Abstract
Since 2013, there have been several alarming influenza‐related events; the spread of highly pathogenic avian influenza H5 viruses into North America, the detection of H10N8 and H5N6 zoonotic infections, the ongoing H7N9 infections in China and the continued zoonosis of H5N1 viruses in parts of Asia and the Middle East. The risk of a new influenza pandemic increases with the repeated interspecies transmission events that facilitate reassortment between animal influenza strains; thus, it is of utmost importance to understand the factors involved that promote or become a barrier to cross‐species transmission of Influenza A viruses (IAVs). Here, we provide an overview of the ecology and evolutionary adaptations of IAVs, with a focus on a review of the molecular factors that enable interspecies transmission of the various virus gene segments.
Keywords: adaptation, pandemic, zoonotic
1. Introduction
Influenza A virus (IAV) has caused significant morbidity and mortality globally in humans, with an estimated 14 pandemics that have occurred since the 1500s.1 Wild aquatic birds are well known to be the natural reservoirs for IAV subtypes harbouring H1–H16 subtypes,2, 3, 4 with the exception of H17 and H18 subtypes that were recently discovered in bats.5, 6 The phylogenetic relationships of all IAV subtypes are displayed in Fig. 1. In addition to its natural reservoir species, influenza viruses infect a wide range of hosts including canids, equids, humans and swine.2 IAVs’ ability to generate novel gene constellations through reassortment between subtypes poses a risk for immune escape in these new hosts.7 Furthermore, IAV undergoes rapid genetic and antigenic evolution, which makes vaccination control difficult in humans and other domestic species.
Figure 1.
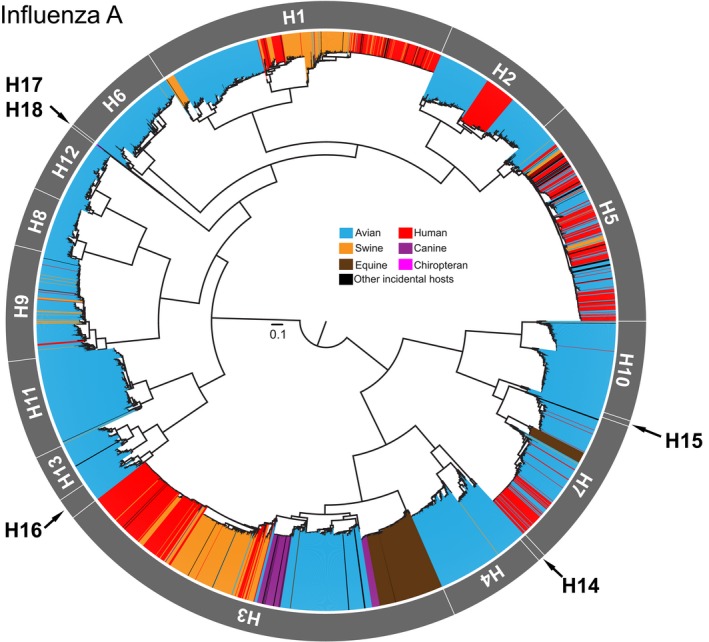
Diversity and host distribution of influenza A viruses (IAVs). Maximum‐likelihood (ML) estimation of the haemagglutinin (HA) gene sequences of all subtypes of IAVs downloaded from the NCBI GenBank database. Overall data set randomly subsampled to include 200 isolates per subtype per host for the tree reconstruction. External branches of tree coloured by the major host groups from which the virus has been isolated from: avian (blue), canine (purple), chiropteran (pink), equine (brown), human (red), swine (amber) and other incidental hosts (black). Scale for branch length represents number of nucleotide substitution per site (subs/site) in the HA alignment
In addition to human pandemics that have emerged from avian and swine hosts, there are also repeated spillover events from domesticated animals, primarily poultry and swine, that pose a significant threat to human health.8, 9, 10, 11, 12, 13, 14 Direct transmission of IAV from a wild avian source to humans is rare, as there has only been a single report of laboratory‐confirmed human infection with H5N1 contracted through close contact with dead and infected wild swan in Azerbaijan.15 However, there is serological evidence of H5N1 infection among Alaskan hunters who handled dead wild avian species,16 indicating that exposure to IAVs from wild birds through close contact can potentially cause infection. More notably, viral genes that are similar to the 1918‐like H1N1 avian virus were recently detected in the influenza gene pools of wild birds, raising the potential for the re‐emergence of a 1918‐like pandemic virus.17 Furthermore, due to increasing human encroachment of wildlife habitats, the potential of a wild‐source threat becomes more relevant, as is seen with the emergence of other pathogens such as human immunodeficiency virus (HIV), severe acute respiratory syndrome coronavirus and the more recent Zaire‐variant Ebola virus in Western Africa.18, 19, 20, 21
In this review, we discuss the current knowledge of ecological and molecular determinants responsible for interspecies transmission of IAV, with specific focus on avian‐derived influenza subtypes involved in zoonotic and epizootic transmission to other hosts (see Fig. 2).
Figure 2.
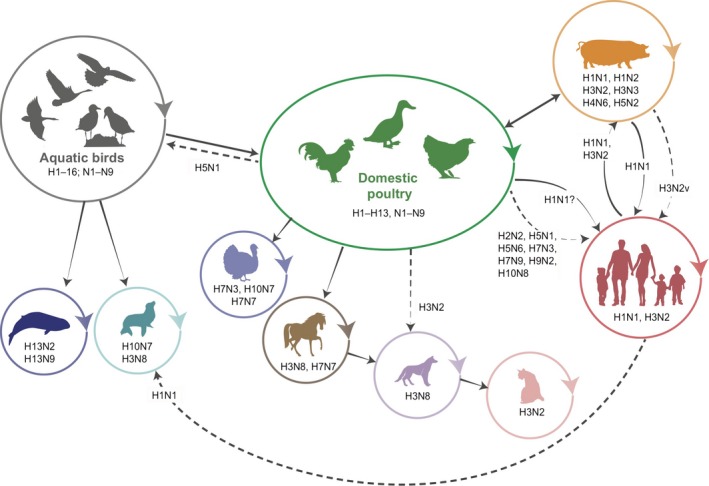
Significant interspecies transmission of influenza A viruses (IAVs). Representative diagram of interspecies transmission events of IAVs and the subtypes involved in these events. Solid arrows represent direct transmission events that have since been established in the host species. Dashed arrows represent sporadic or limited infection of subtypes where sustained transmission in the new host has not been detected
2. Role of wild birds in IAV ecology
Wild birds of the orders Anseriformes (such as ducks and geese) and Charadriiformes (such as shorebirds and gulls) are the primary reservoir of avian influenza viruses.2 The maintenance of this diverse pool of viruses globally is in part due to the migratory nature of the bird species, a mechanism in which the viruses are shed in bird faeces and later acquired by other birds that share the same habitat along migratory flyways.2, 22, 23, 24 Transmission of IAV in wild bird populations is dependent on several factors including virus shedding, virus stability in the environment and the degree of close contact/mixing with other host species.3, 25
Virus shedding by bird species in the environment mainly occurs in fresh water, from where viable viruses of most known subtypes currently circulating in wild birds have been isolated.24 The persistence of infectivity of IAV shed by aquatic birds is dependent on the temperature and pH of the freshwater, where it has been found that shed viruses can remain infectious for 4 days in temperatures of 28°C and >30 days in temperatures below 0°C, with greatest viral recovery noted at a slightly basic pH (7.4–8.2) and temperatures lower than 17°C.26, 27, 28, 29 Prolonged virus shedding, which can be attributed to the development of the avian immune system, facilitates the efficient transmissions of IAVs in their natural hosts.27 In the avian cellular immune response, the maturation of T lymphocytes occurs in the thymus glands that are distributed around the neck and inside the thoracic cavity of most birds. Unlike humans, these thymus glands only fully develop in late juvenile birds and persist throughout adult life.30 In contrast, the plasmacytoid B cells essential for antibody‐mediated response mature in a specialized organ called the bursa of Fabricius (BF), located in the cloaca, but the BF involutes or disappears early in the juvenile bird stage, that is at the onset of feather growth.27, 30 The transition from the loss of the BF to the full functionality of the thymus results in a period of a weakened immunity in these avian species; thus migrating juvenile birds with a high viral load become an infection source of different pathogens.27, 31
The stability of the haemagglutinin (HA) protein, both in the intracellular and extracellular environment, is a key determinant of the influenza virus stability and infectivity in the environment.32 Stability of the HA in low intracellular pH of the early endosome is essential for the intracellular entry of the virus, after which point a threshold pH is reached in the late endosome causing the fusion of the protein to the lipid membrane, enabling the viral genome to escape the late endosome and successfully infect the cell.7 Conversely, the virus must also maintain its stability in variable extracellular pH environments in its transmission between hosts. Thus, it has been observed that the pH stability of IAV subtypes evolves differently depending on host type and the route of transmission.33 As most transmission of avian IAV in aquatic birds is through the oral–faecal route and the replication occurs mostly in the GI tract and cloaca of the birds, these viruses have evolved for stability at higher extracellular pH to accommodate these sites of replication and also maintain viability in the aquatic environment.33, 34, 35, 36
More recently, the incorporation of environmental durability data into a phylogenetic study of avian IAV demonstrated that environmental transmission is correlated with the increase in virus genetic diversity in short‐lived avian hosts, suggesting an environmental reservoir can act as a source of annual outbreaks.37
3. Role of domestic birds in IAV ecology and interspecies transmission to humans and other mammals
AIV can cause low pathogenic infections (LPAI) in domestic birds with an asymptomatic or mild disease, with HA subtypes H1, H3, H5, H6, H7 and H9 most commonly isolated.38, 39 In contrast, certain AIV lineages in subtypes H5 and H7 cause severe disease and rapid mortality in domestic birds and are referred to as highly pathogenic avian influenza (HPAI). While HPAI is phenotypically defined by a viruses lethality for susceptible chickens under strict experimental criteria,40 genetically, the key distinguishing structure of HPAI is the presence of a multibasic cleavage site (MBCS), which contains two to several basic residues (e.g. PQRERRRKR/G) in the HA gene, as opposed to a single basic residue (PQRETR/G) in LPAI strains.41, 42, 43 Furin‐like peptidases, which are expressed ubiquitously in all tissue types of both avian and mammalian species, are responsible for the cleavage of HA at the MBCS site.41, 44 Additionally, HPAI strains have also emerged through non‐homologous recombination with a non‐HA protein nucleotide sequence, introducing a non‐native sequence into the HA gene.45, 46 Although this can enhance the pathogenicity of the virus, systemic replication does not necessarily occur in all species: experimental infection of ferrets and mice with HPAI H5N1 showed systemic virus replication,47, 48 but replication was only observed in the respiratory tract of non‐human primates,49 suggesting that pathogenicity of HPAI viruses is possibly host specific and multifactorial.43 It has been recently understood that domestic birds such as chicken lack the retinoic acid‐inducible gene‐I (RIG‐I), which is essential for signalling in the innate immune response,50, 51 as opposed to presence of RIG‐I in other avian species and mammals as an important sensor of viral RNA leading to activation of type I interferons.52 In experimental infection of chickens and ducks with H7N1 virus, increased viral pathogenesis with clinical signs of disease and death is observed in chickens, but infected ducks do not exhibit clinical signs, highlighting the evolutionary adaption of ducks being a natural host of the influenza virus.53, 54 There has also been back‐transmission of HPAI viruses from domestic poultry to wild bird populations that are implicated in bird die‐offs55 and intercontinental spread.56, 57, 58, 59
Most IAV spillover events to humans have occurred through close contact with domestic poultry.2, 24, 60 Poultry markets have been demonstrated to harbour a variety of influenza subtypes including H5N1, H6N1, H7N9 and H9N2; these viruses were detected from bird cages, drinking water troughs, environmental soils at live poultry markets and farms.38, 61, 62, 63, 64 Furthermore, live poultry markets act as a major source of reassortment of IAV causing the generation of new HA and neuraminidase (NA) subtype combinations and gene constellations, with the H9N2 subtype acting as a major donor of internal gene segments to other subtypes. Those viruses have subsequently caused human infection with H5N1, H5N6, H9N2, and H10N8 subtype viruses.8, 38, 65, 66, 67, 68, 69, 70, 71 The outbreak of H7N9 in China, a novel subtype that also contains H9N2 internal genes, has become one of the more severe zoonotic infections from avian IAV causing high morbidity and case fatality (40.6%) in humans.72, 73, 74 The majority of H7N9 human infection has resulted from close exposure to live poultry markets, whereas human‐to‐human transmission of this virus is limited.74 China is the world's largest producer of domestic poultry, with an estimated 13.1 billion poultry produced in 2015.75 These poultry include mainly chickens, ducks and geese with smaller numbers of a diverse range of species such as guinea fowl, partridge, pheasants and quails.38, 76 Production of domestic poultry ranges from high biosecurity commercial farms to backyard poly‐culture farming, where domestic poultry often come into contact with wild birds.76 Furthermore, the movement and mixing of domestic poultry to live poultry markets, where often multiple species are housed together regardless of origin, enhance the spread and mixing of IAVs.76, 77, 78, 79 Closure of wet markets in China has been shown to effectively reduce the transmission of H5N1, H9N2 and H7N9.80, 81, 82
4. Avian IAV adaptation to human and other mammalian hosts
In a large‐scale analysis of sequence data of IAVs from various hosts, several mutations were reported to be indicative of adaptation of IAVs from avian to other host species.83 However, in the surveillance of swine populations in southern China, many of these sites retain their avian‐like residues despite over 30 years of circulation in these mammalian hosts,84 suggesting these residues are likely founder effects rather than true adaptive mutations. Here, we highlight a few mutations that have been validated experimentally as critical in the adaptation of avian IAVs to humans and other mammalian hosts.
4.1. Avian HA vs mammalian HA
The HA protein is the key mediator of virus infection into the host cell and consists of two domains, HA1 and HA2. Three domains located in the HA1 globular head form the receptor binding site (RBS): the 130‐loop (residues 132–138), the 190 loop (residues 188–195) and 220‐loop (residues 221–228) (HA residues here follow H3 numbering unless otherwise stated).32, 85, 86, 87 Mutations within the RBS determine the specificity of binding to avian‐like α‐2,3‐sialic acid (SA) or mammalian‐like α‐2,6‐SA receptors. Specifically, the widening of the binding pocket through mutation favours α‐2,6‐SA receptor binding, as steric hindrance of the α‐2,6 cis‐linkage SA prevents binding to the narrower avian‐adapted RBS that binds to the smaller α‐2,3 trans‐linkage SA. Two mutations at Gln‐226‐Leu and Gly‐228‐Ser are required to widen the RBS in the human‐adapted H2 and H3 subtypes.88, 89, 90, 91 In comparison, the widening of the RBS in human‐adapted H1 subtype is determined by aspartic acid residues at positions 190 and 225, as they contribute to the conformational change of the 220 loop at the RBS in switching the binding affinity to the α‐2,6‐SA receptor.92, 93 In the case of subtype H5 virus, crystal structure studies show that the Gln‐226‐Leu mutation not only widens the binding pocket but also creates a hydrophobic environment, which alters the binding preference from avian α‐2,3 to the human α‐2,6 receptor.94, 95, 96, 97 Furthermore, mutations in the 130‐loop (Leu‐133‐Val/Ser and Ala‐138‐Val substitutions) of the HA‐H5 enable the virus to bind to both α‐2,3‐SA and α‐2,6‐SA receptors.94, 98, 99 Finally, mutations outside the RBS, specifically in glycosylation sites near the RBS, have also been determinants of receptor binding specificity, as these sites, when glycosylated, cause stearic hindrance to the optimal binding of respective SA linkage to the RBS, thereby restricting binding.100, 101, 102, 103, 104, 105
As mentioned earlier, the stability of the HA protein at various pH is associated with transmission in different hosts, with mammalian viruses having a lower optimum pH of conformational stability than avian viruses.35, 106, 107, 108 However, it was recently discovered that, in the adaptation from avian H1N1 to establishment of the European avian‐like (EA) swine virus lineage, this mammalian virus possessed a higher optimum pH of conformational stability than its avian counterpart, which is hypothesized to be mediated by changes in HA2 residues 72, 75, and 113.109 These findings certainly warrant the increased need to differentially characterize this trait between all susceptible species that can harbour IAVs for pandemic and panzootic preparedness as previously understood host‐specific optimum pH for HA stability may vary between subtypes and hosts.
4.2. Avian NA vs mammalian NA
The primary function of the NA protein is to cleave sialic acid receptors of host cells from where the virion has budded, thereby preventing reinfection into the same cell and promoting viral spread within the host.7 An optimal balance between the HA and NA protein function is required for effective infection and transmission of IAV: excess NA proteins can hinder the binding of HA to host cell receptors, whereas insufficient NA protein results in limited virus spread.110, 111 Avian NA proteins preferentially cleave α‐2,3‐SA residues, whereas mammalian NA proteins can cleave both α‐2,3‐SA and α‐2,6‐SA residues, indicating host‐specific adaptations of the protein.112 In avian IAV subtypes (e.g. H2N2, H7N1 and H9N2), deletions in the NA stalk are crucial for adaptation in chickens and other Galliformes poultry species.113, 114 While the significance of this deletion is yet to be elucidated, the naturally selected deletion serves as a barrier in interspecies transmission, as viruses containing the stalk region deletion were shown to have compromised transmission between ferrets.115
4.3. Avian PB2 vs mammalian PB2
The most widely cited mutation in polymerase PB2 subunit is Glu‐627‐Lys substitution, which is responsible for adaptation from avian to mammalian hosts. This mutation has been shown to mediate replication at a lower temperature (33°C) in the human upper respiratory tract, thereby facilitating efficient aerosol transmission, as opposed to a higher temperature (40°C) in avian intestinal tract.116, 117, 118 Additionally, this PB2 Glu‐627‐Lys mutation enhances the assembly of the viral ribonuclear protein (vRNP) complex in mammalian cells by stabilizing the binding of nucleoprotein (NP) and PB2 proteins during viral replication,119, 120, 121 and this stabilization further contributes in resistance to host RIG‐I sensing.122 Exceptions are found in H5N1 avian viruses as they are able to replicate in mammals successfully even with the absence of PB2 Glu‐627‐Lys mutation; however, a compensatory Asp‐701‐Asn mutation in the PB2 gene is present and has been found to facilitate adaptation to mammalian replication.123 This mutation promotes the transport of the RNA polymerase subunits into the nucleus through NP and PB2 interaction with importin‐α in mammalian cells, thereby promoting transmission in mammalian systems.123, 124
4.4. Avian NP vs mammalian NP
The NP segment plays a key role in interspecies transmission of IAV, particularly the switch from avian to mammalian hosts. The viral NP is associated with the sensitivity of the IAV to host myxovirus resistance A (MxA) proteins—an important intrinsic antiviral factor of mammals known to inhibit IAV as well as other RNA and DNA viral infections.125, 126, 127 It has been experimentally shown that an avian H5N1 virus with a reverse engineered human H1N1 NP protein can restrict the MxA sensitivity to eliminate species barrier and enhance viral replication 10‐fold,125, 126 indicating IAV can overcome the species barrier from an avian host to replicate successfully in the mammalian system through variation in the NP protein. For instance, the avian H7N9 virus has a crucial Asn52 mutation in the NP gene, causing a reduction in MxA sensitivity within human hosts.128 This also suggests that pre‐adaptive Asn‐52 mutation (i.e. resistance to MxA proteins) has been initiated in avian hosts prior to adaptation in mammalian hosts. This was demonstrated experimentally in squirrel monkeys, where the addition of an avian NP segment into a human H3N2 backbone attenuated virus growth.129, 130, 131
4.5. Avian NS vs mammalian NS
The non‐structural protein 1 (NS1) protein has been implicated in variable virulence and pathogenicity within different hosts through its interactions of PDZ‐binding motif at the C terminus of the protein.83, 132, 133, 134 In a large‐scale comparative genomic approach of all IAV, a specific 11 amino acid long sequence feature variant type (SFVT) located between residues 137–147 of the NS1 protein was statistically associated with a host restriction phenotype,135 although this has yet to be tested experimentally. Nonetheless, discovery of this SFVT emphasizes the importance of further studying this protein and its potential role in interspecies species transmission.
Mutations in the non‐structural protein 2 (NS2)/nuclear export protein (NEP) gene has of late received more attention as an important factor in mammalian adaptation of avian IAV, due to its role as a regulator of transcription and replication of influenza virus genome within the host cell.136, 137, 138 Importantly, it was shown that substitution Met‐16‐Ile in the NEP conferred H5N1 avian polymerases the ability to efficiently replicate in mammalian cells by stabilizing the vRNP complex and by enhancing vRNA synthesis.138
4.6. Avian PB1‐F2 vs mammalian PB1‐F2
The complete role of PB1‐F2 in the virus lifecycle still remains to be elucidated, as it has been shown to both aid and disrupt viral virulence. It is generally understood that PB1‐F2's role is subtype and host dependent. It has been demonstrated that within the 1918‐like H1N1 viruses, PB1‐F2 interacts with the PB1 protein to increase replication by retaining the polymerase complex in the nucleus longer.139 In HPAI H5N1, the wild‐type PB1‐F2 protein was found to have no effect on the severity of infection in mice beyond replication in the lung, but replication was detected systemically in multiple organs of experimentally infected ducks, further supporting the existence of a host‐specific function for the protein.140, 141
4.7. Avian CpG vs mammalian CpG non‐coding region
Mutations in the non‐coding region of IAV have been shown to mediate cross‐species transmission. These non‐coding adaptations are due in part to constraints on host mechanisms that restrict the probability of mutational fixation, and consequently, a non‐random pattern of mutation is seen across the virus genome in different host species.142 For instance, many RNA viruses evolve to mimic the CpG dinucleotide frequencies of their hosts.143, 144 Mammalian DNA sequences contain unmethylated CpG motifs responsible for inducing innate immune responses through interactions with Toll‐like receptor 9 (TLR9)144, 145; however, most mammals protect themselves against autoimmunity by maintaining CpG dinucleotides that are methylated and at low frequency.145 To accommodate this, IAV has been observed to evolve lower dinucleotide CpG content following interspecies transmission.143, 146 This phenomenon has also been experimentally confirmed, where the infection with modified human A/WSN/33 influenza virions expressing avian‐like increased CpG content was shown to increase ssRNA recognition and immune activation by human plasmacytic dendritic cells.144 This low CpG preference has been reported in human IAV subtypes such as the H1N1 and H3N2 in comparison with avian IAVs.143 However, this trend was not observed in H2N2 isolates from the 1957 pandemic, likely due to the short duration in which this subtype circulated in humans.147
Another parameter of codon usage bias is relative synonymous codon usage (RSCU), which is commonly applied to IAV genomes as a metric of comparison between different host species.142 A clear distinction in codon usage patterns has been shown in H1N1 and H3N2 subtypes by comparison among different hosts: avian‐derived human and swine viruses demonstrate RSCU values that become closer to their new mammalian hosts over time, and do not show preference for the synonymous codons used in their precursor avian hosts.142, 148, 149, 150 Other IAV subtypes (including H2N2, H5N1 and H9N2) that have transmitted to humans have RSCU values that are similar to those of avian‐like RSCU, as these subtypes have been transmitted directly from an avian source and have not successfully circulated long enough in humans to establish an optimized codon usage in mammals.142, 148, 151
5. Pigs: the mixing vessel?
Pigs are widely recognized as a “mixing vessel” of IAV with the presence of both α‐2,3‐SA and α‐2,6‐SA residues distributed throughout their respiratory tracts, where avian, swine and human IAV strains reassort following co‐infection and give rise to new genetic variants, potentially leading to epidemics and/or epizootics.152 The most remarkable example is the swine‐origin 2009 H1N1 pandemic (H1N1/2009), which emerged from the reassortment of gene segments from the H1N1 European “avian‐like” swine (EA‐swine) lineage, H1N2 triple reassortant swine (TRS) lineage and an Eurasian avian H1N1 lineage.153 Several adaptive changes of specific host residues have been reported that contributed to the establishment of this swine virus in humans, observed especially from viruses isolated during the early phase of the pandemic, with the later circulating viruses showing mutations involved in antigenic escape within human hosts.154 The 2009 pandemic H1N1 (pH1N1) viruses did not contain the Lys‐627 or Asn‐701 substitutions that were considered indicative of mammalian adaptation,43, 155, 156 and it now appears these substitutions are not required for swine to human transmission. Indeed, when the mammalian‐like Lys‐627 and Asn‐701 residues were introduced, a significant increase in polymerase activity and viral pathogenesis was observed,155, 156 but this replicative advantage of the mutation was not necessary for the initial establishment of the avian polymerase in TRS lineage and subsequently human pH1N1 viruses.157 Instead, partial compensatory mutations Gly590Ser and Glu591Arg were observed to aid polymerase activity of in the lack of the mammalian 627 and 701 residues, providing a possible explanation for the successful transmission of the avian‐like residues in mammals.43, 155, 156, 157 Additionally, the H1N1/2009 virus has reduced sensitivity to host MxA proteins though the interaction with the NP gene, as a result of the TRS‐origin NP reassortment.120, 121, 158
There have been several spillover infections of swine IAVs into humans159, 160, 161; however, increasing evidence suggests reintroduction of the human virus into swine populations is more common.162, 163 The susceptibility of swine infection by human virus can be attributed to, among many options, the dynamics of host immune development and farming practices. The immune response to IAV infection in swine is similar to that of the human antiviral responses, with experimental evidence showing similar activation and secretion of cytokines is triggered in response to infection with the same virus.164, 165 Unlike humans, swine are unable to transfer maternal immunoglobulins to porcine embryos due to the existence of a thick placenta comprising six tightly connected tissue layers.166 The first 24 hours after birth of the newborn pigs is exceptionally crucial for the transfer and intestinal absorption of maternal IgG from colostrum.166, 167, 168 Although maternal antibodies through suckling can protect newborn piglets to some extent, the piglets are still highly susceptible to new infections. High‐density commercial farming practices, where piglets are typically sent to market at 6 months of age, can therefore have profound effects on the transmission and diversity of IAVs in various swine populations.162, 163
6. Equine IAV
Two equine IAV subtypes, H7N7 and H3N8, were first detected in the 1950s, although the host origin of the former subtype is not known.169 However, as the H7N7 equine lineage has not been detected in over three decades, it is possible the H7N7 has become extinct in equine hosts.170, 171, 172 Despite its disappearance, the lineage has been studied extensively to elucidate the factors involved in its emergence and potential virulence in mammalian species.173, 174, 175 Of note, it has been observed that the equine H7N7 contains an MBCS in the H7 HA protein,176 which in avian H7 lineages have been implicated in HPAI infections in poultry and have infected humans.177 The equine lineage virus was also found to be highly pathogenic and neurovirulent in mice without prior adaptation.173 However, the intracellular cleavage of the equine H7N7 lineage was found to be due to an 11 amino acid motif adjacent to the MBCS that, when inserted into an LPAI H7N3 virus, increased the pathogenicity of infection.175
In contrast, the H3N8 equine lineage may have originated wholly from an avian IAV source, and these viruses have been shown to undergo frequent intersubtype and intrasubtype reassortments.178, 179, 180 Interestingly, the H3N8 virus in equine hosts preferentially binds to avian‐like α‐2,3‐SA, where the receptors are abundantly present in the upper respiratory tract of horses.181, 182 As such, horses are always considered as dead‐end hosts, but the interspecies transmission of H3N8 virus from horses to dogs and camels raises the question of whether horses can act as mixing vessels for IAV.169, 182
7. Canine IAV
IAV has been detected in canine species since the early 2000s, and there are currently two stably transmitting lineages of IAVs that circulate in dogs: the equine‐derived H3N8 and the avian‐derived H3N2 strains.169 The transmission of the equine H3N8 subtype to dogs seems to have occurred en bloc through close contact with infected horses183 and this virus continues to circulate in dogs with no evidence of reassortment with other subtypes to date.169 This is further supported by recent findings that despite phylogenetic divergence and genetic change from the equine lineage, canine H3N8 was not observed to be phenotypically different from equine H3N8 strains.184 The avian‐derived H3N2 canine IAV has a much broader host range, and unlike the H3N8 lineage, reassortment with other subtypes, such as with H5N1 and H1N1, has been detected.185, 186 Recently, an isolated case of an H6N1 virus was detected a dog in Taiwan, likely through contact with infected chicken, and represents the potential for other avian IAV subtypes to emerge in canid hosts.187 Lineage‐specific mutations have been detected in the canine IAVs that suggest adaptive mutations in various segments towards the new canine hosts,188, 189 but only the role of a leucine residue at position 222 (H3 numbering) has been confirmed as a host‐specific adaptation thus far.190, 191
8. Concluding thoughts
There is a need to understand adaptations in IAV that confer interspecies transmission capabilities to drive active surveillance for these mutations in nature and prevent an impending outbreak. Thus, the continued evolution of influenza viruses in its myriad of hosts necessitates further studies that characterize the risk of human transmission in these viruses, to provide a meaningful assessment of pandemic potential. Furthermore, the benefits of such efforts have been seen in the identification and rapid response to recent H7N9 outbreaks in China,12, 73 while other studies that have characterized mutations that may enable sustained human‐to‐human transmission or describe pathogenesis have been useful for risk assessment of contemporarily circulating IAVs.104, 107, 108 However, as there is continued mixing of viruses normally resident in different species, there is value in the continued experimental characterization of circulating viruses, particularly from subtypes known to have caused pandemics and those viruses present in animals to which humans have high exposure.
Acknowledgements
This study was supported by the Duke‐NUS Signature Research Program funded by the Agency of Science, Technology and Research, Singapore, and the Ministry of Health, Singapore, the National Medical Research Council (NMRC/GMS/1251/2010), and by contract HHSN272201400006C from the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, USA.
Joseph, U. , Su, Y. C. F. , Vijaykrishna, D. and Smith, G. J. D. (2017), The ecology and adaptive evolution of influenza A interspecies transmission. Influenza and Other Respiratory Viruses 11, 74–84. doi: 10.1111/irv.12412
References
- 1. Taubenberger JK, Morens DM. Influenza: the once and future pandemic. Public Health Rep. 2010;125(Suppl 3):16–26. [PMC free article] [PubMed] [Google Scholar]
- 2. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stallknecht DE, Brown JD. Ecology of avian influenza in wild birds. In: Swayne DE, ed. Avian Influenza. Oxford: Blackwell Publishing Ltd; 2008:43–58. [Google Scholar]
- 4. Medina RA, García‐Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Webster RG, Monto AS, Braciale TJ, Lamb RA. Textbook of Influenza. Oxford, UK: John Wiley & Sons, Ltd; 2013. [Google Scholar]
- 8. Peiris M, Yuen KY, Leung CW, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–917. [DOI] [PubMed] [Google Scholar]
- 9. Lin YP, Shaw M, Gregory V, et al. Avian‐to‐human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000;97:9654–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koopmans M, Wilbrink B, Conyn M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. [DOI] [PubMed] [Google Scholar]
- 11. Kandun IN, Wibisono H, Sedyaningsih ER, et al. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med. 2006;355:2186–2194. [DOI] [PubMed] [Google Scholar]
- 12. Gao R, Cao B, Hu Y, et al. Human infection with a novel avian‐origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. [DOI] [PubMed] [Google Scholar]
- 13. Epperson S, Jhung M, Richards S, et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis. 2013;57(Suppl 1):S4–S11. [DOI] [PubMed] [Google Scholar]
- 14. Yang ZF, Mok CK, Peiris JS, Zhong NS. Human infection with a novel avian influenza A(H5N6) virus. N Engl J Med. 2015;373:487–489. [DOI] [PubMed] [Google Scholar]
- 15. Gilsdorf A, Boxall N, Gasimov V, et al. Two clusters of human infection with influenza A/H5N1 virus in the Republic of Azerbaijan, February–March 2006. Euro Surveill. 2006;11:122–126. [PubMed] [Google Scholar]
- 16. Reed C, Bruden D, Byrd KK, et al. Characterizing wild bird contact and seropositivity to highly pathogenic avian influenza A (H5N1) virus in Alaskan residents. Influenza Other Respir Viruses. 2014;8:516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe T, Zhong G, Russell CA, et al. Circulating avian influenza viruses closely related to the 1918 virus have pandemic potential. Cell Host Microbe. 2014;15:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morse SS. The public health threat of emerging viral disease. J Nutr. 1997;127(5 Suppl):951S–957S. [DOI] [PubMed] [Google Scholar]
- 19. Li W, Wong SK, Li F, et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2‐S‐protein interactions. J Virol. 2006;80:4211–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feldmann H. Ebola – a growing threat? N Engl J Med. 2014;371:1375–1378. [DOI] [PubMed] [Google Scholar]
- 22. Slingenbergh JI, Gilbert M, de Balogh KI, Wint W. Ecological sources of zoonotic diseases. Rev Sci Tech. 2004;23:467–484. [DOI] [PubMed] [Google Scholar]
- 23. Bahl J, Krauss S, Kühnert D, et al. Influenza a virus migration and persistence in North American wild birds. PLoS Pathog. 2013;9:e1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Runstadler J, Hill N, Hussein IT, Puryear W, Keogh M. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol. 2013;17:162–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spackman E. The ecology of avian influenza virus in wild birds: what does this mean for poultry? Poult Sci. 2009;88:847–850. [DOI] [PubMed] [Google Scholar]
- 26. Stallknecht DE, Shane SM, Kearney MT, Zwank PJ. Persistence of avian influenza viruses in water. Avian Dis. 1990;34:406–411. [PubMed] [Google Scholar]
- 27. Causey D, Edwards SV. Ecology of avian influenza virus in birds. J Infect Dis. 2008;197(Suppl 1):S29–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shoham D, Jahangir A, Ruenphet S, Takehara K. Persistence of avian influenza viruses in various artificially frozen environmental water types. Influenza Res Treat. 2012;2012:912326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keeler SP, Berghaus RD, Stallknecht DE. Persistence of low pathogenic avian influenza viruses in filtered surface water from waterfowl habitats in Georgia, USA. J Wildl Dis. 2012;48:999–1009. [DOI] [PubMed] [Google Scholar]
- 30. Sharma JM. Overview of the avian immune system. Vet Immunol Immunopathol. 1991;30:13–17. [DOI] [PubMed] [Google Scholar]
- 31. Deibel R, Emord DE, Dukelow W, Hinshaw VS, Wood JM. Influenza viruses and paramyxoviruses in ducks in the Atlantic flyway, 1977–1983, including an H5N2 isolate related to the virulent chicken virus. Avian Dis. 1985;29:970–985. [PubMed] [Google Scholar]
- 32. Mair CM, Ludwig K, Herrmann A, Sieben C. Receptor binding and pH stability – how influenza A virus hemagglutinin affects host‐specific virus infection. Biochim Biophys Acta. 2014;1838:1153–1168. [DOI] [PubMed] [Google Scholar]
- 33. Galloway SE, Reed ML, Russell CJ, Steinhauer DA. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013;9:e1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown JD, Goekjian G, Poulson R, Valeika S, Stallknecht DE. Avian influenza virus in water: infectivity is dependent on pH, salinity and temperature. Vet Microbiol. 2009;136:20–26. [DOI] [PubMed] [Google Scholar]
- 35. Reed ML, Bridges OA, Seiler P, et al. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J Virol. 2010;84:1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DuBois RM, Zaraket H, Reddivari M, Heath RJ, White SW, Russell CJ. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 2011;7:e1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roche B, Drake JM, Brown J, Stallknecht DE, Bedford T, Rohani P. Adaptive evolution and environmental durability jointly structure phylodynamic patterns in avian influenza viruses. PLoS Biol. 2014;12:e1001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pepin KM, Wang J, Webb CT, et al. Multiannual patterns of influenza A transmission in Chinese live bird market systems. Influenza Other Respir Viruses. 2013;7:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peiris M, Yen HL. Animal and human influenzas. Rev Sci Tech. 2014;33:539–553. [DOI] [PubMed] [Google Scholar]
- 40. World Organization for Animal Health (OIE) . Avian Influenza. 2015. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [Internet]. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf. Accessed 7 April 2016.
- 41. Stieneke‐Gröber A, Vey M, Angliker H, et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin‐like endoprotease. EMBO J. 1992;11:2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. [DOI] [PubMed] [Google Scholar]
- 43. Schrauwen EJ, de Graaf M, Herfst S, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Determinants of virulence of influenza A virus. Eur J Clin Microbiol Infect Dis. 2014;33:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bertram S, Glowacka I, Steffen I, Kühl A, Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010;20:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suarez DL, Senne DA, Banks J, et al. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg Infect Dis. 2004;10:693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maurer‐Stroh S, Lee RT, Gunalan V, Eisenhaber F. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virol J. 2013;10:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. [DOI] [PubMed] [Google Scholar]
- 48. Schrauwen EJ, Herfst S, Leijten LM, et al. The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J Virol. 2012;86:3975–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rimmelzwaan GF, Kuiken T, van Amerongen G, Bestebroer TM, Fouchier RA, Osterhaus AD. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J Virol. 2001;75:6687–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barber MR, Aldridge JR, Webster RG, Magor KE. Association of RIG‐I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A. 2010;107:5913–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Magor KE, Miranzo Navarro D, Barber MR, et al. Defense genes missing from the flight division. Dev Comp Immunol. 2013;41:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pichlmair A, Schulz O, Tan CP, et al. RIG‐I‐mediated antiviral responses to single‐stranded RNA bearing 5′‐phosphates. Science. 2006;314:997–1001. [DOI] [PubMed] [Google Scholar]
- 53. Cornelissen JB, Post J, Peeters B, Vervelde L, Rebel JM. Differential innate responses of chickens and ducks to low‐pathogenic avian influenza. Avian Pathol. 2012;41:519–529. [DOI] [PubMed] [Google Scholar]
- 54. Cornelissen JB, Vervelde L, Post J, Rebel JM. Differences in highly pathogenic avian influenza viral pathogenesis and associated early inflammatory response in chickens and ducks. Avian Pathol. 2013;42:347–364. [DOI] [PubMed] [Google Scholar]
- 55. Chen H, Smith GJ, Zhang SY, et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. [DOI] [PubMed] [Google Scholar]
- 56. Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. [DOI] [PubMed] [Google Scholar]
- 57. Winker K, McCracken KG, Gibson DD, et al. Movements of birds and avian influenza from Asia into Alaska. Emerg Infect Dis. 2007;13:547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gilbert M, Slingenbergh J, Xiao X. Climate change and avian influenza. Rev Sci Tech. 2008;27:459–466. [PMC free article] [PubMed] [Google Scholar]
- 59. Abdelwhab EM, Hafez HM. An overview of the epidemic of highly pathogenic H5N1 avian influenza virus in Egypt: epidemiology and control challenges. Epidemiol Infect. 2011;139:647–657. [DOI] [PubMed] [Google Scholar]
- 60. Smith GJ, Bahl J, Vijaykrishna D, et al. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A. 2009;106:11709–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lebarbenchon C, Feare CJ, Renaud F, Thomas F, Gauthier‐Clerc M. Persistence of highly pathogenic avian influenza viruses in natural ecosystems. Emerg Infect Dis. 2010;16:1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Horm SV, Gutiérrez RA, Sorn S, Buchy P. Environment: a potential source of animal and human infection with influenza A (H5N1) virus. Influenza Other Respir Viruses. 2012;6:442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pepin KM, Wang J, Webb CT, et al. Anticipating the prevalence of avian influenza subtypes H9 and H5 in live‐bird markets. PLoS One. 2013;8:e56157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen Z, Li K, Luo L, et al. Detection of avian influenza A(H7N9) virus from live poultry markets in Guangzhou, China: a surveillance report. PLoS One. 2014;9:e107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Claas EC, Osterhaus AD, van Beek R, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. [DOI] [PubMed] [Google Scholar]
- 66. Subbarao K, Klimov A, Katz J, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. [DOI] [PubMed] [Google Scholar]
- 67. Guan Y, Shortridge KF, Krauss S, et al. H9N2 influenza viruses possessing H5N1‐like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Butt KM, Smith GJ, Chen H, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xu KM, Smith GJ, Bahl J, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang T, Bi Y, Tian H, et al. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis. 2014;20:2076–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bi Y, Mei K, Shi W, et al. Two novel reassortants of avian influenza A (H5N6) virus in China. J Gen Virol. 2015;96(Pt 5):975–981. [DOI] [PubMed] [Google Scholar]
- 72. Bao CJ, Cui LB, Zhou MH, Hong L, Gao GF, Wang H. Live‐animal markets and influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2337–2339. [DOI] [PubMed] [Google Scholar]
- 73. Lam TT, Wang J, Shen Y, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qin Y, Horby PW, Tsang TK, et al. Differences in the epidemiology of human cases of avian influenza A(H7N9) and A(H5N1) viruses infection. Clin Infect Dis. 2015;61:563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. USDAFAS . China–People's Republic of, Poultry and Products 2015, GAIN Report:CH 15027, 2015. http://gain.fas.usda.gov/Recent%20GAIN%20Publications/Poultry%20and%20Products%20Annual_Beijing_China%20-%20Peoples%20Republic%20of_8-18-2015.pdf. Accessed March 21, 2016.
- 76. Guan Y, Smith GJ. The emergence and diversification of panzootic H5N1 influenza viruses. Virus Res. 2013;178:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cheung CL, Vijaykrishna D, Smith GJ, et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol. 2007;81:10402–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cardona C, Yee K, Carpenter T. Are live bird markets reservoirs of avian influenza? Poult Sci. 2009;88:856–859. [DOI] [PubMed] [Google Scholar]
- 80. Kung NY, Guan Y, Perkins NR, et al. The impact of a monthly rest day on avian influenza virus isolation rates in retail live poultry markets in Hong Kong. Avian Dis. 2003;47(3 Suppl):1037–1041. [DOI] [PubMed] [Google Scholar]
- 81. Leung YH, Lau EH, Zhang LJ, Guan Y, Cowling BJ, Peiris JS. Avian influenza and ban on overnight poultry storage in live poultry markets, Hong Kong. Emerg Infect Dis. 2012;18:1339–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yu H, Wu JT, Cowling BJ, et al. Effect of closure of live poultry markets on poultry‐to‐person transmission of avian influenza A H7N9 virus: an ecological study. Lancet. 2014;383:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Obenauer JC, Denson J, Mehta PK, et al. Large‐scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. [DOI] [PubMed] [Google Scholar]
- 84. Vijaykrishna D, Smith GJ, Pybus OG, et al. Long‐term evolution and transmission dynamics of swine influenza A virus. Nature. 2011;473:519–522. [DOI] [PubMed] [Google Scholar]
- 85. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. [DOI] [PubMed] [Google Scholar]
- 86. Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. [DOI] [PubMed] [Google Scholar]
- 87. Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell‐surface receptors: structures of five hemagglutinin‐sialyloligosaccharide complexes determined by X‐ray crystallography. Virology. 1997;232:19–31. [DOI] [PubMed] [Google Scholar]
- 88. Keleta L, Ibricevic A, Bovin NV, Brody SL, Brown EG. Experimental evolution of human influenza virus H3 hemagglutinin in the mouse lung identifies adaptive regions in HA1 and HA2. J Virol. 2008;82:11599–11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu J, Stevens DJ, Haire LF, et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009;106:17175–17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol. 2010;84:1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Matrosovich M, Tuzikov A, Bovin N, et al. Early alterations of the receptor‐binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Glaser L, Stevens J, Zamarin D, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X‐ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A. 2001;98:11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lu X, Shi Y, Zhang W, Zhang Y, Qi J, Gao GF. Structure and receptor‐binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell. 2013;4:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xiong X, Coombs PJ, Martin SR, et al. Receptor binding by a ferret‐transmissible H5 avian influenza virus. Nature. 2013;497:392–396. [DOI] [PubMed] [Google Scholar]
- 97. Shi Y, Wu Y, Zhang W, Qi J, Gao GF. Enabling the ‘host jump’: structural determinants of receptor‐binding specificity in influenza A viruses. Nat Rev Microbiol. 2014;12:822–831. [DOI] [PubMed] [Google Scholar]
- 98. Auewarakul P, Suptawiwat O, Kongchanagul A, et al. An avian influenza H5N1 virus that binds to a human‐type receptor. J Virol. 2007;81:9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Stevens J, Blixt O, Chen LM, Donis RO, Paulson JC, Wilson IA. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 1997;71:8377–8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kasson PM, Pande VS. Structural basis for influence of viral glycans on ligand binding by influenza hemagglutinin. Biophys J. 2008;95:L48–L50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yen HL, Aldridge JR, Boon AC, et al. Changes in H5N1 influenza virus hemagglutinin receptor binding domain affect systemic spread. Proc Natl Acad Sci U S A. 2009;106:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jayaraman A, Koh X, Li J, et al. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. Biochem J. 2012;444:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Raman R, Tharakaraman K, Shriver Z, Jayaraman A, Sasisekharan V, Sasisekharan R. Glycan receptor specificity as a useful tool for characterization and surveillance of influenza A virus. Trends Microbiol. 2014;22:632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zaraket H, Bridges OA, Russell CJ. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J Virol. 2013;87:4826–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Linster M, van Boheemen S, de Graaf M, et al. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell. 2014;157:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Baumann J, Kouassi NM, Foni E, Klenk H‐D, Matrosovich M. H1N1 swine influenza viruses differ from avian precursors by a higher pH optimum of membrane fusion. J Virol. 2016;90:1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. [DOI] [PubMed] [Google Scholar]
- 111. Yen HL, Liang CH, Wu CY, et al. Hemagglutinin‐neuraminidase balance confers respiratory‐droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A. 2011;108:14264–14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Munier S, Larcher T, Cormier‐Aline F, et al. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol. 2010;84:940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hoffmann TW, Munier S, Larcher T, et al. Length variations in the NA stalk of an H7N1 influenza virus have opposite effects on viral excretion in chickens and ducks. J Virol. 2012;86:584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Blumenkrantz D, Roberts KL, Shelton H, Lycett S, Barclay WS. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol. 2013;87:10539–10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Van Hoeven N, Pappas C, Belser JA, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yamada S, Hatta M, Staker BL, et al. Biological and structural characterization of a host‐adapting amino acid in influenza virus. PLoS Pathog. 2010;6:e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mehle A, Doudna JA. An inhibitory activity in human cells restricts the function of an avian‐like influenza virus polymerase. Cell Host Microbe. 2008;4:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ng AK, Chan WH, Choi ST, et al. Influenza polymerase activity correlates with the strength of interaction between nucleoprotein and PB2 through the host‐specific residue K/E627. PLoS One. 2012;7:e36415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mänz B, Schwemmle M, Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol. 2013;87:7200–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Weber M, Sediri H, Felgenhauer U, et al. Influenza virus adaptation PB2‐627K modulates nucleocapsid inhibition by the pathogen sensor RIG‐I. Cell Host Microbe. 2015;17:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5:e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dittmann J, Stertz S, Grimm D, et al. Influenza A virus strains differ in sensitivity to the antiviral action of Mx‐GTPase. J Virol. 2008;82:3624–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zimmermann P, Mänz B, Haller O, Schwemmle M, Kochs G. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol. 2011;85:8133–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. The human interferon‐induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol. 2013;87:13053–13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Riegger D, Hai R, Dornfeld D, et al. The nucleoprotein of newly emerged H7N9 influenza A virus harbors a unique motif conferring resistance to antiviral human MxA. J Virol. 2015;89:2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tian SF, Buckler‐White AJ, London WT, Reck LJ, Chanock RM, Murphy BR. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/NY/78 virus and its reassortants in squirrel monkey respiratory tract. J Virol. 1985;53:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Snyder MH, Buckler‐White AJ, London WT, Tierney EL, Murphy BR. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian‐human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987;61:2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Naffakh N, Tomoiu A, Rameix‐Welti MA, van der Werf S. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol. 2008;62:403–424. [DOI] [PubMed] [Google Scholar]
- 132. Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C‐terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Thomas M, Kranjec C, Nagasaka K, Matlashewski G, Banks L. Analysis of the PDZ binding specificities of Influenza A virus NS1 proteins. Virol J. 2011;8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ayllon J, Russell RJ, García‐Sastre A, Hale BG. Contribution of NS1 effector domain dimerization to influenza A virus replication and virulence. J Virol. 2012;86:13095–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Noronha JM, Liu M, Squires RB, et al. Influenza virus sequence feature variant type analysis: evidence of a role for NS1 in influenza virus host range restriction. J Virol. 2012;86:5857–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bullido R, Gómez‐Puertas P, Saiz MJ, Portela A. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J Virol. 2001;75:4912–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol. 2009;90(Pt 6):1398–1407. [DOI] [PubMed] [Google Scholar]
- 138. Mänz B, Brunotte L, Reuther P, Schwemmle M. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat Commun. 2012;3:802. [DOI] [PubMed] [Google Scholar]
- 139. Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, Ludwig S. The proapoptotic influenza A virus protein PB1‐F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol. 2008;10:1140–1152. [DOI] [PubMed] [Google Scholar]
- 140. Zell R, Krumbholz A, Eitner A, Krieg R, Halbhuber KJ, Wutzler P. Prevalence of PB1‐F2 of influenza A viruses. J Gen Virol. 2007;88(Pt 2):536–546. [DOI] [PubMed] [Google Scholar]
- 141. Schmolke M, Manicassamy B, Pena L, et al. Differential contribution of PB1‐F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 2011;7:e1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL. Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol Biol. 2010;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 2008;4:e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jimenez‐Baranda S, Greenbaum B, Manches O, et al. Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. J Virol. 2011;85:3893–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. [DOI] [PubMed] [Google Scholar]
- 146. Greenbaum BD, Rabadan R, Levine AJ. Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PLoS One. 2009;4:e5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Joseph U, Linster M, Suzuki Y, et al. Adaptation of pandemic H2N2 influenza A viruses in humans. J Virol. 2015;89:2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ahn I, Son HS. Comparative study of the nucleotide bias between the novel H1N1 and H5N1 subtypes of influenza A viruses using bioinformatics techniques. J Microbiol Biotechnol. 2010;20:63–70. [PubMed] [Google Scholar]
- 149. Ahn I, Son HS. Evolutionary analysis of human‐origin influenza A virus (H3N2) genes associated with the codon usage patterns since 1993. Virus Genes. 2012;44:198–206. [DOI] [PubMed] [Google Scholar]
- 150. Goñi N, Iriarte A, Comas V, et al. Pandemic influenza A virus codon usage revisited: biases, adaptation and implications for vaccine strain development. Virol J. 2012;9:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Liu X, Wu C, Chen AY. Codon usage bias and recombination events for neuraminidase and hemagglutinin genes in Chinese isolates of influenza A virus subtype H9N2. Arch Virol. 2010;155:685–693. [DOI] [PubMed] [Google Scholar]
- 152. Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med. 2008;3:158–166. [PMC free article] [PubMed] [Google Scholar]
- 153. Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. [DOI] [PubMed] [Google Scholar]
- 154. Su YC, Bahl J, Joseph U, et al. Phylodynamics of H1N1/2009 influenza reveals the transition from host adaptation to immune‐driven selection. Nat Commun. 2015;6:7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106:21312–21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Zhou B, Pearce MB, Li Y, et al. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS One. 2013;8:e67616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Barman S, Krylov PS, Fabrizio TP, et al. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog. 2012;8:e1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Mänz B, Dornfeld D, Götz V, et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 2013;9:e1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Shu B, Garten R, Emery S, et al. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology. 2012;422:151–160. [DOI] [PubMed] [Google Scholar]
- 160. Shinde V, Bridges CB, Uyeki TM, et al. Triple‐reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. [DOI] [PubMed] [Google Scholar]
- 161. Wang DY, Qi SX, Li XY, et al. Human infection with Eurasian avian‐like influenza A(H1N1) virus, China. Emerg Infect Dis. 2013;19:1709–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Nelson MI, Viboud C, Vincent AL, et al. Global migration of influenza A viruses in swine. Nat Commun. 2015;6:6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Nelson MI, Vincent AL. Reverse zoonosis of influenza to swine: new perspectives on the human‐animal interface. Trends Microbiol. 2015;23:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest. 1998;101:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Khatri M, Dwivedi V, Krakowka S, et al. Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: a potential animal model for human H1N1 influenza virus. J Virol. 2010;84:11210–11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Rothkötter HJ. Anatomical particularities of the porcine immune system–a physician's view. Dev Comp Immunol. 2009;33:267–272. [DOI] [PubMed] [Google Scholar]
- 167. Kömüves LG, Heath JP. Uptake of maternal immunoglobulins in the enterocytes of suckling piglets: improved detection with a streptavidin‐biotin bridge gold technique. J Histochem Cytochem. 1992;40:1637–1646. [DOI] [PubMed] [Google Scholar]
- 168. Butler JE, Zhao Y, Sinkora M, Wertz N, Kacskovics I. Immunoglobulins, antibody repertoire and B cell development. Dev Comp Immunol. 2009;33:321–333. [DOI] [PubMed] [Google Scholar]
- 169. Parrish CR, Murcia PR, Holmes EC. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol. 2015;89:2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Morens DM, Taubenberger JK. Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influenza Other Respir Viruses. 2010;4:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Cullinane A, Elton D, Mumford J. Equine influenza – surveillance and control. Influenza Other Respir Viruses. 2010;4:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Murcia PR, Baillie GJ, Daly J, et al. Intra‐ and interhost evolutionary dynamics of equine influenza virus. J Virol. 2010;84:6943–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kawaoka Y. Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol. 1991;65:3891–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Shinya K, Watanabe S, Ito T, Kasai N, Kawaoka Y. Adaptation of an H7N7 equine influenza A virus in mice. J Gen Virol. 2007;88(Pt 2):547–553. [DOI] [PubMed] [Google Scholar]
- 175. Hamilton BS, Sun X, Chung C, Whittaker GR. Acquisition of a novel eleven amino acid insertion directly N‐terminal to a tetrabasic cleavage site confers intracellular cleavage of an H7N7 influenza virus hemagglutinin. Virology. 2012;434:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Gibson CA, Daniels RS, Oxford JS, McCauley JW. Sequence analysis of the equine H7 influenza virus haemagglutinin gene. Virus Res. 1992;22:93–106. [DOI] [PubMed] [Google Scholar]
- 177. Fouchier RA, Schneeberger PM, Rozendaal FW, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Murcia PR, Wood JL, Holmes EC. Genome‐scale evolution and phylodynamics of equine H3N8 influenza A virus. J Virol. 2011;85:5312–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Ito T, Kawaoka Y, Ohira M, et al. Replacement of internal protein genes, with the exception of the matrix, in equine 1 viruses by equine 2 influenza virus genes during evolution in nature. J Vet Med Sci. 1999;61:987–989. [DOI] [PubMed] [Google Scholar]
- 180. Gorman OT, Bean WJ, Kawaoka Y, Webster RG. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990;64:1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Suzuki Y, Ito T, Suzuki T, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Cullinane A, Newton JR. Equine influenza–a global perspective. Vet Microbiol. 2013;167:205–214. [DOI] [PubMed] [Google Scholar]
- 183. Yamanaka T, Nemoto M, Tsujimura K, Kondo T, Matsumura T. Interspecies transmission of equine influenza virus (H3N8) to dogs by close contact with experimentally infected horses. Vet Microbiol. 2009;139:351–355. [DOI] [PubMed] [Google Scholar]
- 184. Feng KH, Gonzalez G, Deng L, et al. Equine and canine influenza H3N8 viruses show minimal biological differences despite phylogenetic divergence. J Virol. 2015;89:6860–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Song D, Moon HJ, An DJ, et al. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J Gen Virol. 2012;93(Pt 3):551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhu H, Hughes J, Murcia PR. Origins and evolutionary dynamics of H3N2 canine influenza virus. J Virol. 2015;89:5406–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lin HT, Wang CH, Chueh LL, Su BL, Wang LC. Influenza A(H6N1) virus in dogs, Taiwan. Emerg Infect Dis. 2015;21:2154–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Hayward JJ, Dubovi EJ, Scarlett JM, Janeczko S, Holmes EC, Parrish CR. Microevolution of canine influenza virus in shelters and its molecular epidemiology in the United States. J Virol. 2010;84:12636–12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Hoelzer K, Murcia PR, Baillie GJ, et al. Intrahost evolutionary dynamics of canine influenza virus in naive and partially immune dogs. J Virol. 2010;84:5329–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Yang G, Li S, Blackmon S, et al. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol. 2013;94(Pt 12):2599–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Collins PJ, Vachieri SG, Haire LF, et al. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A. 2014;111:11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]


