cocozza S. (2008). DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet 12:808–812 [DOI] [PubMed] [Google Scholar]
- 23.Lufino MM, Silva AM, Nemeth AH, Alegre-Abarrategui J, Russell AJ. and Wade-Martins R. (2013). A GAA repeat expansion reporter model of Friedreich's ataxia recapitulates the genomic context and allows rapid screening of therapeutic compounds. Hum Mol Genet 25:5173–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR. and Gottesfeld JM. (2009). Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chem Biol 9:980–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soragni E, Chou CJ, Rusche JR. and Gottesfeld JM. (2015). Mechanism of action of 2-aminobenzamide HDAC inhibitors in reversing gene silencing in Friedreich's ataxia. Front Neurol 6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buganim Y, Faddah DA. and Jaenisch R. (2013). Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 6:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urbach A, Bar-Nur O, Daley GQ. and Benvenisty N. (2010). Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 5:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coffee B, Zhang F, Warren ST. and Reines D. (1999). Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet 1:98–101 [DOI] [PubMed] [Google Scholar]
- 29.Evans-Galea MV, Hannan AJ, Carrodus N, Delatycki MB. and Saffery R. (2013). Epigenetic modifications in trinucleotide repeat diseases. Trends Mol Med 11:655–663 [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharyya A. and Zhao X. (2016). Human pluripotent stem cell models of Fragile X syndrome. Mol Cell Neurosci 73:43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N. and Ben-Yosef D. (2007). Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 5:568–577 [DOI] [PubMed] [Google Scholar]
- 32.Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M. and Gottesfeld JM. (2010). Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell 5:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polak U, Hirsch C, Ku S, Gottesfeld JM, Dent SYR. and Napierala M. (2012). Selecting and isolating colonies of human induced pluripotent stem cells reprogrammed from adult fibroblasts. J Vis Exp, DOI: 10.3791/e3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Polak U, Bhalla AD, Rozwadowska N, Butler JS, Lynch DR, Dent SY. and Napierala M. (2015). Excision of expanded GAA repeats alleviates the molecular phenotype of Friedreich's Ataxia. Mol Ther 6:1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, Gao L, Ye D, Zhou Y, et al. (2013). Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci U S A 8:2858–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Lu Y, Polak U, Lin K, Shen J, Farmer J, Seyer L, Bhalla AD, Rozwadowska N, et al. (2015). Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum Mol Genet 24:6932–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miranda CJ, Santos MM, Ohshima K, Smith J, Li L, Bunting M, Cossee M, Koenig M, Sequeiros J, Kaplan J. and Pandolfo M. (2002). Frataxin knockin mouse. FEBS Lett 1–3:291–297 [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Okita K, Nakagawa M. and Yamanaka S. (2007). Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 12:3081–3089 [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 5:861–872 [DOI] [PubMed] [Google Scholar]
- 40.Hick A, Wattenhofer-Donze M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, Andre C, et al. (2013). Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Dis Model Mech 3:608–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM. and Pandolfo M. (2008). HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One 4:e1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottesfeld JM, Rusche JR. and Pandolfo M. (2013). Increasing frataxin gene expression with histone deacetylase inhibitors as a therapeutic approach for Friedreich's ataxia. J Neurochem 126 Suppl 1: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soragni E, Miao W, Iudicello M, Jacoby D, De Mercanti S, Clerico M, Longo F, Piga A, Ku S, et al. (2014). Epigenetic therapy for Friedreich ataxia. Ann Neurol 4:489–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Candido EP, Reeves R. and Davie JR. (1978). Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 1:105–113 [DOI] [PubMed] [Google Scholar]
- 45.Lee MG, Wynder C, Schmidt DM, McCafferty DG. and Shiekhattar R. (2006). Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol 6:563–567 [DOI] [PubMed] [Google Scholar]
- 46.Kingston WR. (1962). A clinical trial of an antidepressant, tranylcy-promine (“Parnate”). Med J Aust 49:1011–1012 [PubMed] [Google Scholar]
- 47.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. and Shi Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 7:941–953 [DOI] [PubMed] [Google Scholar]
- 48.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R. and Schule R. (2005). LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 7057:436–439 [DOI] [PubMed] [Google Scholar]
- 49.Consortium EP. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 7414:57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, et al. (2001). Regulation of neuronal traits by a novel transcriptional complex. Neuron 3:353–365 [DOI] [PubMed] [Google Scholar]
- 51.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G. and Shiekhattar R. (2002). A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A 11:7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N. and Shiekhattar R. (2006). Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol 17:6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dottori M. and Pera MF. (2008). Neural differentiation of human embryonic stem cells. Methods Mol Biol 438:19–30 [DOI] [PubMed] [Google Scholar]
- 54.Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM. and Pandolfo M. (2010). Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One 1:e8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding VM, Ling L, Natarajan S, Yap MG, Cool SM. and Choo AB. (2010). FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3beta signaling. J Cell Physiol 2:417–428 [DOI] [PubMed] [Google Scholar]
- 56.Du J, Campau E, Soragni E, Ku S, Puckett JW, Dervan PB. and Gottesfeld JM. (2012). Role of mismatch repair enzymes in GAA.TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J Biol Chem 35:29861–29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerhardt J, Bhalla AD, Butler JS, Puckett JW, Dervan PB, Rosenwaks Z. and Napierala M. (2016). Stalled DNA replication forks at the endogenous GAA repeats drive repeat expansion in Friedreich's ataxia cells. Cell Rep 16:1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan PK, Torres R, Yandim C, Law PP, Khadayate S, Mauri M, Grosan C, Chapman-Rothe N, Giunti P, Pook M. and Festenstein R. (2013). Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreich's ataxia can be reduced upon HDAC inhibition by vitamin B3. Hum Mol Genet 13:2662–2675 [DOI] [PubMed] [Google Scholar]
- 59.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 7479:282–286 [DOI] [PubMed] [Google Scholar]
- 60.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, et al. (2014). Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 4:471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.