SUMMARY
Infection with the flavivirus ZIKA (ZIKV) causes neurological, immunological, and developmental defects through incompletely understood mechanisms. We report that ZIKV infection affects viral and human RNAs by altering the topology and function of N6-adenosine methylation (m6A), a modification affecting RNA structure and function. m6A nucleosides are abundant in ZIKV RNA, with 12 m6A peaks identified across full length ZIKV RNA. m6A in ZIKV RNA is controlled by host methyltransferases METTL3 and METTL14 and demethylases ALKBH5 and FTO, and knockdown of methyltransferases increases, while silencing demethylases decreases ZIKV production. YTHDF family proteins, which regulate the stability of m6A-modified RNA, bind to ZIKV RNA and their silencing increases ZIKV replication. Profiling of the m6A methylome of host mRNAs reveals that ZIKV infection alters m6A location in mRNAs, methylation motifs, and target genes modified by methyltransferases. Our results identify a mechanism by which ZIKV interacts with and alters host cell functions.
Keywords: ZIKA virus, m6A methylation, 2′O methylation, methyltransferases
Graphical abstract
INTRODUCTION
Human infection with ZIKA virus (ZIKV), a mosquito-borne flavivirus, has spread rapidly since the 2015 outbreak in Brazil, and the World Health Organization declared ZIKV infection an International Public Health Emergency in 2016 (Fauci and Morens, 2016; Heymann et al., 2016; Petersen et al., 2016). ZIKV was discovered in 1947 (Driggers et al., 2016a) and, although it had previously caused only sporadic disease in Africa and Asia, more recent outbreaks occurred in Micronesia in 2007 and in French Polynesia in 2013 (Broutet et al., 2016). ZIKV infection has been identified as the etiological agent of severe neurological defects, including microcephaly during fetal development (Driggers et al., 2016b) and neuronal injury associated with Guillain-Barré syndrome in adults (Dejnirattisai et al., 2016). New modes of viral transmission, including maternal–fetal (Brasil et al., 2016) and sexual transmission (Hills et al., 2016), have been reported. ZIKV can infect human skin explants, peripheral blood mononuclear cells, human neuroprogenitor cells, and human cerebral organoids (Dang et al., 2016a; Hamel et al., 2015; Tang et al., 2016). In mouse models, ZIKV may be neurotropic (Cugola et al., 2016; Lazear et al., 2016; Li et al., 2016; Mlakar et al., 2016; Sarno et al., 2016).
ZIKV and other members of the Flaviviridae family, such as dengue (DENV), West Nile (WNV), yellow fever (YFV), and Japanese encephalopathy (JEV), are positive (+) single-stranded RNA viruses. The ZIKV genome encodes a single polyprotein precursor that is cleaved by viral and host proteases to produce three structural and seven nonstructural proteins. Although our understanding of the molecular mechanisms involved in ZIKV infection of human cells has increased dramatically in the past few years, key determinants of ZIKV pathogenicity, such as cell-type specificity, mode of entry, and host factors essential for replication, are still largely unknown. In particular, there is a large gap in our understanding of the genetic and epigenetic regulatory mechanisms governing the viral life cycle and viral interactions with host cells.
As is the case with proteins and DNA, chemical modification of RNA affects its metabolism, function, and localization. More than 100 diverse chemical modifications of RNA nucleotides have been identified, most of which affect ribosomal and transfer RNAs. Modifications of mRNAs and long noncoding RNAs include the 5′-cap structure, N6-methylation of adenosine (m6A), and methylation of C5 of cytosine (m5C) (Fu et al., 2014; Squires et al., 2012; Yi and Pan, 2011). m6A is the most prevalent internal modification of eukaryotic mRNA with unique distribution patterns (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2014). While it is becoming increasingly clear that m6A plays an important regulatory role in physiological and pathological processes (Frayling et al., 2007; Jia et al., 2011; Zheng et al., 2013), little is known of the function of m6A in the mammalian immune system or its influence on host–pathogen interactions.
Adenosine methylation is catalyzed by a large RNA methyltransferase complex (MTase), composed of two catalytic subunits (METTL3 and METTL14), a splicing factor (WTAP), a protein (KIAA1429), and other subunits not yet identified (Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014), while removal of methyl groups is catalyzed by two RNA demethylases, FTO and ALKBH5 (Jia et al., 2011; Zheng et al., 2013). m6A is most abundant in translation start sites, stop codons, and 3′-UTRs (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2014), suggesting that it plays important roles in mRNA biology. Indeed, m6A has been shown to contribute to mRNA stability (Geula et al., 2015; Wang et al., 2014a; Xu et al., 2014); RNA structure, with subsequent effects on RNA–protein interactions (Liu et al., 2015); translation (Meyer et al., 2015; Wang et al., 2015); mRNA nuclear export (Zheng et al., 2013); exon splicing (Zhao et al., 2014) by promoting binding of splicing factor SRSF2 (Zhao et al., 2014); circadian gene expression upon METTL3 depletion (Fustin et al., 2013); and embryonic stem cell pluripotency upon modulation of either METTL3 (Batista et al., 2014; Geula et al., 2015) or METTL14 (Wang et al., 2014b) expression. These findings substantiate the notion that RNA methylation is an epitranscriptomic mechanism that regulates gene expression. The precise sites and abundance of m6A are highly regulated under normal conditions, and it might be expected that non-physiological conditions such as stress, inflammation, and infection would perturb m6A homeostasis.
Although the m6A modification was identified in viral RNA several decades ago (e.g., Rous sarcoma virus, (Kane and Beemon, 1985) influenza virus (Krug et al., 1976) SV40 virus, (Finkel and Groner, 1983), its function and relevance to viral replication had remained unclear. Similarly, whether and how viral infections influence the dynamics of the host and viral RNA methylomes was relatively unknown until the recent publication of three reports describing m6A modification of HIV-1 RNA and the mechanisms by which it affects viral gene expression (Kennedy et al., 2016; Lichinchi et al., 2016; Tirumuru et al., 2016). These studies highlight the significance of this post-transcriptional RNA modification in virology.
The epitranscriptomic landscape of flaviviruses, including ZIKV, remains largely unexplored. N-7 and 2′-O ribose methylations (2′-O-Me) in the cap structure by the viral NS5 protein are required for the efficient translation of viral proteins and for evasion from host antiviral responses. NS5 mutation and loss of N7-methylation are lethal for WNV (Kroschewski et al., 2008; Zhang et al., 2008), and defects in 2′-O-Me dramatically decrease WNV fitness due to enhanced restriction by the host factor IFIT (Daffis et al., 2010). 2′-O-Me of internal adenosines have been detected in DENV and WNV RNA, suggesting a further layer of regulation (Dong et al., 2012), but to date, there have been no reports of internal m6A modifications in flaviviral RNA. Here, we investigated the topology and function of the m6A modification of viral and host RNA during ZIKV infection.
RESULTS AND DISCUSSION
To investigate whether ZIKV viral RNA is methylated, human HEK293T cells were infected with MR766 ZIKV for 48 hours and viral RNA was purified from virions released into the supernatant. The presence of internal m6A and 2′-O-Me nucleosides (m6A, Am, Cm, Um, and Gm) in ZIKV RNA was quantified by LC-MS/MS according to published methods (Jia et al., 2011). Interestingly, ZIKV RNA contained a high level of methylation analyzed (Figure 1A). All four nucleotides contained 2′-O-Me groups, with U and C showing the most extensive modification, followed by A and G (Figure 1A). In particular, the abundance of m6A (about 3% of total adenosine) was strikingly high compared with mammalian mRNAs, where m6A accounts for only ~0.4–0.5% of adenosines (Liu et al., 2014).
Figure 1. ZIKV RNA Contains m6A and 2′-O-Me Modifications, and Methylation is Regulated by Host METTL3, METTL14, and ALKBH5.
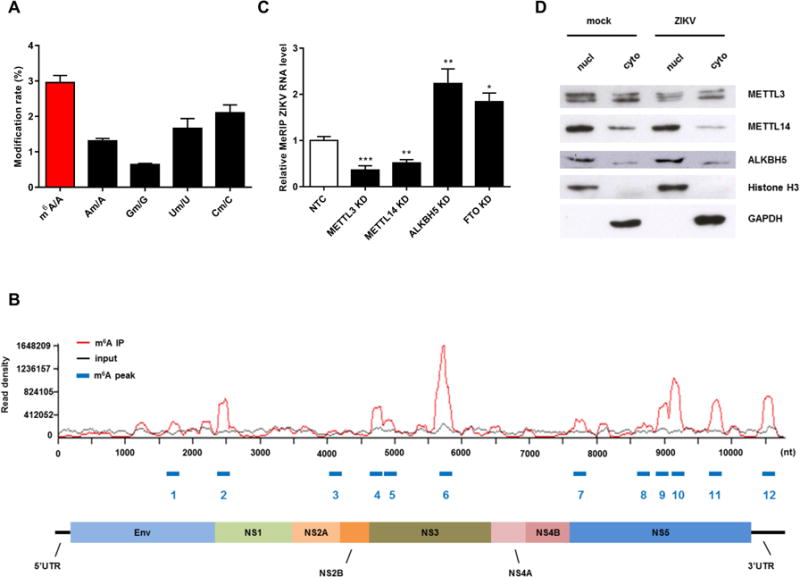
(A) LC-MS/MS quantification of m6A and 2′-O-Me modifications on all four bases of ZIKV genomic RNA (RNA, 50 ng/sample). Data are expressed as the ratio of modified to unmodified bases (m6A/A, Am/A, Gm/G, Um/U, and Cm/C). N = 3.
(B) m6A-seq of ZIKV RNA showing the distribution of m6A reads mapped to the ZIKV genome (red line). The baseline signal from input samples is shown as a black line, and the m6A peaks are shown as blue rectangles along the x-axis. A schematic diagram of the ZIKV genome is shown below to indicate the location of the m6A-enriched sequences. Data are representative of n = 3 determinations.
(C) Modulation of ZIKV RNA methylation by METTL3/METTL14 and ALKBH5. 293T cells were transfected with a non-targeting control shRNA (NTC) or shRNAs targeting METTL3, METTL14, ALKBH5 or FTO (knockdown, KD). RNA was isolated by Me-RIP and quantified by qRT-PCR. N = 3
(D) Localization of METTL3, METTL14, and ALKBH5 in the nucleus and cytoplasm of ZIKV-infected cells. Nuclear and cytoplasmic fractions of mock- or ZIKV-infected 293T cells were subjected to western blot analysis using antibodies against METTL3, METTL14, and ALKBH5 enzymes. Histone H3 and GAPDH were probed as controls for each fraction. Data are the mean ± SEM of the indicated number of replicates. Student’s t-test: * p < 0.05, ** p < 0.005, *** p < 0.0005.
See also Figure S1.
We next examined the topology of the m6A RNA ZIKV methylome by performing methylated RNA immunoprecipitation–sequencing (MeRIP-seq) experiments. For this, ZIKV RNA was purified from Vero cell supernatants, fragmented into 60–200 nucleotide lengths and immunoprecipitated with an m6A-specific antibody (Supplemental Experimental Procedures). The associated RNA was then sequenced and reads were mapped to identify the regions of the ZIKV genome enriched in m6A. We identified 12 discrete m6A peaks spanning the full length of ZIKV RNA ((Figure 1B, Table 1, and Figures S1A and S1B).). We used a stringent peak calling method (FDR<0.01, see supplemental method section) and sequenced the methylome of ZIKV with very high depth leading to the identification of statistically significant peaks. The p values for all identified peaks were < 1E-30, which includes peak 3 shown in Fig 1B. Additionally, correlation test between two biological replicates of vgRNA showed a high correlation (0.998), further indicating a high replicability of our sequencing results (Fig S1A). Of particular note, a cluster of m6A peaks was observed in the NS5 coding region and one peak was present in the 3′-UTR region.
Table 1.
Nucleotide locations and log2 (enrichment) of the 12 m6 A peaks identified in ZIKV RNA by m6A-seq (see Figure 1B).
| m6 A peak number | start | end | log2(enrichment) |
|---|---|---|---|
| 1 | 1652 | 1824 | 1.39 |
| 2 | 2379 | 2531 | 2.52 |
| 3 | 4041 | 4150 | 1.01 |
| 4 | 4658 | 4811 | 1.70 |
| 5 | 4812 | 4999 | 1.11 |
| 6 | 5625 | 5810 | 2.69 |
| 7 | 7673 | 7828 | 1.50 |
| 8 | 8651 | 8800 | 0.98 |
| 9 | 8904 | 9073 | 1.97 |
| 10 | 9080 | 9234 | 3.08 |
| 11 | 9696 | 9849 | 2.32 |
| 12 | 10465 | 10623 | 2.18 |
To determine if m6A sites were conserved among recent strains of ZIKV, we performed m6A consensus motif search within the 12 m6A peaks identified in the MR766 strain and compared these results to four recent ZIKV strains including a Brazilian strain from Paraiba, KX156774 from Panama, KU501215 from Puerto Rico, and FSS13025 from Cambodia. Three common m6A motifs, DRACH, MGACK, and UGAC, were analyzed in this analysis (Fig S1B). Notably, with a few minor exceptions, the majority of the consensus m6A sequences found in MR766 share identity among the other viral strains, suggesting that the m6A landscape of ZIKV RNA is potentially conserved and not dependent on the particular strain under our investigation (Figure S1B).
To date, there have been no reports of flaviviral enzymes with internal m6A MTase activity, and it is possible that ZIKV RNA adenosines are modified by host MTases and demethylases. To investigate this, we transduced 293T cells with shRNAs to knock down the MTases METTL3 METTL14 and the demethylases ALKBH5 and FTO and then examined m6A abundance in ZIKV RNA by MeRIP followed by RT-qPCR. The density of m6A on viral RNA was decreased by silencing of METTL3 and METTL14 and increased by depletion of ALKBH5 and FTO, compared with the abundance in cells expressing the control shRNA (Figure 1C). These results provide evidence that deposition and removal of m6A on ZIKV RNA is mediated by host enzymes.
Since the replication of positive single-stranded flaviviruses takes place in the cytoplasm (Hamel et al., 2015), we examined the subcellular localization of METTL3, METTL14, and ALKBH5 proteins in mock- and ZIKV-infected 293T cells. Although METTL14 and ALKBH5 were more abundant in the nucleus, all three enzymes were readily detectable in both the nuclear and cytoplasmic fractions (Figure 1D). Furthermore, there was no apparent redistribution of the enzymes upon ZIKV infection (Figures 1D and S1). The presence of METTL3, METTL14, and ALKBH5 in the cytoplasm was confirmed by immunofluorescence staining of uninfected and ZIKV-infected cells (Figure S1C). Together, these results show that methylation and demethylation of viral RNA adenosine occurs in the cytoplasm of the host cell.
To evaluate whether perturbation of ZIKV m6A directly or indirectly affects ZIKV replication efficiency, we transduced 293T cells with METTL3, METTL14, ALKBH5, FTO, or control shRNAs. We found that the viral titer, ZIKV RNA levels in cell supernatants, and expression of ZIKV envelope protein were significantly increased by METTL3 and METTL14 knockdown and decreased by ALKBH5 knockdown, respectively (Figures 2A, 2B, and 2C). We confirmed the effects of METTL3, METTL14, and ALKBH5 on the ZIKV life cycle by overexpressing each protein and examining viral replication. Consistent with the results of the knockdown experiments, we found that the viral titer was decreased by METTL3 and METTL14 overexpression and increased by ALKBH5 overexpression without affecting the cell viability (Figures S2B–S2D, S2F–S2G). Thus, modulation of the m6A RNA landscape by host enzymes profoundly influences viral replication.
Figure 2. m6A RNA Methylation Modulates the ZIKV Life Cycle.
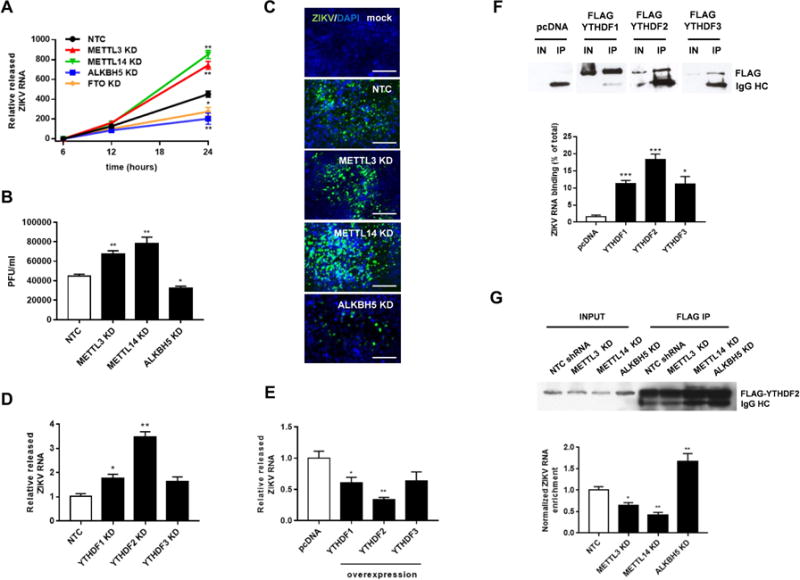
(A) Enhancement of ZIKV replication by METTL3/METTL14 silencing and reduction by ALKBH5/FTO silencing. 293T cells expressing a non-targeting control shRNA (NTC) or shRNAs targeting METTL3, METTL14, ALKBH5 or FTO shRNA (KD) were infected with ZIKV. Supernatants were harvested 6, 12 and 24 h later for quantification of ZIKV RNA by qRT-PCR. N = 3.
(B) Viral titers (PFU/ml) at 24 h post-infection. Cells were treated as described in (A). N = 3
(C) Immunostaining of viral envelope protein in cells treated as described in (A). Scale bars, 100 μm.
(D) Enhancement of ZIKV RNA expression by YTHDF1–3 silencing. 293T cells were transduced with shRNAs targeting YTHDF1–3 or control shRNA. Supernatants were harvested 24 h later for quantification of ZIKV RNA by qRT-PCR. N = 5.
(E) Decrease in ZIKV RNA expression by overexpression of YTHDF 1, 2, and 3 proteins. 293T cells were transfected with FLAG-tagged YTHDF1–3 or control pcDNA vectors. Supernatants were harvested 24 h later for quantification of ZIKV RNA by qRT-PCR. N = 5.
(F) Binding of YTHDF1–3 proteins to ZIKV RNA. 293T cells transfected as described for (E) were immunoprecipitated with an anti-FLAG antibody and immunoblotted for FLAG proteins (top). “IN” (input) lanes contained 5% of the lysate. ZIKV RNA in the FLAG-immunoprecipitates was quantified by qRT-PCR and normalized to percentage of total intracellular ZIKV RNA (bottom). N = 3
(G) Reduction and enhancement of YTHDF2–RNA binding by RNA methylation status. 293T cells were transfected with control or FLAG-YTHDF2 overexpression vector and co-transfected with the indicated shRNAs. Lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted for FLAG protein (top). Input lanes contained 5% of the lysate. ZIKV RNA in YTHDF2 immunoprecipitates was quantified by qRT-PCR and normalized to the level in cells expressing NTC shRNA (bottom). N = 3 All data are the mean ± SEM of the indicated number of replicates. Student’s t-test * p < 0.05, ** p < 0.005, *** p < 0.0005.
See also Figure S2.
Members of the YTH domain protein family (YTHDF) bind to methylated RNA and regulate the stability (Wang et al., 2014a) and translation (Meyer et al., 2015; Wang et al., 2015; Zhou et al., 2015) of cellular m6A-modified transcripts. Recent studies have also shown that YTHDF proteins bind to HIV-1 RNA and regulate gene expression (Kennedy et al., 2016; Tirumuru et al., 2016). Therefore, we analyzed the effect of YTHDF1–3 silencing by RNAi on ZIKV replication. We observed an increase in ZIKV replication when these proteins were depleted (Fig 2D and S2E). Interestingly, YTHDF2 knockdown resulted in the largest increase in ZIKV replication as compared to YTHDF1 and 3. Next, we asked whether YTHDF1, 2, and 3 proteins can bind to ZIKV RNA and, if so, whether they affect RNA metabolism and viral replication. For this, we overexpressed FLAG-tagged YTHDF1–3 proteins in 293T cells and then examined viral RNA abundance and YTHDF protein binding to methylated RNA. Overexpression of all three YTHDF proteins decreased ZIKV RNA expression, with YTHDF2 having the greatest effect (Figure 2E). Furthermore, immunoprecipitation of the YTHDF proteins with anti-FLAG antibodies followed by RT-qPCR revealed that YTHDF1–3 proteins did indeed bind to ZIKV RNA, with YTHDF2 immunoprecipitates containing the greatest amount of viral RNA (Figure 2F).
The results of the YTHDF knockdown and overexpression experiments (Figures 2D and 2E) suggest that host cell YTHDF2 may bind to and destabilize viral RNA (Wang et al., 2014a), possibly as a mechanism to regulate ZIKV infection. To determine whether modulation of m6A abundance on ZIKV RNA affects its binding to YTHDF2, we examined cells expressing control, METTL3, METTL14, or ALKBH5 shRNA. Notably, the association of ZIKV RNA with YTHDF2 was significantly reduced by METTL3 and METTL14 silencing, and conversely, significantly increased by ALKBH5 knockdown (Figure 2G). Thus, taking all these results together, YTHDF2 modulates ZIKV RNA levels by directly binding to m6A-modified viral RNA.
Since viral infection has a profound effect on gene and protein expression in the host cell, we examined the abundance and distribution of m6A on cellular transcripts by performing m6A-seq experiments on mRNA isolated from uninfected and ZIKV-infected 293T cells (Figure S3A–B). Metagene analysis showed that ZIKV infection increased m6A levels in the 5′-UTR regions of the transcriptome and correspondingly decreased the levels in the 3′-UTRs (Figure 3A). This change may represent a cellular response to the stress of viral infection, similar to the previously described change in m6A deposition in response to heat shock (Zhou et al., 2015). To examine the distribution of ZIKV-induced changes in more detail, we compared mRNA m6A peaks that were either newly gained or lost after ZIKV infection. We observed that the newly emerged m6A modifications were preferentially deposited in 5′-UTR and CDS and depleted in exon junctions and 3′-UTR regions, compared to the lost m6A peaks (Figure 3B), suggesting that ZIKV infection might affect gene translation, alternative splicing, and mRNA stability as a consequence of differential deposition of m6A. Gene ontology (GO) analysis of the genes with changed m6A peaks identified a number of immune-related categories among the most enriched pathways in both the newly gained and lost m6A modifications (Figure 3C). Furthermore, quantitative RNA expression analysis of a dozen representative genes showed that the newly methylated genes emerged upon infection had expression levels similar to uninfected conditions suggesting that the selective m6A deposition was not due to an RNA upregulation but rather a consequence of viral infection events (Figure S3C). Thus, ZIKV infection appears to be sensed by the host MTase and demethylase machineries, which then initiate a directed reprogramming of the post-transcriptional landscape of cellular mRNAs (Table S1).
Figure 3. ZIKV Infection Influences RNA Methylation of Host Cell Transcripts.
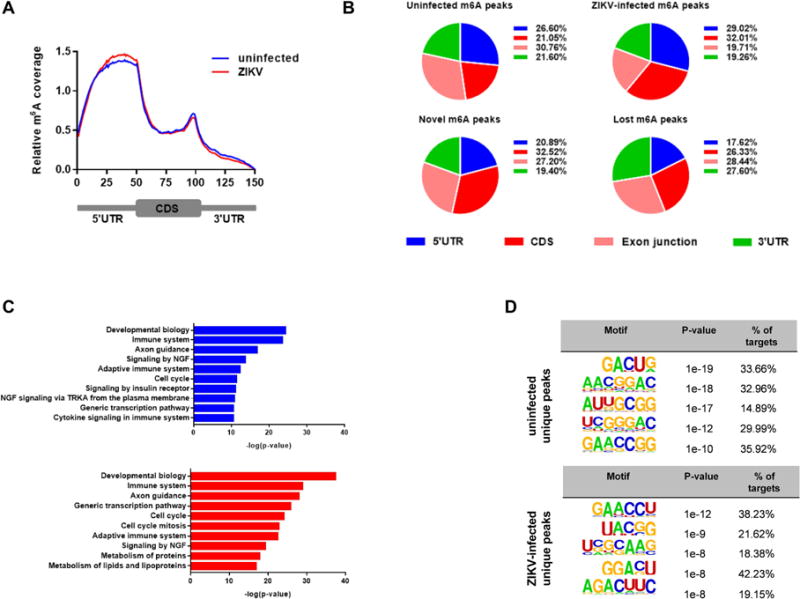
(A) Metagene analysis of normalized m6A peak distribution along a reference mRNA.
(B) Distribution of m6A peaks in the 5′-UTR (blue), coding sequence (CDS, red), exon junction (pink), and 3′-UTR (green) of host cell RNA transcripts. 293T cells were mock- or ZIKV-infected, and m6A peaks in total cellular RNA were analyzed at 24 h after infection. Charts show the proportion of m6A peaks in the indicated regions in uninfected and ZIKV-infected cells (top) and the appearance of newly emerged m6A peaks or loss of existing m6A peaks after ZIKV infection (bottom). Representative of N = 2 determinations.
(C) GSEA analysis of reactome analysis of pathways associated with newly emerged m6A modifications (top, blue) and loss of existing m6A modifications (bottom, red) at 24 h after ZIKV infection of 293T cells. The top 10 enriched categories for each condition are shown.
(D) Motif analysis to identify consensus sequences for m6A methylation in uninfected and ZIKV-infected 293T cells. The top 5 motifs for each are shown.
We have performed a motif analysis of the newly emerged and lost peaks specifically to determine if there was any change in the top motifs between these two groups. The motif usage changes seem to occur on the overall level, with newly emerged ones preferring “AAC” while lost ones preferring “GAC” (Fig 3D). The results showed clear differences in the consensus sequences of m6A, highlighting the possibility that the substrate specificity of m6A-related enzymes may change upon viral infection (Figure 3D).
CONCLUSIONS
Epitranscriptomics has recently been identified as a new layer of regulation of RNA metabolism and function. Several aspects of RNA biology have been shown to be controlled by m6A deposition by METTL3/METTL14 MTases and removal by ALKBH5 and FTO demethylases. Here, we quantified and mapped the internal m6A modifications in ZIKV RNA and showed that the m6A abundance is controlled by host METTL3, METTL14, ALKBH5, and FTO. Subsequently, we investigated the role of m6A during ZIKV infection of human cells. We found that the depletion or over-expression of the central RNA methylation enzymes impact viral replication, demonstrating that the host RNA methyltransferase machinery acts as a negative post-transcriptional regulator of ZIKV virus. Moreover, ZIKV RNA binding to YTHDF proteins indicates another regulatory layer represented by the m6A binding proteins. We also explored the m6A landscape of cellular mRNAs in response to ZIKV infection, and identified two classes of transcripts which undergo specific m6A deposition or loss of the modification. This evidence suggests that RNA methylation machineries (“writers”, “erasers” and finally “readers”) are able to sense and respond to viral infection and modulate gene expression at the post-transcriptional level. The overall effects of m6A on viral replication may be due to a combination of direct events regulating viral RNA metabolism (i.e. YTHDF proteins’ binding to ZIKV RNA) and indirect post-transcriptional regulation of host RNAs, which may act as pro- or anti-viral factors. For example, viral infection triggers RNA modification in host genes to suppress immune surveillance or to perturb host genetic networks for successful replication. In order to delineate the precise contribution of the viral and cell-mediated RNA modification events, further studies will be needed.
EXPERIMENTAL PROCDURES
ZIKV Production and Cell Infection
All studies were conducted in accordance with approved IRB protocols by the University of California, San Diego. All animal work was approved by the Institutional Review Board at the University of California, San Diego and was performed in accordance with Institutional Animal Care and Use Committee guidelines. ZIKV MR766 virus was expanded by inoculation of Vero cells at an MOI of 5 as previously described (Dang et al., 2016b).
Immunofluorescence Staining of Flavivirus Envelope Protein
293T cells previously transduced with NTC, METTL3, METTL14, FTO, or ALKBH5 shRNAs were seeded in 24-well plates pretreated with poly-L-lysine (Sigma) one day before infection, and then infected as described above. At 24 h post-infection, cells were washed three times with PBS and fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were permeabilized in PBS containing 0.2% Triton X-100 for 10 min at room temperature and then blocked with 3% BSA in PBS for 2 h. Cells were stained with primary antibody (1:1000) overnight at 4°C, washed three times with PBS, and then stained with Alexa 488-conjugated secondary antibody (1:1000) for 1 h at room temperature. The nuclei were stained with DAPI, and cells were analyzed using a Leica microscope.
m6A-seq
High-throughput sequencing of the ZIKV methylome was carried out by m6A-seq following the previously published protocol (Dominissini et al., 2012). In brief, total cellular RNA (containing ZIKV RNA) was extracted and purified by the RiboMinus Eukaryote System v2 (Thermo Fisher) to remove contaminating rRNA. Purified RNA was sonicated to ~200 nt lengths, mixed with 2.5 mg of affinity-purified anti-m6A polyclonal antibody (202003; Synaptic Systems, Goettingen, Germany) in IP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl, pH 7.4), and incubated for 2 h at 4°C. The antibody was collected by incubation with protein A beads (Invitrogen) at 4°C for 2 h, and the beads were washed three times and eluted with m6A-free nucleotide solution. RNA in the eluate was purified using RNA Clean & Concentrator kit (Zymo Research) and used for library generation with a Stranded mRNA Library Prep kit (Illumina). Sequencing was performed using an Illumina HiSeq 2000 according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Steve Head and the staff of the Next Generation Sequencing core facility at UCSD for help with the HT-seq and data analysis. We thank Jason Dang for his help with preparation of the artwork, and members of the Rana lab for helpful discussions and advice. This work was supported in part by grants from the National Institutes of Health. C.H. is an Investigator of the Howard Hughes Medical Institute (HHMI). B.S.Z. is an HHMI International Student Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The GEO accession number for the RNA-seq data reported in this paper is GEO number GSE87516.
AUTHOR CONTRIBUTIONS STATEMENTS
G.L. designed and performed the experiments, analyzed the data, and wrote the manuscript; B.S.Z. performed the experiments, analyzed the data, and contributed to manuscript writing; Y.W. designed and performed the experiments and analyzed the data; Z.L. analyzed the data; Y.Q. analyzed the data; Q. Z. performed the experiments; C.H. contributed to the experimental design, data analysis, and interpretation; and T.M.R. contributed to the experimental design, data analysis and interpretation, and manuscript writing. All authors approved the final version of this manuscript.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
References
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Jr, Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro – Preliminary Report. The New England journal of medicine. 2016 doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutet N, Krauer F, Riesen M, Khalakdina A, Almiron M, Aldighieri S, Espinal M, Low N, Dye C. Zika Virus as a Cause of Neurologic Disorders. The New England journal of medicine. 2016;374:1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016 doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016b;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016 doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, et al. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS pathogens. 2012;8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. The New England journal of medicine. 2016a:10. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. The New England journal of medicine. 2016b;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- Fauci AS, Morens DM. Zika Virus in the Americas–Yet Another Arbovirus Threat. The New England journal of medicine. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- Finkel D, Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann DL, Hodgson A, Sall AA, Freedman DO, Staples JE, Althabe F, Baruah K, Mahmud G, Kandun N, Vasconcelos PF, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387:719–721. doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills SL, Russell K, Hennessey M, Williams C, Oster AM, Fischer M, Mead P. Transmission of Zika Virus Through Sexual Contact with Travelers to Areas of Ongoing Transmission – Continental United States, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Molecular and cellular biology. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. The Journal of biological chemistry. 2008;283:19410–19421. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]
- Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. Journal of virology. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.1003.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.1004.1017. [DOI] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nature Microbiology. 2016;1 doi: 10.1038/nmicrobiol.2016.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. The New England journal of medicine. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno M, Sacramento GA, Khouri R, do Rosario MS, Costa F, Archanjo G, Santos LA, Nery N, Jr, Vasilakis N, Ko AI, et al. Zika Virus Infection and Stillbirths: A Case of Hydrops Fetalis, Hydranencephaly and Fetal Demise. PLoS Negl Trop Dis. 2016;10:e0004517. doi: 10.1371/journal.pntd.0004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and noncoding RNA. Nucleic acids research. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016;5 doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nature chemical biology. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- Yi C, Pan T. Cellular dynamics of RNA modification. Acc Chem Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Zhou Y, Shi PY. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5′ stem-loop of genomic RNA. Journal of virology. 2008;82:7047–7058. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


